Abstract
Background
The dramatic increase of intervertebral disc degeneration (IDD) is considered to be a major cause of discogenic low back pain. The current study focused on the regulatory function of microRNA-194 (miR-194) on lipopolysaccharide (LPS)-induced inflammatory response in nucleus pulposus (NP) cells.
Material/Methods
LPS was used to treat NP cells to induce inflammatory responses. MiRNA and gene expression were detected by quantitative PCR. Proteins and protein expression levels were detected by Western blot and ELISA kit. Dual luciferase reporter assay was applied to identify the correlation between an miR-194- and TNF receptor-associated factor 6 (TRAF6) and to test NF-κB activity.
Results
MiR-194 expression was reduced in LPS-induced NP cells. Both miR-194 overexpression and miR-194 inhibitor could regulate extracellular matrix (ECM) genes expression (Aggrecan and collagen II), MMP3, MMP13, ADAMTS4, and ADAMTS5, as well as inflammatory cytokines-associated genes (TNF-α, IL-1, IL-6, PGE2). Through a further study of the molecular mechanism, miR-194 was proved to be involved in the regulation of TRAF6 and its downstream signal molecule, nuclear factor-kappa B (NF-κB).
Conclusions
Finding of our study suggest that miR-194 can inhibit LPS-induced inflammatory response in NP cells of the intervertebral disc (IVD) by targeting TRAF6, which may contribute development of IDD biological therapy.
MeSH Keywords: Intervertebral Disc Degeneration, Lipopolysaccharides, MicroRNAs, TNF Receptor-Associated Factor 6
Background
Intervertebral disc degeneration (IDD) is a widely recognized clinical characteristic of chronic low back pain, and is a secondary symptom and characteristic of lumbar disc herniation, spinal stenosis, and spinal deformity [1]. IDD is considered as a global health burden with severe healthcare and socioeconomic consequences, but its pathogenesis remains obscure. With age, intervertebral disc (IVD) will degenerate naturally, and studies have suggested that the degenerative process is accelerated or aggravated by environmental effects and genetic factors [2,3].
Aggrecan is the main proteoglycan in nucleus pulposus (NP) cells, which play a crucial role in the maintenance of IVD height and anti-compression pressure [4]. Collagen II is the major component of fibers in NP cells. Imbalance between anabolic and catabolic gene expression in chondrocytes of NP can result in the occurrence of IDD [5]. Upregulation of some proinflammatory cytokines and extracellular matrix (ECM)- degrading enzymes leads to the homeostasis of catabolism and anabolism in the ECM of IVD, shifting toward a degenerative and catabolic state, and finally causes the breakdown of ECM components, which includes collagen and proteoglycans (PGs) [6,7]. For instance, TIMP3 is an inhibitor of aggrecanase, whose expression is negatively associated with the severity of IDD, and the imbalance between TIMP3 and aggrecanase can have an adverse effect on IVD [8].
Moreover, many inflammatory cytokines, including Interleukin-1β (IL-1β), IL-6, and IL10, and tissue necrosis factor-α (TNF-α), are also involved in the pathogenesis of IDD. Unlike MMPs or ADAMTSs, these inflammatory cytokines aggravate IDD by inducing inflammatory substances instead of degrading ECM of IVD directly. Lipopolysaccharide (LPS) has been identified as an inflammatory mediator that engages in the process of IDD. LPS is capable of inducing variety of proinflammatory cytokines and matrix-degrading enzymes (including MMP-3, MMP-13, ADAMTS-4, and ADAMTS-5), further decreasing PGs content and promoting the occurrence of IDD [9,10].
MicroRNAs (miRNAs) are a type of small noncoding RNA that can bind to target genes through special sequences to repress gene expression. It has been demonstrated that some miRNAs have a key role in inflammatory response of monocyte-macrophages, such as miR-155, miR-146a, and miR-223 [11,12]. In addition, the role of miRNAs in the pathogenesis of IVD has been reported in previous studies [13]. It has been suggested that the expression of miR-194 in cord blood granulocytes of newborn infants was downregulated after LPS induction, which indicates the role of miR-194 in the LPS-stimulated TLR signaling pathway [14]. Another study reported that miR-194 can regulate the palmitic acid-induced TLR4 inflammatory response via targeting TNF receptor-associated factor 6 (TRAF6) in human THP-1 cells [14]. Furthermore, miR-194 is able to regulate the LPS-induced NF-κB signal by mediating TLR4 expression. Collectively, miR-194 is an important miRNA involved in LPS-induced inflammatory responses. However, the role of miR-194 in IDD has not been reported.
In the present study, we aimed to explore the role of miR-194 in LPS-induced inflammatory responses in NP cells and to investigate its regulatory mechanism.
Material and Methods
Cell extraction and culture
Sprague-Dawley (SD) rats aged 6 weeks were purchased from the Experimental Animal Center of Shanghai (Shanghai, China). Animal experiments were approved by the Ethics Committee of Yantai YEDA Hospital. Under sterile conditions, the rat lumber vertebrae were extracted using scalpels, and NP was extracted under a microscope. The NP was washed 3 times, digested with trypsin and collagenase II in turn for 30 min and 3 h, respectively, then filtered with a 100-um filter screen and centrifuged for 5 min at 2000 rpm. NP cells were separated and collected, and cultured to passage 2-3 by DMEM complete medium supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich Chemical Company, St Louis MO, USA), 100 U/ml penicillin (Sigma-Aldrich Chemical Company, St Louis MO, USA), and 100 mg/ml streptomycin (Sigma-Aldrich Chemical Company, St Louis MO, USA).
Cell transfection
NP cells were transfected with miR-194 mimics (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., Shanghai, China), miR-194 inhibitor (Shanghai Sangon Biological Engineering Technology & Services Co., Ltd., Shanghai, China), and TRAF6 over-expressing plasmids. Plasmids over-expressing TRAF6 were constructed as follows: firstly, the open reading frame (ORF) of TRAF6 was cloned from cDNA of NP cells. Then the cloned fragments were combined with pEGFP-N3 vector (Promega Corp., Madison, Wisconsin, USA) to construct pEGFP-N3-TRAF6. Liposome 2000 (Invitrogen Inc., Carlsbad, CA, USA) was used for transient transfection of NP cells. LPS (10 ug/ml) was used to induce NP cells for various lengths of time.
RNA isolation and quantitative polymerase chain reaction (q-PCR)
miRNA and total RNA of NP cells were selected using the mirPremier® microRNA Isolation Kit (Sigma-Aldrich Chemical Company, St Louis MO, USA). Total RNA was reversely transcribed into first-strand cDNA using the First-Strand cDNA Synthesis Kit (Takara Biotechnology Ltd., Dalian, China). To determine the miR-194 expression, synthetase of the First-Strand cDNA Synthesis Kit (Takara Biotechnology Ltd., Dalian, China) was combined with specific reverse transcription primers of miR-194 to generate cDNA of miRNA. The q-PCR was applied to determine gene expression by using the SYBR Premix Ex Taq kit (Takara Biotechnology Ltd., Dalian, China) and ABI Prism 7500 Fast Real-Time PCR system (Applied Biosystems, Grand Island, NY, USA). The experiment was preformed strictly according to the kit instructions. Each reaction system contained: 10 μL SYBR Premix Ex Taq, 0.4 μL 10 μM downstream primers, 0.4 μL 10 μM upstream primers, 2 μL templates, and 7.2 μL sterile solution. For the determination of gene expression, the reaction procedure was: pre-denaturation at 95°C for 30 s and 40 cycles of 95°C for 3 s, and 60°C for 30 s. For the determination of miR-194, the reaction procedure was: pre-denaturation at 95°C for 10 min; denaturation at 95°C for 15 s, annealing at 60°C for 1 min, and extension at 72°C for 2 min (40 cycles). Gene expression level was calculated by 2−ΔΔct method. β-actin and U6 were used as the internal references. All primer sequences are shown in Supplementary Table 1.
ELISA determination
Expression levels of ECM degrading enzymes and inflammatory cytokines-associated genes were measured by use of an ELISA regent kit (R&D Systems, Minneapolis, MN, USA). The experiment was carried out strictly according to the kit instructions.
Western blotting
The obtained NP cells were washed 2–3 times with cold phosphate-buffered saline (PBS) (Sigma-Aldrich, St. Louis, MO, USA), and then lysed with RIPA lysis buffer (Sigma-Aldrich, St. Louis, MO, USA) to extract total protein. The concentration of the total protein was determined by use of a BCA kit (Thermo Fisher Scientific, San Jose, CA, US). A volume of 20 mg protein sample was drawn to the gel, separated by 10% SDS-PAGE, then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore Corp., Bedford, MA, USA). The transfer membrane was sealed for 1 h with 5% skim milk at room temperature, washed once with PBS, and incubated overnight with primary antibodies at 4°C. The primary antibodies were as follows: anti-Aggrecan (ab36861, 1: 200, Abcam, Cambridge, MA, USA), anti-TRAF6 (ab62488, 1: 1000, Abcam, Cambridge, MA, USA), anti-Collage II (ab185430, 1: 200, Abcam, Cambridge, MA, USA), anti-GAPDH (ab9484, 1: 1000, Abcam, Cambridge, MA, USA). Following that, the transfer membrane was washed 3 times by TBST (TBS with Tween20) (Sigma-Aldrich, St. Louis, MO, USA). Secondary antibodies combined with HRP (ab131368, 1: 1000, Abcam, Cambridge, MA, USA) were incubated with the transfer membrane for 1 h. Protein bands were detected with an enhanced chemiluminescence assay kit (Thermo Fisher Scientific, San Jose, CA, USA). Images were captured using a ChemiDoc™ XRS + image analyzer (Bio-Rad Laboratories, Inc. CA, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody served as the internal reference.
Luciferase assay
TargetScanHuman 7.0 (http://www.targetscan.org/) and miRBase (http://www.mirbase.org/) microRNA databases were used to identify the target genes of miR-194. The primers of TRAF6 3′UTR and mutant TRAF6 3′UTR that may contain binding sites of miR-194 were amplified by PCR and then cloned into pMIR-REPORT Luciferase miR Expression Reporter Vector (Applied Biosystems, Grand Island, NY, USA), respectively. Then, the NP cells were transfected with the 2 types of vectors combined with Renilla vector (Promega Corp., Madison, Wisconsin, USA) and miR-194 mimics or negative controls, respectively. The luciferase was detected by use of luciferase reporter assay reagents (Promega Corp., Madison, Wisconsin, USA).
Determination of NF-κB activity
One day before the experiment, the NP cells transfected with miR-194 mimics or miR-194 inhibitor were placed into a 96-well plate at 1×104 cells/well. Co-transfection was performed by liposome 2000 (Invitrogen Inc., Carlsbad, CA, USA) with 20 ng pGL4.32[luc2P/NF-κB-RE/Hygro] vector and 5 ng pRL-TK vector in each well (Promega Corp., Madison, Wisconsin, USA). After that, the cells were cultured for 24 h and then collected. The luciferase was detected by use of the Dual Luciferase Reporter Assay System (Promega Corp., Madison, Wisconsin, USA).
Statistical analysis
Data are expressed as the mean ± standard deviation. All data were processed using SPSS 20.0 software (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance was performed for multiple comparisons, and differences discrepancies between 2 groups were compared using the unpaired t test for parametric data or the Mann-Whitney rank sum test for nonparametric data. P<0.05 was considered to indicate a statistically significant difference.
Results
MiR-194 was decreased in the LPS-induced NP cells
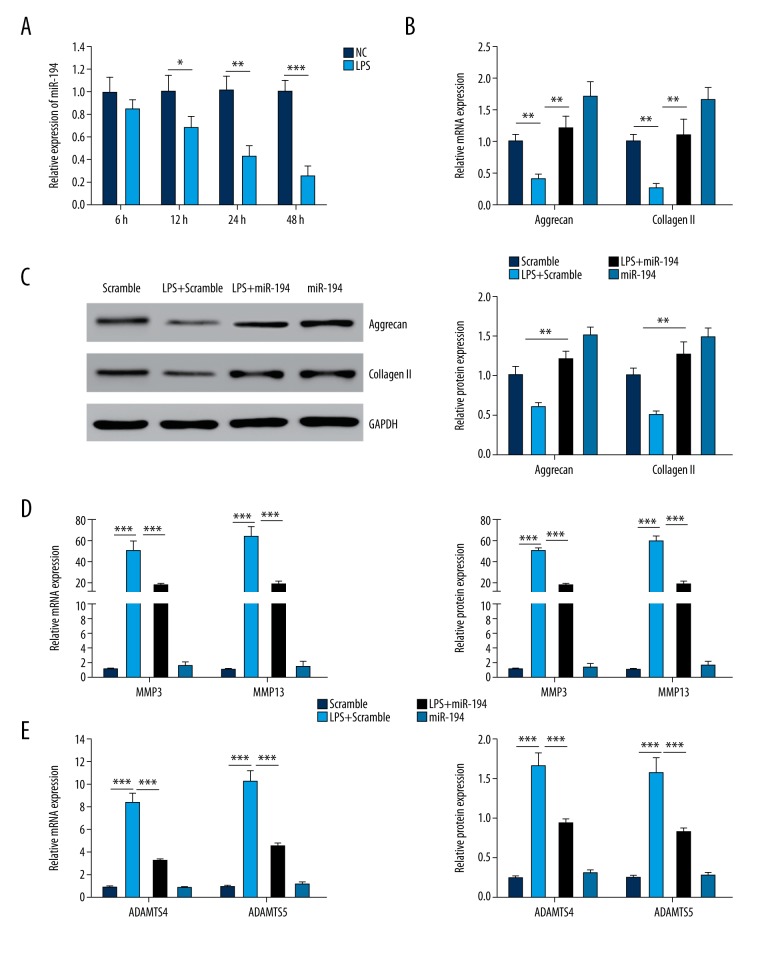
The NP cells were induced by 10 μg/ml LPS. The results demonstrated that miR-194 in the NP cells was significantly lower than that in the control group after 12 h, and was further reduced after 24 and 48 h compared with the control group (P<0.05; Figure 1A).
Figure 1.
The gene expression of miR-194 in NP cells. (A) The gene expression of miR-194 was suppressed by LPS treatment in NP cells (P<0.01). (B) miR-194 stimulated the mRNA expression of Aggrecan and Collagen II (P<0.01). (C) miR-194 stimulated the protein expression of Aggrecan (P<0.01). (D) miR-194 suppressed the expression of MMP3 and MMP13 induced by LPS (P<0.01, P<0.001). (E) miR-194 suppressed the expression of ADAMTS4 and ADAMTS5 induced by LPS (P<0.01, P<0.001). ** P<0.01; *** P<0.001.
Overexpressed miR-194 regulated the expression of ECM genes and synthesis of ECM-degrading enzymes
The experiments revealed that after LPS induction, the gene expression of Aggrecan and Collagen II was notably decreased in the NP cells in the LPS + scramble group, and transfection of miR-194 suppressed the down-regulation of LPS-induced Aggrecan and Collagen II (P<0.01; Figure 1B). We used Western blotting to evaluate the protein levels of Aggrecan and Collagen II. The trend of the result was similar to that of q-PCR. As shown in Figure 1C, LPS induced Aggrecan and Collagen II protein down-expression, and miR-194 significantly reversed the decrease of Aggrecan and Collagen II.
To confirm the regulatory effect of miR-194 on the expression of ECM degrading enzymes (MMP3, MMP13, ADAMTS4, and ADAMTS5) in NP cells, the PCR and ELISA assays were used to measure the mRNA and protein levels, respectively, of various matrix-degrading enzymes. The results showed that the gene expression was significantly increased in the LPS + scramble and LPS + miR-194 groups compared with the scramble group, and was remarkably inhibited in the miR-194 mimics group (P<0.01, P<0.001; Figure 1D, 1E). miR-194 significantly suppressed the increase of these degrading enzymes induced by LPS (P<0.001; Figure 1D, 1E).
Overexpressed miR-194 inhibited the expression of inflammatory cytokines-associated genes
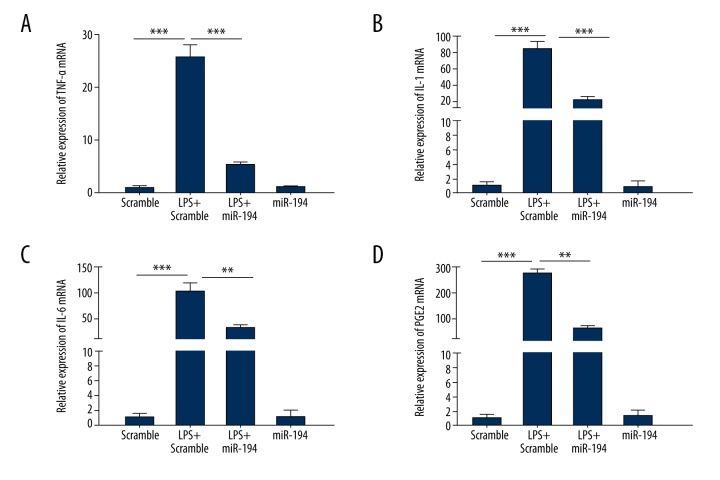
To identify the regulatory effect of miR-194 on inflammatory cytokines-associated genes in LPS-induced NP cells, the expression of related genes was determined. The experiment showed that the presence of LPS (10 ug/ml) markedly increased the mRNA of TNF-α, IL-1, IL-6, and PGE2 compared with the scramble group. Upon transfection of miR-194, the expression levels of inducible TNF-α, IL-1, IL-6, and PGE2 were significantly suppressed by miR-194 (P < 0.01, P < 0.001; Figure 2).
Figure 2.
miR-194 suppressed the LPS-induced inflammatory cytokines-associated gene expression. (A) miR-194 suppressed LPS-induced expression of TNF-α mRNA. (B) miR-194 suppressed LPS-induced expression of IL-1 mRNA. (C) miR-194 suppressed LPS-induced expression of IL-6 mRNA. (D) miR-194 suppressed LPS-induced expression of PGE2 mRNA. ** P<0.01; *** P<0.001.
MiR-194 inhibitor regulated the expression of ECM genes and synthesis of ECM-degrading enzymes
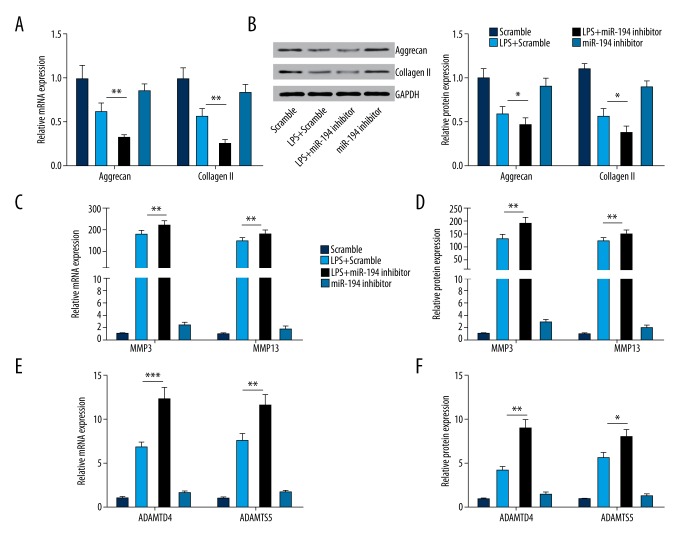
To further affirm the regulatory effect of miR-194 on the inflammatory response in LPS-induced NP cells, NP cells were blocked by transfecting miR-194 inhibitor. NP cells transfected with miRNA scramble served as control. q-PCR results it suggested that miR-194 inhibitor intensifies the inhibitory effect of LPS on the expression of Aggrecan and Collagen II (P<0.01, P<0.001; Figure 3A, 3B). Furthermore, it revealed that miR-194 inhibitor significantly promoted the mRNA and protein expression of ECM-degrading enzymes (including MMP3, MMP13, ADAMTS4, and ADAMT5; Figure 3C–3F) in LPS-induced NP cells.
Figure 3.
miR-194 inhibitor suppressed the expression of extracellular matrix genes and stimulated the expression of ECM degradation enzymes. (A, B) miR-194 inhibitor decreased the mRNA and protein level of Aggrecan and Collagen II (P<0.05, P<0.01). (C, D) miR-194 inhibitor promoted the mRNA and protein level of MMP3 and MMP13 (P<0.05, P<0.01). (E, F) miR-194 inhibitor promoted the mRNA and protein level of ADAMTS4 and ADAMTS5 (P<0.05, P<0.01). * P<0.05; ** P<0.01; *** P<0.001.
MiR-194 inhibitor regulates the expression of inflammatory cytokines-associated genes
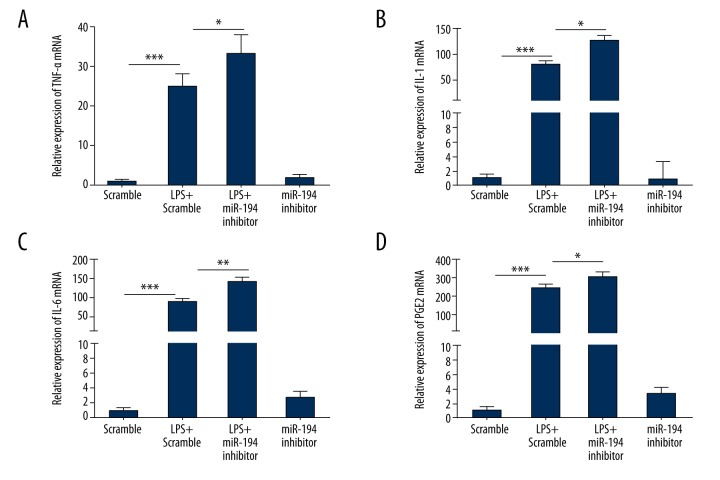
The expressions of inflammatory cytokines-associated genes of TNF-α, IL-1, IL-6, and PGE2 were detected by q-PCR. Through q-PCR assay, we found that the expression levels of these inflammatory cytokines-associated genes notably increased when cells were induced by LPS (P<0.001; Figure 4). As shown in Figure 4, miR-194 inhibitor significantly promoted the gene expressions of TNF-α, IL-1, IL-6, and PGE2 in the LPS + miR-194 inhibitor group in contrast with those in the LPS + scramble group (P<0.05).
Figure 4.
miR-194 inhibitor stimulated the expression of inflammatory cytokines-associated genes. (A) miR-194 inhibitor promoted the mRNA level of TNF-α (P<0.05, P<0.001). (B) miR-194 inhibitor promoted the mRNA level of IL-1 (P<0.05, P<0.001). (C) miR-194 inhibitor promoted the mRNA level of IL-6 (P<0.01, P<0.001). (D) miR-194 inhibitor promoted the mRNA level of PGE2 (P<0.05, P<0.001). * P<0.05; ** P<0.01; *** P<0.001.
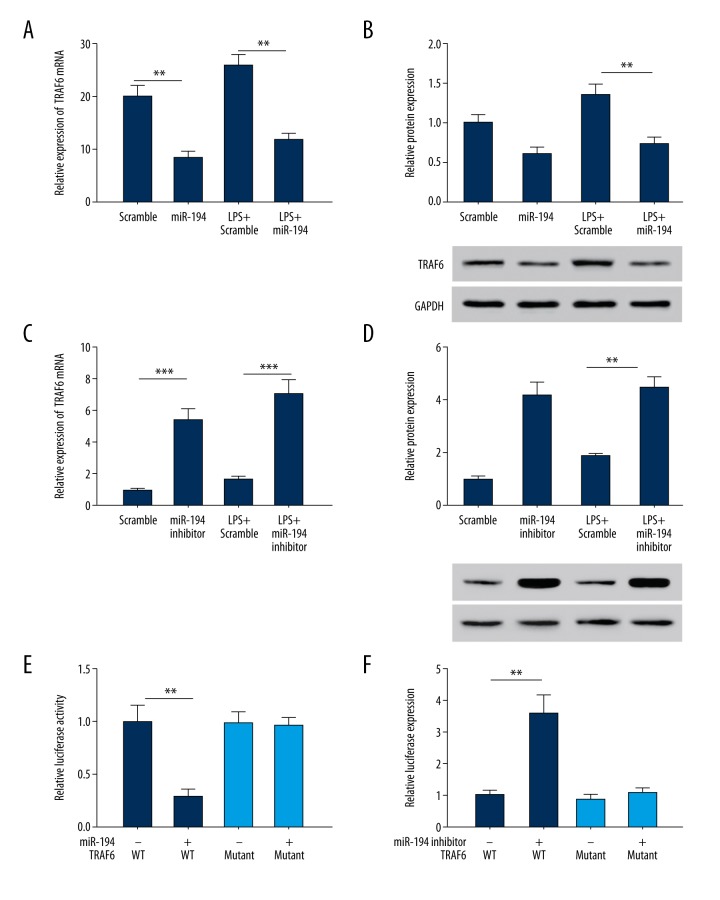
MiR-194 regulates TRAF6 expression
Through target prediction, we found that TRAF6 is a target gene of miR-194. It has been reported that TRAF6 is involved in cellular immunity and inflammatory responses [15,16]. Therefore, we investigated whether miR-194 regulates TRAF6 expression in NP cells. The results showed that overexpression of miR-194 led to a significant decrease in TRAF6 expression in NP cells and LPS-stimulated NP cells (P<0.01; Figure 5A, 5B). In contrast, miR-194 inhibitor led to an increase of TRAF6 expression (P<0.01; Figure 5C, 5D). These findings indicate that miR-194 exerts an inhibitory effect on TRAF6 expression.
Figure 5.
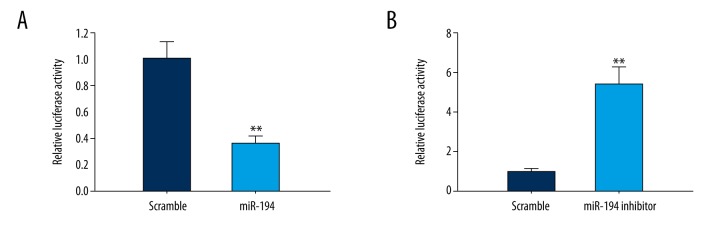
miR-194 regulated the gene expression of TRAF6. (A, B) miR-194 mimic suppressed mRNA and protein levels of TRAF6. (C, D) miR-194 inhibitor stimulated mRNA and protein level of TRAF6. (E) miR-194 suppressed TRAF6 5′UTR derived luciferase activity, but did not affect the luciferase activity of mutant. (F) miR-194 inhibitor stimulated TRAF6 5′UTR derived luciferase activity, but did not affect the luciferase activity of mutant. ** P<0.01; *** P<0.001.
To further assess the direct effect of miR-194 on TRAF6, we established a luciferase reporter plasmid of TRAF6 3′ UTR sequence. The results showed that the TRAF6 3′UTR-mediated luciferase activity was significantly inhibited by miR-194 and it was significantly promoted by miR-194 inhibitor (P<0.01, P<0.001; Figure 5E, 5F). However, there was no significant difference in the mutant TRAF6 3′UTR-mediated luciferase activity.
MiR-194 regulates the inflammation in NP cells via NF-κB signaling pathway
TRAF6 plays a critical role in the ability of the TLR signal transduction pathway to activate NF-κB and mitogen-activated protein kinase (MAPK) pathways [17,18]. Thus, the NP cells were transfected with luciferase reporter plasmid containing NF-κB responsive element, which further determined the regulatory effect of miR-194 on NF-κB transcriptional activity. The result showed that NF-κB-mediated luciferase activity was significantly inhibited by miR-194 and promoted by miR-194 inhibitor (P<0.01; Figure 6).
Figure 6.
miR-194 regulated NF-κB activity. NF-κB response element-derived luciferase activity was suppressed by miR-194 (p<0.01), but was stimulated by miR-194 inhibitor (P<0.01). ** P<0.01.
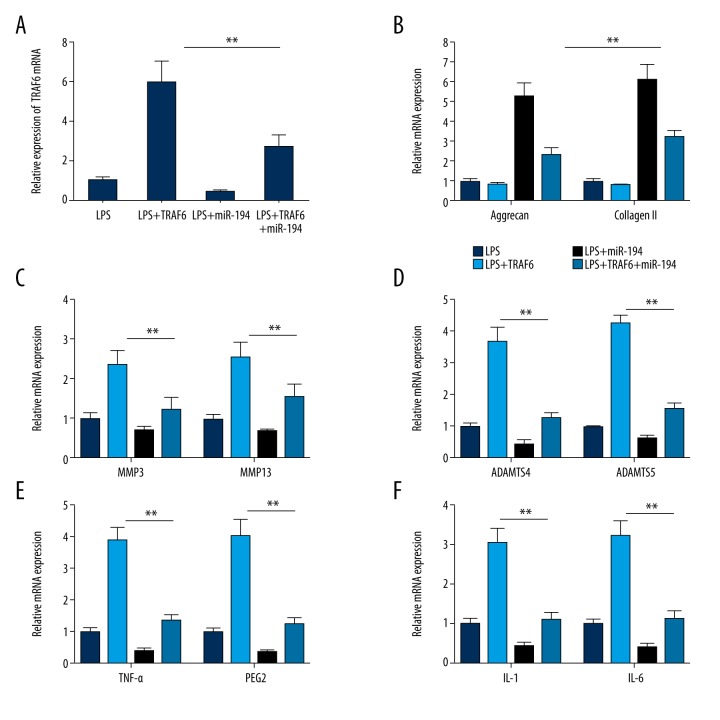
MiR-194 overexpression reverses the regulatory function of TRAF6 on NP cells
To verify whether miR-194 regulates inflammatory responses in LPS-induced NP cells by inhibiting TRAF6, we assessed the effect of miR-194 overexpression on the regulatory function of TRAF6. TRAF6 overexpression improved inflammation and significantly promoted the expression of MMP3, MMP13, ADAMTS4, ADAMTS5, TNF-α, IL-1, IL-6, and PGE2 in LPS-induced NP cells. We also found that TRAF6 overexpression downregulates the expression of Aggrecan and Collagen I. We found that miR-194 overexpression reversed the regulatory effect of TRAF6 on the expression of ECM genes and synthesis of ECM-degrading enzymes, including blocking the inhibitory effect of TRAF6 on Aggrecan and collagen II expression (P<0.01) and blocking the promoting effect of TRAF6 on the gene expression of MMP3, MMP13, ADAMPTS4, ADAMPTS5, TNF-α, IL-1, IL-6, and PGE2 (P<0.01; Figure 7).
Figure 7.
Overexpression of miR-194 reversed the regulatory function of TRAF6 in LPS-induced NP cells. (A) The relative expression levels of TRAF6 in LPS-induced NP cells transfected with miR-194, TRAF6, or miR-194+TRAF6. (B) Overexpression of miR-194 reversed the TRAF6 induced promotion of Aggrecan and Collagen II expression (P<0.05, P<0.01). (C) Overexpression of TRAF6 rescued the expression of MMP3 and MMP13, which were suppressed by miR-194. (D) Overexpression of TRAF6 rescued the expression of ADAMTS4 and ADAMTS5, which were suppressed by miR-194 (P<0.05, P<0.001). (E) Overexpression of TRAF6 rescued the expression of TNF-α and PGE2 (P<0.01). (F) Overexpression of TRAF6 rescued the expression of IL-1 and IL-6 (P<0.01). ** P<0.01; *** P<0.001.
Discussion
IDD is a common chronic disease in clinical practice. It burdens society clinically and economically due to its high rates of disability. About 10% of IDD patients have an obvious physical defect. Currently, relieving clinical symptoms is the main treatment for of IDD. However, biological therapy based on the pathological mechanism has become particularly popular in clinical research on IVD. In recent years, the role of miRNA has also become a focus in the progression of IDD. It has been demonstrated that miRNA is involved in IDD progression through multiple biological processes, mainly including: regulating the proliferation and apoptosis of NP cells [14,19–21], controlling the synthesis and dynamic balance of ECM [22,23], and mediating inflammatory responses [22]. Through exploring the function of miR-194 in NP cells, the present study adds to the theoretical basis for use of miRNA in the treatment of IDD.
In this study, we found that the expression of miR-194 was decreased in LPS-induced NP cells and that the expression of aggrecan and collagen II genes was suppressed by LPS but was promoted by miR-194 overexpression. Results of further experimentation suggest that miR-194 attenuates the inhibitory effect of LPS on aggrecan and collagen II. Additionally, LPS can promote the gene expression of ECM-degrading enzymes (MMP3, MMP13, ADAMPTS4, and ADAMPTS5), which is contrary to the function of miR-194. miR-194 overexpression can mediate the level of inflammatory cytokines induced by LPS, such as TNF-α, IL-1, IL-6, and PGE2. After transfection with miR-194 inhibitor, the expression of ECM and the synthesis of EMC-degrading enzymes induced by LPS were significantly promoted. These findings indicate that miR-194 inhibits the inflammatory responses of NP cells induced by LPS.
Recent research found that inflammatory factors matrix metalloproteinases (MMPs) and ADAMTS-4\ADAMTS-5, which are associated with IDD, were all involved in the NF-κB signaling pathway because they are common target genes of NF-κB [21,24]. NF-κB activation by endogenous or exogenous stimuli through cell antigen leads to NF-κB nuclear translocation, which can elevate the expression of inflammatory factors (such as IL-1/IL-6), MMPs, and ADAMTS. Based on these reports, the present study explored the relationship between miR-194 and NF-κB. Our investigation found that the activity of NF-κB luciferase was inhibited by miR-194 overexpression but promoted by miR-194 inhibitor, which suggests that the regulatory function of miR-194 operates via the NF-κB signaling pathway. Combined with prior conclusions, we conclude that miR-194 regulates the inflammatory responses in LPS-induced NP cells by regulating the NF-κB signaling pathway.
To explore the anti-inflammatory mechanism of miR-194 in NP cells, we used 2 algorithm programs (TargetScan and miRBase) to search for the potential targets of miR-194. We found that TRAF6 is a target of miR-194. In NF-κB pathway stimulated by LPS, activated TRAF6 induced phosphorylation and degradation of NF-κB inhibitor, I kappa B (IκB), further leading to the release of NF-κB from IκB/NF-κB trimer, ultimately activating nuclear factor NF-κB and inducing the expression of inflammatory factors such as TNF-N and IL-1 in some cells [25,26]. Our experimental results demonstrate that miR-194 overexpression downregulates the activity of TRAF6 3′ UTR-mediated luciferase, which is contrary to the effect of miR-194 inhibitor. Notably, both miR-194 overexpression and miR-194 inhibitor have no significant effect on the mutant TRAF6 3′UTR-mediated luciferase activity. Therefore, miR-194 exerts its regulatory function on inflammatory responses of LPS-induced NP cells by targeting TRAF6.
Conclusions
In conclusion, miR-194 inhibits activation of the NF-κB signaling pathway by targeting TRAF6, leading to suppression of the LPS-induced inflammatory responses in NP cells (Figure 8), showing it may be a regulatory mechanism in IDD.
Figure 8.
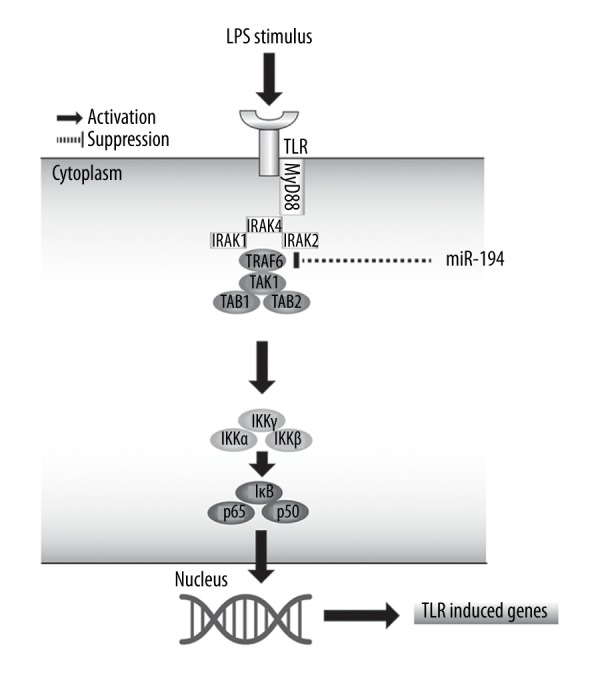
A hypothetical model for the role of miR-194 in LPS-induced inflammation in NP cells. After LPS stimulation in NP cells, TLR4 of cell membranes was polymerized, and then recruited MyD88 inside the cells. Recruited MyD88 bonded with IRAK. Subsequently, TRAF6 was activated by IRAK to induce the phosphorylation and degradation of IκB kinase, leading to the release of NF-κB from IκB/NF-κB complex and transferred into the nucleus to regulate the LPS/TLR-induced genes. In addition, miR-194 inhibits NF-κB signaling and expression of TLR-induced genes by targeting TRAF6.
Supplementary Table
Supplementary Table 1.
Primers for the clone and RT-PCR of genes.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| MiR-194 | GGGTGTAACAGCAACTCCA | CAGTGCGTGTCGTGGAGT |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
| Collagen II | GGCCCTCAGGGACCTGCCGG | CCAGGGGTACCAGGTTCTCC |
| Aggrecan | CAGTGAGTTGGACAGTAGTG | CAGATGTTTCTCCACTGACA |
| IL-6 | GTGTGA AAGCAGCAAAGAGGC | CTGGAGGTACTCTAGGTATAC |
| TNF-α | TCCATGGCCCAGACCCTCAC | CTCAGCCACTCCAGCTGCTC |
| IL-1 | TTGTACAAGGAGAGACAAGC | CAGCTGCAGGGTGGGTGTGC |
| PGE-2 | CGTTGGACTTTTTCTGGGTA | GTGCAACGGTAACCTGTGAC |
| MMP3 | CCTGGACAAGCAGTTCCAAA | TTCACAATCCTGTAGGAGAT |
| MMP13 | CCTGGACAAGCAGTTCCAAA | AGGTATAGATGGGAAACATC |
| ADAMTS4 | GGAGGCGCCCTTAACTCTGC | GGGCTCCCAGAAGGAGCCTT |
| ADAMT5 | GACGTTGGGACCATATGTTC | GAGAGGCCAAGTAGATGTCCC |
| β-actin | GCCTCCCGCGCGCGCACTAG | AAAGGCAAACGCTGGCCCGG |
| TRAF6 | TCATAATGTTAACCTCTCTT | TGTCTTACTAGGCGCCCCTC |
| MiR-194 RT primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACCACA | |
| U6 RT Primer | CGCTTCACGAATTTGCGTGTCAT | |
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–30. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F, Cai F, Shi R, et al. Aging and age related stresses: A senescence mechanism of intervertebral disc degeneration. Osteoarthritis Cartilage. 2016;24:398–408. doi: 10.1016/j.joca.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Virtanen IM, Karppinen J, Taimela S, et al. Occupational and genetic risk factors associated with intervertebral disc disease. Spine (Phila Pa 1976) 2007;32:1129–34. doi: 10.1097/01.brs.0000261473.03274.5c. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe H, Yamada Y, Kimata K. Roles of aggrecan, a large chondroitin sulfate proteoglycan, in cartilage structure and function. J Biochem. 1998;124:687–93. doi: 10.1093/oxfordjournals.jbchem.a022166. [DOI] [PubMed] [Google Scholar]
- 5.Nomura T, Mochida J, Okuma M, et al. Nucleus pulposus allograft retards intervertebral disc degeneration. Clin Orthop Relat Res. 2001;(389):94–101. doi: 10.1097/00003086-200108000-00015. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, Moon CS, Sul D, et al. Comparison of growth factor and cytokine expression in patients with degenerated disc disease and herniated nucleus pulposus. Clin Biochem. 2009;42:1504–11. doi: 10.1016/j.clinbiochem.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 7.Masuda K, Imai Y, Okuma M, et al. Osteogenic protein-1 injection into a degenerated disc induces the restoration of disc height and structural changes in the rabbit anular puncture model. Spine (Phila Pa 1976) 2006;31:742–54. doi: 10.1097/01.brs.0000206358.66412.7b. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Li K, Han X, et al. The imbalance between TIMP3 and matrix-degrading enzymes plays an important role in intervertebral disc degeneration. Biochem Biophys Res Commun. 2016;469:507–14. doi: 10.1016/j.bbrc.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Ellman MB, Kim JS, An HS, et al. Toll-like receptor adaptor signaling molecule MyD88 on intervertebral disk homeostasis: In vitro, ex vivo studies. Gene. 2012;505:283–90. doi: 10.1016/j.gene.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwata M, Ochi H, Asou Y, et al. Variations in gene and protein expression in canine chondrodystrophic nucleus pulposus cells following long-term three-dimensional culture. PLoS One. 2013;8:e63120. doi: 10.1371/journal.pone.0063120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He Y, Sun X, Huang C, et al. MiR-146a regulates IL-6 production in lipopolysaccharide-induced RAW264.7 macrophage cells by inhibiting Notch1. Inflammation. 2014;37:71–82. doi: 10.1007/s10753-013-9713-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Bai X, Song Q, et al. miR-223 inhibits lipid deposition and inflammation by suppressing toll-like receptor 4 signaling in macrophages. Int J Mol Sci. 2015;16:24965–82. doi: 10.3390/ijms161024965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Yu X, Shen J, et al. MicroRNA in intervertebral disc degeneration. Cell Prolif. 2015;48:278–83. doi: 10.1111/cpr.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Liu Z, Yang Y. In vitro screening of LPS-induced miRNAs in leukocytes derived from cord blood and their possible roles in regulating TLR signals. Pediatr Res. 2014;75:595–602. doi: 10.1038/pr.2014.18. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Cao H, Li J, et al. Autophagy induced by DAMPs facilitates the inflammation response in lungs undergoing ischemia-reperfusion injury through promoting TRAF6 ubiquitination. Cell Death Differ. 2017;24:683–93. doi: 10.1038/cdd.2017.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh MC, Lee J, Choi Y. Tumor necrosis factor receptor-associated factor 6 (TRAF6) regulation of development, function, and homeostasis of the immune system. Immunol Rev. 2015;266:72–92. doi: 10.1111/imr.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao R, Fan Y, Mou Y, et al. TAK1 lysine 158 is required for TGF-beta-induced TRAF6-mediated Smad-independent IKK/NF-kappaB and JNK/AP-1 activation. Cell Signal. 2011;23:222–27. doi: 10.1016/j.cellsig.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaisho T, Akira S. Dendritic-cell function in Toll-like receptor- and MyD88-knockout mice. Trends Immunol. 2001;22:78–83. doi: 10.1016/s1471-4906(00)01811-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang HQ, Yu XD, Liu ZH, et al. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol. 2011;225:232–42. doi: 10.1002/path.2931. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Huang X, Liu X, et al. miR-21 promotes human nucleus pulposus cell proliferation through PTEN/AKT signaling. Int J Mol Sci. 2014;15:4007–18. doi: 10.3390/ijms15034007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Z, Yin Z, Liu C, Tian J. The changes in the expression of NF-κB in a degenerative human intervertebral disc model. Cell Biochem Biophys. 2015;72:115–22. doi: 10.1007/s12013-014-0417-3. [DOI] [PubMed] [Google Scholar]
- 22.Gu SX, Li X, Hamilton JL, et al. MicroRNA-146a reduces IL-1 dependent inflammatory responses in the intervertebral disc. Gene. 2015;555:80–87. doi: 10.1016/j.gene.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neidlinger-Wilke C, Boldt A, Brochhausen C, et al. Molecular interactions between human cartilaginous endplates and nucleus pulposus cells: A preliminary investigation. Spine (Phila Pa 1976) 2014;39:1355–64. doi: 10.1097/BRS.0000000000000372. [DOI] [PubMed] [Google Scholar]
- 24.Wuertz K, Vo N, Kletsas D, Boos N. Inflammatory and catabolic signalling in intervertebral discs: The roles of NF-kappaB and MAP kinases. Eur Cell Mater. 2012;23:103–19. doi: 10.22203/ecm.v023a08. discussion 119–20. [DOI] [PubMed] [Google Scholar]
- 25.Verstrepen L, Bekaert T, Chau TL, et al. TLR-4, IL-1R and TNF-R signaling to NF-kappaB: Variations on a common theme. Cell Mol Life Sci. 2008;65:2964–78. doi: 10.1007/s00018-008-8064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber A, Wasiliew P, Kracht M. Interleukin-1 (IL-1) pathway. Sci Signal. 2010;3(105):cm1. doi: 10.1126/scisignal.3105cm1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Primers for the clone and RT-PCR of genes.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| MiR-194 | GGGTGTAACAGCAACTCCA | CAGTGCGTGTCGTGGAGT |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
| Collagen II | GGCCCTCAGGGACCTGCCGG | CCAGGGGTACCAGGTTCTCC |
| Aggrecan | CAGTGAGTTGGACAGTAGTG | CAGATGTTTCTCCACTGACA |
| IL-6 | GTGTGA AAGCAGCAAAGAGGC | CTGGAGGTACTCTAGGTATAC |
| TNF-α | TCCATGGCCCAGACCCTCAC | CTCAGCCACTCCAGCTGCTC |
| IL-1 | TTGTACAAGGAGAGACAAGC | CAGCTGCAGGGTGGGTGTGC |
| PGE-2 | CGTTGGACTTTTTCTGGGTA | GTGCAACGGTAACCTGTGAC |
| MMP3 | CCTGGACAAGCAGTTCCAAA | TTCACAATCCTGTAGGAGAT |
| MMP13 | CCTGGACAAGCAGTTCCAAA | AGGTATAGATGGGAAACATC |
| ADAMTS4 | GGAGGCGCCCTTAACTCTGC | GGGCTCCCAGAAGGAGCCTT |
| ADAMT5 | GACGTTGGGACCATATGTTC | GAGAGGCCAAGTAGATGTCCC |
| β-actin | GCCTCCCGCGCGCGCACTAG | AAAGGCAAACGCTGGCCCGG |
| TRAF6 | TCATAATGTTAACCTCTCTT | TGTCTTACTAGGCGCCCCTC |
| MiR-194 RT primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACCACA | |
| U6 RT Primer | CGCTTCACGAATTTGCGTGTCAT | |