Abstract
The development of antibiotic resistance during treatment is a threat to patients and their environment. Insight in the mechanisms of resistance development is important for appropriate therapy and infection control. Here, we describe how through the application of mass spectrometry-based proteomics, a novel beta-lactamase Axc was identified as an indicator of acquired carbapenem resistance in a clinical isolate of Achromobacter xylosoxidans. Comparative proteomic analysis of consecutively collected susceptible and resistant isolates from the same patient revealed that high Axc protein levels were only observed in the resistant isolate. Heterologous expression of Axc in Escherichia coli significantly increased the resistance towards carbapenems. Importantly, direct Axc mediated hydrolysis of imipenem was demonstrated using pH shift assays and 1H-NMR, confirming Axc as a legitimate carbapenemase. Whole genome sequencing revealed that the susceptible and resistant isolates were remarkably similar. Together these findings provide a molecular context for the fast development of meropenem resistance in A. xylosoxidans during treatment and demonstrate the use of mass spectrometric techniques in identifying novel resistance determinants.
Introduction
Development and spread of antibiotic resistance by pathogenic microorganisms is an increasing healthcare problem. Moreover, certain resistance determinants spread readily1,2, while the introduction of novel antibiotics is lagging behind. Several clinically important classes of antimicrobials such as the beta-lactams, target the bacterial cell wall3. Resistance to beta-lactams can be mediated by beta-lactamases that are capable of hydrolysing the beta-lactam ring. Following the initial introduction of penicillin, second and third generation beta-lactams have been developed which, in turn, triggered the selection of beta-lactamases with broader specificities. Carbapenem treatment is often used as a last resort, since extended-spectrum beta-lactamases (cephalosporinases) are becoming more prevalent in Gram-negative bacteria. The emergence and spread of carbapenemases such as class A KPC4, a number of metallo beta-lactamases5,6 (class B: IMP, VIM, NDM) and class D oxacillinases such as OXA-487, in combination with other resistance mechanisms8, can jeopardize carbapenem efficacy, leaving little or no treatment options for patients.
Achromobacter xylosoxidans is a rod shaped aerobic non-fermentative Gram-negative bacterium. It is widespread in the environment and generally considered as an opportunistic pathogen. Chronic infections with A. xylosoxidans are problematic in cystic fibrosis patients9,10 but reported prevalence numbers vary greatly (3–30%)11,12. Moreover, bacteremia as a result of A. xylosoxidans can occur in immunocompromised patients13. A. xylosoxidans is notorious for its intrinsic high level of resistance, especially towards penicillins and cephalosporins14–16. In general, carbapenem resistance in A. xylosoxidans is not widespread and as a result meropenem treatment is routinely applied, even in the case of recurring infections17,18. Though carbapenem resistance is observed, specifically for meropenem19, there are few reports on the mechanism of carbapenemase resistance in A. xylosoxidans. Notable exceptions are the plasmid-encoded carbapenemase IMP20,21 and the chromosomally encoded class D beta-lactamase OXA-11416, that show low level carbapenemase activity. A comparative genomic exploration of two A. xylosoxidans isolates revealed many genes that could be involved in drug resistance, such as efflux pumps and β-lactamases. However, most of these genes were conserved between carbapenem susceptible and resistant strains, highlighting the difficulty in translating genomic data to the observed resistant phenotypes22.
In this study, two clinical isolates of A. xylosoxidans from an immunocompromised patient with pneumonia were investigated. The initially cultured isolate from the respiratory tract was susceptible to meropenem and treatment was started accordingly. However, a subsequent meropenem resistant isolate was obtained from a blood culture after treatment failure. Since PCR analysis was negative for known carbapenemases, we performed a proteomic analysis which revealed the novel beta-lactamase Axc as highly abundant in the meropenem-resistant, but not in the susceptible isolate. Axc expression led to an increase of minimal inhibitory concentrations for carbapenems when introduced in a susceptible Escherichia coli strain and direct carbapenemase activity of Axc was demonstrated using in vitro imipenem conversion assays. Interestingly, the resistant and the susceptible clinical isolates are genetically almost identical, emphasizing the importance of mass spectrometry as a technique to investigate carbapenem resistance in A. xylosoxidans.
Results
Development of meropenem resistance in Achromobacter xylosoxidans during treatment
An antibiogram of a clinical isolate of Achromobacter xylosoxidans from a patient treated for pneumonia (see Methods for details about patient history and treatment) revealed a multi-resistant character, but it was susceptible to meropenem (Supplemental Table 1). Hence, meropenem treatment was initiated. During treatment, the patient developed a pneumothorax and died from septic shock 6 days later. An antibiogram of a blood culture from a sample taken one day before the patient’s death revealed a meropenem-resistant A. xylosoxidans phenotype (Supplemental Table 1). Subsequently, pure cultures from the first and second clinical isolate were prepared (AchroS and AchroR, respectively (Table 1)). In line with the antibiogram analysis described above, Etests showed that both isolates were resistant to imipenem (MIC > 32 mg/L for both) but differed in their susceptibility towards meropenem (MIC of 0.0094 mg/L (first isolate, AchroS) and 2 mg/L (second isolate, AchroR), respectively). An in-house multiplex PCR assay (based on a published PCR)23 failed to detect common carbapenemases (KPC, IMP, VIM, NDM and OXA-48, data not shown), suggesting that the change in resistance is not mediated by these enzymes. This finding prompted us to perform a comparative proteomic analysis of the two isolates to attempt to identify the meropenem resistance mechanism.
Table 1.
Strains used in this study. Axc: Achromobacter xylosoxidans carbapenemase. PPEP-1: Pro-Pro endopeptidase 131.
| Name | Short description | Full description |
|---|---|---|
| JC188 | AchroS | Meropenem susceptible A. xylosoxidans clinical isolate |
| JC186 | AchroR | Meropenem resistant A. xylosoxidans clinical isolate |
| JC107 | E. coli_Axc | Escherichia coli C43/pET21B-Axc |
| JC108 | E. coli_control | Escherichia coli C43/pET21B-PPEP-1 |
| JC113 | KPC | Carbapenemase positive Klebsiella pneumoniae |
Comparative proteomic analysis shows differential levels of the beta-lactamase Axc
Protein extracts of the meropenem-susceptible and resistant A. xylosoxidans isolates (AchroS and AchroR) were first analysed by SDS-PAGE. Since no major visual differences were observed (Supplemental Fig. S1A), all bands were excised and processed for a bottom-up LC-MS/MS proteomic analysis. In total, 2276 unique proteins were identified, of which 1519 proteins were common to both isolates, while 223 and 534 were only found in the susceptible and resistant isolates, respectively. For a semi-quantitative analysis, the spectra assigned to peptides belonging to a certain protein were counted and compared between the two different isolates (Supplemental Figs S1B and S2, Table S2). Of the uniquely observed proteins, most were proteins with low spectral counts (often single peptide identifications), likely resulting from sampling bias of low abundant proteins. One protein was observed with 102 spectra in the resistant isolate (AchroR) but none in the susceptible isolate (AchroS). This protein, hereafter called Axc (for Achromobacter xylosoxidans carbapenemase, GenBank ID: MF767301), is a putative PenP class A beta-lactamase (COG2367/pfam13354).
To confirm that Axc was highly abundant in the resistant isolate in comparison to the susceptible isolate, a second proteomic analysis was performed on whole cell protein extracts that were digested in-solution and analysed without any prior fractionation. Spectral count analysis resulted in a cumulative quantification of 1342 different proteins, of which 981 were found in both isolates, but 196 and 165 were uniquely quantified in the susceptible and resistant isolates, respectively. The spectral count plot reflects the high similarity of the two clinical isolates, with the vast majority of the proteins distributed along the diagonal (Fig. 1A, Table S2, Supplemental Fig. S2). In accordance with the data described above, Axc was the most prominent outlier in the resistant clinical isolate. A number of Axc tryptic peptides (Fig. 1B) was clearly visible in the LC-MS/MS analysis of the resistant but not the susceptible isolates (Fig. 1C). Of note, cells used for these analyses were grown in the absence of meropenem, so the high level of Axc in the resistant isolate was independent of antibiotic pressure.
Figure 1.
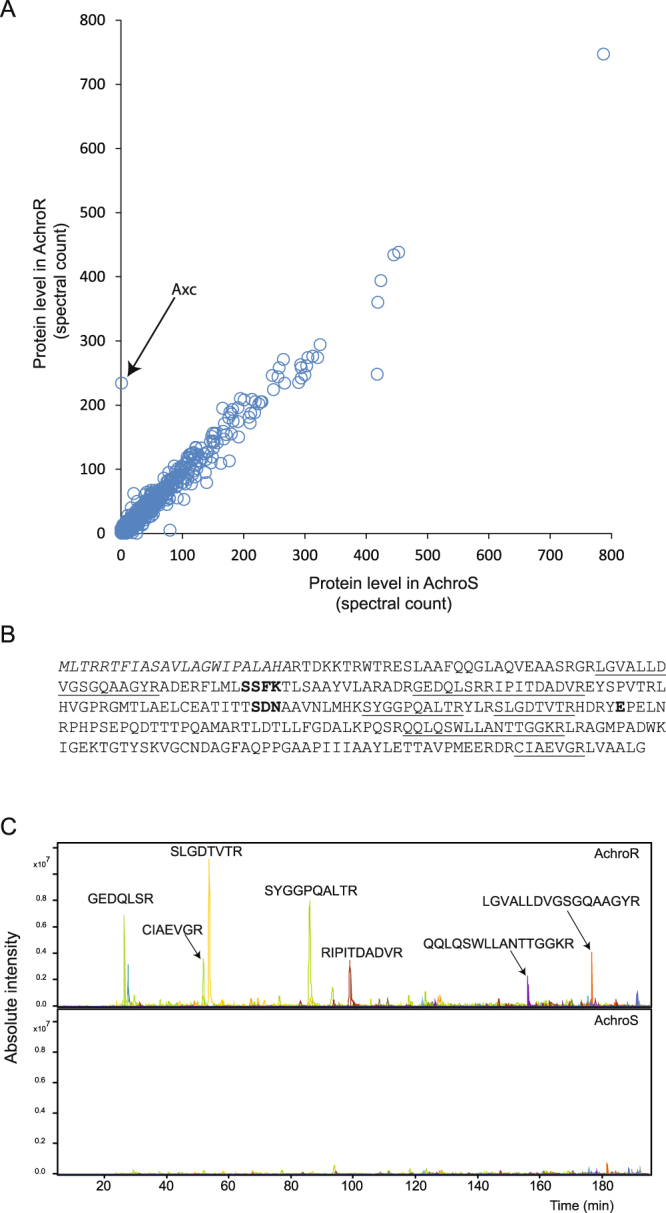
Comparative proteomic analysis of meropenem resistant and susceptible Achromobacter xylosoxidans clinical isolates. (A) Tryptic digests of protein extracts of the meropenem resistant (AchroR, Table 1) and susceptible (AchroS, Table 1) isolate were analysed by LC-MS/MS. Spectra were assigned to peptides based on database searching. Identified spectra were then assigned to the corresponding proteins and the total number of spectra per protein were counted. Each circle represents one protein with the number of spectra observed in the resistant and the susceptible isolate. Hence, proteins on the diagonal were observed in similar counts in both isolates. Axc (arrow), a classA PenP-family beta-lactamase, is the most prominent outlier. See also Table S2 and Supplemental Fig. S2. (B) The full amino acid sequence of Axc, with the peptides identified by LC-MS/MS analysis underlined. Conserved residues from serine beta-lactamases, Ser-X-X-Lys, Ser-Asp-Asn and the active site Glu, are in bold37. (C) Extracted ion chromatograms of m/z values corresponding to tryptic peptides of Axc in the meropenem resistant isolate (AchroR, upper panel) and susceptible isolate (AchroS, lower panel). The corresponding tryptic peptides are indicated above the corresponding peaks.
The axc gene is present in meropenem resistant and susceptible A. xylosoxidans isolates
To investigate whether A. xylosoxidans acquired the axc gene in the course of the treatment, we performed a PCR for the axc open reading frame on both the resistant and susceptible isolates AchroS and AchroR. This analysis demonstrated that axc was present in both clinical isolates (Supplemental Fig. S3A). Moreover, Sanger sequence analysis demonstrated that the sequence of axc was identical in both isolates (data not shown), indicating that the observed meropenem resistance was not likely caused by an alteration of protein function.
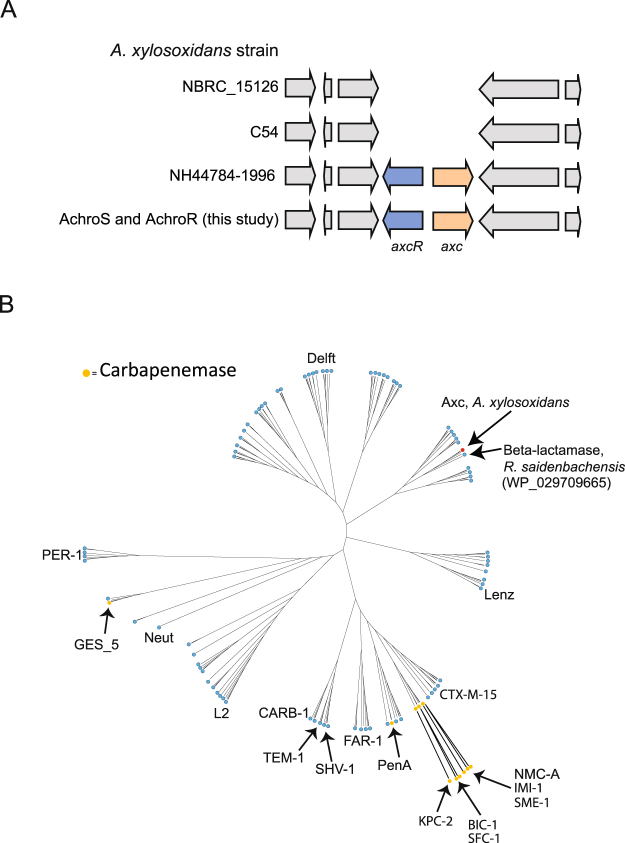
A database search revealed that the axc gene is not present in all A. xylosoxidans strains (Fig. 2A). Like in our clinical isolates, axc is present in the NH44784-1996 strain (ENA assembly GCA_000967095.1)24 but not in the strains NBRC 15126/ATCC 27061 (ENA assembly GCA_000508285.1) and C54 (ENA assembly GCA_000758265.1) for instance. In those strains that contain axc, the gene is located next to a putative LysR-type transcriptional regulator (Pfam 03466), hereafter axcR (for axc-associated regulator). Additional PCR and Sanger sequencing experiments verified that the intergenic regions between axc and axcR in AchroS and AchroR were identical. However, they differed at two positions with the intergenic region between axc and axcR in strain NH44784_1996 (Supplemental Fig. S3B).
Figure 2.
Genomic context of axc in Achromobacter xylosoxidans strains and comparison of Axc with other class A beta-lactamases (A) Axc (a putative PenP class A beta-lactamase, 1.17 e-54) and the gene encoding its putative transcriptional repressor (axcR), were found in both clinical isolates (AchroS and AchroR, Table 1). Three other fully sequenced genomes of Achromobacter xylosoxidans were examined for the presence of axc; NH44784_1996 (ENA assembly GCA_000967095.1), C54 (ENA assembly GCA_000758265.1) and NBRC_15126/ATCC27061 (ENA assembly GCA_000508285.1). Only within the strain NH44784_1996 (used as a reference to search our proteomics data), axc and the putative regulator are also present. (B) Unrooted cladogram obtained for 176 class A beta-lactamases including Axc. The class A β-lactamase protein sequences of Gram negative bacteria were obtained by querying the refseq_protein database using Blastp (<1e-10, http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi) and a consensus β-lactamase-alignment37. Duplicate sequences and sequences causing a strong overrepresentation of branches produced in the tree were removed. Names of known carbapenames (orange) and names of identifier of branches37 are indicated. Axc is indicated with a red dot. The closest homologue to Axc, a β-lactamase from R. saidenbachensis (WP_029709665), is also indicated.
The results showed a high degree of similarity between the meropenem resistant and susceptible strain, raising the possibility that these two strains represented a clonal complex. To further explore the relatedness of both isolates, whole genome sequencing (WGS) analysis was performed. This showed that both patient isolates were highly similar, with only one single-nucleotide polymorphism (SNP) within the gene encoding AxyZ25. This SNP was confirmed by PCR and subsequent Sanger sequencing, ruling out the possibility that this was an artefact of the assembly procedure. Moreover, axc and its putative regulator axcR were found to be located in the same genomic region as in NH44784_1996 strain (Fig. 2A). Overall, the genome sequences suggest that both isolates are clonal, and that the meropenem resistance evolved within the same strain during the course of treatment.
To demonstrate how Axc is related to other class A beta-lactamases, we compared the Axc amino acid sequence with the sequence of another 176 representatives of this family using an alignment consensus based on a report by Walther-Rasmussen and Hoiby26. The resulting unrooted cladogram shows that Axc is most closely related to a class A beta-lactamase of Rhodoferax saidenbachensis (WP_029709665, Fig. 2B). Only a limited number of class A beta-lactamases have activity towards carbapenems, but none of these cluster with the Axc sequence (Fig. 2B).
Functional characterisation of Axc
To establish whether Axc indeed has activity towards carbapenems, Axc was expressed in a heterologous host and hydrolysis of carbapenems was measured indirectly and directly.
The E. coli C43 strain, suitable for the expression of toxic proteins27, is susceptible to carbapenems. We generated a derivative of C43 that allows for IPTG-dependent expression of a plasmid-based copy of axc (E. coli_Axc (JC107)). Susceptibility testing for imipenem and meropenem showed that the MICs for these carbapenems increased 8-fold, following induction of Axc expression (Table 2). Though these levels were lower than for the positive control (KPC) for our assay, they were specific for Axc as the expression of an unrelated protein (PPEP-1)28 did not lead to an increase in MIC values (Table 2). Thus, expression of Axc is correlated to resistance towards carbapenems.
Table 2.
Effect of Axc expression in E. coli on the susceptibility towards imipenem and meropenem.
| Strain | MIC (mg/L) | |||
|---|---|---|---|---|
| Imipenem | Meropenem | |||
| −IPTG | +IPTG | −IPTG | +IPTG | |
| E. coli_ Axca | 0.5 | 4 | 1.563 | 12.5 |
| E. coli_controlb | 0.38 | N.D. | 0.391 | 0.391 |
| KPCc | >32 | >32 | >12.5 | >12.5 |
Minimal inhibitory concentrations of imipenem and meropenem for E.coli C43 expressing Axc (E. coli_ Axc), or PPEP-1 (E. coli_control) and Klesbsiella pneumoniae expressing KPC carbapenemase (KPC). N.D: Not detectable. IPTG: isopropyl β-D-1-thiogalactopyranoside.
aJC107: Escherichia coli C43/pET21B-Axc.
bJC108:Escherichia coli C43/pET21B/PPEP-1 (Pro-Pro endopeptidase 1).
cJC113: Carbapenemase-positive Klebsiella pneumonia.
To directly demonstrate carbapenemase activity, hydrolysis of imipenem was monitored in vitro through colorimetric assays (Fig. 3A) and 1H-NMR (Fig. 3B). Imipenem hydrolysis results in the formation of a carboxylic acid, and monitoring the accompanying pH drop colorimetrically is a well-established method for the detection of carbapenemase activity29,30. Indeed, hydrolysis of imipenem was readily observed using KPC cells (without IPTG). Consistent with our previous results, imipenem hydrolysis was observed for E. coli cells harbouring the Axc expression plasmid (E. coli_Axc) grown in the presence, but not in the absence, of IPTG. As before, these results were specific for Axc, as induction of PPEP-1 expression (E.coli_control) did not result in imipenem hydrolysis (Fig. 3A). In a parallel assay, 1H-NMR was used to directly observe the opening (hydrolysis) of the lactam ring in imipenem (Fig. 3B). The chemical shifts of the peaks change as a result of this hydrolysis, with the protons closest to the ring opening undergoing the largest change. The H-6 proton of imipenem generates an adequately resolved multiplet at 3.42 ppm that decreases in intensity upon hydrolysis. Concomitantly, the doublet generated by the protons of the methyl group (H-9) move upfield, resulting in a decrease of the doublet at 1.3 ppm. After 10 minutes incubation of bacterial cells with imipenem, hydrolysis of imipenem was observed with E. coli_Axc grown in the presence, but not in the absence, of IPTG. Hydrolysis was also apparent for KPC, but not for E. coli cells expressing PPEP-1 (E.coli_control)), even after long incubation times (10 h). Under these conditions, imipenem hydrolysis was also observed for samples with E. coli cells harbouring the Axc expression plasmid grown in absence of IPTG, due to leaky expression from the inducible promoter.
Figure 3.
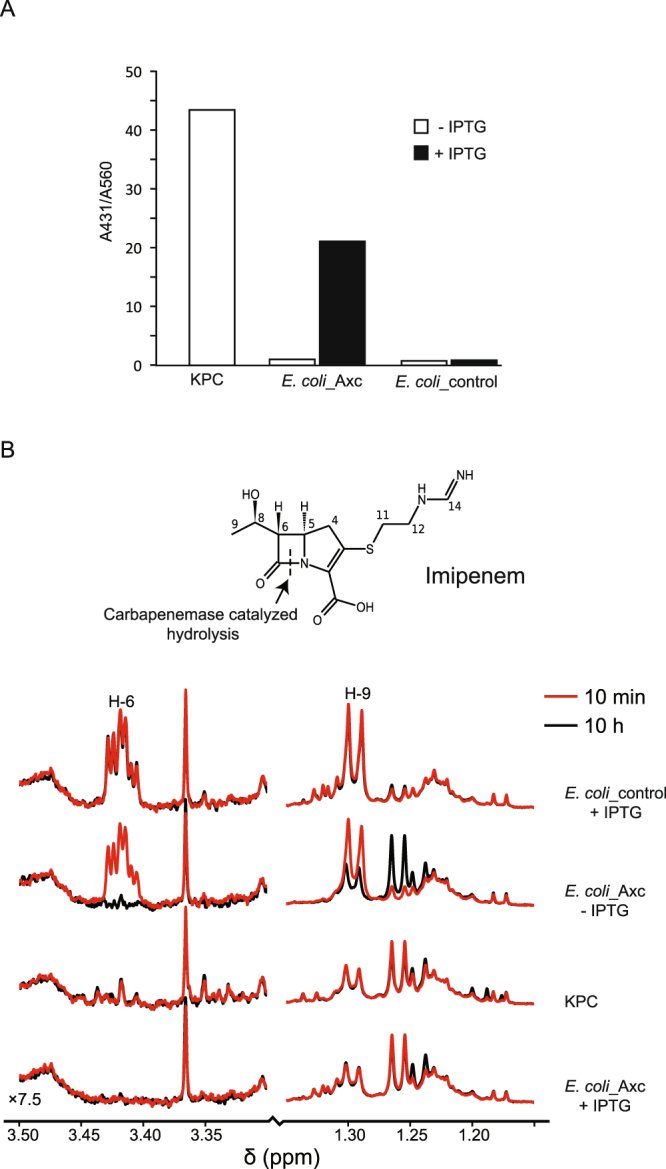
Axc has carbapenemase activity. (A) Axc was expressed in E. coli and cell extracts were tested for the ability to hydrolyse imipenem. Bars show the A431/A560 ratio, which is a measure for the shift in pH due to imipenem hydrolysis. (B) NMR-based identification of imipenem hydrolysis by Axc. The structure of imipenem is shown, with the proton numbering used in the spectrum. The line color indicates the incubation time (red, 10 min at room temperature or black, 10 hrs at 6 °C). Imipenem hydrolysis is accompanied by the loss of the H-6 multiplet at 3.4 ppm, and a shift in the H-9 doublet, resulting in a decrease of the doublet at 1.3 ppm. Strains used (see also Table 1): KPC (JC113): Carbapenem resistant Klebsiella pneumoniae. E. coli Axc (JC107): E. coli strain C43(DE3), containing plasmid pET21-Axc; E. coli_control (JC108): E. coli strain C43, containing plasmid pET21-PPEP-1 (Pro-Pro endopeptidase 1). IPTG: isopropyl β-D-1-thiogalactopyranoside.
Taken together, our data establish that Axc has carbapenemase activity.
Discussion
In this paper we identified a new resistance mechanism that explains the difference in meropenem susceptibility of two clinical isolates of A. xylosoxidans that were collected during treatment. Using a combination of comparative proteomic analyses and functional assays, we have shown that the class A beta-lactamase Axc is highly abundant in the meropenem resistant isolate in comparison to the susceptible isolate and that Axc has carbapenemase activity.
Detection of carbapenemases from sequence data is challenging. First, carbapenemases belong to different subgroups of beta-lactamases which have probably evolved by convergent evolution. Although sequence identities are moderate, most of the class A carbapenemases have a disulphide bridge between Cys-69 and Cys-238, but this is dispensable for activity against carbapenems31 and our data show that Axc does not contain these residues. Next, some studies also revealed a mechanism where beta-lactam trapping, without actual degradation, can be involved in resistance towards carbapenems when levels are sufficiently high. In such cases, concomitant loss of porins is often observed32,33. Thus, it is crucial to determine whether a beta-lactamase actually induces hydrolysis of carbapenems. Our NMR and pH-shift analyses data clearly demonstrate Axc-mediated opening of the beta-lactam ring in imipenem. Finally, many unexplained mechanisms of carbapenem resistance remain. For instance, a recent study demonstrated plasmid derived carbapenem resistance in Klebsiella pneumoniae strains which could not be explained by the most commonly found carbapenemases (KPC-type). Even though none of the plasmid-encoded genes were obvious candidates for the observed resistance towards carbapenems, several TEM-homologs were detected34. Though the prediction of the activity of a certain beta-lactamase against carbapenems is not straightforward, studies such as the present one highlight that mass spectrometry approaches can be used to gain insight in the mechanism of action and role of specific proteins in the observed phenotypes.
We do not know whether the high level of Axc is the only mechanism conferring meropenem resistance to our isolate of A. xylosoxidans. When expressed from an inducible promoter, Axc confers moderate resistance to carbapenems to E. coli; MIC values compared to the KPC strain suggest that Axc has a lower efficiency than KPC, but may also indicate lower overall levels of expression. Differing efficiencies in carbapenemases are well documented, to the point where the activity of a specific class, such as OXA-48, is difficult to detect but of great clinical importance35. Full biochemical characterization of Axc, including kinetic experiments, is subject to further study. Such experiments, in combination with crystallography analysis, will provide more insight in the activity of Axc against different beta-lactams and could resolve the structural characteristics of the binding pocket which facilitates its activity towards carbapenems. We note, however, that two other changes in the antibiogram between the meropenem susceptible and resistant isolates involve beta-lactam antibiotics. Augmentin (amoxicillin/clavulanate) resistance changed from intermediate (8 mg/L) to resistant (>32 mg/L), and piperacilline/tazobactam from susceptible (< = 4 mg/L) to intermediate resistance (8 mg/L). This suggests that Axc may have a broad substrate specificity and is insensitive to inhibition by clavulanate and tazobactam. Strikingly, both the meropenem-susceptible and meropenem-resistant isolates were resistant to imipenem (MIC values higher than the maximum concentration tested (32 mg/L)). This indicates that, notwithstanding the activity of Axc towards imipenem as presented here, imipenem resistance in the clinical isolates is not dependent on Axc. Differences in the sensitivities towards different carbapenems results from the chemical differences between the individual drugs36 and are often linked to the differential permeability of the outer cell membrane37,38.
The regulatory mechanism leading to higher levels of Axc expression are unclear. Sequence analyses showed that the meropenem susceptible and resistant A. xylosoxidans clinical isolates are highly similar, with no differences in the axc promotor and coding sequence, nor in its putative regulator AxcR and the axc-axcR intergenic region. The only SNP we identified is located in the gene encoding AxyZ, the TetR-type repressor of the axyXY-oprZ operon25. This leads to an amino acid substitution (V29G) in a region of AxyZ that is involved in DNA binding in other members of TetR family39. AxyX, AxyY and OprZ form an efflux pump of the resistance-nodulation division (RND) family and are predominantly found in aminoglycoside resistant Achromobacter species40–42. A recent paper showed higher expression levels of axyY in A. xylosoxidans strains containing AxyZ_Gly29, suggesting that this mutation leads to reduced repression of AxyZ43. Closer inspection of our proteomics data indicates that also AxyX and AxyY are more abundant in the resistant isolate (spectral counts of 62 vs 21 for AxyX and 10 vs 2 for AxyY in the results presented in Fig. 1 (see Table S2)). However, the difference is not as pronounced as found for Axc and more accurate quantitative proteomics experiments have to be performed to validate these data.
Mutations in TetR-like repressors have been linked to differences in carbapenem resistance44. For example, a 162 bp deletion in axyZ has been identified in certain carbapenem resistant strains45. However, even though AxyZ_Gly29 leads to higher expression of axyY, resulting in higher MIC values for aminoglycosides, fluoroquinolones and tetracyclines, no correlation between axyY expression and meropenem resistance was observed43. In line with this observation, a deletion of axyY in several A. xylosoxidans strains is reported to result in only a modest increase in the susceptibility towards carbapenems25. Taken together, it is likely that AxyXY-OprZ per se contributes little if any to the meropenem resistance phenotype of our clinical isolate. Instead, our data show a critical role for Axc and suggest that axc expression is regulated by AxyZ. If this is indeed the case, we postulate that the increase in the meropenem MIC for the ACH-CF-911V29G strain43 is accompanied by increased Axc levels.
From the clinical perspective, the development of resistance to meropenem within days following meropenem treatment is remarkable. Previous longitudinal analyses of different A. xylosoxidans isolates from one patient have revealed large phenotypic and genetic differences, for example in the resistance towards different classes of antibiotics but they were generally performed over longer time periods24,44,45. Such changes are believed to be the result of adaptive evolution of the initial strain which infected the patient, but there is also evidence that genetically different strains of A. xylosoxidans can co-exist within the same chronically infected individual46. Though we cannot exclude a co-infection, it is likely that the mutation in axyZ in our case occurred during treatment.
Finally, from a diagnostic point of view, the presence of Axc in both the sensitive and resistant A. xylosoxidans isolates complicates straightforward detection by molecular methods in the future and warrants detection based on protein abundance levels. Even though more elaborate mass spectrometry-based platforms clearly have potential for the detection of resistance and virulence proteins47,48, as also exemplified in our study, it will take more time and effort before they could find their way into the medical microbiology laboratory to complement the now well-established application of MALDI-ToF-MS for bacterial species identification.
Methods
Patient history
All procedures performed in studies involving human participants were in accordance with the ethical standards of LUMC Medical Ethical Committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
A 65-year old patient, diagnosed with chronic lymphocytic leukemia in 1989, underwent a non-myeloablative stem cell transplantation in July 2014. The patient was discharged on July 25th but readmitted on August 1st with a mucositis-associated neutropenic fever and blood cultures positive for Gemella haemolysans. The patient was treated with cefuroxim intravenously but developed pneumonia one week later. Analysis of a sputum sample showed a microscopically and culture confirmed fungal infection and abundant growth of a Gram-negative bacterium. This bacterium was identified as A. xylosoxidans by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. The patient was treated with liposomal amphotericin B for the fungal infection, but this did not result in improvement of the patient’s status. Due to clinical deterioration the patient received ceftazidim (3 dd 1 gram intravenously) between September 3rd and 9th to treat the ceftazidim sensitive A. xylosoxidans isolate. On September 10th, the antibiogram (Supplemental Table 1), revealed a multi-resistant character for the A. xylosoxidans isolate, but it was susceptible to meropenem. Hence, meropenem treatment was initiated (4 dd 1 gram intravenously) on September 10th. During treatment the patient developed a pneumothorax and died from septic shock 6 days later. Because of this, informed consent of the patient was not possible to obtain. We therefore anonymized all patient data and presented the data in such a form that it is not traceable to the patient.
Materials
MilliQ water was obtained from a Q-Gard 2 system (Merck Millipore, Amsterdam the Netherlands). Acetonitrile of LC-MS grade was obtained from Biosolve (Valkenswaard, the Netherlands). Porcine trypsin was purchased from Promega (Madison, WI). If not indicated otherwise, chemicals were from Sigma Aldrich (St Louis, MN, USA).
Susceptibility profiling of clinical isolates
The minimum inhibitory concentrations (MICs) for different antibiotic compounds on the clinical isolates (Clinical IDs: M 14073954-7 (first isolate), M 14076260-2 (second isolate)) were initially determined using a Vitek-2 system (bioMérieux, Marcy-l’Étoile, France). Several colonies of plate grown cultures were inoculated and suspended in 0.45% sterile physiological saline solution. Suspensions for testing had densities between of approx. 0.5 McFarland standards. The testing procedure was performed according to the manufacturer’s instructions.
Pure cultures of the susceptible and resistant isolates (AchroS and AchroR, respectively (Table 1)) were subjected to further susceptibility testing and used for the proteomic and genomic analyses. Etests (bioMérieux) were performed according to the recommendations from EUCAST (http://www.eucast.org/clinical_breakpoints/). EUCAST does not provide recommendations for the interpretation of MIC values or clinical breakpoints for Achromobacter xylosoxidans. Therefore, scoring was performed using locally developed protocols that are based on clinical breakpoints for other non-fermentative Gram-negative rods.
Mass spectrometry-based proteomics
Cells were grown in BHI (Oxoid, Basingstoke, UK). Cells were collected by centrifugation (4000 g, 5 min) from 1 mL cell culture and washed with phosphate- buffered saline (PBS, pH 7.4). Pellets were stored at −80 °C until further use.
Two different proteomics experiments were performed. For the first, cell extracts from A. xylosoxidans strains (AchroS and AchroR, Table 1) were prepared in LDS (Lithium dodecyl sulphate) sample buffer (Novex, Thermo Scientific) and put at 95 °C for 5 minutes for cell lysis and protein extraction. Proteins were separated on Novex precast 4–12% Bis-Tris gels (Thermo Scientific, Waltham, MA, USA) with MOPS (3-(N-morpholino)propanesulfonic acid) running buffer (Thermo Scientific). After overnight staining using a Colloidal Blue Staining Kit (Thermo Scientific), destained gel lanes were processed into 31 slices per lane. Gel pieces were sequentially washed with 25 mM ammonium bicarbonate and acetonitrile. Reduction and alkylation were performed with dithiothreitol (DTT, 10 mM, 30 minutes at 56 °C) and iodoacetamide (IAA, 55 mM, 20 min at room temperature) respectively. Following several washes with 25 mM ammonium bicarbonate and acetonitrile, bands were overnight digested with trypsin (12.5 ng/μl in 25 mM NH4HCO3). Digest solutions were lyophilized and reconstituted in 0.5% trifluoroacetic acid (TFA). Nano-LC separation was carried out using a Ultimate 3000 RSLCnano System equipped with an Acclaim PepMap RSLC column (C18, 75 µm × 15 cm with 2 µm particles, Thermo Scientific) preceded by a 2 cm Acclaim PepMap100 guard column (Thermo Scientific). Peptide elution was performed by applying a mixture of solvents A and B with solvent A being 0.1% formic acid (FA) in water and solvent B 0.1% FA in 80% acetonitrile (ACN). Peptides were eluted from the column with a multi-step gradient from 5% to 55% solvent B in 55 minutes (0–5 min 5%B, then in 15 min to 20%B and finally in 25 min till 55%B), at a constant flow rate of 300 nl min−1. MS analysis was performed employing a maXis Impact UHR-TOF-MS (Bruker Daltonics, Bremen, Germany) in data dependent MS/MS mode in the m/z 150–2200 range. The source settings were as follows: voltage 1500 V, dry gas 3.0 l.min−1, temperature 150 °C. Ten ions were selected at a time, based on relative abundance and subjected to collision-induced-dissociation with helium as collision gas. A 1 minute dynamic exclusion window was applied for precursor selection. The precursor isolation window and collision energy were m/z dependent.
For the second proteomics approach, in-solution digests were prepared as described previously47. In short, following cell disruption proteins were solubilized in 50% trifluoroethanol (TFE). Subsequent reduction with DTT and alkylation with IAA were performed prior to overnight tryptic digestion. Samples were lyophilized and reconstituted in 0.5% TFA for injection. The nano-LC system and solvents were the same as in the above experiment, but using an Acclaim PepMap RSLC column (C18, 75 µm × 50 cm with 2 µm particles, Thermo Scientific) with a 2 cm Acclaim PepMap100 guard column (Thermo Scientific). Peptides were eluted from the column with a multi-step gradient from 5% to 55% of solvent B in 180 minutes (0–5 min 5%B, then in 65 min till 13%B, then in 90 minutes till 35%B and finally in 20 min till 55%B), at a constant flow rate of 300 nl min−1. MS analysis was carried out as described above.
Mass spectrometry data export and spectral count analysis
Conversion of Bruker Impact files into mzXML format using CompassXport version 3.0.9, led to an initial total of 1,207,329 MS/MS and 444,565 MS/MS spectra, for the gel and total lysate based comparisons, respectively (in-solution total lysate digests were analysed twice and the data were merged). These spectra were searched using a concatenated forward and decoy strategy. The forward database was constructed from the 6386 unreviewed sequences from Uniprot for the organism A. xylosoxidans NH44784-1996 (November 2015) together with the cRAP contaminant sequences, as downloaded in January 2015. An in-house developed program, Decoy version V1.0.1-2-gfddc, that preserves homology, amino-acid frequency and peptide length distribution, was used with default flags to construct the decoy search space, which was concatenated to the forward sequences. The search against the resulting database was performed using Comet version 2014.02 rev.2, with precursor mass tolerance equal to 50 ppm and a fragment bin width of 0.05 Da, considering only fully tryptic digests with at most 2 missed cleavages. All cysteines were assumed carbamidomethylated, while methionine oxidation and N-terminal acetylation were regarded as variable modifications. The confidence of these results was assessed by means of Xinteract from the Trans Proteomics Pipeline suite version 4.8.0, retaining peptides longer than 6 amino acids and running in semi-parametric mode. A second in-house developed program, Pepxmltool version 2.5.1, was used to construct a protein quantification table based on spectral counting of only non-degenerate peptides (peptides mapping to a single protein) with corresponding q-value of at most 1%. Plots of the resistant versus susceptible quantifications of all proteins revealed for both methods a predominantly linear relationship, suggesting the applicability of an in-house developed program, Qntdiff version 0.1.1, to assign p-values to deviations from linear behaviour and provide lists of the most significantly differential protein expression levels between the two isolates. The programs Pepxmltool and Qntdiff are available from the authors on request.
Bacterial culture and genomic DNA preparation
AchroS and AchroR (Table 1) were cultured on trypcase soy agar plates (BioMérieux, Marcy l’Etoile, France), inoculated into liquid medium brain-heart-infusion (BHI) broth (Oxoid, Basingstoke, UK) and grown overnight (~16-hrs) at 37 °C. Cells were harvested, washed with phosphate-buffered saline (PBS, pH 7.4), and genomic DNA extraction was performed using a phenol-chloroform extraction as previously described49.
Whole genome sequencing and SNP calling
Paired-end multiplex libraries were created as previously described50. Sequencing was performed on an Illumina Hiseq 2000 platform (Illumina, San Diego, Ca, USA), with a read-length of 100 basepairs. High-throughput de novo assembly of sequenced genome was performed as previously described51,52. The assemblies are then automatically annotated using PROKKA53 with genus-specific databases from RefSeq54. To identify single nucleotide polymorphisms (SNPs), the Illumina sequence data of the meropenem-susceptible A. xylosoxidans isolate (AchroS) was mapped on the assembled genome of the meropenem-resistant isolate (AchroR) using SMALT software (http://smalt.sourceforge.net/), after which SNPs were determined as previously described50.
PCR and heterologous expression of Axc
To corroborate the proteomics results and confirm results from the whole genome sequence analysis, the axc ORF, the axc-axcR intergenic region and part of the axyZ ORF were Sanger sequenced at a commercial provider (Macrogen, Amsterdam, the Netherlands). PCR products (for primers see Table 3) were sequenced using the same primers as those used for generating the product from genomic DNA isolated from the A. xylosoxidans isolates. We have submitted the Axc sequence to Genbank, ID MF767301.
Table 3.
Primers used in this study. Axc: Achromobacter xylosoxidans carbapenemase. axcR: axc-associated regulator. axyZ: TetR-type repressor of the axyXY-oprZ operon36.
| Name | 5′ > 3′ sequence | Description |
|---|---|---|
| AxFor | 5′-GAATGACATGTTGACCCGAAGAACCTTCATTGCC-3′ | axc ORF forward |
| AxRev | 5′-GCCGGATCCCTAGCCCAATGCCGCCACCAGCCTG-3′ | axc ORF reverse |
| Operonfor | 5′-CTGAGCATCAGGAAGCGTT-3′ | axc-axcR intergenic region forward |
| Operonrev | 5′-TCGAAGGATTCGGACAACAC-3′ | axc-axcR intergenic region reverse |
| AC_RR_fw | 5′-AGAAGAATCCCAACGCACCC-3′ | Confirmation of SNP in axyZ, forward |
| AC_RR_rv | 5′-TCGAGGCATACAGCGATTCC-3′ | Confirmation of SNP in axyZ, reverse |
| Axfor2 | 5′-AAACATATGTTGACCCGAAGAACCTTCATTG-3′ | Cloning axc ORF into pET21B forward |
| Axrev3 | 5′-AAACTCGAGGCCCAATGCCGCCACCAGCC-3′ | Cloning axc ORF into pET21B reverse |
To construct an E. coli strain expressing Axc with a C-terminal 6xHis-tag from an IPTG (isopropyl β-D-1-thiogalactopyranoside) inducible promoter, the axc open reading frame was amplified using primers Axfor2 and Axrev3 (Table 3), using Accuzyme polymerase (GC Biotech, Alphen aan den Rijn, The Netherlands) and genomic DNA from A. xylosoxidans (AchroR) as a template. The amplified PCR product was digested using NdeI (Bioké, Leiden, The Netherlands) and XhoI (Roche, Almere, The Netherlands), and cloned into similarly digested pET-21b(+), yielding plasmid pET21B/Axc. The axc expression region was confirmed using Sanger sequencing. Expression of Axc-his6 was carried out in E. coli C43(DE3)27 in Luria-Bertani broth (Affymetrix, Cleveland, OH, USA) with ampicillin (50 µg/mL) and 1 mM IPTG (GC Biotech, Alphen aan de Rijn, the Netherlands) for 3 hrs at 37 °C and verified by immunoblotting using anti-His antibody (Agilent Technologies, Santa Clara, CA, USA).
Susceptibility testing of E. coli expressing Axc
Minimal inhibitory concentration (MIC) values for the carbapenems imipenem (Etest, BioMérieux) and meropenem (microbroth dilution) were established for E. coli C43/pET21B-Axc (strain JC107, Table 1) in the presence and absence of 1 mM IPTG. For imipenem, cells were grown overnight in LB broth at 37 °C in the presence of ampicillin (50 µg/mL). The overnight cultures were diluted 1:100 in LB broth with ampicillin and grown to mid logarithmic phase (OD600nm ~ 0.5). Two hundred μl of bacterial culture was spread on LB-ampicillin (50 µg/mL) plates (with or without 1 mM IPTG) and an Imipenem Etest (BioMérieux) was applied. MIC values were determined after 24 h incubation at 37 °C. The meropenem MIC values were established by microbroth dilution. Bacterial cultures in logarithmic phase (OD600nm ~ 0.5) were diluted into LB-ampicillin medium to an OD600nm of 0.05 in the presence or absence of 1 mM IPTG and subsequently seeded in a 96-well plate. A two-fold serial dilution of meropenem (starting at 12.5 µg/mL) was made by adding equal amounts of meropenem (25 µg/mL) to the first row, from which a two-fold dilution series was made in the rest of the plate. Samples were investigated for growth by measuring the OD600nm after 24 hrs incubation at 37 °C, while shaking. The MIC was the lowest concentration of meropenem at which no growth was observed. As controls for our assays, a carbapenem resistant Klebsiella pneumoniae clinical isolate (KPC; JC113) and an unrelated expression construct (JC108; which expresses PPEP-128 in an IPTG-dependent manner in the same E. coli C43 background, Table 2) were included.
Colorimetric imipenem hydrolysis assay
Overnight bacterial cultures were diluted 100 fold in LB-ampicillin (50 µg/mL) medium and grown to exponential growth phase at 37 °C while shaking. At the time of induction, the OD600nm was determined, the cultures were split in two and 1 mM IPTG was added to one of the cultures, followed by a further incubation for 3 hrs at 37 °C while shaking. At T = 3 h, the OD600nm was determined and cells were harvested by centrifugation (4000 g, 5 min) and stored at −20 °C overnight. Cells were resuspended in water to yield equal densities based on measured OD600nm values. Then, 7.5 µL of bacterial suspension was mixed with 25 µL of imipenem/phenol red/ZnSO4 solution (3 mg/mL imipenem, 0.35% (wt/vol) phenol red, pH 7.8, 70 µM ZnSO4) and incubated at 37 °C for 1 hr. Conversion of imipenem leads to a pH drop that can be visualized by the color change of the buffer from red to yellow29. To quantify this effect, the UV-Vis spectrum was determined with a Nanodrop ND1000 spectrophotometer (Thermo Scientific) and the ratio between the absorption peaks at 431 and 560 nm was taken as a measure of imipenem hydrolysis.
NMR spectroscopy of imipenem conversion
All proton nuclear magnetic resonance (1H-NMR) experiments were performed on a 600 MHz Bruker Avance II spectrometer (Bruker BioSpin, Karlsruhe, Germany) equipped with a 5-mm triple resonance inverse (TCI) cryogenic probe head with a Z-gradient system and automatic tuning and matching. All experiments were recorded at 310 K. Temperature calibration was done before each batch of measurements55. The duration of the π/2 pulses was automatically calibrated for each individual sample using a homonuclear-gated nutation experiment on the locked and shimmed samples after automatic tuning and matching of the probe head. The samples were prepared by adding 70 µL imipenem aqueous solution (5 mg/mL) to 280 µL milliQ water. This solution was mixed with 350 µL 75 mM phosphate buffer (pH 7.4) in water/deuterium oxide (80/20) containing 4.6 mM sodium 3-[trimethylsilyl] d4-propionate. Twenty µL of bacterial cell suspension were added and the sample was mixed. Samples were manually transferred into 5-mm SampleJet NMR tubes. The cell suspension samples were kept at 6 °C on a SampleJet sample changer while queued for acquisition. For water suppression, presaturation of the water resonance with an effective field of γB1 = 25 Hz was applied during the relaxation delay. A 1D-version of the NOESY (Nuclear Overhauser effect spectroscopy) experiment was performed with a relaxation delay of 4 seconds. A NOESY mixing time of 10 ms was used during which the water resonance was irradiated with the presaturation RF field. After applying 4 dummy scans, a total of 98,304 data points covering a spectral width of 18,029 Hz were collected using 16 scans. The Free Induction Decay was zero-filled to 131,072 complex data points, and an exponential window function was applied with a line broadening factor of 0.3 Hz before Fourier transformation. The spectra were automatically phased and baseline corrected.
Bioinformatic analysis
Comparison of Axc with other class A beta lactamases was performed by multiple alignment using the Geneious 9.0 (Biomatters Ltd, Auckland, New Zealand) software algorithm for Global alignment with free end gaps, cost Matrix Blosum62. The tree was then built using Jukes-Cantor genetic distance model with the Neighbor Joining tree build method.
Data availability
Illumina raw reads were deposited at the European Nucleotide Archive (ENA). Study ID: PRJEB19781. Sample IDs: ERS1575148 (AchroR) and ERS1575149 (AchroS).
Electronic supplementary material
Acknowledgements
This research was financially supported by The Netherlands organisation of scientific research (NWO, ZonMW grant number 50-51700-98-142).
Author Contributions
F.F., H.C.v.L., S.T.N., E.J.K. and P.J.H. designed the research. F.F., J.C., C.W.K., I.D., N.K., A.V., H.C.v.L. and P.J.H. performed experiments. F.F., A.A.H., J.C., C.W.K., W.K.S., M.G., N.K., T.D.L. A.V. and P.J.H. analyzed data. F.F., J.C., W.K.S., H.C.v.L., E.J.K. and P.J.H. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26079-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.van der Bij AK, Pitout JD. The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother. 2012;67:2090–2100. doi: 10.1093/jac/dks214. [DOI] [PubMed] [Google Scholar]
- 2.Marston HD, Dixon DM, Knisely JM, Palmore TN, Fauci AS. Antimicrobial Resistance. Jama. 2016;316:1193–1204. doi: 10.1001/jama.2016.11764. [DOI] [PubMed] [Google Scholar]
- 3.Jovetic S, Zhu Y, Marcone GL, Marinelli F, Tramper J. beta-Lactam and glycopeptide antibiotics: first and last line of defense? Trends Biotechnol. 2010;28:596–604. doi: 10.1016/j.tibtech.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 4.da Silva RM, Traebert J, Galato D. Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae: a review of epidemiological and clinical aspects. Expert Opin Biol Ther. 2012;12:663–671. doi: 10.1517/14712598.2012.681369. [DOI] [PubMed] [Google Scholar]
- 5.Hong DJ, et al. Epidemiology and Characteristics of Metallo-beta-Lactamase-Producing Pseudomonas aeruginosa. Infect Chemother. 2015;47:81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zmarlicka MT, Nailor MD, Nicolau DP. Impact of the New Delhi metallo-beta-lactamase on beta-lactam antibiotics. Infect Drug Resist. 2015;8:297–309. doi: 10.2147/IDR.S39186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Potron A, Nordmann P. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother. 2012;67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 8.Findlay J, Hamouda A, Dancer SJ, Amyes SG. Rapid acquisition of decreased carbapenem susceptibility in a strain of Klebsiella pneumoniae arising during meropenem therapy. Clin Microbiol Infect. 2012;18:140–146. doi: 10.1111/j.1469-0691.2011.03515.x. [DOI] [PubMed] [Google Scholar]
- 9.Hansen CR, et al. Inflammation in Achromobacter xylosoxidans infected cystic fibrosis patients. J Cyst Fibros. 2010;9:51–58. doi: 10.1016/j.jcf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 10.De Baets F, Schelstraete P, Van Daele S, Haerynck F, Vaneechoutte M. Achromobacter xylosoxidans in cystic fibrosis: prevalence and clinical relevance. J Cyst Fibros. 2007;6:75–78. doi: 10.1016/j.jcf.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 11.LiPuma JJ. The Changing Microbial Epidemiology in Cystic Fibrosis. Clinical Microbiology Reviews. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkins MD, Floto RA. Emerging bacterial pathogens and changing concepts of bacterial pathogenesis in cystic fibrosis. Journal of Cystic Fibrosis. 2015;14:293–304. doi: 10.1016/j.jcf.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Cerezo J, et al. Achromobacter xylosoxidans bacteremia: a 10-year analysis of 54 cases. Eur J Clin Microbiol Infect Dis. 2003;22:360–363. doi: 10.1007/s10096-003-0925-3. [DOI] [PubMed] [Google Scholar]
- 14.Lambiase A, et al. Achromobacter xylosoxidans respiratory tract infection in cystic fibrosis patients. Eur J Clin Microbiol Infect Dis. 2011;30:973–980. doi: 10.1007/s10096-011-1182-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saiman L, et al. Identification and antimicrobial susceptibility of Alcaligenes xylosoxidans isolated from patients with cystic fibrosis. J Clin Microbiol. 2001;39:3942–3945. doi: 10.1128/JCM.39.11.3942-3945.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi Y, Poirel L, Paterson DL, Nordmann P. Characterization of a naturally occurring class D beta-lactamase from Achromobacter xylosoxidans. Antimicrob Agents Chemother. 2008;52:1952–1956. doi: 10.1128/AAC.01463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amoureux L, et al. Detection of Achromobacter xylosoxidans in Hospital, Domestic, and Outdoor Environmental Samples and Comparison with Human Clinical Isolates. Applied and Environmental Microbiology. 2013;79:7142–7149. doi: 10.1128/AEM.02293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nichols KR, Knoderer CA, Jackson NG, Manaloor JJ, Christenson JC. Success With Extended-Infusion Meropenem After Recurrence of Baclofen Pump-Related Achromobacter Xylosoxidans Meningitis in an Adolescent. J Pharm Pract. 2015;28:430–433. doi: 10.1177/0897190015585757. [DOI] [PubMed] [Google Scholar]
- 19.Amoureux L, et al. Epidemiology and resistance of Achromobacter xylosoxidans from cystic fibrosis patients in Dijon, Burgundy: first French data. J Cyst Fibros. 2013;12:170–176. doi: 10.1016/j.jcf.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, et al. IMP-1 encoded by a novel Tn402-like class 1 integron in clinical Achromobacter xylosoxidans, China. Sci Rep. 2014;4:7212. doi: 10.1038/srep07212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto M, et al. Molecular characterization of IMP-type metallo-beta-lactamases among multidrug-resistant Achromobacter xylosoxidans. J Antimicrob Chemother. 2012;67:2110–2113. doi: 10.1093/jac/dks179. [DOI] [PubMed] [Google Scholar]
- 22.Hu Y, et al. Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrobial agents and chemotherapy. 2015;59:1152–1161. doi: 10.1128/AAC.04260-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Zee A, et al. Multi-centre evaluation of real-time multiplex PCR for detection of carbapenemase genes OXA-48, VIM, IMP, NDM and KPC. BMC Infect Dis. 2014;14:27. doi: 10.1186/1471-2334-14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakobsen TH, et al. Complete genome sequence of the cystic fibrosis pathogen Achromobacter xylosoxidans NH44784-1996 complies with important pathogenic phenotypes. PLoS One. 2013;8:e68484. doi: 10.1371/journal.pone.0068484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bador J, Amoureux L, Blanc E, Neuwirth C. Innate aminoglycoside resistance of Achromobacter xylosoxidans is due to AxyXY-OprZ, an RND-type multidrug efflux pump. Antimicrob Agents Chemother. 2013;57:603–605. doi: 10.1128/AAC.01243-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walther-Rasmussen J, Hoiby N. Class A carbapenemases. J Antimicrob Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 27.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 28.Hensbergen PJ, et al. Clostridium difficile secreted Pro-Pro endopeptidase PPEP-1 (ZMP1/CD2830) modulates adhesion through cleavage of the collagen binding protein CD2831. FEBS Lett. 2015;589:3952–3958. doi: 10.1016/j.febslet.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 29.Nordmann P, Poirel L, Dortet L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2012;18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamma PD, et al. Comparison of 11 Phenotypic Assays for Accurate Detection of Carbapenemase-Producing Enterobacteriaceae. J Clin Microbiol. 2017;55:1046–1055. doi: 10.1128/JCM.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith CA, et al. Role of the Conserved Disulfide Bridge in Class A Carbapenemases. J Biol Chem. 2016;291:22196–22206. doi: 10.1074/jbc.M116.749648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goessens WH, et al. Antibiotic trapping by plasmid-encoded CMY-2 beta-lactamase combined with reduced outer membrane permeability as a mechanism of carbapenem resistance in Escherichia coli. Antimicrob Agents Chemother. 2013;57:3941–3949. doi: 10.1128/AAC.02459-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Boxtel R, Wattel AA, Arenas J, Goessens WH, Tommassen J. Acquisition of Carbapenem Resistance by Plasmid-Encoded-AmpC-Expressing Escherichia coli. Antimicrob Agents Chemother. 2017;61:01413–01416. doi: 10.1128/AAC.01413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerqueira GC, et al. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci USA. 2017;114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koroska F, et al. Comparison of phenotypic tests and an immunochromatographic assay and development of a new algorithm for OXA-48-like detection. J Clin Microbiol. 2016;55:877–883. doi: 10.1128/JCM.01929-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Gamal MI, et al. Recent updates of carbapenem antibiotics. Eur J Med Chem. 2017;131:185–195. doi: 10.1016/j.ejmech.2017.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Bornet C, et al. Imipenem and expression of multidrug efflux pump in Enterobacter aerogenes. Biochem Biophys Res Commun. 2003;301:985–990. doi: 10.1016/S0006-291X(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 38.Yang FC, Yan JJ, Hung KH, Wu JJ. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J Clin Microbiol. 2012;50:223–226. doi: 10.1128/JCM.01263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos JL, et al. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anes J, McCusker MP, Fanning S, Martins M. The ins and outs of RND efflux pumps in Escherichia coli. Front Microbiol. 2015;6:587. doi: 10.3389/fmicb.2015.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bador J, et al. First description of an RND-type multidrug efflux pump in Achromobacter xylosoxidans, AxyABM. Antimicrob Agents Chemother. 2011;55:4912–4914. doi: 10.1128/AAC.00341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bador J, et al. Distribution of innate efflux-mediated aminoglycoside resistance among different Achromobacter species. New Microbes New Infect. 2016;10:1–5. doi: 10.1016/j.nmni.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bador J, et al. Role of AxyZ Transcriptional Regulator in Overproduction of AxyXY-OprZ Multidrug Efflux System in Achromobacter Species Mutants Selected by Tobramycin. Antimicrob Agents Chemother. 2017;61:e00290–00217. doi: 10.1128/AAC.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ormerod KL, George NM, Fraser JA, Wainwright C, Hugenholtz P. Comparative genomics of non-pseudomonal bacterial species colonising paediatric cystic fibrosis patients. PeerJ. 2015;3:e1223. doi: 10.7717/peerj.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ridderberg W, Nielsen SM, Norskov-Lauritsen N. Genetic Adaptation of Achromobacter sp. during Persistence in the Lungs of Cystic Fibrosis Patients. PLoS One. 2015;10:e0136790. doi: 10.1371/journal.pone.0136790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dupont C, Jumas-Bilak E, Michon AL, Chiron R, Marchandin H. Impact of High Diversity of Achromobacter Populations within Cystic Fibrosis Sputum Samples on Antimicrobial Susceptibility Testing. J Clin Microbiol. 2017;55:206–215. doi: 10.1128/JCM.01843-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleurbaaij F, et al. Capillary-electrophoresis mass spectrometry for the detection of carbapenemases in (multi-)drug-resistant Gram-negative bacteria. Anal Chem. 2014;86:9154–9161. doi: 10.1021/ac502049p. [DOI] [PubMed] [Google Scholar]
- 48.Charretier Y, et al. Rapid Bacterial Identification, Resistance, Virulence and Type Profiling using Selected Reaction Monitoring Mass Spectrometry. Sci Rep. 2015;5:13944. doi: 10.1038/srep13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knetsch CW, et al. Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 2014;19:20954. doi: 10.2807/1560-7917.ES2014.19.45.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Page AJ, et al. Robust high-throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microb Genom. 2016;2:e000083. doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 54.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Findeisen M, Brand T, Berger S. A 1H-NMR thermometer suitable for cryoprobes. Magn Reson Chem. 2007;45:175–178. doi: 10.1002/mrc.1941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina raw reads were deposited at the European Nucleotide Archive (ENA). Study ID: PRJEB19781. Sample IDs: ERS1575148 (AchroR) and ERS1575149 (AchroS).