Abstract
We established a light microscopy-based assay that reconstitutes the binding of phagosomes purified from mouse macrophages to preassembled F-actin in vitro. Both endogenous myosin Va from mouse macrophages and exogenous myosin Va from chicken brain stimulated the phagosome–F-actin interaction. Myosin Va association with phagosomes correlated with their ability to bind F-actin in an ATP-regulated manner and antibodies to myosin Va specifically blocked the ATP-sensitive phagosome binding to F-actin. The uptake and retrograde transport of phagosomes from the periphery to the center of cells in bone marrow macrophages was observed in both normal mice and mice homozygous for the dilute-lethal spontaneous mutation (myosin Va null). However, in dilute-lethal macrophages the accumulation of phagosomes in the perinuclear region occurred twofold faster than in normal macrophages. Motion analysis revealed saltatory phagosome movement with temporarily reversed direction in normal macrophages, whereas almost no reversals in direction were observed in dilute-lethal macrophages. These observations demonstrate that myosin Va mediates phagosome binding to F-actin, resulting in a delay in microtubule-dependent retrograde phagosome movement toward the cell center. We propose an “antagonistic/cooperative mechanism” to explain the saltatory phagosome movement toward the cell center in normal macrophages.
INTRODUCTION
Phagocytosis, the mechanism by which certain cells take up large particles such as bacteria, is a complex and dynamic process (reviewed in Greenberg and Silverstein, 1993; Aderem and Underhill, 1999). After binding to receptors on the cell surface, the particle is enclosed by a specialized domain of the plasma membrane. Oxygen and nitrogen-derived radicals are then released into the phagosome, to kill microorganisms. Subsequently, the phagosome fuses with compartments of the endocytic pathway, allowing digestion of the microorganism by lysosomal hydrolases.
The engulfment and subsequent intracellular transport of phagocytic particles requires a highly regulated and dynamic interaction of the phagosome membrane with cytoskeletal elements. The initial uptake step of phagocytosis requires extensive remodeling of the cortical actin cytoskeleton (Greenberg and Silverstein, 1993; Aderem and Underhill, 1999; May and Machesky, 2001). The newly formed phagosome bears a dense coat of actin and actin binding proteins (ABPs), which is usually lost with time (Greenberg et al., 1990), although in some systems it may persist and even accumulate additional F-actin (Stockem et al., 1983). Recently, an in vitro assay was established that monitors the de novo assembly of F-actin on phagosomes loaded with latex beads (Defacque et al., 2000). It was shown that mature phagosomes also have the capacity to assemble F-actin, and that this process requires the presence of ezrin and/or moesin on the phagosomal membrane (Defacque et al., 2000). At later stages in the pathway, the interaction of phagosomes with cytoskeletal elements is important to facilitate their fusion with endocytic organelles and their transport to the perinuclear region of cells. Treatment of cells with drugs that depolymerize either F-actin or microtubules blocks this fusion (Pitt et al., 1992; Jahraus et al., 1994, 2001; Desjardins et al., 1994b).
The later events of phagocytosis, such as phagosome transport from the cell periphery toward the perinuclear region and the acquisition by phagosomes of late endocytic organelle markers, is dependent on microtubules (Toyohara and Inaba, 1989; Desjardins et al., 1994b; Blocker et al., 1996, 1997). Analysis of phagosome motility in living cells revealed that drugs that induce microtubule depolymerization inhibit the movement of phagosomes along linear elements (Blocker et al., 1997, 1998). Additional studies showed that the movement of phagosomes along microtubules in vitro is driven in one direction by kinesin and in the other by cytoplasmic dynein. A role for microtubule dynamics in slow movements of newly formed phagosomes in peripheral regions was also shown (Blocker et al., 1998). The latter study showed that phagosomes preferentially bind in vitro to the microtubule plus-ends; in vivo, these are localized near the plasma membrane.
The machinery responsible for F-actin involvement in both the uptake as well as later steps of phagocytosis is not well defined. Because conventional myosin (myosin II) appears not to be involved in phagocytic uptake (Fukui et al., 1990; de Lanerolle et al., 1993), it has been proposed that uptake is driven by actin polymerization, a process that is controlled by actin binding proteins on the plasma membrane (Silverstein et al., 1989; Chimini and Chavrier, 2000). It also seems likely that myosin IC is involved in the production of a contractile activity that allows the phagosomes to separate from the plasma membrane (Swanson et al., 1999). Despite the fact that several myosins are associated with fully formed phagosomes (Desjardins et al., 1994a; Swanson et al., 1999; Garin et al., 2001), the role of myosins in the later events of phagocytosis, such as phagosome maturation and transport from the periphery to the center of cells, is an open question.
Evidence that myosins play a role in membrane trafficking is most compelling for the class V myosins (reviewed in Langford and Molyneaux, 1998; Mermall et al., 1998; Reck-Peterson et al., 2000; Wu et al., 2000). It was shown both in vitro and in vivo that myosin Va mediates the transport of such organelles as melanosome granules (Rogers and Gelfand, 1998), brain vesicles (Evans et al., 1998; Bridgman, 1999), and the endoplasmic reticulum (Tabb et al., 1998). Recently, myosin Va was shown to colocalize with fully internalized phagosomes (Swanson et al., 1999).
Here, we investigated the role of myosin Va in later events of phagocytosis with the use of two different approaches. First, we devised an in vitro phagosome–F-actin binding assay and used this to demonstrate that phagosome-associated myosin Va is involved in ATP-regulated binding of phagosomes to F-actin. Second, we analyzed phagosome movement in normal and dilute-lethal (myosin Va null) macrophages and observed a paradoxical acceleration of phagosome accumulation at the cell center in dilute-lethal macrophages. This analysis shows that myosin Va motor activity is not necessary for rapid, long-range retrograde transport of phagosomes to the perinuclear region of macrophages, but rather appears to be necessary for short-range transport of phagosomes at the cell periphery. We suggest that these myosin Va-mediated motility events might be important for phagosome maturation and fusion with endocytic organelles.
MATERIALS AND METHODS
Cell Culture
J774A.1 mouse macrophages were obtained from American Type Culture Collection (Manassas, VA) and maintained as described in Blocker et al. (1996). For cytosol preparation, cells were grown in suspension in Joklik's modified Eagle's medium (Biochrom Beteiligungs, Berlin, Germany), 5% newborn calf serum, 5% fetal calf serum (Biochrom) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Biochrom).
Mouse bone marrow macrophages from normal mice and mice homozygous for the dilute-lethal spontaneous mutation MyoVa d-l (strain DLS/Le a/a Myo5ad-l +/+ Bmp5se; Jackson Laboratories, Bar Harbor, ME) were grown in macrophage media [100 ml RPMI 1640, 50 ml of L929 cell supernatant, 1 ml of nonessential amino acids, 1 ml of l-glutamine (Gibco Life Technologies GmbH, Karlsruhe, Germany), supplemented with 1 ml of penicillin/streptomycin (Biochrom), 0.5 ml of 2-mercaptoethanol, 10 ml of fetal calf serum [Gibco Life Technologies]) on uncoated Petri dishes for bacteria. After harvesting, bone marrow cells were resuspended in 10 ml of phosphate-buffered saline (PBS) by pipetting and then centrifuged at 1500 × g for 3 min at 4°C. The pelleted cells were then resuspended in 10 ml of macrophage media and transferred to Petri dishes at 3 × 106 cells/150-mm dish. Three days after the initial plating, the medium was changed. Five days after initial plating, cells were cooled down in a −20°C freezer until they rounded up (1–2 min). Cells were detached by scraping and then replated onto sterile coverslips in a new Petri dish and cultured until use. Normally, cells were used for experiments 6–7 d after initial plating.
Preparation of Latex Beads
Carboxylate-modified nile red fluorescent (535/575 nm) or blue fluorescent (350/440 nm) latex beads, 1 μm in diameter (FluoSpheres; Molecular Probes Europe B.V., Leiden, The Netherlands) were covalently coupled to fish skin gelatin (FSG) (1 mg/ml in 50 mM MES buffer, pH 6.7) with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide according to the manufacturer's recommendations.
Fixed-time Movement Analysis In Vivo
Bone marrow macrophages were fed a 0.0066% (wt/vol) solution of blue fluorescent latex beads coupled to FSG for 15 min at 37°C in macrophage media, chased for 15 min in macrophage media at 37°C to wash out excess beads, and then chased in macrophage media containing no additions, or 5 μM nocodazole (Sigma Chemie GmbH, Deisenhofen, Germany). At 0 time or after 15, 30, 45, 60, or 240 min of chase, cells were fixed in 3% paraformaldehyde and permeabilized with 0.1% Triton X-100. Fixed cells were labeled with polyclonal DIL2 anti-myosin Va, increased against a glutathione S-transferase fusion protein that contained myosin Va heavy chain residues 910-1106 corresponding to the first segment of the α helical coiled-coil in the central rod domain (Wu et al., 1997) at 1:500 dilution, followed by fluorescein isothiocyanate-conjugated goat anti-rabbit Ig (Calbiochem Novabiochem GmbH, Bad Soden/Ts., Germany) at 1:50 dilution.
To quantify phagosome localization inside cells, fixed cells were observed by video-enhanced contrast differential interference contrast (VEC-DIC) microscopy (Allen et al., 1985; Weiss et al., 1999). The total number of phagosomes loaded with latex beads within 20 cells at each time point was determined. In the same 20 cells, the number of phagosomes in the central region, defined as the area including the cell nucleus and a zone 4–5 μm from the nucleus, was determined. The extent of phagosome accumulation in the central region was calculated as the percentage of total phagosomes that were localized in the perinuclear region.
Video Microscopy
Slides with macrophages were mounted onto microscope chambers containing macrophage media, with latex beads at a concentration of 0.003% (wt/vol) and observed by VEC-DIC microscopy (Weiss et al., 1999) at 37°C.
A Nikon Diaphot 300 inverted microscope (Nikon GmbH, Düsseldorf, Germany) equipped with an oil immersion condenser (NA 1.4), a 100× DIC PlanApo oil objective (NA 1.4) and a Xenon lamp (XBO 100) was used. A Hamamatsu C2400-07 Newvicon camera was used for acquiring DIC images (Hamamatsu Photonics Deutschland GmbH, Herrsching, Germany). The analog and digital processing of VEC-DIC signals was performed with the use of an ARGUS 20 real time image processor (Hamamatsu Photonics). Single-frame images were captured from videotape or directly from the ARGUS 20-image processor with the use of a Power Macintosh 9600 (Apple Computer, Cupertino, CA) equipped with an LG-3 frame grabber (Scion, Frederick, MD), with the use of IPLab Spectrum software (Signal Analytics, Vienna, VA). Movements were analyzed either by hand with the use of IPLab Spectrum software or more precisely with the use of the MicroTrack FG32 path software (HaSoTec GmbH, Rostock, Germany).
Phagosome Purification
Phagosomes loaded with latex beads were purified from J774A.1 mouse macrophages pulsed with red or blue fluorescent latex beads (Molecular Probes Europe) coupled to FSG for 1 h and chased for 1 h in bead-free media as described by Desjardins et al. (1994b) and Blocker et al. (1996). This procedure yields phagosomes of high purity with a defined protein composition (Desjardins et al., 1994a,b). For some experiments, the phagosomes were isolated by flotation in a discontinuous sucrose gradient as described in Blocker et al. (1996) with the use of postnuclear supernatant diluted 1:10 with HB (HEPES buffer: 25 mM HEPES, pH 7.4 [KOH], 2 mM MgCl2, 2 mM EGTA, 0.1 mM EDTA, 2 mM diothiothreitol, and protease inhibitors [1 μg/ml aprotinin, leupeptin, pepstatin, N-tosyl-l-phenylalanine chloromethyl ketone, all from Sigma]) containing 0.3 mg/ml casein (Sigma). For salt-stripping, the phagosomes were treated with 2 M NaCl for 30 min at 4°C and then reisolated by flotation in a discontinuous sucrose gradient as described in Blocker et al. (1996). Unless stated otherwise, salt-stripped phagosomes were used in all experiments. The number of phagosomes in any preparation was determined by measuring the optical density of the preparations at 600 nm with the use of the extinction coefficient for 1% of latex beads (0.01 g/ml) E600 = 1000.
Where indicated, phagosome membrane proteins were digested with 100 μg/ml trypsin (Sigma) for 30 min at 37°C. Trypsin was inactivated with 3,4-dichloroisocoumarin (Boehringer Mannheim GmbH, Mannheim, Germany). Phagosomes were reisolated as previously described (Blocker et al., 1996) and used immediately.
Preparation of Cytosol
Cytosol was prepared from J774A.1 cells cultured in suspension at a maximum density of 1 × 106 cells/ml. Cells were collected by centrifugation at 2500 × g for 3 min and washed twice with ice-cold PBS. The cell pellet was resuspended in HBKS (25 mM HEPES, pH 7.4 [KOH], 50 mM KCl, 2 mM MgCl2, 2 mM EGTA, 0.1 mM EDTA, 2 mM dithiothreitol, 10% sucrose) and repelleted by centrifugation for 5 min at 2500 × g, 4°C. Cells were resuspended in 1 volume HBKS plus protease inhibitors and homogenized by passage through a 24-guage syringe needle until 90–95% of the cells were lysed. The homogenate was then centrifuged for 30 min at 40,000 × g. The supernatant was removed and centrifuged for 1 h at 150,000 × g, 4°C, to generate cytosol. Cytosol preparations from J774A.1 cells typically contained 30–35 mg/ml protein as measured by the method of Bradford (1976). Cytosol from scraped bone marrow macrophages isolated from normal and dilute-lethal mice was prepared as for J744A.1 cells. Due to the limited amount of starting material, these cytosol preparations had a protein concentration <1 mg/ml. All cytosol preparations were flash frozen in liquid nitrogen and stored at −80°C.
For some experiments, the cytosol was further fractionated by gel filtration with the use of a Superose 12 column on a SMART system (Amersham Pharmacia Biotech Europe GmbH, Freiburg, Germany). The column was equilibrated with HB and 50-μl fractions were collected.
Preparation of Actin Gel and Isolation of ABPs
Actin gels were prepared from cytosol by a modification of the extract gelation procedure of Hartwig and Stossel (1982). Endogenous actin was polymerized by incubation of cytosol at 25°C for a total of 1 h. After 30 min, 10 μM phalloidin (Sigma) was added to stabilize the filaments. The entire incubation mixture was loaded onto a 20% sucrose cushion in HB and centrifuged at 100,000 × g, 4°C for 40 min, producing a pellet of the gel lattice.
The actin gel pellet was washed in HBKS for 10 min at room temperature then centrifuged at 150,000 × g for 30 min, 4°C. ATP-independent actin binding proteins (KCl-ABPs) were then isolated from the actin pellet by resuspension in HBKS containing 0.5 M KCl, followed by centrifugation at 150,000 × g for 2 h, 4°C. The supernatant of ATP-independent ABPs was collected and the pellet was washed again as described above with the addition of 2 mM ATP to isolate ATP-dependent ABPs (ATP-ABPs). ABP fractions were desalted into HB by dialysis. All samples were frozen in liquid nitrogen and stored at −80°C.
ATP-dependent ABPs were further fractionated by chromatography on a hydroxyapatite (HAP) column (Bio-Gel HTP; Bio-Rad Laboratories GmbH, Munich, Germany) according to Gyoeva et al. (1983) with some modifications. The column was equilibrated with HB containing 0.5 M KCl. After loading ABPs, the column was washed with equilibration buffer and the proteins were eluted with a potassium phosphate step gradient of 75, 150, and 300 mM. Collected fractions were desalted into HB by dialysis.
Binding Assays
F-Actin was polymerized from rabbit skeletal muscle G-actin, prepared by the method of Spudich and Watt (1971) by the addition of 2 mM MgCl2. After overnight polymerization, F-actin was stabilized and labeled with rhodamine-phalloidin (Sigma) with the use of a 1:1 M ratio.
Microscope chambers were built from a glass microscope slide (Menzel Super Frost; Gerhard Menzel GmbH, Braunschweig, Germany) and an 11-mm circular glass coverslip (Gerhard Menzel GmbH) sealed onto two pieces of double-sided tape (3M Scotch), forming a 2–3-μl chamber. All incubations were carried out in a moist chamber at room temperature. Filamentous actin (0.5 μM), stabilized and labeled with rhodamine-phalloidin, was perfused into the chamber and incubated for 5 min. Nonspecific binding was blocked by perfusion of the chamber with 3 mg/ml casein (Sigma) in HB. Excess F-actin and casein were washed away by perfusion with three chamber volumes of HBS, (HB containing 10% sucrose). One volume of phagosome binding reaction mixture, containing phagosomes (working concentration 0.001% [wt/vol]), 0.3 mg/ml casein, and binding factors to be tested, was perfused into the chamber and incubated for 20 min. Unbound phagosomes were washed away by perfusion with 3 volumes of HBS. Binding was analyzed by fluorescence microscopy with the use of a Nikon Diaphot 300 microscope with a 10× eyepiece and Nikon 100× PlanApo oil objective (field surface area of 22,000 μm2). The bound phagosomes were counted by eye, and in each experiment, values from at least 10 fields from two separate, but identical, reactions were averaged. The errors reported are the SDs of these 20 fields.
In some cases, the phagosome-binding reaction mixture, containing phagosomes at a concentration of 0.160% (wt/vol), 0.3 mg/ml casein, and one of the following components: cytosol at different concentrations, 0.1 mg/ml ABPs, or 50 μg/ml myosin Va isolated from chicken brain according Cheney (1998), was preincubated with or without 2 mM ATP for 20 min. Phagosomes were reisolated and used in the binding assay.
Immunofluorescence Microscopy
For immunofluorescence microscopy, phagosome-binding mixtures in HB were layered onto a 1.5-ml HBS cushion and centrifuged onto coverslips at 20,000 × g for 20 min at 20°C. Coverslips were fixed in 3% paraformaldehyde for 10 min and then washed with PBS and processed for immunofluorescence microscopy. Fixed phagosomes were labeled with DIL2 anti-myosin Va at 1:500 dilution or with polyclonal antibody against chicken myosin Va head domain (Nascimento et al., 1997) at 1:100 dilution followed by fluorescein isothiocyanate-conjugated goat anti-rabbit Ig (Calbiochem Novabiochem GmbH) at 1:50 dilution. Images were acquired with Nikon microscope equipped with a chilled charge-couple device camera system (SenSys; Photometrics, Munich, Germany).
Antibodies and Electrophoresis
SDS-PAGE was performed according to Laemmli (1970) on 7.5 and 10% polyacrylamide gels. Immunoblotting was performed according to Towbin et al. (1979), with the use of horseradish peroxidase-labeled goat anti-rabbit secondary antibodies and the enhanced chemiluminescence detection system (Amersham Pharmacia Biotech Europe GmbH). Myosin II was probed with the monoclonal antibody m2.42 (IgG1) against the head domain of Acanthamoeba myosin II heavy chain (r179–289) (a gift of D. Kaiser, Johns Hopkins University, Baltimore, MD) or rabbit polyclonal antibody against skeletal myosin II (Sigma). Myosin Va was detected with a rabbit polyclonal antibody against the tail domain of chicken myosin Va (a gift of M.S. Mooseker, Yale University, New Haven, CT), a rabbit polyclonal antibody against the head domain of chicken myosin Va (a gift of R.E. Larson, University of Sao Paulo, Sao Paulo, Brazil), or the DIL2 rabbit polyclonal antibody (see above). The rabbit polyclonal antibody to annexin III was a gift of J. Ernst (University of California, San Francisco, CA).
RESULTS
Binding of Phagosomes to F-Actin Is Stimulated by Cytosolic Factors
Previous studies have shown that after their formation, phagosomes move within cells toward the cell center (Toyohara and Inaba, 1989; Blocker et al., 1997, 1998). The latter studies established that microtubules are critical for this transport. To study the possible involvement of myosin-based motility in this process, we first asked whether phagosomes interact with F-actin in vitro. For this, we established an in vitro binding assay. Rhodamine-phalloidin–labeled F-actin was perfused into microscopy chambers prepared as described in MATERIALS AND METHODS and nonspecific binding sites were blocked with casein (Figure 1A). In parallel, phagosomes enclosing fluorescent latex beads were prepared by internalization into J774 cells for 1 h, followed by 1 h of chase. After isolation on sucrose step-gradients, the phagosomes were treated with 2 M NaCl to remove peripheral membrane proteins (Defacque et al., 2000). These salt-stripped phagosomes were then allowed to bind to the coverslip-adsorbed F-actin in the presence or absence of macrophage cytosol. After unbound phagosomes had been washed out with buffer, the number of bound phagosomes was analyzed by fluorescence microscopy (Figure 1B).
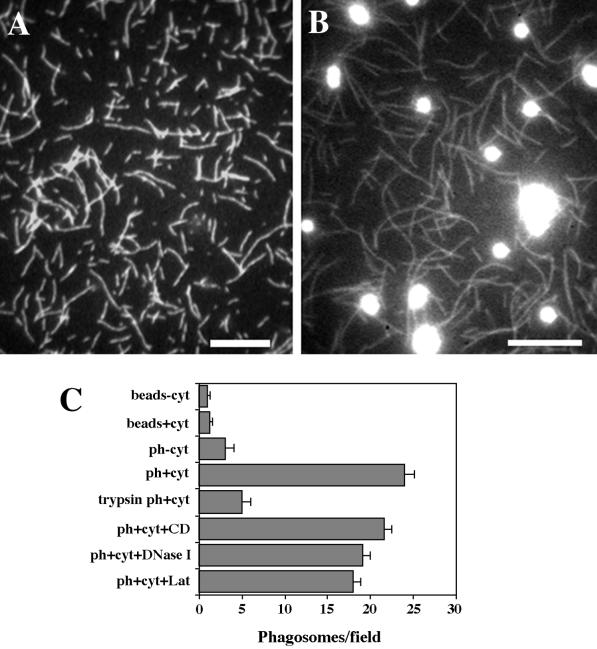
Figure 1.
Reconstitution of salt-stripped phagosome binding to preassembled F-actin in vitro. (A and B) Fluorescence images of typical fields of the binding assay. (A) Rhodamine-phalloidin–labeled F-actin was absorbed on the coverslip surface. (B) Phagosomes were bound to F-actin in the presence of 1 mg/ml cytosol and unbound phagosomes were washed out. Bars, 10 μm. (C) Stimulation of phagosome binding to F-actin by cytosolic (cyt) factor(s). Binding to the F-actin lawn of uninternalized latex beads (beads−cyt; beads+cyt). Phagosome (ph) binding to F-actin in the presence and absence of 1 mg/ml cytosol (ph+cyt, ph−cyt). Binding of phagosomes treated with 100 μg/ml trypsin (trypsin ph+cyt) (see MATERIALS AND METHODS). Binding of phagosomes in the presence of 5 μM cytochalasin D (ph+cyt+CD), 15 μM DNase I (ph+cyt+DNase I), or 5 μM latrunculin (ph+cyt+Lat).
Binding of phagosomes to F-actin was minimal in the absence of cytosol (Figure 1C). However, when cytosol was added, numerous bound phagosomes were observed (Figure 1C). No binding was observed in the absence of an F-actin network absorbed on the coverslip surface. The number of phagosomes bound per field varied only slightly from experiment to experiment and was essentially linear with the number of added phagosomes within the concentration range from 0.0015 to 0.0125% solids (OD600 = 1.5–12.5). The addition of cytosol at a final protein concentration of 1–4 mg/ml stimulated the binding of phagosomes to F-actin, typically by 10–15-fold.
Phagosome membrane proteins were required for the stimulation of binding, because F-actin did not bind free FSG beads in the presence or absence of cytosol (Figure 1C). Moreover, phagosome–F-actin interaction in the presence of cytosol was inhibited by pretreatment of phagosomes with trypsin (Figure 1C), or after heating to 95°C for 5 min (not shown).
Actin found on isolated phagosomes by immunoblotting (Desjardins et al., 1994a; Rezabek et al., 1997) was seen to be removed after the salt-stripping procedure (Defacque et al., 2000). It nevertheless remained possible that small, but functionally significant, amounts of phagosome-bound actin might resist this treatment and contribute to the binding that we measure, via actin–actin interactions (for example, mediated by actin cross-linkers in the cytosol). To test for this possibility, we investigated the effects of reagents that block actin polymerization or actively depolymerize F-actin on the ability of salt-stripped phagosomes to bind to F-actin. For this, cytochalasin D (5 μM), latrunculin (5 μM), and DNase I (15 μM) were tested. None of these agents had a significant effect on phagosome binding to F-actin in the presence of cytosol (Figure 1C). Under the conditions used, rhodamine-phalloidin-stabilized F-actin adsorbed on the glass surface was not depolymerized by these agents. This argues that actin–actin interactions were not responsible for phagosome binding to F-actin in our binding assay.
The ability of macrophage cytosol to stimulate the binding of salt-stripped phagosomes to F-actin indicated that phagosomes could acquire cytosolic factor(s) that promote this binding. To test this, salt-stripped phagosomes were preincubated with cytosol and then repurified by flotation in a sucrose gradient. The binding activity of the reisolated phagosomes that had been preincubated with 1 mg/ml cytosol, and tested in the absence of cytosol was about the same as that of the phagosomes coincubated in the presence of cytosol (Table 1). Thus, proteins from cytosol that facilitate phagosome–F-actin interaction must bind to salt-stripped phagosomes in a relatively stable manner that withstands gradient purification.
Table 1.
Characterization of phagosome F-actin-binding activity and myosin Va association in different phagosome preparations
| Type of phagosome preparation | Binding
activitya (phagosomes/field) of different
phagosome preparations in the presence of
|
Detection of
myosin Va in different phagosome preparations by
|
|||
|---|---|---|---|---|---|
| No additions | 2 mM ATP | DIL2 anti-myosin V | Immunoblottingb | Microscopyc | |
| 1. Isolated salt-stripped phagosomes | 1–3 | 1–3 | 1–3 | No | No |
| 2. Salt-stripped phagosomes reisolated after preincubation with 1 mg/ml cytosol | 20–30 | 15–20 | 15–20 | Yes | Yes |
| 3. Salt-stripped phagosomes reisolated after preincubation with 20 mg/ml cytosold | 2–5 | 2–5 | 2–5 | Traces | No |
| 4. Salt-stripped phagosomes reisolated after preincubation with 0.1 mg/ml ATP-ABPs | 20–30 | 5–8 | 5–8 | Yes | Yes |
Binding activity typically observed in 3–5 experiments.
Preparations were probed with DIL2 anti-myosin Va antibody.
Phagosomes were stained with DIL2 anti-myosin Va antibody and observed by fluorescence microscopy.
High concentrations of cytosol inhibited F-actin—phagosome binding activity and prevented myosin Va from binding to phagosomes due to low molecular mass inhibitor (Kuznetsov, unpublished data).
Two Different Classes of ABPs Facilitate Interaction between Phagosomes and F-Actin
To examine whether the cytosolic binding factor(s) that bind phagosomes are ABPs that could then directly interact with F-actin, the F-actin lawn was preincubated for 20 min with 1 mg/ml cytosol in the absence of phagosomes. Unbound cytosolic proteins were washed out and the phagosomes were added in the absence of additional cytosol. The number of salt-stripped phagosomes that bound was as high as that found after simultaneous addition of phagosomes and cytosol (data not shown). Because the binding factors interact with F-actin, they must represent ABPs.
To assess more directly the role of ABPs in phagosome–F-actin interactions, two types of ABP preparations were carried out by extract gelation as described in MATERIALS AND METHODS. First, the ATP-independent ABPs (KCl-ABPs) were eluted from the actin gel with 0.5 M KCl (Figure 2 A, lane KCl-ABPs). The actin gels were then resedimented in the buffer with 0.5 M KCl plus 2 mM ATP to elute ABPs that bind F-actin in an ATP-dependent manner (ATP-ABPs) (Figure 2A, lane ATP-ABPs). Phagosomes were then tested for their ability to bind F-actin in the presence of either KCl-ABPs (nonmyosin ABPs) or ATP-ABPs (putative myosins). Both fractions stimulated phagosome binding to F-actin (Figure 2B). As expected, only the binding supported by ATP-ABPs was inhibited by the addition of ATP to the assay mixture (Figure 2B). The addition of both fractions together did not result in a synergistic stimulation of binding (Figure 2B). In fact, at saturation, the stimulation of binding by the combined ABP fractions was about the same as that found for KCl-ABPs or ATP-ABPs alone. These data therefore suggest that the two classes of ABPs compete for a limited number of binding sites on the phagosomal membrane.
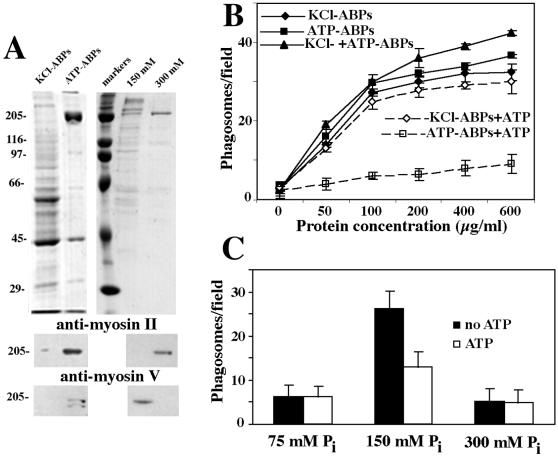
Figure 2.
Involvement of ABPs in salt-stripped phagosome–F-actin interaction. (A) Analysis of ABP fractions by SDS-PAGE and immunoblotting. (Top) ABPs eluted from actin gel in the presence of 0.5 M KCl (KCl-ABPs), ABPs eluted from actin gel in the presence of 0.5 M KCl plus 2 mM ATP (ATP-ABPs), molecular mass markers (markers), and ATP-ABP fraction eluted from the HAP column in the presence of 150 mM Pi (150 mM) and in the presence of 300 mM Pi (300 mM) were run on 10% gel and stained with Coomassie blue. (Middle) Blot of the fractions described above probed with a monoclonal anti-myosin II heavy chain. (Bottom) Blot of corresponding fractions probed with a polyclonal antibody against the tail domain of chicken myosin V heavy chain. (B) Phagosome binding to F-actin in the presence of ABPs eluted from an actin gel without ATP (KCl-ABPs), with ATP (ATP-ABPs), and in the presence of both ABPs fractions (KCl+ATP-ABPs). ATP (2 mM) effects on phagosome binding in the presence of KCl-ABPs (KCl-ABPs+ATP), ATP-ABPs (ATP-ABPs+ATP). The protein concentrations for each ABP fraction are indicated. (C) Binding activity of ATP-ABPs after fractionation on the HAP column. Fractions were eluted from the column in the presence of 75, 150, and 300 mM Pi. For all fractions, the binding activity was tested in the presence (ATP) and absence (no ATP) of 2 mM ATP.
The effect of ADP (1 mM) and the nonhydrolyzable ATP analog AMP-PNP (1 mM) on phagosome binding in the presence of ATP-ABPs was then tested. ADP caused no reduction in phagosome binding, whereas AMP-PNP caused a slight reduction (not shown). The results indicate that ATP effectively decreases the phagosome binding activity only of the ATP-ABPs, which is consistent with the known nucleotide dependence of active myosins (Stossel and Hartwig, 1975; Pollard, 1982; Hartwig and Stossel, 1982; Cheney et al., 1993).
High Molecular Mass Myosin, but not Myosin II, Is Involved in Phagosome–F-Actin Interaction
To gain a better understanding of the type of myosin that could be responsible for phagosome binding to F-actin, we first estimated the molecular mass of the relevant proteins by gel filtration on a Superose 12 column. The fractionation of both cytosol and ATP-ABPs yielded a single peak of binding activity, corresponding to a molecular mass (assuming a globular protein) of >670 kDa (not shown). Thus, the peak of activity was in the range expected for high molecular mass myosins, like myosin II (Hartwig and Stossel, 1982) and myosin Va (Espindola et al., 1992). We therefore performed immunoblotting of the macrophage cytosol and the ABP fractions with a monoclonal antibody m2-42 to myosin II and a polyclonal antibody to myosin Va. Both antibodies cross-reacted specifically with polypeptides from the ATP-ABP fraction (Figure 2A, lane ATP-APBs). As expected, little or no immunoreactivity for myosin II or myosin Va was seen in the KCl-ABP fraction (Figure 2A, lane KCl-ABPs).
Because the ATP-ABP fraction contained both myosin II and myosin Va, we separated the two on an HAP column and then tested their relative abilities to stimulate phagosome binding to F-actin. After HAP chromatography of the ATP-ABPs, the fraction that eluted with 75 mM phosphate contained almost no proteins and had no activity in the phagosome binding assay (Figure 2C). The fraction that eluted with 150 mM phosphate contained several polypeptides (Figure 2A, lane 150 mM) and had a high level of phagosome-binding activity that was inhibited by 50% in the presence of ATP (Figure 2C). The presence of myosin Va, but not myosin II, was detected in this fraction by immunoblotting (Figure 2A, lane 150 mM). Myosin II was eluted from the column in 300 mM phosphate (Figure 2A, lane 300 mM), however this fraction did not influence binding (Figure 2C).
Based on these results, we concluded that myosin II does not stimulate phagosome binding to F-actin. However, myosin Va was a strong candidate for mediating ATP-dependent phagosome–F-actin interactions.
Localization of Myosin Va on Isolated Phagosomes
We further examined the association of myosin Va with phagosomes with the use of the DIL2 rabbit polyclonal antibody raised against the rod domain of mouse myosin Va heavy chain. As seen in Figure 3A, lane 2, this antibody detected a single band at ∼190 kDa, consistent with the known molecular mass of myosin Va.
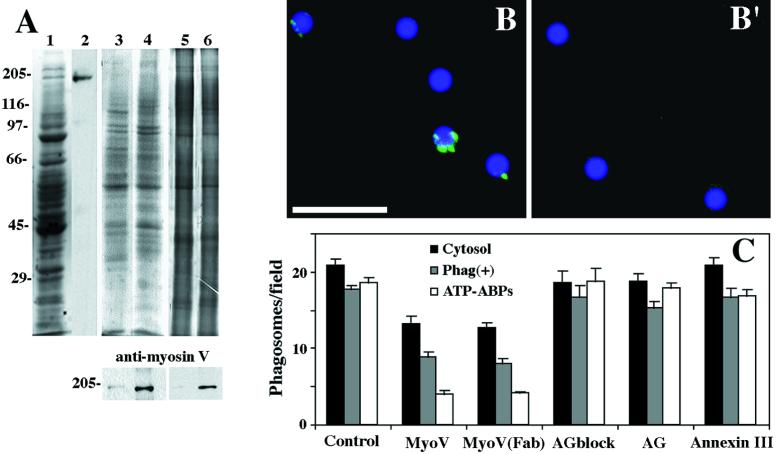
Figure 3.
Myosin Va detection in phagosome preparations with associated F-actin–binding activity. (A) DIL2 antibody strongly reacted with myosin V in macrophage cytosol. (Lane 1) SDS-PAGE of cytosol (Coomassie staining). (Lane 2) Immunoblot probed with DIL2 antibody. Salt-stripped phagosomes were reisolated by flotation after incubation in the presence of cytosol at 20 mg/ml (lane 3) or 1 mg/ml (lane 4). The upper panel shows SDS-PAGE of phagosomes (Coomassie staining). The lower panel shows an immunoblot probed with DIL2 antibody. Reisolated (mock) salt-stripped phagosomes (lane 5) and phagosomes reisolated by flotation after incubation in the presence of ATP-ABPs (lane 6). The upper panel shows SDS-PAGE of phagosomes (silver staining). The lower panel shows an immunoblot probed with DIL2 antibody. (B and B′) Immunofluorescence labeling of isolated phagosomes with DIL2 antibody against myosin Va. (B) Phagosomes with F-actin–binding activity reisolated after preincubation with 1 mg/ml cytosol. Fluorescent patches were found associated with phagosomes. (B′) Phagosomes without F-actin–binding activity reisolated after preincubation with 20 mg/ml cytosol. No fluorescence signal was found associated with these phagosomes. Bar, 5 μm. (C) Inhibition of phagosome binding to F-actin with DIL2 antibody against myosin Va. Polyclonal DIL2 antibody (bars MyoV), Fab fragments [bars Myo(Fab)], and antigen-blocked DIL2 antibody (bars AGblock) were tested for effects on phagosome binding to F-actin when added directly to the binding reaction mixture in the presence of 2 mg/ml cytosol and salt-stripped phagosomes (cytosol), in the presence of reisolated phagosomes with F-actin-binding activity after preincubation with 2 mg/ml cytosol [Phag(+)], and in the presence of 0.1 mg/ml ATP-ABPs. Control, AG and annexin III show the effects on phagosome binding of no additions, DIL2 antigen alone, or control polyclonal antibody to annexin III (a protein normally associated with isolated phagosomes; Diakonova et al., 1997), respectively.
Immunofluorescence labeling of isolated phagosomes yielded positive labeling of discrete patches around the phagosomes that displayed F-actin–binding activity (Figure 3B and Table 1). After preincubation of salt-stripped phagosomes, which are devoid of labeling, with 1 mg/ml cytosol, 76% of reisolated phagosomes were labeled with anti-myosin Va antibody. No labeling was detected on phagosomes reisolated after preincubation with 0 mg/ml or 20 mg/ml cytosol, both of which showed no binding activity (Figure 3B′ and Table 1).
The fluorescence microscopy data were confirmed by immunoblot analysis. Myosin Va was not detected in salt-stripped phagosome preparations (Figure 3A, lane 5) and only traces of myosin Va were found in phagosome preparations reisolated after incubation with 20 mg/ml cytosol (Figure 3A, lane 3). However, immunoblotting analysis revealed the presence of myosin Va in reisolated phagosome preparations after preincubation of salt-stripped phagosomes with 1 mg/ml cytosol or 0.1 mg/ml ATP-ABP fraction, both conditions that support F-actin binding (Figure 3A, lanes 4 and 6, and Table 1). Although myosin II was abundant in cytosol, and enriched in the ATP-ABP fraction, no myosin II was detected in reisolated phagosome preparations (not shown). Thus, both immunoblotting analysis and fluorescence microscopy data revealed a specific binding of myosin Va to phagosomes and a strong correlation between the presence of actin binding activity and myosin Va association with phagosomes (Table 1).
Anti-Myosin Va Antibody Inhibits Phagosome Binding to F-Actin
We also tested the effect of DIL2 anti-myosin Va antibody on phagosome binding to F-actin in the presence of either cytosol or the ATP-ABP fraction, and on the binding of reisolated phagosomes after preincubation with 1 mg/ml cytosol. The addition of DIL2 antibody reduced the binding of salt-stripped phagosomes to F-actin in the presence of cytosol by ∼40%, in the presence of ATP-ABPs by ∼80%, and in preparations of reisolated phagosomes (with bound myosin Va and binding activity; see above) by 50% (Figure 3C). Importantly, the inhibitory effect of the DIL2 antibody in all three different reaction mixtures was about the same as the inhibitory effect of ATP (Table 1). To investigate whether the inhibition of binding was due to the cross-linking of myosin Va by bivalent DIL2 antibody molecules, Fab fragments were tested. These Fab fragments had the same inhibitory effect on phagosome binding as that observed for intact antibody (Figure 3C).
As controls for the specificity of inhibition by DIL2 antibody, we tested the effects of including in the binding assay a rabbit polyclonal anti-myosin II antibody (not shown), a rabbit polyclonal antibody against annexin III, a known phagosome protein (Diakonova et al., 1997), or the DIL2 antibody that had been pretreated with purified antigen. None of the control treatments had any significant effect on phagosome binding to F-actin (Figure 3C).
Chicken Brain Myosin Va Stimulates ATP-dependent Binding of Phagosomes to F-Actin
To show directly that myosin Va was able to bind to phagosomes and stimulate their interaction with F-actin, we preincubated the salt-stripped phagosomes with myosin Va isolated from chicken brain and, after reisolation, tested their F-actin–binding activity. As shown on Figure 4, A and B, chicken myosin Va tightly associated with phagosomes and stimulated their binding to F-actin. The binding was strongly dependent on ATP and it was specific, because it was inhibited by the polyclonal antibody against chicken myosin Va “head” domain (Figure 4B). Moreover, immunofluorescence labeling with this antibody of salt-stripped phagosomes that had been incubated with chicken myosin Va yielded the same positive labeling of discrete patches around the phagosomes as was shown for endogenous mouse myosin Va (Figure 4C). This establishes that myosin Va is the essential factor in the ATP-regulated binding of phagosomes to F-actin.
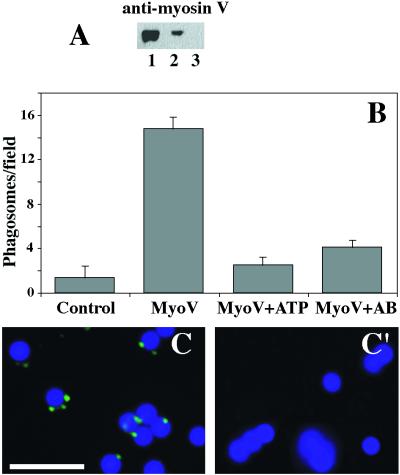
Figure 4.
ATP-dependent interaction between phagosomes and F-actin in the presence of myosin Va isolated from chicken brain. (A) Immunoblot probed with polyclonal antibody against the head domain of chicken myosin Va (anti-myosin V). (Lane 1) Purified myosin Va from chicken brain. (Lane 2) Salt-stripped phagosomes reisolated by flotation after incubation in the presence of chicken myosin V at 50 μg/ml. (Lane 3) Reisolated (mock) salt-stripped phagosomes. (B) Phagosome binding to F-actin in the presence of chicken myosin Va. Mock phagosomes without binding activity (bar control). Stimulation of phagosome binding in the presence of 50 μg/ml chicken myosin Va (bar MyoV). Inhibition of phagosome binding to F-actin with 2 mM ATP (bar MyoV+ATP) and with polyclonal antibody against the head domain of chicken myosin Va (bar MyoV+AB). (C and C′) Immunofluorescence labeling of isolated phagosomes with polyclonal antibody against chicken myosin Va. (C) Phagosomes with F-actin–binding activity reisolated after preincubation with 50 μg/ml chicken myosin Va. (C′) Phagosomes without F-actin–binding activity reisolated by flotation after preincubation with buffer. Bar, 5 μm.
Phagosome Movement in Normal and dilute-lethal Macrophages
In macrophages, phagosomes loaded with latex beads move toward the cell center (Figure 5, A and B). To determine whether myosin Va is involved in phagosome–F-actin interaction in vivo, we examined this centripetal phagosome movement in mouse bone marrow macrophages isolated from normal and dilute-lethal mice. The absence of myosin Va in dilute-lethal macrophages was confirmed by immunoblotting and binding assay (Figure 5, C and D). Moreover, the cytosol isolated from dilute-lethal macrophages stimulated phagosome binding to F-actin only to 65% of the level seen with the cytosol from normal macrophages (Figure 5D). This is in good agreement with studies with the use of the anti-myosin Va DIL2 antibody to inhibit phagosome–F-actin interaction in the presence of cytosol containing myosin Va, where binding was reduced to ∼60% of control (Figure 3C and Table 1).
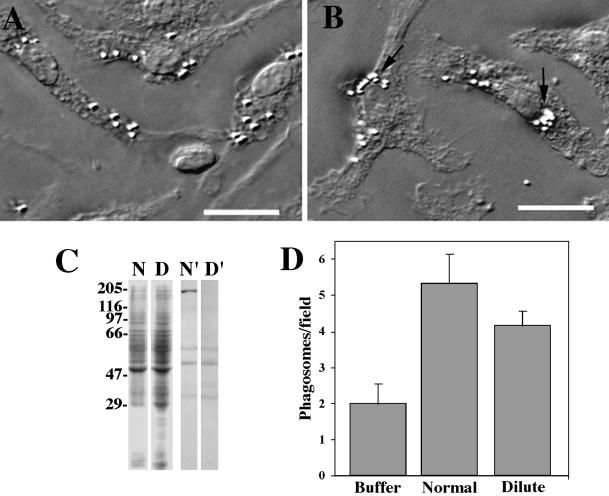
Figure 5.
(A and B) Accumulation of the phagosomes in the perinuclear region of macrophages. Cells were loaded with latex beads for 15 min (A) and then chased for 4 h (B). Accumulation of phagosomes is seen at the cell centers (arrows). The same accumulation was observed in dilute-lethal macrophages (not shown). Bars, 10 μm. (C) Detection of myosin Va in normal and dilute-lethal macrophages. SDS-PAGE of lysates of normal (N) and dilute-lethal (D) macrophages; immunoblot of lysates of normal (N′) and dilute-lethal (D′) macrophages probed with DIL2 antibody. No myosin Va was detected in dilute-lethal macrophages. (D) Binding activity of phagosomes to F-actin in the absence of cytosol (buffer) and in the presence of 0.2 mg/ml cytosol isolated from normal macrophages (normal) or from dilute-lethal macrophages (dilute). The difference of stimulation between the two types of cytosol is ∼35%.
To examine centripetal phagosome movement, normal and dilute-lethal macrophages were allowed to internalize latex beads, fixed at different chase times, and the cells were then visualized with VEC-DIC microscopy. The extent of phagosome movement toward the cell center was quantified by determining the relative number of phagosomes in the perinuclear region. In both types of macrophages, essentially all internalized latex beads were accumulated near the nucleus at 4 h after internalization (Figure 6A). Also, the number of internalized latex beads per cell was about the same in normal and dilute-lethal macrophages. In normal macrophages that had internalized latex beads, immunolabeling for myosin Va showed a positive labeling of discrete patches around the phagosomes (Figure 6B) that were similar to the patches observed on isolated phagosomes with bound myosin Va (Figure 3B).
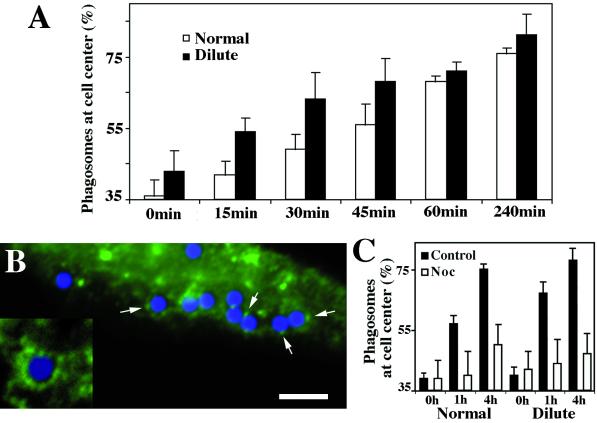
Figure 6.
(A) Time course of phagosome accumulation near the cell center in normal and dilute-lethal macrophages based on three independent experiments. Cells were loaded for 15 min, washed for 15 min (0 time), and then chased for 15, 30, 45, 60, and 240 min. For each time point, 20 cells were analyzed (see MATERIALS AND METHODS). An increased rate of accumulation was found in dilute-lethal macrophages (see time points 15, 30, and 45 min). (B) Immunofluorescence labeling with DIL2 antibody of normal macrophages engaged in phagocytosis. Fluorescent patches were found associated with some phagosomes enclosing blue latex beads in normal macrophages (arrows), but not in dilute-lethal cells (not shown). Bar, 10 μm. (Inset) Twofold-higher magnification. (C) Time course of phagosome accumulation near the cell center in normal (normal) and dilute-lethal (dilute) macrophages in the presence of nocodazole. The cells were treated as in A and chased for 1 h and 4 h in media with 5 μM nocodazole (Noc).
Surprisingly, despite no apparent difference in final phagosome uptake, the accumulation of phagosomes at the perinuclear region occurred twofold faster in dilute-lethal than in normal macrophages (Figure 6A). These data indicate that myosin Va is not required for uptake or the movement to the perinuclear regions, but seems to be involved in phagosome functions subsequent to their formation, soon after their internalization, by delaying their microtubule-dependent movement to the cell center. This directed centripetal phagosome movement is indeed mainly microtubule-dependent, because microtubule depolymerization by nocodazole strongly reduced the perinuclear phagosome accumulation in both normal and dilute-lethal macrophages (Figure 6C).
Motion Analysis of Phagosome Movement within Normal and dilute-lethal Macrophages
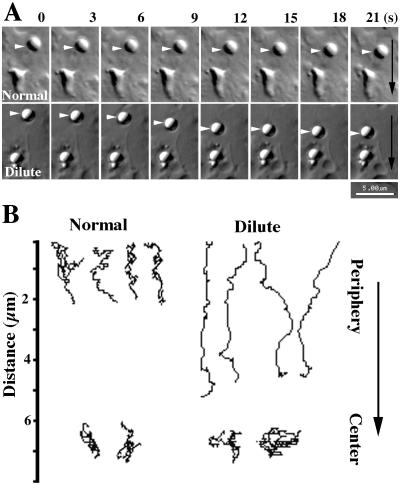
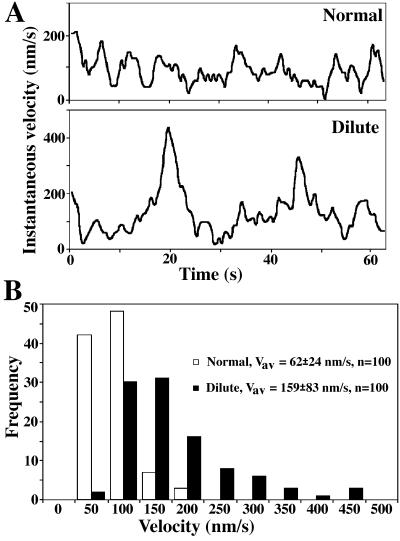
Real time VEC-DIC microscopy confirmed that all latex bead-loaded phagosomes moved toward the cell center, both in normal and dilute-lethal macrophages (Figure 7A). A more detailed analysis of 20 phagosomes in 10 cells showed that phagosome movement in normal macrophages was saltatory; phagosomes paused and temporarily reversed direction but progressed generally in a single direction (Figure 7B). Pauses and direction reversals appeared much more numerous in normal macrophages than in dilute-lethal macrophages. Remarkably, almost no reversals in direction were observed for centripetal phagosome movements in dilute-lethal macrophages (Figure 7B). However, when phagosomes reached the perinuclear region their movements were identical in normal and dilute-lethal macrophages; phagosome movement at this stage was mostly unidirectional, and predominantly via short saltatory movements (Figure 7B). In the inward movement from the periphery, phagosomes in dilute-lethal macrophages tended to make larger “jumps” between time points compared with those in normal cells (Figure 8A). The average velocities and maximum rates achieved for some phagosomes in macrophages from dilute-lethal mice were significantly greater (2.5-fold) than those observed in macrophages from normal mice (Figure 8B). Based on these observations, we propose a new antagonistic/cooperative mechanism that could be involved in saltatory phagosome movement toward the cell center (Figure 9).
Figure 7.
Phagosome movement in normal and dilute-lethal macrophages. (A) Successive video frames (3-s intervals) of centripetal phagosome movements (arrowheads) in normal (normal) and dilute-lethal (dilute) macrophages. The black arrows indicate the direction of movement toward the nucleus. (B) Typical paths for phagosome movements that occurred over a 2-min period inward from the periphery of normal macrophages and dilute-lethal macrophages. In the lower part, typical paths in the perinuclear region (center) of normal and dilute-lethal macrophages are shown. The arrow indicates the directionality of movement toward the nucleus. Download movies at http://www.biologie.uni-rostock.de/abt/tierphys/kuznet/movies.htm.
Figure 8.
Velocity of phagosome movement in the periphery of normal and dilute-lethal macrophages. (A) Distribution of instantaneous velocities attained by one phagosome in normal (normal) and in dilute-lethal (dilute) macrophages. The sampling intervals were 0.5 s. (B) Distribution of the frequencies of instantaneous velocities attained during 2 min by 10 phagosomes in five normal macrophages (normal) and by 10 phagosomes in five dilute-lethal macrophages (dilute). The sampling intervals were 1 s.
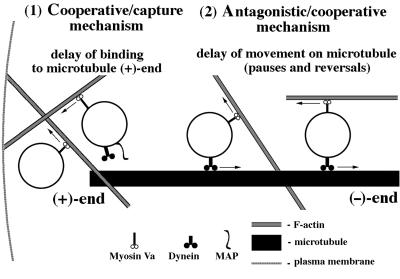
Figure 9.
Model depicting how myosin Va-based motility on F-actin and dynein-based motility on microtubules could compete and influence phagosome transport from the periphery to the cell center in macrophages. (1) According to the “cooperative/capture mechanism” proposed by Wu et al. (1998), after uptake, myosin Va serves to capture phagosomes, so that short-range movement on F-actin results in a delay of phagosome binding to (+)-ends of microtubules via dynein–dynactin complex (a microtubule-associated protein [MAP] could also be required for this binding; Blocker et al., 1998). (2) According to the “antagonistic/cooperative mechanism” proposed in this report, phagosomes move after interaction with microtubules by dynein–dynactin complex toward microtubule (−)-ends. Kinesin seems not to be involved in this process because almost no movements toward microtubule (+)-ends were observed in dilute-lethal macrophages. Myosin Va does not dissociate from the phagosomes moving on microtubules, so that it may interact with adjacently located actin filaments of random polarity. Such interactions would result in myosin Va-dependent phagosome movement on F-actin in different directions, including a direction opposite to the dynein-based movement. The myosin Va-dependent interactions and short-range movements would reduce the average and maximum rates of microtubule-dependent movement and lead to the more saltatory manner of phagosome movement, with numerous pauses and reversals. We therefore suggest that cooperation of both actin- and microtubule-based motility systems participate not only in phagosome capture but also in long-distance translocation.
DISCUSSION
The loading of phagosomes with latex beads provides a unique system for the study of the molecular interactions between a defined membranous organelle and the cytoskeleton. Because these organelles are generated by the engulfment of individual fluorescent latex beads, they are easy to detect by microscopy, and the high buoyant density of latex makes them easy to purify after different stages of organelle internalization and biogenesis (Desjardins, 1994a,b). These organelles have ∼500 proteins, of which many have already been identified (Garin et al., 2001).
In this study, we used this phagosome system to investigate the molecular basis for the in vitro interactions between mature phagosomes and F-actin and to show the involvement of myosin Va in centripetal phagosome movement in macrophages. Although the involvement of cortical F-actin in the initial uptake step of phagocytosis is well documented (Axline and Reaven, 1974; Silverstein et al., 1989; Greenberg et al., 1990, May and Machesky, 2001), the idea that F-actin and myosin-based motility could play a role in later stages of phagocytosis was until recently not often discussed (Jahraus et al., 2001).
To determine which myosins might be involved in the interaction between phagosomes and F-actin, we developed an in vitro light microscopy assay to measure the binding of phagosomes to F-actin. With the use of this binding assay, we have demonstrated that the interactions between phagosome and F-actin depend on specific cytosolic ABPs, which can bind to phagosomes and facilitate their subsequent binding to F-actin. At least two stimulatory ABPs were shown to be involved. The first factor could bind to phagosomes and then to F-actin in manner that was independent of ATP. This factor remains to be identified. The second factor, which we identified in this report as myosin Va, behaved differently. When bound to phagosomes, this myosin could not subsequently tightly bind F-actin in the presence of ATP. This behavior is typical of myosins. Our data indicate that the two classes of ABPs share the same or overlapping binding sites for F-actin on the phagosome membrane.
Our results demonstrate that among the high molecular weight myosins detected in macrophages (myosin II, myosin Ixb, and myosin Va; Swanson et al., 1999), myosin Va, but not myosin II, is involved in ATP-dependent phagosome binding to F-actin in vitro. However, we cannot yet exclude possible roles for other high molecular weight myosins, e.g., myosin IXb, in this binding. Nevertheless, all our in vitro and in vivo data show that myosin Va is one essential factor responsible for phagosome–F-actin interaction.
Recently, myosin Va was shown to colocalize with fully internalized phagosomes (Swanson et al., 1999). We confirmed the colocalization of myosin Va with phagosomes in macrophages and showed the presence of discrete patches of myosin Va associated with phagosomes. Furthermore, our data suggest that myosin Va is not associated with the entire phagosome surface but rather with defined regions of the phagosomal membrane; such a domain-restricted localization of myosin V was previously seen on membranes of the endoplasmic reticulum from squid axon (Tabb et al., 1998).
Because myosin Va associates with phagosomes in vitro and in vivo, we investigated the role of myosin Va in the process of phagosome accumulation near the nucleus in macrophages. We analyzed phagosome movement from the periphery to the cell center in normal and dilute-lethal (myosin Va null) macrophages and found that in dilute-lethal macrophages, the accumulation of phagosomes at the perinuclear region occurred significantly faster than in normal cells (Figure 6A). Moreover, the average and maximum rates of phagosome movement were higher in dilute-lethal macrophages (Figure 8). Therefore, in vivo, myosin Va seems to play an inhibitory role, delaying the ability of phagosomes to undergo the long-range microtubule-dependent motility that brings them toward the macrophage cell center. Unfortunately, our attempts to rescue dilute-lethal macrophages by microinjection of purified chicken brain myosin V were not successful. Even the microinjection of macrophages with control fluorescence proteins (tubulin or actin) led immediately to accumulation of the injected proteins inside membrane organelles, most probably due to autophagy.
The microtubule-dependent motor dynein is likely to play the key role in net, long-range, centripetal phagosome movement. First, microtubule depolymerization by nocodazole strongly inhibits phagosome motility in vivo (Blocker et al., 1998; this report, Figure 6C). Second, the majority (70%) of phagosomes moving in vitro along microtubules was found to be minus-end directed and dynein-dependent (Blocker et al., 1997). Third, in vitro, phagosomes preferentially interacted with microtubule plus-ends (Blocker et al., 1998), which are mainly located in the cell periphery.
Taken together, our observations suggest that the “cooperative/capture model” proposed for melanosome transport within melanocytes by Wu et al. (1998) might help us to explain some aspects of phagosome transport from the periphery to the cell center in macrophages. According to this model, a myosin Va-dependent interaction of phagosomes with F-actin in the peripheral regions of macrophages would temporarily prevent the phagosomes from docking at the plus-end of microtubules, a process necessary for the fast retrograde, microtubule-dependent phagosome transport toward the perinuclear region. This means that the myosin Va-dependent phagosome interaction with F-actin in the cell periphery competes with the ability of phagosomes to bind microtubules.
However, the cooperative/capture model alone is not able to explain why in normal macrophages the inward phagosome movement was always saltatory with frequent reversals in direction, whereas in dilute-lethal macrophages almost no reversals in direction were observed. Therefore, we are forced to postulate that an additional mechanism must operate in conjunction with the cooperative/capture mechanism (Wu et al., 1998). In particular, we propose that an antagonistic interaction between the F-actin and microtubule systems is involved not only in deciding when a peripheral phagosome starts its journey toward the cell center but also in defining how fast this journey will be (Figure 9). According to our model, simultaneous and antagonistic interactions of phagosomes with both F-actin, via myosin Va, and with microtubules, via dynein and dynactin, would be responsible for the numerous pauses and reversals in direction that we observed in normal macrophages. A phagosome already moving inwards in the cell via dynein could interact with F-actin and even switch to the F-actin system via myosin Va. These actin-dependent “digressions” would thus antagonize the dynein-dependent motility. This possibility would also be consistent with our previous observation that, in axoplasm preparations from squid giant axons, a given organelle can move on both F-actin and microtubules, visibly switching from one system to the other (Kuznetsov et al., 1992). The “antagonistic/cooperative model” as proposed here is also supported by the recent finding that myosin Va is a processive motor (Mehta et al., 1999; Walker et al., 2000; Rief et al., 2000), suggesting that myosin Va activity can indeed compete with activity of the dynein motor complex.
We think that the antagonistic/cooperative mechanism could be involved not only in the saltatory movement of phagosomes in macrophages but also in the saltatory movement of other organelles that has been observed in different cell types for many years (Rebhun, 1972). Indeed, when Weiss et al. (1986) compared various types of intracellular organelle motility by quantitative motion analysis, they found that “interrupted motion type II” (pauses and direction reversals) is typical for larger organelles, such as lysosomes in cultured cells.
If myosin Va associated with phagosomes is not responsible for net centripetal phagosome movement, and in fact seems to delay their arrival at the cell center, what role could this motor play for phagosomes in macrophages? We suggest that one role of myosin Va is to delay the passage of phagosomes to the microtubular machinery, as well as to antagonize already started microtubule motility events. We speculate that the role of this delay in the peripheral regions may facilitate the fusion of phagosomes with early endosomal organelles, an idea we are currently testing.
The association of myosin Va with fully internalized phagosomes, as well as the involvement of F-actin in phagosome fusion with endocytic organelles in vitro (Jahraus et al., 2001), supports our hypotheses that the interaction of phagosomes with F-actin, including the myosin Va-dependent process, is an integral part of the life of a phagosome. This is clearly a complex system in which the actin cytoskeleton interacts with the microtubule motor-dependent machinery. The latter is responsible for the long-range transport to the perinuclear region of cells. The work presented here describes yet another novel aspect of phagosomes in their ability to interact with the cytoskeleton. Besides their ability to bind F-actin, these organelles can also nucleate actin polymerization (Defacque et al., 2000), interact with microtubules (Blocker et al., 1996), move along microtubules (Blocker et al., 1997), and fuse with early and late endocytic organelles (Rabinowitz et al., 1992; Desjardins et al., 1994b; Claus et al., 1998; Jahraus et al., 1998, 2001). A major challenge now is to link these different processes involving the cytoskeleton to one another and to the events that control docking and membrane fusion.
ACKNOWLEDGMENTS
We thank D. Kaiser for supplying the antibody against myosin II, M.S. Mooseker and R.E. Larson for that against myosin Va, and J. Ernst for providing us with the anti-annexin III antibody. We are grateful to V.I. Gelfand, J. Hodgkinson, R. Kjeken, and R. Palazzo for critical comments on the manuscript. This work was supported by a W.J. Fulbright Commission fellowship to M.A.S.; a Human Frontiers Science Program grant to H.Y, J.K.B., G.G., and S.A.K.; and DFG Innovationskolleg “Komplexe und Zelluläre Sensorsysteme.”
Abbreviations used:
- ABP
actin binding protein
- FSG
fish skin gelatin
- HAP
hydroxyapatite
- HB
HEPES buffer
- HBS
HB plus 10% sucrose
- HBKS
HBS plus 50 mM KCl
- VEC-DIC
video-enhanced contrast differential interference contrast microscopy
REFERENCES
- Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- Allen RD, Weiss DG, Hayden JH, Brown DT, Fujiwake H, Simpson M. Gliding movement of and bidirectional transport along single native microtubules from squid axoplasm: evidence for an active role of microtubules in cytoplasmic transport. J Cell Biol. 1985;100:1736–1752. doi: 10.1083/jcb.100.5.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axline SG, Reaven EP. Inhibition of phagocytosis and plasma membrane mobility of the cultivated macrophage by cytochalasin B. Role of subplasmalemmal F-actin. J Cell Biol. 1974;62:647–659. doi: 10.1083/jcb.62.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Griffiths G, Olivo J-C, Hyman AA, Severin FF. A role for microtubule dynamics in phagosome movement. J Cell Sci. 1998;111:303–312. doi: 10.1242/jcs.111.3.303. [DOI] [PubMed] [Google Scholar]
- Blocker A, Severin FF, Burkhardt JK, Bingham JB, Yu H, Olivo J-C, Schroer TA, Hyman AA, Griffiths G. Molecular requirements for bi-directional movement of phagosomes along microtubules. J Cell Biol. 1997;137:113–129. doi: 10.1083/jcb.137.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Severin FF, Habermann A, Hyman AA, Griffiths G, Burkhardt JK. MAP-dependent binding of phagosomes to microtubules. J Biol Chem. 1996;271:3803–3811. doi: 10.1074/jbc.271.7.3803. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bridgman PC. Myosin Va movements in normal and dilute-lethal axons provide support for a dual filament motor complex. J Cell Biol. 1999;146:1045–1060. doi: 10.1083/jcb.146.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney RE. Purification and assay of myosin V. Methods Enzymol. 1998;298:3–18. doi: 10.1016/s0076-6879(98)98003-x. [DOI] [PubMed] [Google Scholar]
- Cheney RE, O'Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, Forscher P, Larson RE, Mooseker MS. Brain myosin V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- Chimini G, Chavrier P. Function of Rho family proteins in actin dynamics during phagocytosis and engulfment. Nat Cell Biol. 2000;10:E191–E196. doi: 10.1038/35036454. [DOI] [PubMed] [Google Scholar]
- Claus V, Jahraus A, Tjelle T, Berg T, Kirschke H, Faulstich H, Griffiths G. Lysosomal enzyme trafficking between phagosomes, endosomes, and lysosomes in J774 macrophages: enrichment of cathepsin H in early endosomes. J Biol Chem. 1998;273:9842–9851. doi: 10.1074/jbc.273.16.9842. [DOI] [PubMed] [Google Scholar]
- de Lanerolle P, Gorgas G, Li X, Schluns K. Myosin light chain phosphorylation does not increase during yeast phagocytosis by macrophages. J Biol Chem. 1993;268:6883–16886. [PubMed] [Google Scholar]
- Defacque H, Egeberg M, Habermann A, Diakonova M, Roy C, Mangeat P, Voelter W, Marriott G, Pfannstiel J, Faulstich H, Griffiths G. Involvement of erzin/moesin in de novo actin assembly on phagosomal membranes. EMBO J. 2000;19:199–212. doi: 10.1093/emboj/19.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjardins M, Celis JE, van Meer G, Dieplinger H, Jahraus A, Griffiths G, Huber LA. Molecular characterization of phagosomes. J Biol Chem. 1994a;269:32194–32200. [PubMed] [Google Scholar]
- Desjardins M, Huber AM, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994b;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakonova M, Gerke V, Ernst J, Liautard JP, van der Vusse G, Griffiths G. Localization of five annexins in J774 macrophages and on isolated phagosomes. J Cell Sci. 1997;110:1199–1213. doi: 10.1242/jcs.110.10.1199. [DOI] [PubMed] [Google Scholar]
- Espindola FS, Espreafico EM, Coelho MV, Martins AR, Costa FR, Mooseker MS, Larson RE. Biochemical and immunological characterization of p190-calmodulin complex from vertebrate brain: a novel calmodulin-binding myosin. J Cell Biol. 1992;118:359–368. doi: 10.1083/jcb.118.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LL, Lee AJ, Bridgman PC, Mooseker MS. Vesicle-associated brain myosin V can be activated to catalyze actin-based transport. J Cell Sci. 1998;111:2055–2066. doi: 10.1242/jcs.111.14.2055. [DOI] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S, Burridge K, Silverstein SC. Co-localization of F-actin and talin during Fc receptor-mediated phagocytosis in mouse macrophages. J Exp Med. 1990;172:1853–1856. doi: 10.1084/jem.172.6.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S, Silverstein SC. Phagocytosis. In: Paul WE, editor. Fundamental Immunology. 3rd ed. New York: Raven Press; 1993. pp. 137–191. [Google Scholar]
- Gyoeva FK, Gelfand VI, Rosenblat VA. Spleen protein forming gel with actin. Dokladi Academii Nauk USSR. 1983;268:246–248. (in Russian). [PubMed] [Google Scholar]
- Hartwig JH, Stossel TP. Macrophages: their use in elucidation of the cytoskeletal roles of actin. Methods Cell Biol. 1982;25:201–225. doi: 10.1016/s0091-679x(08)61426-0. [DOI] [PubMed] [Google Scholar]
- Jahraus A, Egeberg M, Hinner B, Habermann A, Sackman E, Pralle A, Faulstich H, Rubin VV, Defacque H, Griffiths G. ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol Cell Biol. 2001;12:155–170. doi: 10.1091/mbc.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahraus A, Storrie B, Griffiths G, Desjardins D. Evidence for retrograde traffic between terminal lysosomes and the prelysosomal/late endosome compartment. J Cell Sci. 1994;107:145–157. doi: 10.1242/jcs.107.1.145. [DOI] [PubMed] [Google Scholar]
- Jahraus A, Tjelle TE, Berg T, Habermann A, Storrie B, Ullrich O, Griffiths G. In vitro fusion of phagosomes with different endocytic organelles from J774 macrophages. J Biol Chem. 1998;273:30379–30390. doi: 10.1074/jbc.273.46.30379. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Langford GM, Weiss DG. Actin-dependent organelle movement in squid axoplasm. Nature. 1992;356:722–725. doi: 10.1038/356722a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. Cleavage of structural proteins during the assembly of the bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langford GM, Molyneaux BJ. Myosin V in the brain: mutations lead to neurological defects. Brain Res Rev. 1998;28:1–8. doi: 10.1016/s0165-0173(98)00020-4. [DOI] [PubMed] [Google Scholar]
- May RC, Machesky LM. Phagocytosis and the actin cytoskeleton. J Cell Sci. 2001;114:1061–1077. doi: 10.1242/jcs.114.6.1061. [DOI] [PubMed] [Google Scholar]
- Mehta AD, Rock RS, Rief M, Spudich JA, Mooseker MS, Cheney RE. Myosin-V is a processive actin-based motor. Nature. 1999;400:590–593. doi: 10.1038/23072. [DOI] [PubMed] [Google Scholar]
- Mermall V, Post PL, Mooseker MS. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science. 1998;279:527–533. doi: 10.1126/science.279.5350.527. [DOI] [PubMed] [Google Scholar]
- Nascimento AAC, Amaral RG, Bizario JCS, Larson RE, Espreafico EM. Subcellular localization of myosin V in the B16 melanoma cells, a wild-type cell line for the dilute gene. Mol Biol Cell. 1997;8:1971–1988. doi: 10.1091/mbc.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt A, Mayorga LS, Schwartz AL, Stahl PD. Transport of phagosomal components to an endosomal compartment. J Biol Chem. 1992;267:126–132. [PubMed] [Google Scholar]
- Pollard TD. Myosin purification and characterization. Methods Cell Biol. 1982;24:333–371. doi: 10.1016/s0091-679x(08)60665-2. [DOI] [PubMed] [Google Scholar]
- Rabinowitz S, Horstmann H, Gordon S, Griffiths G. Immunocytochemical characterization of the endocytic and phagolysosomal compartments in peritoneal macrophages. J Cell Biol. 1992;116:95–112. doi: 10.1083/jcb.116.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhun LI. Polarized intracellular particle transport: saltatory movements and cytoplasmic streaming. Int Rev Cytol. 1972;32:93–137. doi: 10.1016/s0074-7696(08)60339-3. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Provance DW, Jr, Mooseker MS, Mercer JA. Class V myosins. Biochem Biophys Acta. 2000;1496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- Rezabek BL, Rodriguez-Paris JM, Cardelli JA, Chia CP. Phagosomal proteins of Dictyostelium discoideum. J Eukaryot Microbiol. 1997;44:284–292. doi: 10.1111/j.1550-7408.1997.tb05668.x. [DOI] [PubMed] [Google Scholar]
- Rief M, Rock RS, Mehta AD, Mooseker MS, Cheney RE, Spudich JA. Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci USA. 2000;97:9482–9486. doi: 10.1073/pnas.97.17.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Gelfand VI. Myosin cooperates with microtubule motors during organelle transport in melanophores. Curr Biol. 1998;8:161–164. doi: 10.1016/s0960-9822(98)70063-6. [DOI] [PubMed] [Google Scholar]
- Silverstein SC, Greenberg S, Di Virgilio F, Steinberg T. Phagocytosis. In: Paul WE, editor. Fundamental Immunology. New York: Raven Press; 1989. pp. 703–720. [Google Scholar]
- Spudich JA, Watt SJ. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Stockem W, Hoffmann HU, Gruber B. Dynamics of the cytoskeleton in Amoeba proteus. I. Redistribution of microinjected fluorescein-labeled actin during locomotion, immobilization and phagocytosis. Cell Tissue Res. 1983;232:79–96. doi: 10.1007/BF00222375. [DOI] [PubMed] [Google Scholar]
- Stossel TP, Hartwig JH. Interactions between actin, myosin, and an actin-binding protein from rabbit alveolar macrophages. Alveolar macrophage myosin Mg2+-adenosine triphosphatase requires a cofactor for activation by actin. J Biol Chem. 1975;250:5706–5712. [PubMed] [Google Scholar]
- Swanson JA, Johnson MT, Beningo K, Post P, Mooseker M, Araki N. A contractile activity that closes phagosomes in macrophages. J Cell Sci. 1999;112:307–316. doi: 10.1242/jcs.112.3.307. [DOI] [PubMed] [Google Scholar]
- Tabb JS, Molyneaux BJ, Cohen DL, Kuznetsov SA, Langford GM. Transport of ER vesicles on actin filaments in neurons by myosin V. J Cell Sci. 1998;111:3221–3234. doi: 10.1242/jcs.111.21.3221. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyohara A, Inaba K. Transport of phagosomes in mouse peritoneal macrophages. J Cell Sci. 1989;94:143–153. doi: 10.1242/jcs.94.1.143. [DOI] [PubMed] [Google Scholar]
- Walker ML, Burgess SA, Sellers JR, Wang F, Hammer III JA, Trinick J, Knight PJ. Two-headed binding of a processive myosin to F-actin. Nature. 2000;405:804–807. doi: 10.1038/35015592. [DOI] [PubMed] [Google Scholar]
- Weiss DG, Keller F, Gulden J, Maile W. Towards a new classification of intracellular particle movements based upon quantitative analysis. Cell Motil Cytoskeleton. 1986;6:128–135. doi: 10.1002/cm.970060210. [DOI] [PubMed] [Google Scholar]
- Weiss DG, Maile W, Wick RA, Steffen W. Video microscopy. In: Lacey AJ, editor. Light Microscopy in Biology. A Practical Approach. 2nd ed. Oxford, UK: Oxford University Press; 1999. pp. 73–105. [Google Scholar]
- Wu X, Bowers B, Rao K, Wei Q, Hammer III JA. Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function in vivo. J Cell Biol. 1998;28:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bowers B, Wei Q, Kocher B, Hammer III JA. Myosin V associates with melanosomes in mouse melanocytes: evidence that myosin V is an organelle motor. J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- Wu X, Jung G, Hammer III JA. Functions of unconventional myosins. Curr Opin Cell Biol. 2000;12:42–51. doi: 10.1016/s0955-0674(99)00055-1. [DOI] [PubMed] [Google Scholar]