Abstract
The signal recognition particle (SRP) enables cotranslational delivery of proteins for translocation into the endoplasmic reticulum (ER), but its full in vivo role remains incompletely explored. We combined rapid auxin-induced SRP degradation with proximity-specific ribosome profiling to define SRP’s in vivo function in yeast. Despite the classic view that SRP recognizes amino-terminal signal sequences, we show that SRP was generally essential for targeting transmembrane domains regardless of their position relative to the amino-terminus. By contrast, many proteins containing cleavable amino-terminal signal peptides were efficiently cotranslationally targeted in SRP’s absence. We also reveal an unanticipated consequence of SRP loss: Transcripts normally targeted to the ER were mistargeted to mitochondria, leading to mitochondrial defects. These results elucidate SRP’s essential roles in maintaining the efficiency and specificity of protein targeting.
Main Text
Regardless of their ultimate destination, nuclear-encoded genes are translated in the cytosol. In non-plant eukaryotes, two cellular locations serve as the major sites of translocation across lipid bilayers: the mitochondrial outer membrane and the surface of the endoplasmic reticulum (ER), the latter being the entry point for proteins in the secretory pathway. The role of the signal recognition particle (SRP) in mediating cotranslational ER targeting is well established. SRP recognizes N-terminal hydrophobic signals of nascent polypeptide chains and, through interaction with the ER-localized SRP receptor, directs them to the translocon (1).
This classical model, however, fails to capture the full diversity of SRP targeting strategies and SRP function, leaving open several unanswered questions. A substantial fraction of proteins containing N-terminal targeting sequences do not require SRP for translocation (2, 3), and, in vitro, these are capable of posttranslational ER translocation (4, 5). However, SRP was recently shown to be comparably associated with SRP-independent and -dependent nascent chains (6); whether this engagement is important for cotranslational targeting of SRP-independent proteins (6, 7) is unknown. Recent studies have also pointed to several nonconventional modes of SRP interaction with nascent chains in vivo. For example, SRP can be recruited to mRNAs before the targeting signal is accessible (6, 8, 9), and in bacteria, SRP has been found to bind internal transmembrane domains (TMDs) (8). However, a general role of SRP in enabling cotranslational targeting of eukaryotic proteins lacking N-terminal targeting sequences is not established. Furthermore, targeting to either the ER or mitochondria is highly specific, with minimal mistargeting or dual targeting of proteins (10). In metazoans, the nascent polypeptide-associated complex (NAC) plays a critical role in this specificity by preventing the mistargeting of mitochondrial proteins to the ER (11). It is not understood how cells prevent mistargeting of ER proteins to mitochondria.
To address these issues, we sought to monitor cotranslational protein targeting after rapid SRP depletion. We used the plant-based auxin-induced degradation (AID) system (12) to induce acute and robust loss of one of the subunits of SRP, Srp72 (Fig. 1, A and B). The depletion of Srp72 was rapid, with a half-life of about 5 minutes (Fig. 1C). We used proximity-specific ribosome profiling (7), which utilizes a biotin ligase (BirA) localized to a subcellular location, such as the surface of the ER or the mitochondrial outer membrane, to label ribosomes at this location. Subsequent affinity purification of labeled ribosomes and sequencing of ribosome-protected mRNA fragments provide a snapshot of translation at that subcellular location.
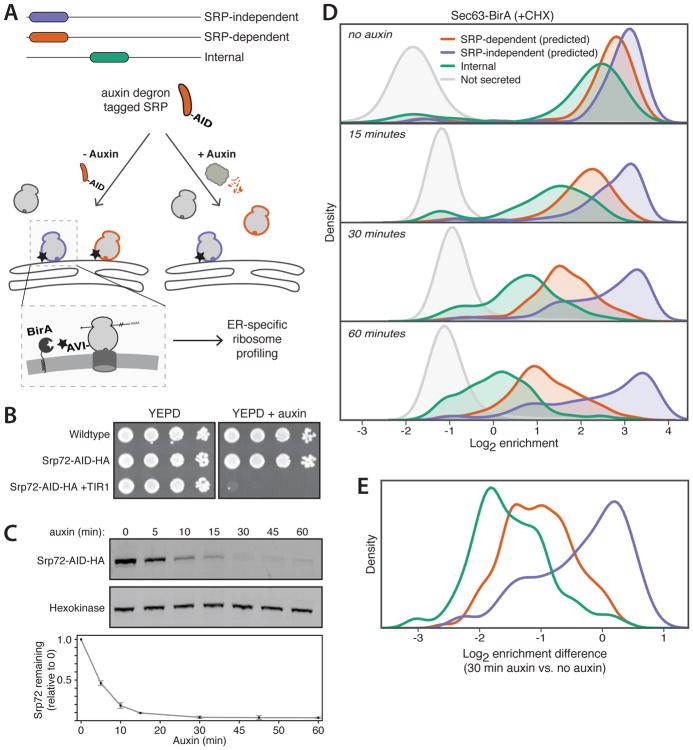
Fig. 1. A strategy to functionally probe acute loss of SRP.
(A) Experimental setup. Ribosomes (round gray shapes) translating SRP-independent (purple outline) or -dependent (orange outline) proteins at the ER are labeled with biotin (star) on the AVI tag. Experiments are also performed with auxin treatment (right) to induce degradation of SRP by the proteasome. (B) Growth of auxin-degron tagged Srp72 (Srp72-AID) yeast in the absence or presence of auxin. YEPD, yeast extract peptone dextrose medium; HA, hemagglutinin; TIR1, Oryza sativa TIR1. (C) Immunoblots and quantification of Srp72-AID depletion upon auxin addition (mean ± SD, N=3). (D) Density distributions of Sec63-BirA log2 enrichments with Srp72-AID after indicated times of auxin treatment. Genes are separated and colored by predictions of SRP-dependence from (3). (E) Density distributions of log2 enrichment differences comparing 30 min auxin to no auxin for each gene. Genes are categorized as in (D).
To identify proteins that rely on SRP for efficient ER targeting in yeast, we performed a time course of proximity-specific ribosome profiling in the presence of cycloheximide (CHX) after Srp72 depletion using two ER-localized BirA fusions: Sec63-BirA (Fig. 1D) and BirA-Ubc6 (fig. S1A). Upon loss of SRP, transcripts of proteins previously predicted to be SRP-dependent (3) showed time-dependent depletion at the ER, validating our system. Transcripts of genes encoding internal TMDs, but lacking N-terminal targeting signals, also depleted from the ER upon loss of SRP on a similar timescale (Fig. 1D and fig. S1A) and at a similar magnitude (Fig. 1E and fig. S1B). Thus, with the prominent exception of tail-anchored (TA) proteins and substrates of the SRP-independent targeting (SND) proteins whose TMDs are at or near the C-terminus, respectively (13–15), SRP is essential for targeting TMDs to the ER regardless of their distance to the N-terminus.
By contrast, a prominent subset of mRNAs predicted to be SRP-independent remained at the ER throughout the 1-hour auxin treatment (Fig. 1D and fig. S1A). A small group of mRNAs is known to be cotranslationally targeted to the ER in the presence of CHX, which uncouples the kinetics of translation and ER targeting (7). To ensure that the observed cotranslational targeting in the absence of SRP was not due to this effect, we performed a time course of Srp72 depletion and proximity-specific ribosome profiling without CHX treatment (Fig. 2A). In the absence of auxin treatment, the extent of ER enrichment for secretory-protein transcripts was reduced as compared to that observed in CHX-treated cells (fig. S2), as expected (7). Proteins containing targeting TMDs were nearly universally SRP-dependent, whereas a prominent group of proteins containing cleavable signal peptides with low hydrophobicity remained efficiently cotranslationally targeted after prolonged SRP depletion (fig. S3).
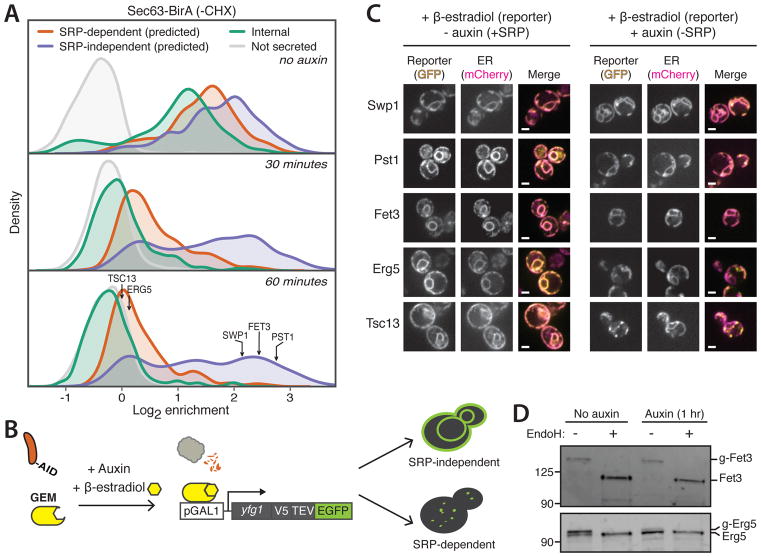
Fig. 2. Efficient cotranslational ER targeting in the absence of SRP.
(A) Density distributions of Sec63-BirA log2 enrichments with Srp72-AID after indicated times of auxin treatment. Arrows indicate enrichments of reporter genes. (B) Schematic of β-estradiol-inducible reporter system. GEM is a fusion protein consisting of GAL DNA-binding domain, β-estradiol-binding domain, and Msn2 activation domain. pGAL1, GAL1 promoter; yfg1, reporter gene; V5, V5 epitope tag; TEV, tobacco etch virus protease site; EGFP, enhanced green fluorescent protein. (C) Single-plane confocal fluorescence microscopy images of yeast with Srp72-AID expressing reporter-GFP (yellow) and ER-mCherry-HDEL (magenta) in the absence (left) or presence (right) of auxin. Scale bar, 2 μm. (D) Immunoblots of reporters Fet3 and Erg5 in the absence or presence of auxin. Where indicated, lysates were treated with endoglycosidase H (EndoH). g, glycosylated.
The ER enrichment of transcripts encoding SRP-independent proteins observed at later time points theoretically could be due to polysome tethering of mRNAs, although the 1-hour duration of our time course was considerably longer than typical yeast mRNA half-lives (~10 min) (16). To examine the effect of SRP depletion on de novo mRNA targeting, we induced expression of reporter mRNAs with β-estradiol under conditions of SRP depletion (Fig. 2B), focusing on five genes representing a range of targeting signals (fig. S4). In our ribosome profiling experiments, transcripts of three genes (SWP1, PST1, and FET3) displayed efficient ER targeting in SRP’s absence (Fig. 2A). However, FET3 is predicted to be SRP-dependent, and its localization is independent of SEC72 (3), a factor implicated in translocation of some SRP-independent substrates (17, 18). All three proteins showed ER localization by fluorescence microscopy, even when SRP was depleted (Fig. 2C). By contrast, two proteins whose targeting was observed to be SRP-dependent, Erg5 and Tsc13, which have an N-terminal or an internal targeting TMD, respectively, failed to localize to the ER when SRP was depleted (Fig. 2C). Consistent with the microscopy, in cells depleted of SRP, Fet3, but not Erg5, was glycosylated, indicating translocation into the ER (Fig. 2D). Thus, SRP-independent proteins can be de novo cotranslationally targeted and translocated into the ER, even in the prolonged absence of SRP.
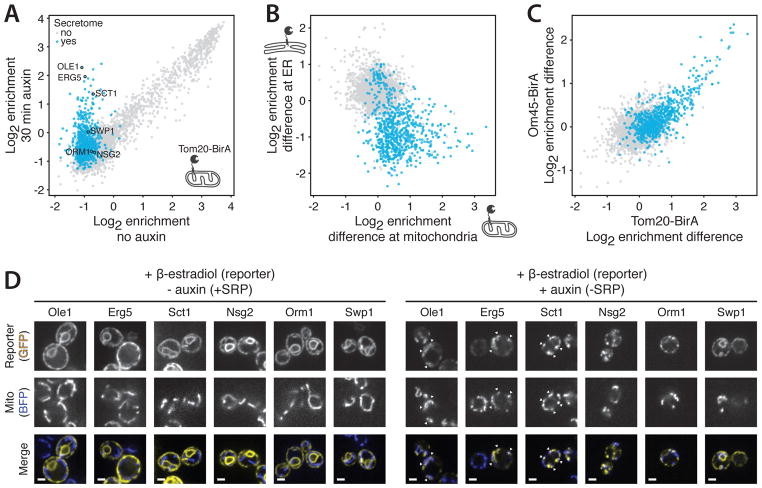
We next examined if loss of SRP can lead to mistargeting of proteins. Although the ER and mitochondria are connected (19), targeting to these organelles is exquisitely specific (10). To explore if SRP contributes to this specificity, we performed the proximity-specific ribosome profiling experiment using a mitochondrially localized BirA fusion (Fig. 3A). Mitochondrial protein targeting was unaffected by loss of SRP (fig. S5). By contrast, a number of mRNAs were newly targeted to mitochondria, and most encoded for proteins normally cotranslationally targeted to the ER (Fig. 3A). Transcripts that remained efficiently targeted to the ER in the absence of SRP were generally not mistargeted to mitochondria (Fig. 3B). We observed the same set of mistargeted mRNAs in multiple replicates and with two different mitochondrially localized BirA fusions (Tom20 and Om45) and strain backgrounds (Fig. 3C). Thus, a prominent subset of SRP-dependent substrates, including proteins with N-terminal and internal targeting sequences, were cotranslationally targeted to the mitochondrial surface in SRP’s absence.
Fig. 3. Secretory proteins are mistargeted to mitochondria in absence of SRP.
(A) Scatter plot of Tom20-BirA log2 enrichments with Srp72-AID comparing no treatment or treatment with auxin. Genes encoding secretory proteins are shown in blue. (B) Scatter plot comparing log2 enrichments differences of 30-min auxin treatment relative to no auxin treatment. Biotinylation was carried out at mitochondria (Tom20-BirA, x axis) in the presence of CHX or at the ER (Sec63-BirA, y axis) in the absence of CHX. Colors as in (A). (C) Scatter plot comparing log2 enrichment differences of 30-min auxin treatment relative to no auxin treatment. Experiments were performed in W303 (x axis) or BY4741 (y axis) strain backgrounds. Colors as in (A). (D) Single-plane confocal fluorescence microscopy images of yeast with Srp72-AID expressing reporter-GFP (yellow) and mitochondrial Su9-TagBFP (blue; BFP, blue fluorescent protein) in the absence (left) or presence (right) of auxin. Scale bar, 2 μm.
We next used our β-estradiol-inducible system (Fig. 2B) to visualize protein localization of six proteins (fig. S4) consisting of five (Ole1, Erg5, Sct1, Orm1, and Nsg2) that displayed SRP-dependent ER targeting and one (Swp1) that was SRP independent. In our ribosome profiling experiments, OLE1, ERG5, and SCT1 transcripts were consistently mistargeted to mitochondria, whereas ORM1 and NSG2 transcripts were not. In SRP-depleted cells, Ole1, Erg5, and Sct1 colocalized with mitochondria (Fig. 3D and fig. S6), although whether these proteins cross the mitochondrial membranes is an open question. By contrast, Swp1, Nsg2, and Orm1 were predominantly co-localized with the ER or were observed in focal structure adjacent to the ER (Fig. 3D and fig. S6), which might represent cytosolic aggregates.
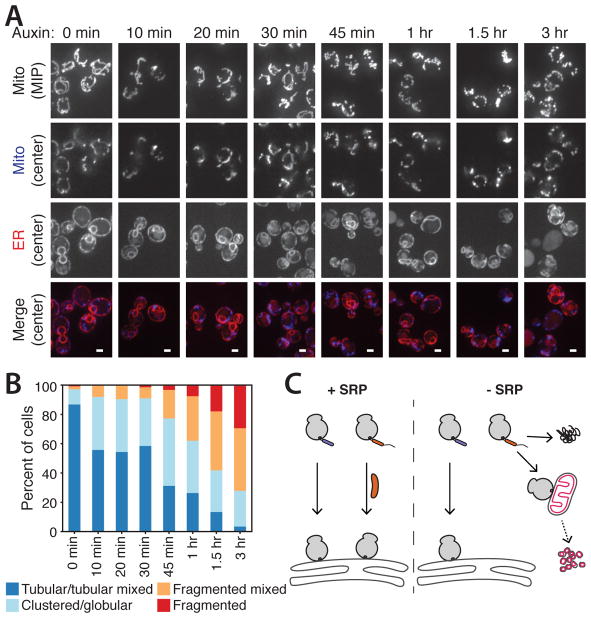
In cells depleted of SRP for 1 hour, mitochondria lost their normal tubular structure and, instead, were more fragmented (Fig. 3D). To examine how proximal this morphological defect is to SRP depletion, we imaged cells at various time points after auxin treatment (Fig. 4A and fig. S7A) and categorized mitochondrial morphology (fig. S7B). Mitochondrial morphology defects were observed at early time points (Fig 4B), before detection of ER stress (fig. S7C) and while ER morphology appeared normal (fig. S8). The rapid emergence of mitochondrial morphology defects in cells suggests that they are not a secondary consequence of ER morphological defects; however, it is possible that the later, severe mitochondrial defects are affected by the ER defects. The observed mitochondrial fragmentation required synthesis of new proteins, because cells depleted of SRP and treated with CHX possessed predominantly tubular mitochondria (fig. S9). Thus, mistargeting of SRP-dependent proteins to mitochondria disrupts mitochondrial structure.
Fig. 4. Loss of SRP leads to rapid mitochondrial defects.
(A) Confocal fluorescence microscopy of mitochondrial and ER morphology with Srp72-AID after indicated times of auxin treatment. Mitochondrial Su9-TagBFP is shown as a maximum intensity projection (MIP, top) or a single plane (blue, below) with ER-mCherry-HDEL (red). Scale bar, 2 μm. (B) Quantification of mitochondrial morphologies from (A). (C) Summary of physiological consequences of loss of SRP. Left: In the presence of SRP (orange rod), ribosomes translating SRP-independent (purple) and -dependent (orange) proteins are targeted to the ER. Right: Without SRP, SRP-independent proteins are targeted to the ER, and proteins not targeted to the ER can form aggregates (top) or be mistargeted to mitochondria (pink), leading to fragmentation.
Three important principles of SRP action now emerge (Fig. 4C). First, a prominent subset of proteins with cleavable N-terminal signal peptides is efficiently cotranslationally targeted to the ER in the complete and extended absence of SRP. Second, with the exception of proteins only containing TMDs at or near to the C-terminus (13–15), targeting and translocation of proteins with anchoring TMDs, regardless of their position relative to the N-terminus, are fully dependent on SRP. Together with work in bacteria (8), our findings suggest that a universal feature of SRP’s action is to engage TMDs across the length of the nascent chain. Third, our results reveal an unanticipated role for SRP in maintaining the specificity of organelle targeting. Without SRP, a subset of proteins becomes susceptible to aberrant cotranslational targeting to mitochondria reminiscent of the mistargeting of some TA proteins to mitochondria in the absence of Get3/TRC40 ER targeting factors (13). This may reflect a more general tendency of hydrophobic or aggregation-prone proteins to be recruited to mitochondria (20). Mistargeting in the absence of SRP triggered rapid mitochondrial fragmentation, an indicator of mitochondrial dysfunction. Consistent with SRP playing a role in maintaining mitochondrial integrity, yeast adapted for SRP deletions acquire rho− (respiration-deficient) phenotypes (21, 22). This role of SRP parallels that of NAC, which can prevent mistargeting of mitochondrial proteins to the ER in metazoans (11). Together, these observations illustrate the critical role of maintaining specificity of protein targeting for proper organelle function.
Supplementary Material
Acknowledgments
We thank members of the Weissman and Nunnari labs for discussion; C. Jan, M. Jost, and M. Shurtleff for critical reading of the manuscript; and the University of California, San Francisco, Center for Advanced Technology for technical assistance. This work was supported by the Howard Hughes Medical Institute (J.S.W.) and NIH grants: R01AG041826 (J.S.W), P50GM102706 (J.S.W.), R01GM062942 (J.N.), R01GM097432 (J.N.), and R01GM106019 (J.N.). E.A.C. was supported by NSF Graduate Research Fellowship 1144247. J.N. serves on the scientific advisory board of Mitobridge, Inc. Data are deposited in the Gene Expression Omnibus under accession number GSE106329.
References and Notes
- 1.Zhang X, Shan SO. Fidelity of cotranslational protein targeting by the signal recognition particle. Annu Rev Biophys. 2014;43:381–408. doi: 10.1146/annurev-biophys-051013-022653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. The Journal of Cell Biology. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ast T, Cohen G, Schuldiner M. A Network of Cytosolic Factors Targets SRP-Independent Proteins to the Endoplasmic Reticulum. Cell. 2013;152:1134–1145. doi: 10.1016/j.cell.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Waters MG, Blobel G. Secretory protein translocation in a yeast cell-free system can occur posttranslationally and requires ATP hydrolysis. The Journal of Cell Biology. 1986;102:1543–1550. doi: 10.1083/jcb.102.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen W, Walter P. Prepro-carboxypeptidase Y and a truncated form of pre-invertase, but not full-length pre-invertase, can be posttranslationally translocated across microsomal vesicle membranes from Saccharomyces cerevisiae. The Journal of Cell Biology. 1988;106:1075–1081. doi: 10.1083/jcb.106.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartron JW, Hunt KCL, Frydman J. Cotranslational signal-independent SRP preloading during membrane targeting. Nature. 2016;536:224–228. doi: 10.1038/nature19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jan CH, Williams CC, Weissman JS. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521–1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schibich D, et al. Global profiling of SRP interaction with nascent polypeptides. Nature. 2016;536:219–223. doi: 10.1038/nature19070. [DOI] [PubMed] [Google Scholar]
- 9.Voorhees RM, Hegde RS. Structures of the scanning and engaged states of the mammalian SRP-ribosome complex. eLife. 2015;4:1485. doi: 10.7554/eLife.07975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams CC, Jan CH, Weissman JS. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamerdinger M, Hanebuth MA, Frickey T, Deuerling E. The principle of antagonism ensures protein targeting specificity at the endoplasmic reticulum. Science. 2015;348:201–207. doi: 10.1126/science.aaa5335. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 13.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 15.Aviram N, et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature. 2016;540:134–138. doi: 10.1038/nature20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller C, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458–458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feldheim D, Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. The Journal of Cell Biology. 1994;126:935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:561–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- 19.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruan L, et al. Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature. 2017;543:443–446. doi: 10.1038/nature21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felici F, Cesareni G, Hughes JM. The most abundant small cytoplasmic RNA of Saccharomyces cerevisiae has an important function required for normal cell growth. Mol Cell Biol. 1989;9:3260–3268. doi: 10.1128/mcb.9.8.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 23.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods in Enzymology. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 24.Morawska M, Ulrich HD. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast. 2013;30:341–351. doi: 10.1002/yea.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ares M. Isolation of total RNA from yeast cell cultures. Cold Spring Harb Protoc. 2012;2012:1082–1086. doi: 10.1101/pdb.prot071456. [DOI] [PubMed] [Google Scholar]
- 26.Di Santo R, Aboulhouda S, Weinberg DE. The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA. eLife. 2016;5:e2573. doi: 10.7554/eLife.20069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murley A, et al. ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. eLife. 2013;2:e00422. doi: 10.7554/eLife.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGlincy NJ, Ingolia NT. Transcriptome-wide measurement of translation by ribosome profiling. Methods. 2017;126:112–129. doi: 10.1016/j.ymeth.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn JG, Weissman JS. Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics. 2016;17:958. doi: 10.1186/s12864-016-3278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Käll L, Krogh A, Sonnhammer ELL. A combined transmembrane topology and signal peptide prediction method. Journal of Molecular Biology. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 31.Elstner M, Andreoli C, Klopstock T, Meitinger T, Prokisch H. The mitochondrial proteome database: MitoP2. Methods in Enzymology. 2009;457:3–20. doi: 10.1016/S0076-6879(09)05001-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.