Abstract
Humans have a close phylogenetic relationship with nonhuman primates (NHPs) and share many physiological parallels, such as highly similar immune systems. Importantly, NHPs can be infected with many human, or related simian viruses. In many cases, viruses replicate in the same cell types as in humans and infections are often associated with the same pathologies. In addition, many reagents that are used to study the human immune response cross-react with NHP molecules. As such, NHPs are often used as models to study viral vaccine efficacy, antiviral therapeutic safety and efficacy, and to understand aspects of viral pathogenesis. With several emerging viral infections becoming epidemic, NHPs are proving to be a very beneficial benchmark for investigating human viral infections.
ToC blurb
Nonhuman primates (NHPs) are increasingly used as models of human viral infections. Here, Estes and colleagues introduce different NHP models, summarize the similarities and differences of human and NHP immune systems and discuss important examples of human viruses that can be modeled in NHP.
Introduction
Animal models of human viral infections are critical for understanding disease pathogenesis, vaccine modalities, and therapeutic interventions in vivo. Ideally, studies of human viral infections would benefit from the ability to perform experiments using humans and samples from humans. Indeed, this is sometimes possible, as immune responses in humans can be measured before and after vaccination in peripheral blood, broncheoalveolar lavage, peripheral lymph nodes, and the upper and lower gastrointestinal tract (via endoscopy). Moreover, vaccine development studies for certain infectious diseases can directly investigate the efficacy of the vaccine with pathogen challenge after vaccination (for example, Zika virus). However, there are many important research questions which cannot directly be answered with experiments using human subjects, thus animal models are required. Indeed, some infections result in high morbidity or mortality in humans, therefore vaccine and/or challenge experiments in humans are not possible. Moreover, comparative observations that require substantial tissue volumes from multiple anatomical sites can negate the possibility of performing experiments in humans, especially if tissues are difficult to access. Finally, the outbred nature of the human population, coupled with demographic differences creates substantial amounts of variation, which considerably complicates many research projects. Therefore, the development and detailed study of animal models for human diseases are absolutely critical.
Small animals, particularly rodents, are a mainstay for biological and immunological research. Indeed, studies in mice offer specific advantages including: the ability to perform adoptive transfers of immune cells; the selective deletion of genes, embryonically or in specific cell types; the ability to control for MHC types1; selective colonization with controlled microbiota or removal of the microbiota2; the ability to intravitally image lymphocytes responding to antigens3,4; and the ability to create transgenic animals that only express specific antigen recognition receptors1. Moreover, small animals such as mice are fairly inexpensive to house, mature to adulthood rapidly, have short gestation periods, and give birth to multiple offspring simultaneously. Indeed, the vast majority of our current knowledge of the immune system has, at some level, involved experiments with mice. Thus, Mus musculus is a phenomenal animal model whose utility cannot be overstated.
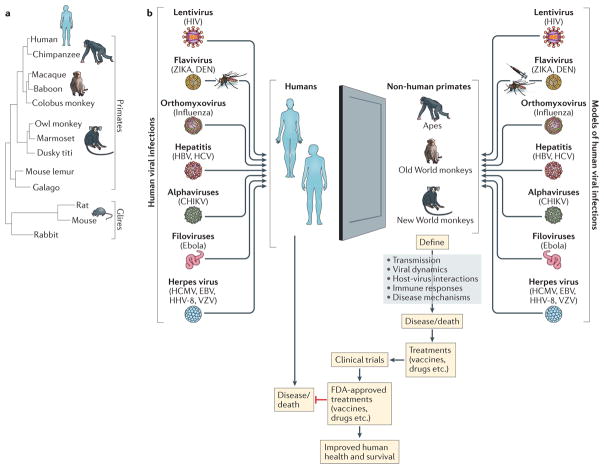
Unfortunately, there are numerous biological phenomena that occur in humans, which cannot be perfectly modeled in mice. Specifically, many viruses that cause disease in humans do not replicate in mice. Furthermore, human viruses that do replicate in mice may not result in the same types of pathologies that occur in humans. However, human viruses that are not well modeled in mice often do cause human-like disease in nonhuman primates (NHPs). Moreover, many of the reagents that are used to study immune responses in humans cross react with NHPs (Table 1); as such, NHPs have become important animal models to study human viral infections (Figure 1).
Table 1.
Nonhuman primate resources and web databases
| Database | Website address | Information |
|---|---|---|
| NIH Office of Research Infrastructure Programs – Comparative Medicine: Vertebrate Models | https://orip.nih.gov/comparative-medicine/programs/vertebrate-models | Contains information pertaining to available vertebrate models that have support through the NIH Office of Research Infrastructure Programs (ORIP). |
| National Primate Research Centers | https://nprcresearch.org/primate/ | A national resource for the scientific research community. |
| Biomedical Primate Research Centre | http://www.bprc.nl/en/home/ | A primate research center in The Netherlands, which is committed to using NHP for critical research when there are no suitable alternatives. |
| NIH NHP Reagent Resource | http://www.nhpreagents.org/NHP/default.aspx | An NIH supported resource to facilitate the optimal use of NHP models in biomedical research. The resource identifies and can provide specific validated reagents for in vitro and in vivo analysis. |
| NHP MHC web portal | https://dholk.primate.wisc.edu/_webdav/dho/mhc_contract/web_portal/%40files/prototype/index.html | Contains information and protocols for genotyping and allele discovery in macaques |
| Immuno Polymorphism Database (IPD) - MHC | https://www.ebi.ac.uk/ipd/mhc/group/NHP | NHP database for major histocompatibility complex (MHC) genes |
| The Macaque Genotype And Phenotype Database | https://mgap.ohsu.edu/ | The Macaque Genotype and Phenotype Database (mGAP) provides access to genotype data collected on a large, pedigreed, rhesus macaque colony housed at the Oregon National Primate Research Center (ONPRC). |
| NHP Genome Sequences | https://www.hgsc.bcm.edu/non-human-primates | The Baylor College of Medicine-Human Genome Sequencing Center is sequencing genomes from representative species from most of the major branches of the phylogeny: hominoids, Old World monkeys, New World monkeys and lemurs. |
| NHP Reference Transcriptome Resource | http://nhprtr.org/ | Phase I: RNAseq analysis of tissue pools for individual NHP species Phase II: RNAseq analysis of NHP tissue-specific transcriptomes Phase III: Tissue-specific small RNA data for rhesus macques |
| Virus Pathogen Database and Analysis Resource | https://www.viprbrc.org | Database of genomic sequences of virus pathogen strains and analysis tools |
Figure 1. Nonhuman primate models to study viral diseases in humans.
A. A phylogenetic tree demonstrating that nonhuman primates are the closest related animals to humans, whereas mice are highly genetically distant. B. Many viruses which cause disease in humans also cause diseases in nonhuman primates (Old world, New World, or apes); alternatively, there are similar viruses which recapitulate the salient aspects of viral disease in humans. Thus, nonhuman primates provide a valuable window into mechanisms of viral diseases and in developing novel therapeutic interventions to treat or vaccine modalities to prevent viral diseases. Figure 1A is adapted from REF177.
In this Review, we introduce species of NHP that are experimentally used to recapitulate salient features of viral diseases in humans. We then discuss the immunological similarities and differences between NHPs and humans, and the advantages and disadvantages of using NHP models of viral diseases (Box 1). Finally, we consider several key examples of human viruses that have been experimentally modeled in NHP species.
Box 1. The advantages and disadvantages of non-human primates over other models.
Viral infections have been modeled in mice and men. However, nonhuman primate (NHP) models present an alternative approach with unique advantages and challenges.
The advantages of nonhuman primates versus mouse models
Mice are not suitable for many viral diseases due to lack of replication or inappropriate pathology
NHP biology and physiology is the closest to humans
Human-specific reagents can be used
Many human viruses are overly sensitive to mouse type I IFN
Disparate genotypes of NHPs better mirror the outbred human population
The disadvantages of nonhuman primates versus mouse models
Increased cost and increased gestation period
It is not possible to create gene knock out or transgenic NHPs
It is not possible to create germ free NHPs or to perform parabiosis. Experiments are limited to specific pathogen free animals.
It is not possible to perform congenic animal studies
The advantages of nonhuman primates versus human models
NHP and human immune systems are highly similar
It is possible to perform longitudinal studies
Environmental factors can be controlled
Any anatomic site can be sampled
The effects of additional infections can be studied
Preventative vaccine or therapeutic efficacy can be studied with lethal viruses
It is possible to deplete lymphocyte populations from NHPs in vivo
The disadvantages of nonhuman primates versus human models
NHPs are not suitable for many viral diseases due to lack of replication or inappropriate pathology
Some NHP genes exert antiviral activities differently to human equivalents.
NHPs express a greater variety of MHC genes than humans
NHP studies are limited to smaller numbers of animals
Studies in NHPs are costly
The genotypes of interferon stimulated genes (TRIM5α for example) can influence antiviral responses
Nonhuman primates as models
For many different viruses, NHPs offer a window into the pathogenesis of human disease, and in developing vaccine and therapeutic interventions aimed to treat and prevent viral infections (Figure 1). Importantly, the innate and adaptive immune responses that NHPs elicit against viruses are, for the most part, very similar to human responses, meaning these animals are more appropriate for some viral infections that cannot be modelled in mice. Moreover, many of the immunological reagents that are routinely used to identify human immunological molecules also cross-react with NHP molecules, which makes the transition to NHP studies less problematic as information specific for NHPs is available on databases on the worldwide web (Table 1).
Old world monkey species
Old World macaque NHP species are, by far, the most commonly used primate species for biomedical research. There are three species of Asian macaques that are commonly used for viral and/or immunological studies: Macaca mulatta (rhesus macaque), Macaca fascicularis (cynomolgus macaque), and Macaca nemestrina (pigtail macaque)5–8. A wide variety of reagents, most of which are monoclonal antibodies against human proteins that also cross react with NHP antigens, are available to interrogate immunological responses to viruses, overt inflammation, and immunological abnormalities. Moreover, viral epitope-loaded MHC tetramers [G] exist for rhesus macaques9–11 (at least three different MHC alleles) and pigtail macaques12 (at least one MHC allele) to study virus-specific CD8+ T cells (Table 1). Although there are no viral epitope-loaded MHC tetramers that are widely used for studies in cynomolgus macaques, the relatively limited MHC diversity of these animals make them particularly attractive for studies involving allogenic responses and/or adoptive transfer experiments13–15.
Although rhesus, pigtail and cynomolgus macaques are very similar in genetic sequences and susceptibility to most of the viral infections we discuss herein, important differences exist that create particular nuances across these three species. For example, the type I interferon-induced, antiviral protein TRIM5α has multiple genetic isoforms, which have differing antiviral activity in each NHP species16. Rhesus macaques express heterogeneous TRIM5α genotypes, which differentially restrict simian immunodeficiency virus (SIV); however, pigtail macaques only express one TRIM5α genotype, which does not restrict SIV17. Of the three Asian macaque species typically used to study host and virus interactions, rhesus macaques are the most often studied, largely due to their availability.
In addition to Asian macaques, several other old world NHP species are employed for virology studies. In Africa, there are more than 50 species of NHPs that are natural hosts for SIV18. These species are often experimentally studied to understand how SIV has co-evolved with the natural hosts such that these NHP species do not develop simian AIDS (discussed below). The two most commonly used NHP SIV natural host species are sooty mangabeys (Cercocebus atys) and African green monkeys (Chlorocebus pygerythrus and Chlorocebus sabeus)18. However, ethical issues have limited the use of sooty mangabey subspecies (Box 2), which confines the types of experiments that can be performed in all subspecies of Cercocebus atys. Therefore, the African green monkey Chlorocebus species have become a mainstay in natural host studies of SIV. In addition, African green monkeys can be infected with orthomyxoviruses19 (discussed below). Baboons (Papio sp.) are one of the more rarely used species of the old world primates to study host and virus interactions20. These animals can be infected with many of the viruses discussed below, however their availability and increased size (relative to other species) can prove limiting.
Box 2. Special ethical considerations for nonhuman primate research.
All experiments that involve live animals require special ethical considerations. Given the close phylogenic similarities of nonhuman primates (NHPs) to humans, special concerns arise. Indeed, most animal care and use committees (ACUC) set up specific review procedures to gauge the scientific validity of experiments that use NHPs, in addition to reviewing concerns related to safety and well-being of animals enrolled in animal protocols.
Recently the sooty mangabey subspecies Cercocebus atys lunatus (the white crowned mangabey) was added to the United States Fish and Wildlife list of endangered species. Cercocebus atys are natural hosts for certain strains of simian immunodeficiency viruses (SIV). These animals exhibit nonprogressive infections with SIV and were the source of cross-species transmission of the sooty mangabey strain of SIV (SIVsmm) into humans, which gave rise to HIV-2. However, owing to the fact the lunatus subspecies was put on the endangered list, it has considerably limited the types of experiments that can be performed in all subspecies of Cercocebus atys. This is despite the fact that one of the major atys subspecies (atys) is thriving in its natural environment and there are colonies of these animals at primate centers.
Chimpanzees also present specific considerations. These animals are the most closely genetically related NHPs to humans (Figure 1), and as such these animals are highly relevant for studying viral diseases that affect humans. For example, a strain of SIV infects chimpanzees in the wild (SIVcpz); cross-species transmission of SIVcpz gave rise to the HIV-1 epidemic and chimpanzees are the only NHPs wherein HIV-1 replicates efficiently in vivo. Moreover, chimpanzees were the gold standard model for vaccine and pathogenesis studies of hepatitis C virus (HCV). Indeed, HCV does not replicate in other species of NHPs. However, in 2015 the US Fish and Wildlife Service (FWS) announced that it is categorizing captive chimpanzees as an endangered species, which are subject to legal protections. This has almost negated research involving chimpanzees, substantially slowing progress on vaccine and antiviral therapeutic development.
New world monkey species
Old World primates represent the most frequently employed experimental primates for virology studies, however, New World primates, when appropriate, have distinct advantages. In particular, the marmosets (Calilthrix jacchus) are less expensive than the larger Old World primates and can be experimentally infected with several viruses that cause disease in humans (herpes viruses in particular, discussed below). Thus, it is more feasible to conduct studies which require larger numbers of animals compared to studies involving Old World primates. However, New World primate specific reagents are not as abundantly available to interrogate host responses to viruses or viral-mediated pathogenesis.
Ape species
Ape species are more closely evolutionarily related to humans than any other primate species (Figure 1), thus, they are ideally suited as animal models aimed at studying phenomena that occur in humans. Indeed, experiments with chimpanzees were crucial for understanding the ontogeny of HIV, and chimpanzees are the only NHP species that support viral replication of HIV and hepatitis C virus (HCV). However, particular ethical concerns have dramatically limited ape species as experimental animal models (Box 2). In summary, NHPs can be appropriate animals to model many viral infections which cause diseases in humans. However, NHPs should be employed only when appropriate, and careful attention should be given to understanding the particular nuances and advantages that each NHP and viral species pair contain so that experiments may be designed appropriately.
Immune responses in nonhuman primates
In NHPs, immune responses to antigens, from both the adaptive and innate arms of the immune system, are incredibly similar to those in humans. Although innate immune responses (which are commonly measured by expression patterns of interferon stimulated genes) to viral infections are fairly widely conserved between NHPs and humans21–24, the most commonly measured immunological parameters in viral infections of primates are the magnitude and functionalities of virus-specific T and B cells.
The MHC region maps to chromosome 6 of humans; in rhesus macaques the MHC-I region is expanded and seemingly more complex. Humans contain one copy of the HLA-A, HLA-B, and HLA-C gene per haplotype, whereas rhesus macaque and other macaque species can have multiple MHC-A and MHC-B genes per chromosome. Macaques can possess up the three MHC-A genes per haplotype. More is known about macaque MHC-A genes than the MHC-B genes. The MHC-A1 gene is the most polymorphic, whereas the other MHC-A genes are more conserved and show lower levels of transcription. The MHC-A1 gene is thought to encode MHC molecules that perform classical peptide presentation whereas the other MHC-A genes are thought to share features with the nonclassical MHC-I genes25,26. A similar increased diversity exists for NHP MHC-II alleles, compared to the equivalent human HLA-DR, HLA-DP and HLA-DQ genes 27.
Importantly, the canonical flow cytometric and intracellular cytokine staining modalities that are used for measuring antigen-specific T cell responses in humans are also meaningful for the analysis of virus-specific T cell responses in NHPs. Nucleated cells which express surface MHC from NHPs can be loaded with overlapping peptide pools that span pathogen open reading frames. Pathogen-specific T cells then become stimulated and produce effector cytokines that can be measured by standard Enzyme-Linked ImmunoSpot (ELISPOT) assay or intracellular cytokine staining and flow cytometry22,28. Indeed, the magnitude and variability of antigen-specific T cells which recognize individual MHC–peptide complexes are quite similar between NHPs and humans, providing additional value to the NHP models.
The analysis of virus-specific B cells is also now possible in many species of NHPs. Traditionally, B cell responses were evaluated by removing B cells from peripheral blood by negative selection and then assessing antibody production after antigen stimulation using ELISPOT technology29. However, recent advances have used fluorochrome-conjugated viral antigens to isolate and analyze virus-specific B cells from multiple anatomical sites using flow cytometry. Moreover, the immunoglobulin locus of macaque species has recently been sequenced and studied in great detail30,31. Similar to the increased diversity of NHP MHCs, macaque immunoglobulin loci are substantially more diverse than in humans, with higher sequence diversity and copy-number variation. Although, the antibody isotypes of NHP antibodies broadly reflect those found in both humans and mice, there are subtle differences, with NHPs having four IgG isotypes. Moreover, the different isotypes bind individual Fc receptors (such as CD16, CD32, CD64) and complement similarly32. Thus, NHPs are a good model to understand antibody development against viruses in vivo.
RNA Viruses
Numerous RNA virus families cause diseases in humans. RNA viruses can have a double stranded or single stranded genome, which may be negative or positive sense. Many of the RNA viruses that cause disease in humans induce similar diseases in NHPs, making NHPs phenomenal animal models.
Lentiviruses
HIV is responsible for the AIDS pandemic, and infection results in the progressive loss of CD4+ T cells and lymphoid tissue pathology.
The HIV-1 pandemic in humans resulted from cross-species transmissions of a strain of SIV (SIVcpz) that infects central African chimpanzees (Pan troglodytes troglodytes). SIVcpz is associated with increased mortality and AIDS-like immunopathology in wild-living chimpanzee communities and, in one case, a captive chimpanzee (reviewed in REF33). Importantly, chimpanzees are the only NHP species that can be persistently infected with unmodified HIV-1; however, owing to their endangered status, the high costs of upkeep, and the long duration of infection that is required to definitively assess AIDS development, chimpanzees are not a practical model for HIV infection and AIDS (Box 2).
An extremely useful primate model of HIV is SIVmac, (otherwise known as SIVsmm), which was first isolated from captive rhesus macaque monkeys with an AIDS-like illness in US primate centers (Table 1, Table 2)34–36. SIVmac was discovered to have arisen from a cross-species transmission between SIVsmm infected sooty mangabeys (the natural host, which do not develop disease) and rhesus macaques in captivity37,38. Importantly, SIVsmm from infected sooty mangabeys was also discovered to have established HIV-2 infection in humans39. Asian macaques are not natural hosts for primate lentiviruses and when infected with certain strains of SIV develop a disease that resembles HIV-1 infection in humans. This discovery has led to the development of many SIV NHP models of HIV-1 infection. Collectively, these models have provided exceptional insight into fundamental aspects of HIV-1 transmission, prevention, and pathogenesis.
Table 2.
Key NHP models of human virus infections.
| Human virus | Equivalent NHP virus* |
|---|---|
| NHP species: Asian macaques | |
| HIV-1 | SIV SHIV simian tropic HIV |
| Dengue | Dengue |
| Zika | Zika |
| Ebola | Ebola |
| Chikunguyna | Chikungunya |
| Varicella Zoster Virus | Simian Varicella Virus |
| Cytomegalovirus (CMV) | Rhesus CMV |
| Epstein-Barr Virus | Rhesus lymphocryptovirus |
| Kaposi’s sarcoma-associated herpesvirus | Rhesus rhadinovirus |
| Pox viruses | Monkeypox |
| NHP species: Marmosets | |
| Hepatitis C | GB Virus-B |
All of the listed NHP models have been used to study disease pathogenesis and the development of vaccines and therapeutics, apart from the NHP HIV models, which have also been used to study transmission.
Mucosal transmission
NHPs closely resemble humans in their physiology and immune system development40, thus, these animal models have been utilized extensively for lentiviral transmission studies. NHPs, primarily rhesus macaques, have been used for intravaginal, intrarectal, penile, oral and mother-to-child transmission studies41; these studies have imparted a better understanding of viral transmission across mucosal surfaces, the early host-viral dynamics within mucosal tissues, the first cells to be infected, and the process of viral dissemination and establishment42–47.
Vaginal transmission studies in NHPs suggest that SIV initially infects antigen-presenting cells in the vagina, with virus subsequently disseminating into genital draining lymph nodes before spreading to proximal lymphoid nodes and finally systemically48. Subsequent vaginal challenge studies demonstrated that although macrophages and dendritic cells can be infected (vRNA+), the vast majority of infected cells (~90%) at the initial site of infection, in the lymph nodes, and in other sites of dissemination, were CD4+ T cells49. Importantly, the predominant infected (vRNA+) CD4+ T cell population within the female genital tract at the early time points was demonstrated to be a population of HLA-DR− and Ki67− “resting” effector memory T cells49. This study also suggested that expanding viral replication in local CD4+ T cells at the mucosal portal of entry, which is fueled by a robust innate inflammatory response to the virus, is required for the establishment of productive infection50. In addition, NHP vaginal transmission studies have highlighted the importance of resident and recruited CD4+ T cells in mucosal tissues for HIV mucosal transmission49,51.
NHP mucosal transmission experiments that use a high dose inoculum have demonstrated the importance of the mucosal barrier as a “bottleneck” to sexual transmission. In these studies, the initial high amount of viral RNA in the inoculum was reduced to very low levels of tissue-associated viral RNA shortly after challenge, suggesting that virus may initially gain access to a small number of susceptible target cells through vulnerable mucosal sites. Thus, there may only be small populations of founder infected cells at the portal of entry51. Importantly, this finding was recapitulated using low-dose, repeated challenge transmission models via the rectal, vaginal or penile routes in rhesus macaques using distinct SIV isolates (SIVmac251 and SIVsmE660)52. Many preclinical NHP vaccine and prevention studies now utilize the evaluation of the numbers of transmitted/founder viruses [G] to determine efficacy, thus showing the important role NHP models have played in delineating and recapitulating HIV-1 transmission53–57.
Prevention
NHP models have been at the forefront of efforts to identify the types of immune responses needed for protection against HIV-1 and have become an accepted essential pre-clinical step for assessing vaccine strategies58. Currently the only HIV vaccine clinical trial that has demonstrated any protective efficacy was the RV144 trial, in which HIV-1 gp120 variable regions 1 and 2 (V1/V2)-specific IgG antibody titers were associated with some protection against HIV-1 acquisition59,60. Importantly, this finding was supported by studies in NHPs, as in macaques that were vaccinated with an ‘RV144-like’ immunization regimen, vaccine-induced protection from SIV acquisition was associated with non-neutralizing antibodies against gp120, which mapped to the V1/V2 region of SIV envelope61. Additional evidence for the critical role of antibodies in protecting from HIV-1 transmission came from proof-of-concept passive immunization NHP SIV studies62,63. Subsequent passive transfer experiments have demonstrated that certain HIV-specific neutralizing antibodies can provide complete protection against transmission of chimeric simian human immunodeficiency viruses (SHIVs), [G] which express HIV-1 envelope glycoproteins64–66.
The induction of broadly neutralizing antibodies (bNAbs) remains a major goal of HIV-1 vaccine development67. However, none of the previously tested HIV-1 vaccine candidates have been effective in eliciting bNAbs67. Owing to the challenges in designing vaccine candidates that could induce bNAbs, alternative vaccine approaches were pursued, which focused on generating CD8+ cytotoxic T lymphocyte (CTL) responses68. This approach was based, in part, on studies of “elite” controllers[G]; there was initial hope that powerful prime-boost vaccine approaches, which could greatly increase the magnitude and function of CD8+ T cell responses, would provide meaningful efficacy69.
Disease pathogenesis
Although SIV infection of Asian macaques (rhesus, cynomolgus, and pigtail) is not identical to HIV-1 infection of humans, SIV infection of macaques does recapitulate key pathological features of HIV-1 infection, including: CD4+ T cell tropism; progressive CD4+ T cell depletion; lymphoid tissue pathology; establishment of the latent reservoir; neuropathology; and progression to AIDS70. The array of both nonpathogenic and pathogenic NHP models utilized for HIV and AIDS research have authoritatively contributed to the discovery and establishment of key hallmarks of HIV-1 pathogenesis71.
Chronic immune activation and inflammation has emerged as one of the central features in HIV-1 pathogenesis72,73. Importantly, chronic immune activation is a hallmark of pathogenic HIV-1 infection in humans and SIV infection in macaques, whereas SIV chronically-infected natural hosts (sooty mangabeys and African green monkeys) show an absence of immune activation74. Studies in HIV-infected humans and SIV-infected NHP demonstrate that HIV or SIV-associated inflammation is a complex and multifactorial phenomenon that ultimately plays a major role in disrupting T cell homeostasis and driving lymphoid tissue pathology75,76. However, many studies have demonstrated that the elevation of microbial products (such as lipopolysaccharide (LPS), flagellin, bacterial DNA) that translocate from the damaged gastrointestinal tract into the systemic circulation of HIV-infected individuals may be a cause of chronic immune activation during progressive HIV-1 infection77–79. Moreover, studies in SIV-infected Asian macaques that capitalized on large tissue collections at necropsy demonstrated direct in situ evidence for microbial translocation into the gut parenchyma and subsequently into draining lymph nodes, liver and distal peripheral lymph nodes80, which does not occur in SIV-infected natural hosts77,80,81. Notably, a novel SIV-negative NHP model of experimental colitis demonstrated that gastrointestinal tract barrier damage was sufficient to drive local and systemic microbial translocation, inflammation, and AIDS virus-like pathology, independently of SIV infection82. Collectively, human and NHP studies support a model of early structural and immunological damage to the gastrointestinal tract in progressive HIV-1 or SIV infections in humans and macaques, which leads to microbial translocation that contributes to persistent chronic immune activation.
Soon after initial studies described numerous histopathological changes to lymphoid tissue in HIV-infected humans, studies in SIV-infected NHPs also demonstrated similar pathology to HIV-infected humans, with parallel histological features such as lymphadenopathy with follicular hyperplasia to severe lymphoid depletion83. Since these early reports, NHP models have proven to be an invaluable tool to elucidate the timing and mechanisms driving the progressive immunopathology of lymphoid tissue in HIV infection76. Studies in HIV-infected patients have demonstrated considerable lymphoid tissue damage associated with the deposition of fibrotic collagen84; an observation that was subsequently confirmed in an NHP model of SIV infection, which linked fibrosis to immune activation and the induction of lymphocytes expressing tumour growth factor-β (TGFβ)76. In addition, the rapid induction of a heterogeneous immunosuppressive regulatory T cell (Treg) response shortly after SIV infection in rhesus macaques recapitulates the reported increases and accumulation of Treg cells and TGFβ expression in peripheral lymph nodes in HIV-infected humans85,86. Taken together, SIV infection of Asian macaques has recapitulated salient pathologies which occur in HIV-infected humans and has provided insights into novel therapeutic interventions and vaccine modalities to prevent HIV infection and improve the prognosis of HIV-infected humans.
Hepatitis viruses
Hepatitis viruses are important human pathogens that infect cells of the liver and cause liver damage (Table 2). The following section summarizes important NHP research on Hepatitis C virus (HCV) and Hepatitis E virus (HEV).
Hepatitis C virus
HCV is a blood borne virus that can cause both acute and chronic hepatitis. Although there is currently no vaccine to prevent HCV infection, new FDA approved direct-acting antivirals that target the HCV replication cycle have been demonstrated to cure more than 95% of HCV-infected individuals. However, access to diagnosis and treatment remains low worldwide87,88. One substantial barrier to developing new, cheaper and more potent treatments and an efficacious vaccine is the lack of a suitable animal model. Currently, chimpanzees are the only known animal species that it is possible to infect with HCV. However, due to high costs, ethical concerns and regulatory restrictions (Box 2) this NHP model has a very limited future89–91.
The NHP chimpanzee model played an instrumental role in the identification of HCV in 1989, and in advancing our understanding of HCV infection and disease, the study of immune responses to HCV, and the pathogenesis of the disease92. Numerous reports have demonstrated the importance of the innate (for example, type I IFNα and IFNβ responses) and adaptive (for example CD4+ and CD8+ T cell responses and B cell responses) arms of the immune system in the spontaneous resolution of acute HCV infection in chimpanzees, which confers long-lived immunity that considerably reduces the risk of re-infection92. One important caveat of the chimpanzee NHP model is that the natural course of HCV infection in chimpanzees is substantially different from that in humans93. Typically, HCV-induced hepatitis in chimpanzees is milder than in human HCV infection, however, severe hepatitis has been reported94. In addition, the progression to chronic hepatitis is much less common in chimpanzees, and in contrast to humans, liver cirrhosis is extremely rare95. Currently, it is unclear why the pathogenic course of HCV infection differs between chimpanzees and humans, but it may be owing, in part, to the lack of liver disease contributing cofactors (for example, alcohol consumption, obesity, a high-iron diet) in HCV-infected chimpanzees92. Alternatively, this difference may be a reflection of the low number of animals that have been followed in long term studies to effectively evaluate the pathological consequences of long-term HCV infection; however, given the current NIH policy to stop funding research in chimpanzees, the U.S. Fish and Wildlife policy to restrict research in chimpanzees, and the limited resources available at national primate centers and Chimp Haven, where most HCV infected chimpanzees are located, it is unlikely that the differences in HCV infection between chimpanzees and humans will ever be fully understood 92.
Although an alternative NHP HCV model remains elusive, the GB virus (GBV)-Bvirus, which is closely related to HCV, and efficiently replicates in marmosets, represents a surrogate model for the study of HCV infection and disease96. The sequencing of GBV-B and the generation of a GBV-B molecular clone revealed important differences and similarities between GBV-B and HCV that has allowed for the construction of chimeric viruses96. Even though GBV-B/HCV chimeric viruses do not perfectly recapitulate HCV biology, they do infect and replicate in New World NHPs and may serve as an important model for the development of HCV-targeting drugs that target specific regions of these chimeric viruses.
Hepatitis E virus
HEV is in the genus Orthohepevirus in the Hepeviridae family97, and shares many features with Hepatitis A virus (HAV); however, HEV causes disease in humans and NHPs, whereas HAV does not cause disease in NHPs. The inoculation of NHPs with material from patients infected with water-borne non-A hepatitis was instrumental in the identification of HEV98,99. A number of NHP species (for example, chimpanzees, marmosets, macaques) are susceptible to experimental infection with HEV; however, cynomolgus macaques and rhesus macaques are the most commonly used species for studying HEV infection and disease100–105. The severity of disease appears dependent on both the strain and dose of virus used; however, cynomolgus macaques may be more sensitive to HEV infection, demonstrating more severe acute hepatitis compared to rhesus macaques or chimpanzees103,106,107.
As with HAV infection, damage to liver cells is thought to result more from the host immune response rather than from the direct effects of viral infection of hepatocytes108. Histological changes in the liver of NHPs and patients are characteristic of acute hepatitis, including focal necrosis with lesions found in all regions of the lobule [G] 108. Similar to infection in humans, HEV infection in NHPs often leads to inflammation, consisting of Kupffer cells [G] and polymorphonuclear leukocytes in focal lesions that are typical of hepatitis108. In contrast to humans, pregnancy does not exacerbate the disease course in cynomolgus macaques104. Importantly, NHP models have been extensively used for the pre-clinical testing of vaccines against HEV, and a vaccine to prevent HEV infection has been developed but is not yet available worldwide109–114.
Flaviviruses
Pathogenic viruses belonging to the family Flaviviridae are transmitted to humans principally by arthopods and include yellow fever virus (YFV), dengue virus, West Nile virus (WNV), Japanese encephalitis virus (JEV) and zika virus (ZIKV)115. Mice can be infected with many species of flaviviruses, however, these viruses are incredibly sensitive to type I interferons. Consequently, most mouse models of flavivirus infection and pathogenesis use mice deficient in type I interferon signaling116–118. Thus, the mouse model of flavivirus infection may not recapitulate all of the salient aspects of virus and host interactions that occur in vivo in humans. As such, NHPs have become an important model to study these important viral infections.
Dengue
Given the huge public health concerns associated with dengue virus (DENV) infection (Table 2), it is important to develop animal models that recapitulate key features of human infection119. Wild type mice are not susceptible to DENV infection, thus, considerable effort has been put into identifying appropriate NHP models119. There are at least four serotypes of DENV, each of which shares around 65% genetic similarity with the other three, which further complicates the modeling of DENV infection in NHPs120. Moreover, there is substantial evidence that preinfection with one serotype of DENV can result in more severe disease upon challenge with another DENV serotype, which may be facilitated by weakly cross-reacting antibodies binding the different serotypes120. Thus, ideally, several serotypes of DENV would need to be capable of replicating and causing disease in NHPs. Many strains of dengue virus, routes of infections, sources of virus, and species of NHPs have been tested119,121. The most commonly studied NHPs for DENV infections are macaque species, old world patas monkeys, and African green monkeys119,121. All of the serotypes of DENV that have been tested (DENV 1–4) replicate to high levels (up to 100 million copies per milliliter of plasma) and viremia lasts for around a week, which is similar to infection in humans. However, NHPs do not seem to manifest the same types of symptoms that are observed in DENV-infected humans119,121.
NHPs have been employed to measure vaccine-induced immunity against DENVs and to ensure that attenuated viral vaccines lack the ability to replicate to high levels in vivo121,122. Many vaccine modalities that have been shown to be safe in NHPs and that induce high titers of neutralizing antibodies against DENV have been advanced to human clinical trials121,123,124, and one has even shown protection after human challenge with DENV124.
Zika virus
Zika virus (ZIKV) was first identified as a human pathogen in Africa in 1952, but the first human epidemic was recognized in the Yap Island in 2007, which spread to a larger epidemic in French Polynesia in 2013–2014125. Subsequently, ZIKV has spread widely in warmer climates, with infections numbering in the tens of thousands and coinciding with mosquito life cycles125. ZIKV infections are generally not life-threatening, however, infection is associated with a rare chance of Guillian-Barre syndrome [G]. In addition ZIKV infection of pregnant women has been linked with birth defects.
Several studies have employed NHPs (cynomologus macaques, rhesus macaques, and pigtail macaques) to study salient features of ZIKV pathogenesis (Table 2)126–129. In general, ZIKV infection of NHPs is facilitated by subcutaneous injection, which models infection via mosquito bite, andrecapitulates many features of human infection including mild weight loss, slight fever, and elevated liver enzymes126–129. NHPs infected with ZIKV became viremic and plasma viremia peaks between 2–6 days post infection; subsequently these animals appear to clear plasma viremia within 10 days126–129. ZIKV can be detected in semen, vaginal fluid, urine, saliva, cerebral spinal fluid, lymphoid tissue, gastrointestinal tract, and central nervous system (CNS) after the resolution of viremia in blood127,128. Finally, NHPs infected with ZIKV mount virus-specific humoral and cellular immune responses, which protect against subsequent homologous and heterologous challenges, suggesting that NHPs are an appropriate animal model to study vaccine-mediated protection22,127,128. Indeed, multiple vaccine modalities have been tested in NHPs, which induce neutralizing antibodies against ZIKV and are protective against subsequent challenge130–132. Having shown protection in NHPs, many of these vaccines are moving forward into human clinical trials.
One of the most concerning aspects of ZIKV infection is the apparent ability of ZIKV to cross the placenta during pregnancy and infect the developing fetus. Indeed, ZIKV infection during pregnancy is associated with fetus developmental abnormalities leading to microcephaly. An animal model which recapitulates this phenomenon would be important for developing therapeutic interventions. Pregnant rhesus macaques infected with ZIKV had viremia lasting up to 55 days, which was far longer than nonpregnant animals and similar to what has been reported in pregnant women127,133. Importantly, in pregnant pigtail macaques infected with ZIKV the fetus showed reduced brain growth, neuronal damage, white matter deficiency and gliosis, and the virus was detected in the placenta and fetal brain and liver134. Finally, recent work has suggested that ZIKV RNA can persist in the tissues of NHPs for prolonged periods of time after the resolution of viremia22,135. Thus, NHPs may represent an appropriate animal model to understand the mechanisms of maternal–fetal ZIKV transmission, pathogenesis and persistence, and to develop vaccines and/or therapeutic interventions.
West Nile virus
Multiple NHP species have been experimentally infected with West Nile virus (WNV). WNV is spread by mosquitoes and infection in humans is usually asymptomatic, however, ~20% may have fevers, aches, vomiting, diarrhea, or rash. Moreover, ~0.5% of WNV-infected humans have more severe disease with central nervous system (CNS) involvement and paralysis or death are possible136. As WNV is a biohazard level 3 pathogen [G], special precautions are required and as such, not all primate facilities are capable of working with the virus. Rhesus macaques and marmosets can be experimentally infected with WNV and both replicate the virus in vivo to high titers, however, marmosets show higher viral titers and more extensive tissue penetrance137. Although both species demonstrated adaptive and innate immune responses to WNV, neither had any clinical signs of WNV infection, which is similar to the majority of WNV infected humans137. Notably, intrathalmic infection of rhesus macaques led to viral spread throughout the CNS without any clinical symptoms of disease138. It is unclear whether WNV fever or neurological disease occurs in NHPs and very large numbers of animals would likely be required to ascertain this. However, NHPs may represent an appropriate model to study vaccine-mediated protection against WNV infection considering that they replicate the virus and mount antibody and cellular immune responses. Correspondingly, several vaccine modalities against WNV have already been employed in NHPs; some of these seem to protect NHPs from subsequent WNV challenge and have entered trials in humans139–142.
DNA viruses
There are five families of DNA viruses that infect humans: adenoviridae, herpesviridae, papoviridae, parvoviridae and poxviridae. Of these, the herpesviridae and poxviridae are associated with many important diseases in humans, whereas the papoviridae are associated with rare pathological conditions, and the adenoviridae and parvoviridae, which are rarely associated with human disease, are being developed for heterologous gene delivery.
Herpesvirus
Herpesviruses are a family of large enveloped DNA viruses with linear double stranded DNA genomes, which infect almost all animal species. Here, we describe studies of simian herpesviruses (which are closely related to human herpesviruses) that aim to elucidate mechanisms of pathogenesis, to define the host immune response to control latent herpesvirus infections [G], and to identify the viral and host factors that lead to virus reactivation.
Alpha herpesvirus
Human varicella zoster-virus (VZV, Table 2) is an alpha-herpesvirus that causes chickenpox and herpes zoster in humans. An attenuated strain of VZV has been licensed as a vaccine and is effective at preventing chickenpox and but less effective at preventing VZV reactivation (herpes zoster) in elderly and immunocompromised individuals. Unfortunately, VZV exhibits strict species-specificity, which prevents animal infection studies to elucidate the mechanisms of herpes zoster disease. Thus, an animal model that closely parallels key features of VZV infection and pathogenesis is required.
Simian varicella virus (SVV) is a related virus that was isolated in the 1960s; SVV is morphologically and genetically similar to VZV, and several groups have reported that inoculation of Old World monkeys (African green monkeys and Cynomolgus macaques) can lead to varicella with or without the establishment of latency (reviewed in REF143). Importantly, intra-bronchial inoculation of rhesus macaques with SVV appears to recapitulate all the essential features of VZV infection in humans, including latency. Furthermore, this model has enabled researchers to determine how the antibody-mediated depletion of specific immune cells impacted primary SVV infection. One group found that depletion of CD4+ T cells during a primary SVV infection led to widely disseminated varicella accompanied with higher and prolonged viral loads, compared to animals depleted of CD8+ T cells and B cells144. This implies that CD4+ T cell responses may be important for the control of varicella or herpes zoster disease in humans.
Several attempts have been made to reproducibly induce SVV reactivation in macaques that have an established latent infection. The most recent study utilized full body irradiation, in addition to treatment with prednisone and tacrolimus (both are immunosuppressive drugs) and was successful in reactivation in four of four rhesus macaques. Surprisingly, a control animal also developed a rash, implying the stress associated with the transport of animals to the site of irradiation was sufficient to induce reactivation145. Taken together, the NHP model of VZV infection has provided important information related to immunological pressure and mechanisms of viral reactivation from latency, including how immunodeficiency can lead to viral reactivation. Moreover, this model will be useful in advancing vaccines aimed at reducing incidence of herpes zoster.
Beta herpesvirus infections: rhesus cytomegalovirus
Human cytomegalovirus (HCMV; strictly tropic to humans) is an important pathogen with widespread seropositivity in humans. NHPs are hosts to homologues of HCMV (Table 2) and several CMV isolates have been obtained and sequenced from numerous NHPs, including chimpanzees, baboons, cynomolgus macaques and other Old World monkeys. The morphology and genome sequences of NHP CMV isolates are quite similar across species and reveal a close evolutionary relationship with their hosts. The most widely studied NHP CMV is rhesus CMV (RhCMV), of which two isolates are utilized for pathogenesis and vaccine studies (reviewed in REF146).
Our current fundamental understanding of rhesus macaque cellular immunity originated from studies characterizing ex vivo CD4+ and CD8+ T cell subsets from RhCMV-infected animals147. These early studies defined the key immunological markers on CD4+ and CD8+ naïve and memory (central and effector subsets) T cells and performed comprehensive functional analyses of these populations. This immunological information led to a continued and expanded use of the rhesus macaque model for investigations into viral pathogenesis and vaccine development.
Some of the most intriguing studies of RhCMV in rhesus macaques involve using RhCMV as a vaccine vector to prevent SIV infection and disease. Here, investigators found that RhCMV vectors, particularly rhCMV68.1, which encoded SIV antigens, were capable of infecting CMV naïve and CMV seropositive rhesus macaques, and eliciting persistent SIV-specific CD4+ and CD8+ T effector memory (TEM) cell responses148. SIV-specific CD8+ T cells were demonstrated to be MHC-I restricted, and were also restricted by MHC class II or non-polymorphic MHC-E, implying that the SIV RhCMV vaccine elicited responses from unconventional CD8+ T cells28,149. Importantly, TEM cell responses were present at extra-lymphoid sites in vaccinated animals and were effective at preventing progressive SIV infection after challenge with SIVmac239. Moreover, an analysis of vaccinated animals revealed that protection from progressive SIV infection was correlated with the magnitude of the peak SIV-specific CD8+ T cell response. Interestingly, these protected animals exhibited effective SIV control even after antibody-mediated depletion of CD4+ and CD8+ T cells, implying the vaccine was capable of clearing SIV infection150. That RhCMV based vaccines against SIV have led to such impressive control against pathogenic SIV infection has led to ongoing studies using similar hCMV-vectored approaches being developed for human trials.
Gamma-1 herpesviruses: lymphocryptoviruses
Epstein-Barr virus (EBV; Table 2) is a gamma-1 herpesvirus or lymphocryptovirus (LCV) that infects most humans by the time they reach adulthood. EBV is strictly human-tropic, which makes studies of viral pathogenesis extremely difficult in animals.
Multiple NHP species from Old World and New World monkeys harbor related LCVs, with each LCV capable of immortalizing B cells from their natural host, which is a key feature of human EBV infection (reviewed in REF151). The most widely studied NHP LCV is rhesus LCV (rhLCV), which was originally isolated from a lymphoblastoid cell line that was established from a malignant lymphoma that developed in a rhesus macaque. Importantly, several publications have reported that rhLCV is associated with diseases that parallel EBV, including B cell lymphomas and hairy leukoplakia [G] in SIV-infected macaques. In addition, primary rhLCV infection in rhesus macaques parallels primary EBV infection in humans151. The utility of the rhLCV rhesus macaque model is further underscored by the creation of a novel rhLCV BACmid [G], which can produce infectious virus that is capable of immortalizing B cells from rhesus macaques152. The rhLCV BACmid system enables the interrogation of EBV-expressed genes involved in viral pathogenesis and for development of novel vaccine approaches.
Immunologically, rhLCV infection of rhesus macaques closely parallels EBV infection in humans. A detailed study of naturally rhLCV-infected animals revealed that the humoral response develops against multiple viral antigens. Animals exhibited hierarchical responses, with the more robust humoral response to late viral proteins and less frequent responses to early and immediate-early proteins. Although a direct comparison could not be made between rhesus macaque humoral responses and human sera responses, the macaque responses still correlated well with EBV-infected human samples153.
T cell specific immunity to rhLCV has also been interrogated. An exhaustive study compared naturally rhLCV-infected rhesus macaques and EBV-infected humans and challenged both species with a recombinant vaccinia virus encoding individual EBV antigens; the results revealed similar T cell responses develop to late antigens in both humans and macaques as persistent infection progresses over time. The close correlations in humoral and T cell specific responses that are shared between rhesus macaques and humans will allow future vaccine evaluation trials using the rhLCV rhesus macaque model. For example: the phase II clinical trial in humans vaccinated with soluble EBV gp350154,155. Related studies in rhesus macaque inoculated with a recombinant rhLCV (encoding EBV gp350 in place of rhLCV gp350) revealed that EBV gp350 could functionally substitute for rhLCV gp350, as the recombinant could infect animals as effectively as wild type rhLCV156. This demonstrates that the rhLCV rhesus macaque model is an excellent system to develop and evaluate novel vaccine approaches to prevent EBV-associated diseases, including infectious mononucleosis156–158.
Gamma-2 herpesviruses: rhadinoviruses
Kaposi’s sarcoma-associated herpesvirus (KSHV; Table 2) is a gamma-2 herpesvirus or rhadinovirus159 and the etiological agent of several rare cancers: Kaposi’s sarcoma, primary effusion lymphoma (PEL)160, and multi-centric Castleman’s disease (MCD)161. Unfortunately, KSHV displays strict tropism for humans, with the exception of one report of KSHV infection and Kaposi’s sarcoma-like lesions in common marmosets162, a model which is being further developed. The lack of a direct animal model of KSHV has led to the development of an NHP model that uses the closely related rhesus macaque rhadinovirus (RRV; a gamma-2 herpesvirus)163. Two independent strains of RRV have been isolated163,164, which exhibit different pathogenic potential. Strain H26–95 is less pathogenic than strain 17577, as strain H26–95 has not been associated with disease, even in SIV-infected animals165. By contrast, strain 17577 is associated with B cell lymphoma and a mesenchymal proliferative lesion, referred to as retroperitoneal fibromatosis, that resembles Kaposi’s sarcoma in SIV-infected rhesus macaque166. The difference in pathogenic potential has led investigators in different research directions as to how to best utilize the RRV rhesus macaque model for human virus infection studies (reviewed in REF167).
Immunologically, RRV infection can induce robust innate and virus-specific humoral and cell-mediated responses in rhesus macaque. This was demonstrated when naïve RM were experimentally infected with a recombinant RRV lacking the eight viral interferon regulatory factors (vIRFs, RRVvIRF-ko) or wild-type (WT) RRV. Animals infected with RRVvIRF-ko had earlier and sustained production of proinflammatory cytokines and earlier induction of RRV-specific T cell responses compared to the WT RRV-infected animals. Humoral responses were essentially identical. The altered host response to RRVvIRF-ko infection was associated with decreased viral loads and diminished B cell hyperplasia, implying the vIRFs function to slow the innate and adaptive immune responses in animals168. That KSHV also expresses IRFs, these data point to the importance of innate immunity against the human virus and point to potential therapeutic interventions that might inhibit vIRFs.
Conclusions and perspective
Here we provide an overview of many of the important human viruses that can be effectively modeled in NHPs. It is outside the scope of this review to cover all human viral pathogens and several other notable examples are briefly mentioned in Box 3. NHPs, represent essential animal models to study many viral infections that impact on human health, in a controlled setting, where strict adherence to protocols and/or drug regimens is enforced to ensure reproducibility. These studies include virological and immunological analyses, and interventional vaccine and therapeutic strategies aimed at treatment and prevention. Longitudinal analyses, coupled with the ability to sample any anatomical site in animal models provides distinct capabilities not possible in humans. NHP studies are highly regulated at the local and federal levels, which ensures animal welfare and well-being. In summary, NHPs are a valuable and reliable animal model.
Box 3. Emerging viruses that can be modeled in nonhuman primates.
Filoviruses
Filoviruses are filamentous single-stranded negative sense RNA viruses. Some members of this family (Ebola virus and Marburg virus in particular) can cause severe viral hemorrhagic fevers, with a very high mortality rate. Viral-induced inflammation is thought to result in much of the pathology observed in infected individuals, with vascular permeability and organ failure occurring in upwards of 70% of infected individuals. Asian macaques can be infected with Ebola and Marburg viruses and the pathology closely resembles that found in humans, although the mortality rate might be slightly higher in NHPs compared to humans, with near 100% mortality in NHPs169. NHPs have been used to test the therapeutic capacity of antiviral compounds against filoviruses and the ability of different vaccine modalities to protect against subsequent viral challenge170–173. Many of these modalities are being tested in humans to treat and prevent future filovirus epidemics.
Pox viruses
Although smallpox virus (Table 1) has been eradicated, there are concerns that it could be weaponized. In addition, monkeypox (MPXV) virus infections of humans lead to symptoms that are similar to smallpox, with fever, weight loss, lesions and up to 10% mortality. Thus, there is interest in developing animal models of pox virus infection. Monkeypox infection of rhesus macaques also results in disease pathology that closely resembles smallpox infection of humans; the degree of symptoms and the overall lethality varies by dose and route of virus challenge, and NHPs have been used to test vaccine modalities and antivirals174–176.
Chikungunya virus
Chikungunya virus (CHIKV) can infect both rhesus macaques and cynomolgus macaque. Moreover, similar to humans, aged macaques have higher and more prolonged viremia compared to younger animals. In addition, Asian macaques become febrile during acute infection and some animals also experience rash and joint swelling (especially if higher doses of virus are used for infections), which is also similar to human infection. After infection CHIKV can be detected in liver, lymphoid tissue, synovial and skeletomuscular tissues, lung, and kidney. These sites can also manifest some degree of structural abnormalities and lymphocyte infiltration. CHIKV infection of Asian macaques can also lead to neurological abnormalities with symptoms of meningoencephalitis and leukocytes in cerebrospinal fluid. Adaptive immune responses are evident in NHPs with proliferation of T cells and induction of T cell responses directed against multiple CHIKV protein epitopes. During infection NHPs also induce B cell responses to CHIKV against numerous viral proteins, the antibodies produced include multiple isotypes, and by 30 days post infection, antibodies are able to neutralize CHIKV in vitro. Thus, Asian macaques represent a viable animal model to study vaccines and therapeutics to prevent CHIKV infection and/or reduce symptoms associated with infection.
Data from NHP studies have provided invaluable insights into human viral infection with a better understanding of mechanisms of disease pathogenesis, understanding of innate and adaptive immune responses, and vaccine and thereapeutic modalities. Studies from NHPs have guided numerous vaccine and therapeutic trials in humans including Zika virus vaccine trials. Future work might involve: determinations as to how modulation of commensal microbiota influences viral pathogenesis; novel strategies for viral prevention and eradication; direct impact of how immune responses and/or host genetics impact susceptibility and outcome to viral infections; novel approaches to diagnose and treat viral diseases, including virus-mediated cancers; testing novel combination (viral and host targeted), approaches to treatment; and development of multi-pathogen models to better reflect certain regions in the world with high pathogen burdens.
Acknowledgments
Funding for this work was provided in part with federal funds from the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health (NIH) and the National Cancer Institute, NIH under Contract number HHSN261200800001E and NIH grants P51-OD011092 (SWW), R01-CA75922 (SWW), and R01-CA206404 (SWW). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Glossary
- MHC tetramers
Biotinylated, peptide loaded, Major Histocompatibility Class I proteins made in bacteria that are tetramerized with fluorophore-conjugated streptavidin. MHC tetramers can be used to identify epitope-specific CD8 T cells.
- Transmitted/founder viruses
The specific viral clone that initiates infection from a polyclonal infectious challenge swarm.
- Simian human immunodeficiency viruses (SHIVs)
Chimeric SIV viruses that encode portions of the HIV (generally envelope) genome.
- “Elite” controllers
Individuals with protective MHC-I alleles that exert CD8+ T cell control of HIV or SIV viral replication.
- Kupffer cells
Specialized liver-resident macrophages involved in recycling iron from dying red blood cells and phagocytosis of microbial antigens that drain into the liver from the portal vein.
- Lobule
Small division of the liver defined at a histological scale.
- Guillian-Barre syndrome
Rare autoimmune disease associated with neurological symptoms, which can lead to paralysis.
- Biohazard level 3 pathogen
Pathogen which requires biosafety level 3 precautions, including use of biological safety cabinets, solid-front protective clothing, and a laboratory entrance separated from areas that have unrestricted traffic flow.
- Latent herpesvirus infections
A state of infection in which the viral genome persists in an infected cell without any viral replication and occasional limited viral gene transcription.
- Hairy leukoplakia
Occasional symptom of Epstein-Barr virus infection in immunocompromised patients, involving a white patch on the side of the tongue with a corrugated appearance.
- rhLCV BACmid
Bacterial artificial chromosome encoding rhesus lymphocryptovirus, which can be used to immortalize rhesus macaque B cells.
Footnotes
Further information box
NIH Office of Research Infrastructure Programs – Comparative Medicine: Vertebrate Models [https://orip.nih.gov/comparative-medicine/programs/vertebrate-models]
National Primate Research Centers [https://nprcresearch.org/primate/]
Biomedical Primate Research Centre [http://www.bprc.nl/en/home/]
NIH NHP Reagent Resource [http://www.nhpreagents.org/NHP/default.aspx]
NHP MHC web portal [https://dholk.primate.wisc.edu/_webdav/dho/mhc_contract/web_portal/%40files/prototype/index.html]
Immuno Polymorphism Database (IPD) - MHC [https://www.ebi.ac.uk/ipd/mhc/group/NHP]
The Macaque Genotype And Phenotype Database [https://mgap.ohsu.edu/]
NHP Genome Sequences [https://www.hgsc.bcm.edu/non-human-primates]
NHP Reference Transcriptome Resource [http://nhprtr.org/]
Virus Pathogen Database and Analysis Resource [https://www.viprbrc.org Database of genomic sequences of virus pathogen strains and analysis tools]
Author contributions
The authors wrote the article together and all contributed to research, discussions, review and editing.
CFI
The authors declare no significant competing interests
Subject terms
Biological sciences / Biological techniques / Experimental organisms [URI /631/1647/334]
Biological sciences / Microbiology / Pathogens [URI /631/326/421]
Biological sciences / Immunology / Infectious diseases [URI /631/250/255]
Biological sciences / Biological techniques / Biological models / Immunological models [URI /631/1647/767/1972]
Bibliography
- 1.Zehn D, Wherry EJ. Immune Memory and Exhaustion: Clinically Relevant Lessons from the LCMV Model. Adv Exp Med Biol. 2015;850:137–152. doi: 10.1007/978-3-319-15774-0_10. [DOI] [PubMed] [Google Scholar]
- 2.Belkaid Y, Harrison OJ. Homeostatic Immunity and the Microbiota. Immunity. 2017;46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hickman HD. New insights into antiviral immunity gained through intravital imaging. Curr Opin Virol. 2017;22:59–63. doi: 10.1016/j.coviro.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Mueller SN, Hickman HD. In vivo imaging of the T cell response to infection. Curr Opin Immunol. 2010;22:293–298. doi: 10.1016/j.coi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Rimmelzwaan GF, et al. Induction of protective immunity against influenza virus in a macaque model: comparison of conventional and iscom vaccines. J Gen Virol. 1997;78(Pt 4):757–765. doi: 10.1099/0022-1317-78-4-757. [DOI] [PubMed] [Google Scholar]
- 6.Klatt NR, et al. Compromised gastrointestinal integrity in pigtail macaques is associated with increased microbial translocation, immune activation, and IL-17 production in the absence of SIV infection. Mucosal Immunol. 2010;3:387–398. doi: 10.1038/mi.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baskin CR, et al. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina) J Virol. 2004;78:10420–10432. doi: 10.1128/JVI.78.19.10420-10432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ericsen AJ, et al. Microbial Translocation and Inflammation Occur in Hyperacute Immunodeficiency Virus Infection and Compromise Host Control of Virus Replication. PLoS Pathog. 2016;12:e1006048. doi: 10.1371/journal.ppat.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mudd PA, et al. Escape from CD8(+) T cell responses in Mamu-B*00801(+) macaques differentiates progressors from elite controllers. J Immunol. 2012;188:3364–3370. doi: 10.4049/jimmunol.1102470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price DA, et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med. 2009;206:923–936. doi: 10.1084/jem.20081127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minang JT, et al. The Mamu B 17-restricted SIV Nef IW9 to TW9 mutation abrogates correct epitope processing and presentation without loss of replicative fitness. Virology. 2008;375:307–314. doi: 10.1016/j.virol.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klatt NR, et al. Dynamics of simian immunodeficiency virus SIVmac239 infection in pigtail macaques. J Virol. 2012;86:1203–1213. doi: 10.1128/JVI.06033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karl JA, et al. Major histocompatibility complex haplotyping and long-amplicon allele discovery in cynomolgus macaques from Chinese breeding facilities. Immunogenetics. 2017;69:211–229. doi: 10.1007/s00251-017-0969-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ericsen AJ, et al. Whole genome sequencing of SIV-infected macaques identifies candidate loci that may contribute to host control of virus replication. Genome Biol. 2014;15:478. doi: 10.1186/s13059-014-0478-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene JM, et al. Allogeneic lymphocytes persist and traffic in feral MHC-matched mauritian cynomolgus macaques. PLoS One. 2008;3:e2384. doi: 10.1371/journal.pone.0002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, et al. TRIM5alpha restriction affects clinical outcome and disease progression in simian immunodeficiency virus-infected rhesus macaques. J Virol. 2015;89:2233–2240. doi: 10.1128/JVI.02978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu CQ, et al. The TRIMCyp genotype in four species of macaques in China. Immunogenetics. 2013;65:185–193. doi: 10.1007/s00251-012-0670-9. [DOI] [PubMed] [Google Scholar]
- 18.Sodora DL, et al. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat Med. 2009;15:861–865. doi: 10.1038/nm.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuoka Y, et al. African green monkeys recapitulate the clinical experience with replication of live attenuated pandemic influenza virus vaccine candidates. J Virol. 2014;88:8139–8152. doi: 10.1128/JVI.00425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locher CP, et al. Baboons as an animal model for human immunodeficiency virus pathogenesis and vaccine development. Immunol Rev. 2001;183:127–140. doi: 10.1034/j.1600-065x.2001.1830111.x. [DOI] [PubMed] [Google Scholar]
- 21.Harris LD, et al. Downregulation of robust acute type I interferon responses distinguishes nonpathogenic simian immunodeficiency virus (SIV) infection of natural hosts from pathogenic SIV infection of rhesus macaques. J Virol. 2010;84:7886–7891. doi: 10.1128/JVI.02612-09. JVI.02612-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aid M, et al. Zika Virus Persistence in the Central Nervous System and Lymph Nodes of Rhesus Monkeys. Cell. 2017;169:610–620 e614. doi: 10.1016/j.cell.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquelin B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–3555. doi: 10.1172/JCI40093. 40093 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson BA, Estep RD, Messaoudi I, Rogers KS, Wong SW. Viral interferon regulatory factors decrease the induction of type I and type II interferon during rhesus macaque rhadinovirus infection. J Virol. 2012;86:2197–2211. doi: 10.1128/JVI.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Groot N, Doxiadis GG, Otting N, de Vos-Rouweler AJ, Bontrop RE. Differential recombination dynamics within the MHC of macaque species. Immunogenetics. 2014;66:535–544. doi: 10.1007/s00251-014-0783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doxiadis GG, et al. Haplotype diversity generated by ancient recombination-like events in the MHC of Indian rhesus macaques. Immunogenetics. 2013;65:569–584. doi: 10.1007/s00251-013-0707-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otting N, et al. The orthologs of HLA-DQ and -DP genes display abundant levels of variability in macaque species. Immunogenetics. 2017;69:87–99. doi: 10.1007/s00251-016-0954-6. [DOI] [PubMed] [Google Scholar]
- 28.Hansen SG, et al. Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science. 2016;351:714–720. doi: 10.1126/science.aac9475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klatt NR, et al. SIV infection of rhesus macaques results in dysfunctional T- and B-cell responses to neo and recall Leishmania major vaccination. Blood. 2011;118:5803–5812. doi: 10.1182/blood-2011-07-365874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramesh A, et al. Structure and Diversity of the Rhesus Macaque Immunoglobulin Loci through Multiple De Novo Genome Assemblies. Front Immunol. 2017;8:1407. doi: 10.3389/fimmu.2017.01407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu GY, et al. Cynomolgus macaque (Macaca fascicularis) immunoglobulin heavy chain locus description. Immunogenetics. 2016;68:417–428. doi: 10.1007/s00251-016-0921-2. [DOI] [PubMed] [Google Scholar]
- 32.Sundling C, et al. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol. 2014;192:3637–3644. doi: 10.4049/jimmunol.1303334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Groot NG, Heijmans CMC, Bontrop RE. AIDS in chimpanzees: the role of MHC genes. Immunogenetics. 2017;69:499–509. doi: 10.1007/s00251-017-1006-6. [DOI] [PubMed] [Google Scholar]
- 34.Letvin NL, et al. Acquired immunodeficiency syndrome in a colony of macaque monkeys. Proc Natl Acad Sci U S A. 1983;80:2718–2722. doi: 10.1073/pnas.80.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel MD, et al. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 36.King NW, Hunt RD, Letvin NL. Histopathologic changes in macaques with an acquired immunodeficiency syndrome (AIDS) Am J Pathol. 1983;113:382–388. [PMC free article] [PubMed] [Google Scholar]
- 37.Hirsch VM, et al. Molecular cloning of SIV from sooty mangabey monkeys. J Med Primatol. 1989;18:279–285. [PubMed] [Google Scholar]
- 38.Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–392. doi: 10.1038/339389a0. [DOI] [PubMed] [Google Scholar]
- 39.Sharp PM, Robertson DL, Hahn BH. Cross-species transmission and recombination of ‘AIDS’ viruses. Philos Trans R Soc Lond B Biol Sci. 1995;349:41–47. doi: 10.1098/rstb.1995.0089. [DOI] [PubMed] [Google Scholar]
- 40.Makori N, et al. Functional and morphological development of lymphoid tissues and immune regulatory and effector function in rhesus monkeys: cytokine-secreting cells, immunoglobulin-secreting cells, and CD5(+) B-1 cells appear early in fetal development. Clin Diagn Lab Immunol. 2003;10:140–153. doi: 10.1128/CDLI.10.1.140-153.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abel K. The rhesus macaque pediatric SIV infection model - a valuable tool in understanding infant HIV-1 pathogenesis and for designing pediatric HIV-1 prevention strategies. Curr HIV Res. 2009;7:2–11. doi: 10.2174/157016209787048528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller CJ, et al. Genital mucosal transmission of simian immunodeficiency virus: animal model for heterosexual transmission of human immunodeficiency virus. J Virol. 1989;63:4277–4284. doi: 10.1128/jvi.63.10.4277-4284.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sodora DL, Gettie A, Miller CJ, Marx PA. Vaginal transmission of SIV: assessing infectivity and hormonal influences in macaques inoculated with cell-free and cell-associated viral stocks. AIDS Res Hum Retroviruses. 1998;14(Suppl 1):S119–123. [PubMed] [Google Scholar]
- 44.Li Q, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keele BF, et al. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med. 2009;206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma ZM, et al. SIVmac251 is inefficiently transmitted to rhesus macaques by penile inoculation with a single SIVenv variant found in ramp-up phase plasma. AIDS Res Hum Retroviruses. 2011;27:1259–1269. doi: 10.1089/aid.2011.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barouch DH, et al. Rapid Inflammasome Activation following Mucosal SIV Infection of Rhesus Monkeys. Cell. 2016;165:656–667. doi: 10.1016/j.cell.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller CJ. Localization of Simian immunodeficiency virus-infected cells in the genital tract of male and female Rhesus macaques. J Reprod Immunol. 1998;41:331–339. doi: 10.1016/s0165-0378(98)00069-2. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Z, et al. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 50.Miller CJ, et al. Propagation and dissemination of infection after vaginal transmission of simian immunodeficiency virus. J Virol. 2005;79:9217–9227. doi: 10.1128/JVI.79.14.9217-9227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haase AT. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu Rev Med. 2011;62:127–139. doi: 10.1146/annurev-med-080709-124959. [DOI] [PubMed] [Google Scholar]
- 52.Fennessey CM, Keele BF. Using nonhuman primates to model HIV transmission. Curr Opin HIV AIDS. 2013;8:280–287. doi: 10.1097/COH.0b013e328361cfff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J, et al. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol. 2010;84:10406–10412. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee FH, et al. Breakthrough Virus Neutralization Resistance as a Correlate of Protection in a Nonhuman Primate Heterologous Simian Immunodeficiency Virus Vaccine Challenge Study. J Virol. 2015;89:12388–12400. doi: 10.1128/JVI.01531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gach JS, et al. Relationship between Vaccine-Induced Antibody Capture of Infectious Virus and Infection Outcomes following Repeated Low-Dose Rectal Challenges with Simian Immunodeficiency Virus SIVmac251. J Virol. 2016;90:8487–8495. doi: 10.1128/JVI.00812-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roederer M, et al. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature. 2014;505:502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keele BF, et al. Adenovirus prime, Env protein boost vaccine protects against neutralization-resistant SIVsmE660 variants in rhesus monkeys. Nat Commun. 2017;8:15740. doi: 10.1038/ncomms15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evans DT, Silvestri G. Nonhuman primate models in AIDS research. Curr Opin HIV AIDS. 2013;8:255–261. doi: 10.1097/COH.0b013e328361cee8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 60.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pegu P, et al. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol. 2013;87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Putkonen P, et al. Prevention of HIV-2 and SIVsm infection by passive immunization in cynomolgus monkeys. Nature. 1991;352:436–438. doi: 10.1038/352436a0. [DOI] [PubMed] [Google Scholar]
- 63.Van Rompay KK, et al. Passive immunization of newborn rhesus macaques prevents oral simian immunodeficiency virus infection. J Infect Dis. 1998;177:1247–1259. doi: 10.1086/515270. [DOI] [PubMed] [Google Scholar]
- 64.Shibata R, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 65.Mascola JR, et al. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nat Med. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 66.Hessell AJ, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- 67.Kelsoe G, Haynes BF. Host controls of HIV broadly neutralizing antibody development. Immunol Rev. 2017;275:79–88. doi: 10.1111/imr.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shiver JW, et al. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature. 2002;415:331–335. doi: 10.1038/415331a. [DOI] [PubMed] [Google Scholar]
- 69.McMichael AJ, Koff WC. Vaccines that stimulate T cell immunity to HIV-1: the next step. Nat Immunol. 2014;15:319–322. doi: 10.1038/ni.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar N, Chahroudi A, Silvestri G. Animal models to achieve an HIV cure. Curr Opin HIV AIDS. 2016;11:432–441. doi: 10.1097/COH.0000000000000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brenchley JM, Douek DC. Microbial translocation across the GI tract. Annu Rev Immunol. 2012;30:149–173. doi: 10.1146/annurev-immunol-020711-075001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deeks SG, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104:942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 73.Giorgi JV, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 74.Silvestri G, Paiardini M, Pandrea I, Lederman MM, Sodora DL. Understanding the benign nature of SIV infection in natural hosts. J Clin Invest. 2007;117:3148–3154. doi: 10.1172/JCI33034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Douek DC, Roederer M, Koup RA. Emerging concepts in the immunopathogenesis of AIDS. Annu Rev Med. 2009;60:471–484. doi: 10.1146/annurev.med.60.041807.123549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Estes JD. Pathobiology of HIV/SIV-associated changes in secondary lymphoid tissues. Immunol Rev. 2013;254:65–77. doi: 10.1111/imr.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 78.Ciccone EJ, et al. Cycling of gut mucosal CD4+ T cells decreases after prolonged anti-retroviral therapy and is associated with plasma LPS levels. Mucosal Immunol. 2010;3:172–181. doi: 10.1038/mi.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang W, et al. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-Treated HIV infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Estes JD, et al. Damaged intestinal epithelial integrity linked to microbial translocation in pathogenic simian immunodeficiency virus infections. PLoS Pathog. 2010;6:e1001052. doi: 10.1371/journal.ppat.1001052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pandrea IV, et al. Acute Loss of Intestinal CD4+ T Cells Is Not Predictive of Simian Immunodeficiency Virus Virulence. J Immunol. 2007;179:3035–3046. doi: 10.4049/jimmunol.179.5.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hao XP, et al. Experimental colitis in SIV-uninfected rhesus macaques recapitulates important features of pathogenic SIV infection. Nat Commun. 2015;6:8020. doi: 10.1038/ncomms9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chalifoux LV, et al. Lymphadenopathy in macaques experimentally infected with the simian immunodeficiency virus (SIV) Am J Pathol. 1987;128:104–110. [PMC free article] [PubMed] [Google Scholar]
- 84.Schacker TW, et al. Collagen deposition in HIV-1 infected lymphatic tissues and T cell homeostasis. J Clin Invest. 2002;110:1133–1139. doi: 10.1172/JCI16413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nilsson J, et al. HIV-1-driven regulatory T-cell accumulation in lymphoid tissues is associated with disease progression in HIV/AIDS. Blood. 2006;108:3808–3817. doi: 10.1182/blood-2006-05-021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andersson J, et al. The prevalence of regulatory T cells in lymphoid tissue is correlated with viral load in HIV-infected patients. J Immunol. 2005;174:3143–3147. doi: 10.4049/jimmunol.174.6.3143. [DOI] [PubMed] [Google Scholar]
- 87.Lawitz E, et al. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet. 2014;383:515–523. doi: 10.1016/S0140-6736(13)62121-2. [DOI] [PubMed] [Google Scholar]
- 88.Rockstroh JK, et al. Ledipasvir-sofosbuvir for 6 weeks to treat acute hepatitis C virus genotype 1 or 4 infection in patients with HIV coinfection: an open-label, single-arm trial. Lancet Gastroenterol Hepatol. 2017;2:347–353. doi: 10.1016/S2468-1253(17)30003-1. [DOI] [PubMed] [Google Scholar]
- 89.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 90.Houghton M, Abrignani S. Prospects for a vaccine against the hepatitis C virus. Nature. 2005;436:961–966. doi: 10.1038/nature04081. [DOI] [PubMed] [Google Scholar]
- 91.Catanese MT, Dorner M. Advances in experimental systems to study hepatitis C virus in vitro and in vivo. Virology. 2015;479–480:221–233. doi: 10.1016/j.virol.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 92.Lanford RE, Walker CM, Lemon SM. The Chimpanzee Model of Viral Hepatitis: Advances in Understanding the Immune Response and Treatment of Viral Hepatitis. ILAR J. 2017:1–18. doi: 10.1093/ilar/ilx028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Farci P, et al. The natural history of infection with hepatitis C virus (HCV) in chimpanzees: comparison of serologic responses measured with first- and second-generation assays and relationship to HCV viremia. J Infect Dis. 1992;165:1006–1011. doi: 10.1093/infdis/165.6.1006. [DOI] [PubMed] [Google Scholar]
- 94.Farci P, et al. Experimental transmission of hepatitis C virus-associated fulminant hepatitis to a chimpanzee. J Infect Dis. 1999;179:1007–1011. doi: 10.1086/314653. [DOI] [PubMed] [Google Scholar]
- 95.Major ME, et al. Hepatitis C virus kinetics and host responses associated with disease and outcome of infection in chimpanzees. Hepatology. 2004;39:1709–1720. doi: 10.1002/hep.20239. [DOI] [PubMed] [Google Scholar]
- 96.Manickam C, Reeves RK. Modeling HCV disease in animals: virology, immunology and pathogenesis of HCV and GBV-B infections. Front Microbiol. 2014;5:690. doi: 10.3389/fmicb.2014.00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith DB, et al. Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol. 2016;97:537–542. doi: 10.1099/jgv.0.000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bradley DW, et al. Enterically transmitted non-A, non-B hepatitis: serial passage of disease in cynomolgus macaques and tamarins and recovery of disease-associated 27- to 34-nm viruslike particles. Proc Natl Acad Sci U S A. 1987;84:6277–6281. doi: 10.1073/pnas.84.17.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kane MA, et al. Epidemic non-A, non-B hepatitis in Nepal. Recovery of a possible etiologic agent and transmission studies in marmosets. JAMA. 1984;252:3140–3145. doi: 10.1001/jama.252.22.3140. [DOI] [PubMed] [Google Scholar]
- 100.Arankalle VA, Chobe LP, Chadha MS. Type-IV Indian swine HEV infects rhesus monkeys. J Viral Hepat. 2006;13:742–745. doi: 10.1111/j.1365-2893.2006.00759.x. [DOI] [PubMed] [Google Scholar]
- 101.van Cuyck-Gandre H, et al. Experimental African HEV infection in cynomolgus macaques (Macaca fascicularis) J Med Virol. 1998;55:197–202. doi: 10.1002/(sici)1096-9071(199807)55:3<197::aid-jmv3>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 102.McCaustland KA, et al. Hepatitis E virus infection in chimpanzees: a retrospective analysis. Arch Virol. 2000;145:1909–1918. doi: 10.1007/s007050070065. [DOI] [PubMed] [Google Scholar]
- 103.Tsarev SA, et al. Variation in course of hepatitis E in experimentally infected cynomolgus monkeys. J Infect Dis. 1993;167:1302–1306. doi: 10.1093/infdis/167.6.1302. [DOI] [PubMed] [Google Scholar]
- 104.Tsarev SA, et al. Experimental hepatitis E in pregnant rhesus monkeys: failure to transmit hepatitis E virus (HEV) to offspring and evidence of naturally acquired antibodies to HEV. J Infect Dis. 1995;172:31–37. doi: 10.1093/infdis/172.1.31. [DOI] [PubMed] [Google Scholar]
- 105.Zhang J, et al. Evaluation of antibody-based and nucleic acid-based assays for diagnosis of hepatitis E virus infection in a rhesus monkey model. J Med Virol. 2003;71:518–526. doi: 10.1002/jmv.10523. [DOI] [PubMed] [Google Scholar]
- 106.Aggarwal R, Kamili S, Spelbring J, Krawczynski K. Experimental studies on subclinical hepatitis E virus infection in cynomolgus macaques. J Infect Dis. 2001;184:1380–1385. doi: 10.1086/324376. [DOI] [PubMed] [Google Scholar]
- 107.Purcell RH, et al. Pathobiology of hepatitis E: lessons learned from primate models. Emerg Microbes Infect. 2013;2:e9. doi: 10.1038/emi.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Purcell RH, Emerson SU. Animal models of hepatitis A and E. ILAR J. 2001;42:161–177. doi: 10.1093/ilar.42.2.161. [DOI] [PubMed] [Google Scholar]
- 109.Li SW, et al. A bacterially expressed particulate hepatitis E vaccine: antigenicity, immunogenicity and protectivity on primates. Vaccine. 2005;23:2893–2901. doi: 10.1016/j.vaccine.2004.11.064. [DOI] [PubMed] [Google Scholar]
- 110.Purcell RH, et al. Pre-clinical immunogenicity and efficacy trial of a recombinant hepatitis E vaccine. Vaccine. 2003;21:2607–2615. doi: 10.1016/s0264-410x(03)00100-2. [DOI] [PubMed] [Google Scholar]
- 111.Tsarev SA, et al. Successful passive and active immunization of cynomolgus monkeys against hepatitis E. Proc Natl Acad Sci U S A. 1994;91:10198–10202. doi: 10.1073/pnas.91.21.10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tsarev SA, et al. Recombinant vaccine against hepatitis E: dose response and protection against heterologous challenge. Vaccine. 1997;15:1834–1838. doi: 10.1016/s0264-410x(97)00145-x. [DOI] [PubMed] [Google Scholar]
- 113.Zhang M, et al. Immunogenicity and protective efficacy of a vaccine prepared from 53 kDa truncated hepatitis E virus capsid protein expressed in insect cells. Vaccine. 2001;20:853–857. doi: 10.1016/s0264-410x(01)00399-1. [DOI] [PubMed] [Google Scholar]
- 114.Shrestha MP, et al. Safety and efficacy of a recombinant hepatitis E vaccine. N Engl J Med. 2007;356:895–903. doi: 10.1056/NEJMoa061847. [DOI] [PubMed] [Google Scholar]
- 115.Pierson TC, Kielian M. Flaviviruses: braking the entering. Curr Opin Virol. 2013;3:3–12. doi: 10.1016/j.coviro.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J Virol. 2005;79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meier KC, Gardner CL, Khoretonenko MV, Klimstra WB, Ryman KD. A mouse model for studying viscerotropic disease caused by yellow fever virus infection. PLoS Pathog. 2009;5:e1000614. doi: 10.1371/journal.ppat.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Morrison TE, Diamond MS. Animal Models of Zika Virus Infection, Pathogenesis, and Immunity. J Virol. 2017;91 doi: 10.1128/JVI.00009-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Clark KB, Onlamoon N, Hsiao HM, Perng GC, Villinger F. Can non-human primates serve as models for investigating dengue disease pathogenesis? Front Microbiol. 2013;4:305. doi: 10.3389/fmicb.2013.00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fox A, et al. Immunological and viral determinants of dengue severity in hospitalized adults in Ha Noi, Viet Nam. PLoS Negl Trop Dis. 2011;5:e967. doi: 10.1371/journal.pntd.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sariol CA, White LJ. Utility, limitations, and future of non-human primates for dengue research and vaccine development. Front Immunol. 2014;5:452. doi: 10.3389/fimmu.2014.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Durbin AP, Kirkpatrick BD, Pierce KK, Schmidt AC, Whitehead SS. Development and clinical evaluation of multiple investigational monovalent DENV vaccines to identify components for inclusion in a live attenuated tetravalent DENV vaccine. Vaccine. 2011;29:7242–7250. doi: 10.1016/j.vaccine.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Whitehead SS, et al. In a randomized trial, the live attenuated tetravalent dengue vaccine TV003 is well-tolerated and highly immunogenic in subjects with flavivirus exposure prior to vaccination. PLoS Negl Trop Dis. 2017;11:e0005584. doi: 10.1371/journal.pntd.0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kirkpatrick BD, et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med. 2016;8:330ra336. doi: 10.1126/scitranslmed.aaf1517. [DOI] [PubMed] [Google Scholar]
- 125.Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. An update on Zika virus infection. Lancet. 2017 doi: 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- 126.Li XF, et al. Characterization of a 2016 Clinical Isolate of Zika Virus in Non-human Primates. EBioMedicine. 2016;12:170–177. doi: 10.1016/j.ebiom.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dudley DM, et al. A rhesus macaque model of Asian-lineage Zika virus infection. Nat Commun. 2016;7:12204. doi: 10.1038/ncomms12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Osuna CE, et al. Zika viral dynamics and shedding in rhesus and cynomolgus macaques. Nat Med. 2016;22:1448–1455. doi: 10.1038/nm.4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nguyen SM, et al. Highly efficient maternal-fetal Zika virus transmission in pregnant rhesus macaques. PLoS Pathog. 2017;13:e1006378. doi: 10.1371/journal.ppat.1006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abbink P, et al. Protective efficacy of multiple vaccine platforms against Zika virus challenge in rhesus monkeys. Science. 2016;353:1129–1132. doi: 10.1126/science.aah6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dowd KA, et al. Rapid development of a DNA vaccine for Zika virus. Science. 2016;354:237–240. doi: 10.1126/science.aai9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Larocca RA, et al. Vaccine protection against Zika virus from Brazil. Nature. 2016;536:474–478. doi: 10.1038/nature18952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Driggers RW, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- 134.Koide F, et al. Development of a Zika Virus Infection Model in Cynomolgus Macaques. Front Microbiol. 2016;7:2028. doi: 10.3389/fmicb.2016.02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hirsch AJ, et al. Zika Virus infection of rhesus macaques leads to viral persistence in multiple tissues. PLoS Pathog. 2017;13:e1006219. doi: 10.1371/journal.ppat.1006219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yeung MW, Shing E, Nelder M, Sander B. Epidemiologic and clinical parameters of West Nile virus infections in humans: a scoping review. BMC Infect Dis. 2017;17:609. doi: 10.1186/s12879-017-2637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Verstrepen BE, et al. Experimental infection of rhesus macaques and common marmosets with a European strain of West Nile virus. PLoS Negl Trop Dis. 2014;8:e2797. doi: 10.1371/journal.pntd.0002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Maximova OA, Bernbaum JG, Pletnev AG. West Nile Virus Spreads Transsynaptically within the Pathways of Motor Control: Anatomical and Ultrastructural Mapping of Neuronal Virus Infection in the Primate Central Nervous System. PLoS Negl Trop Dis. 2016;10:e0004980. doi: 10.1371/journal.pntd.0004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lieberman MM, et al. Immunogenicity and protective efficacy of a recombinant subunit West Nile virus vaccine in rhesus monkeys. Clin Vaccine Immunol. 2009;16:1332–1337. doi: 10.1128/CVI.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Verstrepen BE, et al. Vaccine-induced protection of rhesus macaques against plasma viremia after intradermal infection with a European lineage 1 strain of West Nile virus. PLoS One. 2014;9:e112568. doi: 10.1371/journal.pone.0112568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Arroyo J, et al. ChimeriVax-West Nile virus live-attenuated vaccine: preclinical evaluation of safety, immunogenicity, and efficacy. J Virol. 2004;78:12497–12507. doi: 10.1128/JVI.78.22.12497-12507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Widman DG, et al. Evaluation of RepliVAX WN, a single-cycle flavivirus vaccine, in a non-human primate model of West Nile virus infection. Am J Trop Med Hyg. 2010;82:1160–1167. doi: 10.4269/ajtmh.2010.09-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ouwendijk WJ, Verjans GM. Pathogenesis of varicelloviruses in primates. J Pathol. 2015;235:298–311. doi: 10.1002/path.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Haberthur K, et al. CD4 T cell immunity is critical for the control of simian varicella virus infection in a nonhuman primate model of VZV infection. PLoS Pathog. 2011;7:e1002367. doi: 10.1371/journal.ppat.1002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Traina-Dorge V, et al. Simian Varicella Virus Is Present in Macrophages, Dendritic Cells, and T Cells in Lymph Nodes of Rhesus Macaques after Experimental Reactivation. J Virol. 2015;89:9817–9824. doi: 10.1128/JVI.01324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Itell HL, Kaur A, Deere JD, Barry PA, Permar SR. Rhesus monkeys for a nonhuman primate model of cytomegalovirus infections. Curr Opin Virol. 2017;25:126–133. doi: 10.1016/j.coviro.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pitcher CJ, et al. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 148.Hansen SG, et al. Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med. 2009;15:293–299. doi: 10.1038/nm.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fruh K, Picker L. CD8+ T cell programming by cytomegalovirus vectors: applications in prophylactic and therapeutic vaccination. Curr Opin Immunol. 2017;47:52–56. doi: 10.1016/j.coi.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Muhe J, Wang F. Non-human Primate Lymphocryptoviruses: Past, Present, and Future. Curr Top Microbiol Immunol. 2015;391:385–405. doi: 10.1007/978-3-319-22834-1_13. [DOI] [PubMed] [Google Scholar]
- 152.Ohashi M, Orlova N, Quink C, Wang F. Cloning of the Epstein-Barr virus-related rhesus lymphocryptovirus as a bacterial artificial chromosome: a loss-of-function mutation of the rhBARF1 immune evasion gene. J Virol. 2011;85:1330–1339. doi: 10.1128/JVI.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Orlova N, Fogg MH, Carville A, Wang F. Antibodies to lytic infection proteins in lymphocryptovirus-infected rhesus macaques: a model for humoral immune responses to epstein-barr virus infection. Clin Vaccine Immunol. 2011;18:1427–1434. doi: 10.1128/CVI.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Moutschen M, et al. Phase I/II studies to evaluate safety and immunogenicity of a recombinant gp350 Epstein-Barr virus vaccine in healthy adults. Vaccine. 2007;25:4697–4705. doi: 10.1016/j.vaccine.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 155.Sokal EM, et al. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J Infect Dis. 2007;196:1749–1753. doi: 10.1086/523813. [DOI] [PubMed] [Google Scholar]
- 156.Herrman M, Muhe J, Quink C, Wang F. Epstein-Barr Virus gp350 Can Functionally Replace the Rhesus Lymphocryptovirus Major Membrane Glycoprotein and Does Not Restrict Infection of Rhesus Macaques. J Virol. 2015;90:1222–1230. doi: 10.1128/JVI.02531-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Leskowitz R, et al. Adenovirus-based vaccines against rhesus lymphocryptovirus EBNA-1 induce expansion of specific CD8+ and CD4+ T cells in persistently infected rhesus macaques. J Virol. 2014;88:4721–4735. doi: 10.1128/JVI.03744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Leskowitz RM, et al. CD4+ and CD8+ T-cell responses to latent antigen EBNA-1 and lytic antigen BZLF-1 during persistent lymphocryptovirus infection of rhesus macaques. J Virol. 2013;87:8351–8362. doi: 10.1128/JVI.00852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Russo JJ, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci U S A. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 161.Soulier J, et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 162.Chang H, et al. Non-human primate model of Kaposi’s sarcoma-associated herpesvirus infection. PLoS Pathog. 2009;5:e1000606. doi: 10.1371/journal.ppat.1000606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Desrosiers RC, et al. A herpesvirus of rhesus monkeys related to the human Kaposi’s sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wong SW, et al. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi’s sarcoma-associated herpesvirus. J Exp Med. 1999;190:827–840. doi: 10.1084/jem.190.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mansfield KG, et al. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol. 1999;73:10320–10328. doi: 10.1128/jvi.73.12.10320-10328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Orzechowska BU, et al. Rhesus macaque rhadinovirus-associated non-Hodgkin lymphoma: animal model for KSHV-associated malignancies. Blood. 2008;112:4227–4234. doi: 10.1182/blood-2008-04-151498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Estep RD, Wong SW. Rhesus macaque rhadinovirus-associated disease. Curr Opin Virol. 2013;3:245–250. doi: 10.1016/j.coviro.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Robinson BA, et al. Viral interferon regulatory factors are critical for delay of the host immune response against rhesus macaque rhadinovirus infection. J Virol. 2012;86:2769–2779. doi: 10.1128/JVI.05657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nakayama E, Saijo M. Animal models for Ebola and Marburg virus infections. Front Microbiol. 2013;4:267. doi: 10.3389/fmicb.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Warren TK, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Misasi J, et al. Structural and molecular basis for Ebola virus neutralization by protective human antibodies. Science. 2016;351:1343–1346. doi: 10.1126/science.aad6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Corti D, et al. Protective monotherapy against lethal Ebola virus infection by a potently neutralizing antibody. Science. 2016;351:1339–1342. doi: 10.1126/science.aad5224. [DOI] [PubMed] [Google Scholar]
- 173.Mire CE, et al. Therapeutic treatment of Marburg and Ravn virus infection in nonhuman primates with a human monoclonal antibody. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Earl PL, et al. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature. 2004;428:182–185. doi: 10.1038/nature02331. [DOI] [PubMed] [Google Scholar]
- 175.Edghill-Smith Y, et al. Smallpox vaccine-induced antibodies are necessary and sufficient for protection against monkeypox virus. Nat Med. 2005;11:740–747. doi: 10.1038/nm1261. [DOI] [PubMed] [Google Scholar]
- 176.Stittelaar KJ, et al. Antiviral treatment is more effective than smallpox vaccination upon lethal monkeypox virus infection. Nature. 2006;439:745–748. doi: 10.1038/nature04295. [DOI] [PubMed] [Google Scholar]
- 177.Margulies EH, et al. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–774. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]