Abstract
Understanding brain function requires technologies that can control the activity of large populations of neurons with high fidelity in space and time. We developed a new multiphoton holographic approach to activate or suppress the activity of ensembles of cortical neurons with cellular resolution and sub-millisecond precision. Since existing opsins were inadequate, we engineered new soma-targeted (ST) optogenetic tools, ST-ChroME and IRES-ST-eGtACR1, optimized for multiphoton activation and suppression. Employing a three-dimensional all-optical read/write interface, we demonstrate the ability to photo-stimulate up to 50 neurons simultaneously distributed in three dimensions in a 550 × 550 × 100 μm volume of brain tissue. This new approach allows the synthesis and editing of complex neural activity patterns needed to gain insight into the principles of neural codes.
Introduction
Neural circuits can encode information in the rate1, timing2, number3, and synchrony of action potentials4, and in the identity of active neurons5. Yet the technical inability to create or edit custom patterns of spatiotemporal neural activity is a key impediment to understanding the logic and syntax of the neural code6. Experimental approaches that allow high fidelity temporal control of neural activity7 from specific groups of neurons would make it possible to systematically vary spike rate, timing, synchrony, and number, and permit definitive tests for how neural ensembles encode information. Similarly, deleting action potentials from functionally defined neurons will allow experimenters to probe the elements of endogenous activity patterns that contribute to neural computations and behaviors with unprecedented precision.
Optogenetics offers the basis for such a technology, but many neural computations rely on genetically similar, yet functionally distinct neurons that are physically intermixed8,9, and are thus beyond the reach of conventional approaches. Two-photon (2P) optogenetics10–13 allows experimenters to stimulate neurons based on their precise spatial location as well as their genetic identity. Combined with 2P calcium imaging, this allows activation of specific neurons on the basis of any desired feature14–18. However, in vivo all-optical approaches in mice have suffered from low temporal precision (>10 ms jitter)14–16, or could only photo-stimulate several neurons simultaneously15,19, and thus could not create precise neural activity patterns. Furthermore, in vivo multiphoton suppression of neural activity was not previously possible20, critically limiting the ability of experimenters to assess the necessity of spikes originating from specific, functionally defined neurons.
Several optical methods can stimulate neurons using 2P excitation, although all have limitations. A standard 2P optogenetic technique is to scan a small laser spot over neurons that express variants of slow red-shifted opsins like C1V1 T/T10,13. While this approach effectively drives spiking, the slow kinetics of the scanning laser and the opsin preclude precise control of the specific sequence of neural activity, resulting in uncertain trial-to-trial reproducibility12,14,16. In contrast, scanless multiphoton stimulation using computer generated holography (CGH) and temporal focusing (TF) allows simultaneous illumination of the entire somatic membrane, and can provide higher temporal fidelity when used with a fast opsin19,21–25. However, existing excitatory opsins are too weak or too slow to drive precise neural activity patterns with scanless 2P optogenetics in vivo15,20. Additionally, only recent innovations have allowed scanless holographic optogenetics to function with high axial resolution in three dimensions (3D)26,27.
Therefore, to synthesize and edit custom distributed patterns of neural ensemble activity, we engineered powerful new opsins optimized for multiphoton optogenetics. We developed ChroME, a new ultra-fast and highly potent opsin with 3–5 times larger photocurrents than opsins commonly used for multiphoton optogenetics12,13,28. This allows high fidelity sub-millisecond control of pyramidal neuron spiking. In vivo, we activated ensembles of neurons expressing soma targeted (ST-) ChroME using 3D-SHOT (3D-Scanless Holographic Optogenetics with Temporal Focusing)27, synthesizing precise sequences of neural activity with cellular resolution and millisecond precision. By combining 3D-SHOT with volumetric 2P calcium imaging, we obtain all-optical control of distributed neural ensembles, simultaneously stimulating up to 50 neurons with high temporal precision. Furthermore, to achieve all-optical suppression, we improved and employed the extremely potent inhibitory anion opsin, GtACR129, which exhibits 80 fold increases in photocurrent over previously employed pump-based opsins for multiphoton optogenetic silencing13. Critically, we identified a strategy to prevent the cellular toxicity we observed when the transgene was expressed conventionally. Using this new construct, IRES-ST-eGtACR1, with 3D-SHOT, we provide electrophysiological and all-optical demonstration of high fidelity silencing of neural activity from identified neurons in vivo. Together, these data represent a novel technological approach for precise multimodal control of neural ensemble activity with high fidelity.
Results
Requirements for controlling neural activity with millisecond precision
To control neural activity with sub-millisecond precision, we sought an opsin and a 2P stimulation paradigm that can generate large currents with rapid kinetics30. Injecting current into patched neurons (layer 2/3 (L2/3) pyramidal cells, used throughout this study unless otherwise noted) in brain slices, we could reliably evoke precise spike trains with brief, high amplitude current steps. In contrast, long current injections, analogous to some spiral scanning approaches14,16, resulted in variable spike number and timing, but required lower current amplitudes (Fig 1a–b, Supplementary Fig. 1).
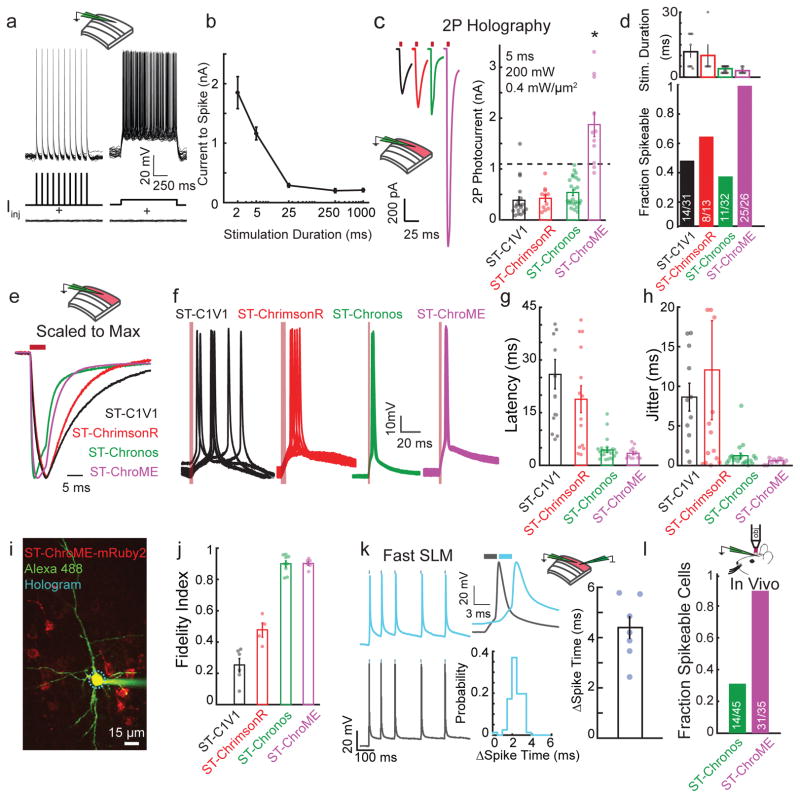
Figure 1. ST-ChroME allows precise high-fidelity 2P activation.
a) Overlay of 25 trials from a representative L2/3 pyramidal neuron showing the Vm response to 5 ms current pulses or sustained current injection near the neuron’s rheobase.
b) Current needed to induce action potentials as a function of stimulus duration (n = 8 L2/3 pyramidal neurons).
c) Left, grand average photocurrent traces from neurons expressing ST-C1V1T/T (black, n = 19), ST-ChrimsonR (red, n =11), ST-Chronos (green, n =25), or ST-ChroME (magenta, n=11), via IUE. Right, photocurrent amplitudes elicited by CGH stimulation. Dashed line represents mean rheobase for 5 ms stimulation of L2/3 pyramidal neurons (ST-ChroME vs others: p<0.0008, Kruskal-Wallis test with multiple comparisons correction, all others comparisons p>0.13).
d) Top, duration of CGH stimulation needed to elicit action potentials in neurons expressing each opsin (n = 8 ST-C1V1, n = 5 ST-ChrimsonR, n=25 ST-Chronos, n= 8 ST-ChroME). Bottom, fraction of electroporated L2/3 neurons that could be driven at 1 Hz with best CGH stimulation.
e) Traces shown in (c) scaled to the peak current amplitude for each.
f) 10 overlaid traces from representative L2/3 neurons expressing ST opsins during 1 Hz CGH stimulation (red line indicates light pulses).
g) Spike latency for 1 Hz CGH stimulation of L2/3 neurons expressing ST-Opsins. ST-C1V1 T/T 25.9±4 ms (n=15), ST-ChrimsonR 18.8±3.8 ms (n=14), ST-Chronos 4.4±0.65 ms (n=23), ST-ChroME 3.48±0.49 ms (n = 12). ST-ChroME vs ST-C1V1 T/T, p=0, vs ST-ChrimsonR p=0.0028; vs ST-Chronos p = 0.95, by Kruskal-Wallis test with multiple comparisons correction.
h) Jitter for 1 Hz CGH stimulation of neurons expressing ST-Opsins. ST-C1V1 T/T 8.6±1.8 ms (n=11), ST-ChrimsonR 12±6.3 ms (n=14), ST-Chronos 1.2±0.36 ms (n=20), ST-ChroME 0.54±0.1 ms (n=10). ST-ChroME vs ST-C1V1 T/T, p=0.0011, vs ST-ChrimsonR p=0.048; vs ST-Chronos p = 0.7 by Kruskal-Wallis test with multiple comparisons correction.
i) Two-photon image of whole cell recording from L2/3 pyramidal neuron expressing ST-ChroME-mRuby2 (image representative of n=10 ST-ChroME-mRuby2 neurons).
j) Fidelity index in response to Poisson-like stimulation. ST-ChroME (n=7) vs ST-C1V1 T/T (n = 6), p = 0, vs ST-ChrimsonR (n=4), p= 0, vs ST-Chronos (n=9), p = 0.99, by Kruskal-Wallis test with multiple comparisons corrections.
k) Left, representative traces of two simultaneously recorded ST-ChroME expressing neurons stimulated with an identical Poisson train for 2.5 ms with a temporal offset of 3 ms. Top middle, example light-evoked spikes in the two neurons. Middle bottom, distribution of the difference in spike times from an example pair of neurons. Right, difference in mean spike times for n = 7 pairs.
l) Bar graph showing the fraction of neurons expressing ST-Chronos (green) or ST-ChroME (magenta) that could be optogenetically driven in vivo (p=0.0089, two-sided Fisher Exact Test). All data represent mean and s.e.m.
To achieve fast currents using 2P excitation, we adopted scanless holographic paradigms, CGH and 3D-SHOT, that can simultaneously illuminate the entire soma. We developed an experimental setup that combines a standard 2P imaging system with a custom photostimulation laser path. The photostimulation path features a high power (20W or 40W) 2 MHz 1040 nm laser with a spatial light modulator (SLM) placed in Fourier space to simultaneously target neurons in 3D. For targeting neural ensembles in vivo with high spatial resolution, the CGH path was replaced with 3D-SHOT27, a new form of 3D holography with temporal focusing that we recently developed and further improved (Supplementary Fig. 2, see Supplementary Table 1–2, and methods).
We first sought to identify the best opsin for precise temporal control of neural activity using scanless approaches. We rejected ChR2, as its 2P excitation peak is centered at 920 nm, and would be strongly activated by GCaMP imaging, resulting in undesirable optical cross-talk10. Instead we tested several red-shifted opsins: C1V113 T/T, ChrimsonR, and Chronos28 (Supplementary Fig. 3a). CGH stimulation (5 ms, 0.4 mW/μm2) of Chronos-expressing neurons elicited small photocurrents that were typically unable to generate action potentials (205±50 pA). To improve spatial resolution and photocurrent amplitude, we employed the Kv2.1 sequence tag to synthesize soma-targeted31,32 (ST-) variants of Chronos, C1V1 T/T, and ChrimsonR that increased photocurrents elicited by CGH stimulation (ST-Chronos: 460±60 pA, Supplementary Fig. 3b). Neurons expressing ST-opsins had photocurrent kinetics similar to published reports28 and normal intrinsic properties (Supplementary Fig. 3c–i).
Despite this improvement, photocurrents were insufficient to reliably spike pyramidal neurons (Fig. 1a). We therefore optimized the pulse parameters of the stimulation laser. We tested the effect of peak power, pulse energy, and average power on photocurrents by systematically varying the laser repetition rate (2 – 40 MHz) and pulse dispersion. We could saturate photocurrents across a range of powers, but high-peak powers (i.e. low rep-rates) saturated more efficiently (Supplementary Fig. 4a–d). Stimulation powers used in experiments did not damage cells33 (Supplementary Fig. 4e–f). For all subsequent experiments, we employed 250–300 fs laser pulses at a repetition rate of 2 MHz with power varying from 0.1–0.4 mW/μm2.
Even after optimization, average maximal photocurrents elicited by CGH stimulation (5 ms, 0.4 mW/um2) of pyramidal neurons expressing ST-opsins remained relatively weak (ST-C1V1 T/T: 380±80 pA, ST-ChrimsonR: 430±60 pA, ST-Chronos: 530±50 pA, Fig. 1c). A simple integrate and fire model of typical L2/3 neurons suggested that these photocurrents were unlikely to generate spikes (Supplementary Fig. 5a). Even using long light pulses and high light powers, only a minority of neurons expressing these opsins could be activated by CGH stimulation (Fig 1d, Supplementary Fig. 5).
The new opsin ChroME allows high fidelity replay of complex activity patterns
Since none of these opsins could reliably spike neurons in response to brief holographic stimulation, we engineered a stronger opsin with the goal of holographically stimulating large ensembles of neurons. We focused on mutating ST-Chronos, aiming to develop a variant that would preserve its fast kinetics but would generate sufficiently large photocurrents with brief light pulses. Guided by homology modeling to the crystal structure of C1C234 (Supplementary Fig. 6a), we mutated the pore region of ST-Chronos, identifying a neutral putative pore residue (M140) in Chronos that is negatively charged in other opsins (Supplementary Fig. 6b). We reasoned that mutating this methionine to a negatively charged residue might increase the flux of positive ions through the pore and therefore increase current amplitudes. We tested several mutations via 1P stimulation in Chinese hamster ovary (CHO) cells against a panel of ST-opsins and identified several mutants with larger photocurrent amplitudes than any other opsin that we tested (Supplementary Fig. 6c–d). One of these mutants, ST-Chronos-M140E, or ‘ChroME’, exhibited rapid decay kinetics while exhibiting photocurrents more than 10 times higher than ChR2 (Supplementary Fig. 6e). Neurons electroporated with ST-ChroME exhibited photocurrent amplitudes 3–5 times larger than ST-C1V1T/T or ST-Chronos in response to CGH stimulation (5ms 0.4 mW/μm2 evoked 1.8±0.2 nA, Fig. 1c). ST-ChroME retained ST-Chronos’s excitation spectrum and rapid rise time (Fig 1e, Supplementary Fig. 3c), but its decay time constant (3.0±0.4 ms) was slightly slower than ST-Chronos (1.7±0.6 ms, Supplementary Fig. 3d).
In contrast to other ST-opsins (and as predicted by modeling, Supplementary Fig 5a), 96% of ST-ChroME-expressing neurons were activated by CGH stimulation (Fig. 1d), requiring lower laser powers and shorter light pulses to evoke spikes than the other opsins (Supplementary Fig. 5b). This was true whether opsins were delivered via in utero electroporation (IUE) or by viral infection (Supplementary Fig. 6f–k).
We next examined the temporal precision of action potentials evoked from ST-ChroME-expressing neurons and the minority of neurons expressing other ST-opsins that could be activated. At 1 Hz, light-evoked spikes from neurons expressing ST-ChroME or ST-Chronos occurred with short latency and low jitter, whereas the timing of spikes from ST-C1V1T/T or ST-ChrimsonR-expressing neurons was more variable (Fig. 1f–h). To test temporal precision while eliciting naturalistic sequences of action potentials, we stimulated neurons with Poisson trains of holographic light pulses. Neurons expressing ST-ChroME and ST-Chronos followed these patterns with high fidelity, exhibiting high spike probability and low jitter across a wide range of stimulation frequencies throughout the stimulus train (fidelity index score: ST-ChroME: 0.87±0.03; ST-Chronos: 0.90±0.02, see methods). However, neurons expressing ST-ChrimsonR or ST-C1V1 T/T could not follow complex stimulus patterns (fidelity index score: ST-ChrimsonR: 0.48±0.05, ST-C1V1 T/T: 0.25±0.04, Fig. 1i–j, Supplementary Fig. 5c–g).
Since ST-ChroME allowed fast and reliable responses with brief stimulation, we reasoned that we could employ high speed SLMs to spike different sets of neurons at high rates. To test the speed at which we could generate spike patterns in two different neurons, we recorded two ChroME-expressing neurons and used a fast SLM (Supplementary Fig. 7, Supplementary Movie 1) to interleave holographic stimulation of each cell at the maximum SLM rate. We generated a Poisson train of light pulses on each trial and delivered the same sequence to both neurons separated by 3 ms. This experiment showed we could generate naturalistic spike trains in multiple neurons offset by brief periods (Fig. 1k).
To test if ST-ChroME drives reliable spiking under more relevant in vivo conditions, we performed 2P guided loose-patch in anesthetized animals. While only 31% of ST-Chronos-expressing could be made to spike with 5 ms CGH pulses, over 89% ST-ChroME-expressing neurons could be activated in vivo (Fig. 1l). Together, these data demonstrate that ST-ChroME can reliably generate the rapid, large photocurrents necessary to drive the temporally precise, short-latency spikes needed to replicate naturalistic neural activity patterns.
Anion opsins permit rapid and potent silencing of neural activity
We next asked if we could identify or engineer an optogenetic silencer to suppress neural activity with high efficacy and temporal precision. We synthesized and tested a suite of ST-inhibitory opsins with ER export motifs35 (‘e’) including pumps (eNpHR3 and eArch3)36,37 and anion channels (GtACR1, psuACR29,38, and iC++39). ST-eGtACR1 generated the largest outward photocurrents while retaining moderately fast kinetics (rise time 1.5 ± 0.7 ms, decay time 12.5 ± 0.7 ms, Fig. 2a–b, Supplementary Fig. 8a–b). GtACR1 photocurrents were near saturation in normal conditions and not improved by the ‘e’ signal (Supplementary Fig. 8c–g). Furthermore, ST-eGtACR1 was more sensitive to 1040 nm light than 930 nm (Supplementary Fig. 8h).
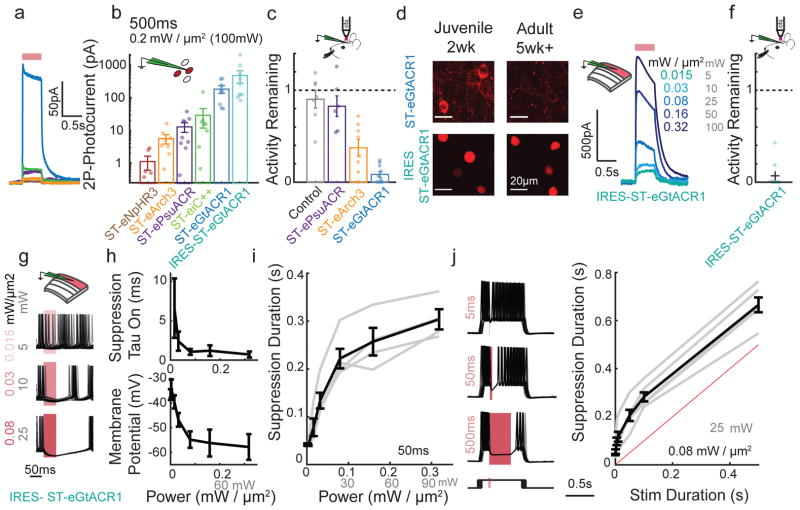
Figure 2. Fast and potent holographic suppression of neural activity.
a) Example average traces of whole cell photocurrents elicited by 500 ms (100 mW, 0.2 mW/μm2) CGH stimulation from CHO cells held at 0 mV expressing inhibitory ST-opsins color-coded as in (b).
b) Mean photocurrent elicited during a 500 ms stimulation, as in (a), plotted on a log scale. (n=5 cells expressing ST-eNpHR3, n=8 ST-eArch3, n=9 ST-ePsuACR, n=8 ST-eiC++, n=5 ST-eGtACR1, n=10 IRES-ST-eGtACR1)
c) In vivo firing activity that persists during optogenetic suppression of L2/3 neurons expressing ST-opsins. Each dot represents mean activity for a single neuron (5–60 sweeps per cell). n=7 no opsin control, n=6 ST-ePsuACR, n=8 ST-eArch3, n=10 ST-eGtACR1.
d) Example confocal images from juvenile (14–15 days old) and adult (35+ days old) mice expressing ST-eGtACR1-mRuby2 or H2b-mRuby3 IRES-ST-eGTACR1. Imaging conditions are matched within an opsin. Representative image from 3 mice each condition.
e) Example whole cell voltage clamp recording of a L2/3 neuron expressing IRES-ST-eGtACR1 held at 0 mV and stimulated for 500 ms with varying illumination powers.
f) In vivo activity that persists during optogenetic suppression with IRES-ST-eGtACR1 using 3D-SHOT (0.32mW/μm2, 100 mW), presented as in (c). (n=9 IRES-ST-eGtACR1 cells)
g) Overlay of 30 current clamp traces from a L2/3 pyramidal neuron expressing IRES-ST-eGtACR1 during current injection, aligned to the onset of a 50 ms stimulation at three different power levels.
h) Top, as in (g), the time for suppression to take effect calculated for a 50 ms light stimulation. Reported as the tau of a fit to the observed number of action potentials after light onset in 1ms bins, assuming a Poisson noise model (n=6 neurons).
Bottom, membrane potential of each neuron during the last 10 ms of a 50 ms stimulus as a function of stimulus intensity (n=6 neurons).
i) Duration of suppression, defined as the mean time until the next action potential as a function of stimulus intensity. Grey bars indicate individual replicates, black mean and s.e.m. (n=6 neurons).
j) Left, overlay of 30 whole cell current clamp traces during light stimulation of different durations. Bottom, schematic of current injection where onset of current injection was varied with respect to the light stimulus. Right, quantification of the duration of suppression as a function of stimulus duration. Grey bars indicate individual replicates, black mean (n=6 neurons). All data represent mean and s.e.m.
Since these silencers function through different biophysical mechanisms, it was possible that the opsin with the largest photocurrent might not be the most effective suppressor of endogenous neural activity. We therefore tested 2P holographic suppression in vivo by performing targeted loose patch recordings from cells expressing inhibitory opsins. Of the opsins that we tested, ST-eGtACR1 was the most efficient silencer, reducing activity to 8.4±3% of normal firing rate with 0.2 mW/μm2 of 2P stimulation. In contrast, at the same laser power, ST-eArch3 only reduced activity to 37±8 %, whereas ST-ePsuACR or light alone did not significantly alter firing rates (82±4% and 90±9 %, respectively, Fig. 2c, Supplementary Fig. 8i).
However, unlike all other opsins we expressed in vivo, we had difficulty identifying ST-eGtACR1-mRuby2 positive neurons. This seemed to be a problem only in adult animals but was partially mitigated by the ‘e’ signal (Fig 2d, Supplementary Fig. 9a). We suspected this might be related to aggregation of GtACR1 protein when highly expressed, possibility leading to degradation or toxicity. To address this problem, we generated a bicistronic construct with a nuclear localized fluorophore (H2B-mRuby3-IRES-ST-eGtACR1) that lowers expression levels of the opsin and spares the fluorophore from degradation. IRES-ST-eGtACR1 exhibited large photocurrents in CHO cells and neurons (460 ± 200 pA CHO cells, 920 ± 140 pA neurons, Fig. 2b,e). Antibody staining to a FLAG epitope on the GtACR1 protein confirmed that it remained soma-targeted (Supplementary Fig. 9b). Importantly, unlike ST-eGtACR1, IRES-ST-eGtACR1-expressing cells were easily identified into adulthood (Fig. 2d, Supplementary Fig. 9a). IRES-ST-eGtACR1-expressing neurons had normal intrinsic properties (Supplementary Fig. 3e–j) and spontaneous in vivo firing rates (Supplementary Fig. 9c) even in older mice. Targeted in vivo loose patch revealed that IRES-ST-eGtACR1-expressing neurons reduced their firing to 6.8 ± 5% of nominal rate in response to CGH stimulation (0.3 mW/um2), suggesting that lowering expression levels did not affect the efficacy of silencing (Fig 2f).
To measure the timing of suppression, we induced spiking in brain slices through current injection in cells electroporated with IRES-ST-eGtACR1. We varied the onset time of holographic suppression so that spike timing was randomized trial-to-trial, and we varied the stimulation intensity and duration in separate experiments (Fig. 2g–j). We found that onset of suppression was rapid, with spiking eliminated within 1.5±0.3 ms after light onset. Similar to photocurrents, the onset time of suppression was power-dependent (Fig. 2g–h, Supplementary Fig. 8e). Despite current injection, cells hyperpolarized to near the reversal potential of GtACR1 when stimulated with <0.1mW/μm2, indicating potent suppression (−54 ± 3 mV at 0.08 mW/μm2 stimulation, Fig. 2h). Although the onset of suppression was rapid, suppression of neural activity persisted for 50 – 250 ms after the cessation of photostimulation, due to the decay kinetics of the GtACR1 channel. This suppression was dependent on both the intensity and duration of the light stimulus (Fig. 2h–i). Together, these data validate IRES-ST-eGtACR1 as a new tool for stable, rapid suppression of neural activity using 2P optogenetics.
Creating and editing spatiotemporal sequences of neural activity in vivo
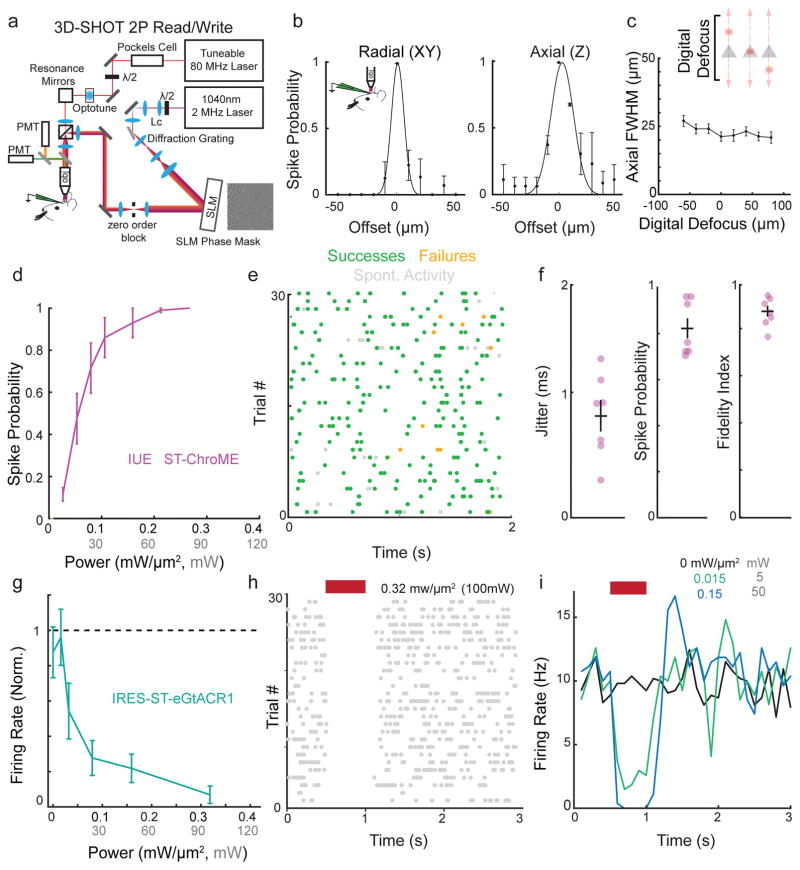
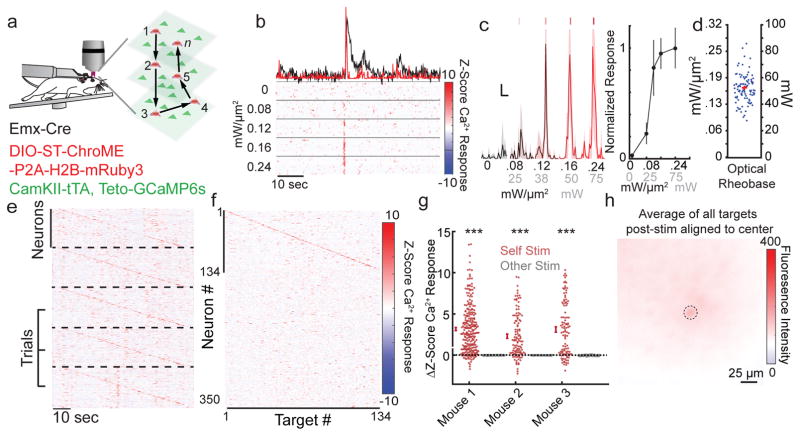
Next, we employed ST-ChroME and IRES-ST-eGtACR1 in the intact brain to create and edit spatiotemporal patterns of neural activity. For this, we employed 3D-SHOT27 (Fig. 3a, Supplementary Fig. 2,11) to enable 3D holographic stimulation with high axial resolution in vivo. To validate spatial resolution, we recorded the physiological point spread function (PPSF) using targeted loose patch recordings from ST-ChroME-expressing neurons in anesthetized mice at multiple focal planes (Fig. 3b–c, spot diameter = 20 μm; radial full-width half-max (FWHM): 11±3 μm, axial FWHM: 28±4 μm).
Figure 3. Creating and editing spatiotemporal neural activity in vivo.
a) Simplified schematic of light path allowing simultaneous 2P imaging and 3D-SHOT photostimulation.
b) Physiological point spread function (PPSF) of 3D-SHOT stimulation of neurons measured by in vivo loose patch. Left, spike probability for radial (XY) axis; right for axial (Z) axis (n=3 neurons).
c) In vivo recording of 3D-SHOT’s axial PPSF as a function of distance from the system’s zero order. PPSFs were measured as a function of depth by testing the spiking response to digital defocusing of the hologram while mechanically offsetting the objective varying distances from the focal plane (n=3 neurons).
d) Spike probability as a function of stimulation power in vivo for 1Hz stimulation in L2/3 pyramidal neurons expressing ST-ChroME-mRuby2 via IUE (n = 10 neurons).
e) Representative experiment showing in vivo Poisson stimulation of a L2/3 neuron expressing ST-ChroME.
f) Jitter, spike probability, and fidelity index score for Poisson stimulation of L2/3 neurons expressing ST-ChroME (n = 7 neurons).
g) Firing rate of neurons during stimulation normalized to pre-stimulation rate and measured through in vivo loose patch recordings from cells expressing IRES-ST-eGtACR1 (n=9 neurons).
h) Representative raster plot from a neuron suppressed with 500 ms stimulation.
i) Representative histogram of firing rate during IRES-ST-eGtACR1 suppression at several stimulation powers for the same neuron as in (h). Each line is the mean of 25+ stimulations binned at 100ms. All data indicate mean and s.e.m.
The majority of ST-ChroME-expressing neurons fired reliable, temporally precise action potentials in response to brief 3D-SHOT stimulation using powers less than 0.2 mW/μm2. This was true when electroporated with ST-ChroME-mRuby2 or virally transduced with AAV DIO-ST-ChroME-P2A-H2B-mRuby3 (Fig 3d, Supplementary Fig 12). We then stimulated with a naturalistic Poisson pattern, varying the pattern each trial to generate unique sequences of evoked activity (Fig. 3e). Quantifying these experiments revealed that ST-ChroME-expressing neurons reliably spiked with sub-millisecond jitter, allowing the production of spatiotemporal activity patterns with high fidelity (Fig 3f, Supplementary Fig 13a).
Conversely, to remove spikes from endogenous neural activity, we recorded from IRES-ST-eGtACR1-expressing neurons. 3D-SHOT stimulation at 0.32 mW/μm2 produced at least a 95% reduction in firing in >75% of IRES-ST-eGtACR1-expressing cells (Fig. 3g). The efficacy of holographic suppression increased with stimulation power, allowing us to either completely silence the activity of a neuron during a defined time window at high power, or titrate a neuron’s average firing rate with lower powers (Fig. 3h–I, Supplementary Fig 10a–b). Suppression appeared constant over the entire stimulation period, consistent with the observation that GtACR1-evoked photocurrents did not substantially desensitize (Fig. 3h–I). Suppression was repeatable over many trials without loss of efficacy or an apparent change in spontaneous firing rates of stimulated neurons (Fig. 3h Supplementary Fig 10c). This demonstration of single neuron suppression using 3D-SHOT represents the second element in a bidirectional toolbox to control spatiotemporal patterns of neural activity.
Holographic spatiotemporal control of cortical inhibitory neurons
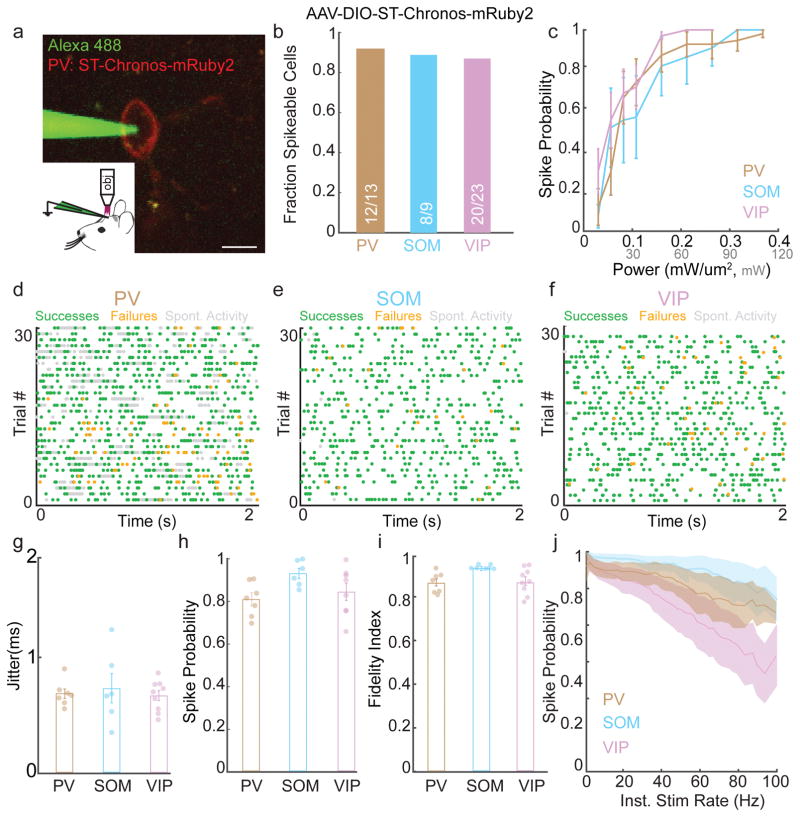
Whereas L2/3 neurons typically fire sparsely, cortical inhibitory neurons are heterogeneous and many fire at much higher frequencies40. We therefore combined spatial and genetic selectivity by stimulating specific subsets of GABAergic neurons (PV, SOM, or VIP)41,42 expressing Cre recombinase transgenically and infected with AAV-DIO-ST-Chronos-mRuby2. Inhibitory neurons are typically more excitable than pyramidal neurons, and ST-Chronos was sufficient to generate reliable action potentials in these cells24 (Fig 4a–b). We identified power levels needed to elicit reliable spiking at 1 Hz (<0.3 mW/μm2, Fig. 4c) and performed Poisson 3D-SHOT stimulation (Fig. 4d–f). Stimulation of each GABAergic cell type drove reliable, short latency spikes with sub-millisecond jitter across many stimulation frequencies, allowing these neurons to follow stimulus trains with high fidelity (Fig. 4g–j, Supplementary Fig. 13b–d). Unlike L2/3 pyramidal neurons and VIP neurons, PV and SOM cells were able to follow stimuli with instantaneous frequencies up to 100 Hz (Fig. 4j). Additionally, we replayed several unique patterns of action potentials with identical mean rates, demonstrating our ability to reliably generate precise activity patterns over many trials (Supplementary Fig. 13e–g).
Figure 4. Spatiotemporal activation of cortical inhibition.
a)Example image of in vivo loose patch recording of a PV-neuron expressing AAV-ST-Chronos-mRuby2 (scalebar = 10 μm).
b) Fraction of PV, SOM, and VIP neurons expressing AAV-DIO-ST-Chronos-mRuby2 that could be induced to fire action potentials in vivo using 3D-SHOT.
c) Line plots showing mean spike probability at 1 Hz as a function of stimulation power for inhibitory neurons expressing ST-Chronos (PV: n = 10, SOM: n = 7, VIP: n= 14 cells).
d–f) Representative experiments showing in vivo 3D-SHOT Poisson stimulation of PV (30 Hz), SOM (10 Hz), or VIP (10 Hz) neurons expressing ST-Chronos (green: light-evoked action potentials, orange: failures, gray: spontaneous activity).
g–i) Bar graphs indicating the jitter, spike probability, and fidelity index score for 3D-SHOT Poisson stimulation of PV, SOM, or VIP neurons expressing ST-Chronos (PV: n = 7, SOM: n = 7, VIP: n = 9 cells).
j) Spike probability as a function of instantaneous stimulation frequency for PV (brown), SOM (blue), or VIP (purple) neurons undergoing Poisson-stimulation in vivo (PV: n = 7, SOM: n = 7, VIP: n = 9 cells). All data represent mean and s.e.m.
Addressing multiple forms of optical crosstalk
To edit spatiotemporal activity patterns while simultaneously reading out network activity, we sought to combine our approach with 2P calcium imaging. To accomplish this, we addressed two forms of optical crosstalk that we encountered. First, the photostimulation laser can directly excite GCaMP6 adding severe artifacts to the imaging data. To overcome this problem, we synchronized the stimulation laser pulse gating with the resonant galvomirrors using a custom thresholded RC circuit (Supplementary Fig. 14a–d). This resulted in a stimulation duty cycle of ~16 kHz, providing stimulation on either side of each imaging line. Because this circuit is tunable, it provides a customizable tool to trade average stimulation power for effective field of view along the x-axis. Typically, we sacrifice ~50% of our stimulation power, resulting in light artifacts in <240 μm of imaging area along the x-axis (Supplementary Fig. 14d: imaging window normally: 550 × 550 μm; with gate synchronization: 310 × 550 μm free of stimulation artifact). However, due to the extremely fast duty cycle compared to the kinetics of the opsins, we observed that this loss of stimulation power results in only a 10–20% reduction in photocurrents when using the laser gate (Supplementary Fig. 14e–f). All reported estimates of illumination densities account for losses from use of this circuit.
Secondly, the imaging laser can excite opsins and thereby modulate neural activity independently of the stimulation laser. To characterize the effect of the imaging laser on photocurrents, we patched opsin-expressing neurons in brain slices and imaged at different powers, window sizes, and volumes (i.e. frame rate). During two photon imaging of GCaMP6, the imaging laser induced brief photocurrents as the laser contacted the cell. These currents decayed between frames and were substantially smaller than holographic currents (Supplementary Fig 15a,d). Imaging volumetrically reduced the effective frame rate, decreasing the imaging-induced photocurrents. When reducing the size of the imaging window, thus increasing the dwell time of the imaging laser on the opsin-expressing neuron, photocurrents increased (Supplementary Fig. 15a–f).
Together, these data indicate that photocurrents caused by the imaging laser under standard wide-field volumetric imaging conditions are unlikely to influence firing rates. Nevertheless, to directly test 2P imaging-induced crosstalk in vivo, we performed loose patch recordings from all four classes of cortical neurons. Pyramidal neurons were electroporated with ST-ChroME or IRES-ST-eGtACR1 or virally transduced with AAV DIO-ST-ChroME, while PV, SOM, and VIP neuron were virally transduced with AAV DIO-ST-Chronos. We did not observe detectable modulation of firing rates by the imaging laser when scanning with 50 mW at 30 Hz over a 400 × 400 μm window, at depths between 100 and 270 μm (Supplementary Figure 15g–h). However, ChroME-expressing neurons increased their firing rates when the size of the imaging window was below 400×400 μm (Supplementary Figure 15i). These data indicate that wide-field volumetric or video-rate 2P imaging is compatible with these optogenetic tools if care is taken to minimize cross-talk.
Next, to enable 3D all-optical read/write experiments (Fig. 5a), we created a custom suite of software to co-align 3D-SHOT and 3D calcium imaging (Supplementary Fig. 16). This alignment was facilitated by an improved version of 3D-SHOT that employs a rotating diffuser instead of a lens to shape the phase of the temporally focused disc. Using this approach, phase is randomly encoded spatially and temporally rather than shaped into a static spherical pattern. This increases available power through the objective and eliminates the secondary geometric focus27, further enhancing axial confinement (Supplementary Figure 11).
Figure 5. All-optical read/write with high spatiotemporal fidelity.
a) Schematic illustrating single-cell 3D all-optical read/write experiments performed in head-fixed mice freely running on a circular treadmill.
b) Example optical rheobase experiment (10 × 5ms pulses at 30 Hz) with varying light intensity using 3D-SHOT. Top, example dF/F calcium trace (black) or deconvolution (red). Below, raster plots of z-scored deconvolved calcium traces.
c) Left, average deconvolved calcium traces from an optical rheobase experiment (shaded area indicates 95% CI, scale bars represent 10% max response and 1 second). Right, the example neurons’ all-optical response function.
d) Power intensity needed to approach saturation of the all-optical response function (80% of maximum, n = 96 neurons, representative experiment from N = 3 mice).
e) Five consecutive trials of sequential stimulation of n=134 neurons from a representative experiment. Each panel corresponds to one trial (separated by dashed lines), and each line shows the trial-wise z-scored deconvolved calcium response for each neuron (see colorbar on f). Neurons were stimulated at 2 Hz with 10 × 5 ms pulses at 30 Hz.
f) Mean z-scored deconvolved dF/F for each neuron in response to 3D-SHOT stimulation of each holographically targeted neuron. A neuron’s response to its own stimulation is plotted on the diagonal. Data represent the mean z-scored deconvolved calcium response from 12 trials from a representative experiment (N=3 mice).
g) Each point represents the mean change in z-scored calcium response of a stimulated neuron upon stimulation (red) or the mean change in response to stimulation of other cells (gray). Mouse 1: n = 255 neurons, p=3.76×10−51, Mouse 2: n = 115 neurons, p<4.3×10−17 Mouse 3: n = 106 neurons, p<1.95×10−18, two-sided paired t-test).
h) Mean fluorescence of all stimulated neurons aligned so the targeted neuron is centered (two post-stimulation frames per neuron). Image is the mean response of 134 targets. Dashed black lines shows the size of the stimulation area, r = 10 μm. Data is from a representative experiment (N = 3 mice). All data represent mean and s.e.m. unless otherwise noted.
All-optical control of neural activity with high spatiotemporal fidelity
Using these technical advances, we tested our ability to perform all-optical read/write using 3D-SHOT stimulation to generate spikes with high fidelity, sub-millisecond temporal precision, and cellular resolution in full 3D. Experiments were performed in primary somatosensory cortex (S1) of awake, head-fixed mice on a treadmill. Mice expressed both GCaMP6s43 and ST-ChroME in excitatory neurons (See methods). To avoid failures or extra spikes, we determined the minimum laser power needed for each cell to reliably drive spiking with short pulses (Fig. 5b–c). The all-optical data matched in vivo physiology measurements, as neurons’ optical response function reached 80% of saturation with 0.16±0.02 mW/μm2 (Fig. 5d).
We then rapidly activated neurons located throughout a 550 μm × 550 μm × 100 μm volume (3 imaging planes spaced 50 μm apart using an electro-tunable lens44, as used throughout the study), well within the accessible volume of our stimulation (Supplementary Figure 11h–i). Neurons were stimulated one-by-one with a series of ten light pulses (5 ms, 30 Hz), and we read out the effects via GCaMP fluorescence. Generation of action potentials in this manner elicited large increases in GCaMP6s fluorescence. OASIS deconvolution of the calcium signal (see methods) revealed that the temporal sequence of activation was reliable across many trials and repeatable in multiple animals (Fig. 5e–g). On average, spatial resolution remained high even in awake in vivo conditions (Fig. 5h), but failures and off-target activation could occur during rare episodes of brain motion (Supplementary Fig 17a). Such motion was easily identified post-hoc and trials where motion coincided with photo-stimulation were excluded from analysis (Supplementary Fig. 17b). Holographic stimulation did not affect the animals’ running behavior (Supplementary Fig. 17c).
All-optical suppression of neurons
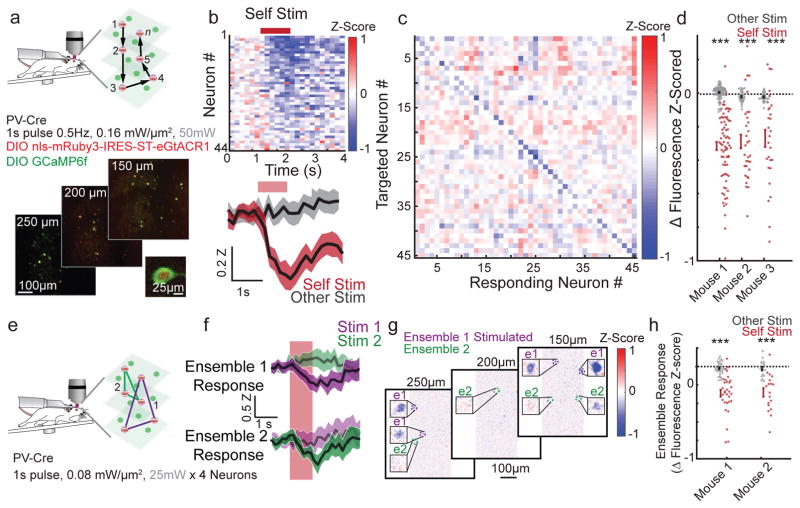
Next, we performed all-optical suppression of activity in awake mice. As L2/3 pyramidal neurons fire sparsely, we focused on PV-expressing interneurons, which have high tonic firing rates. To accomplish this, we created a Cre-dependent viral version of IRES-ST-eGtACR1 (AAV9 DIO-NLS-mRuby3-IRES-ST-eGtACR1), and confirmed its efficacy in vitro via whole cell recordings and in PV cells in vivo via cell attached recordings (Supplementary Fig 18a–c). We co-infected PV-Cre mice with viral DIO-IRES-ST-eGtACR1 and DIO-GCaMP6f. As in Figure 5 we imaged calcium activity while the animal was awake and head-fixed on a treadmill (Fig 6a). We sequentially suppressed individual PV cells (1s illumination, 0.16mW/μm2, Fig 6b, Supplementary Fig 18d–e); most (90.6%) cells exhibited reduced fluorescence when targeted but showed no consistent change when other neurons were targeted (Fig 6b–d). We observed no correlation between the direction or magnitude of a response and its distance from the targeted cell (Supplementary Fig 18f).
Figure 6. All optical suppression.
a) Schematic of experimental design as in (Fig 5). Mice expressed GCaMP6f and IRES-ST-eGtACR1 in PV interneurons via viral infection of PV-cre mice. Individual neurons are suppressed sequentially at 0.5 Hz, with 1 second of illumination. Bottom, representative image of 3 plane FOV (550 × 550 × 100 μm). Inset, enlargement of a PV-cell expressing both GtACR1 and GCaMP6f (Representative image of 16 recordings, 3 mice).
b) Trial averaged Z-scored fluorescence response of each targeted neuron during suppression of that neuron (Top, and Bottom, red) and during stimulation of a different neuron in the field of view (Bottom, gray). Bottom, mean and 95% CI (shaded), of all neurons from this recording (n=45, 24 trials each). Red bar indicates period of stimulation. Targeted cells that were obscured by the optical artifact were excluded from analysis.
c) Averaged fluorescence response of each targeted neuron to stimulation of each targeted neuron during the reporting window (0.5–1.5s after light onset). The trial immediately after self-targeting was ignored and blanked. Each box is the average of all 24 trials from this experiment.
d) Summary data from 3 mice (n=78, 32, and 28 cells stimulated and recorded each mouse). Each dot is the mean Z-Scored fluorescence of a cell not located in the optical artifact in response to self-targeting (red) or in response to all other stimulations (gray). Bars indicate mean and s.e.m. of the population response (Mouse 1: p<2.6×10−19, Mouse 2: p<2.3×10−7, Mouse 1: p<6.3×10−5, Paired two-sided T-Test, ‘***’ denotes p<0.001).
e) Schematic of experimental design as in (a) but depicting simultaneous suppression of multiple neurons in consecutive ensembles. 4 neurons per ensemble 0.5Hz 1s stimulation.
f) Representative responses of two ensembles (trial average z-score of 4 cells) response of ensemble 1 (top) and ensemble 2 (bottom) to suppression of ensemble 1 (purple), and ensemble 2 (green). Shaded region is the trial wise 95% CI.
g) Mean Z-score image of entire field of view during ensemble 1 stimulation with cells from ensemble 1 and ensemble 2 enlarged (insets). The area of the optical artifact was blanked. Mean of 24 Trials.
h) Summary data from 2 mice (n=31 and 18 ensembles of 4 cells, each mouse repeated twice in different areas). Each dot represents the mean Z-scored fluorescence of an ensemble of 4 cells in response to suppression of that ensemble (red) or all other ensembles (gray). Bars indicate mean and s.e.m. of the population response. Both mice showed increased suppression when recording from a targeted ensemble compared to when recording from a non-targeted ensemble (Mouse 1 p<2.0×10−5, Mouse 2 p<5.2×10−8, paired two-sided T-Test, ‘***’ denotes p<0.001).
Next, we suppressed groups of 4 randomly selected PV cells while simultaneously imaging (Fig 6e–h. 6–12 groups per experiment, 1s illumination 0.08 mW/μm2). Suppression of ensembles was also selective, as the laser caused suppression in only the cells targeted by the holographic pattern (Fig 6h). These data demonstrate all-optical suppression of neural activity in multiple neurons across a large working volume.
All-optical spatiotemporal control of neural ensembles
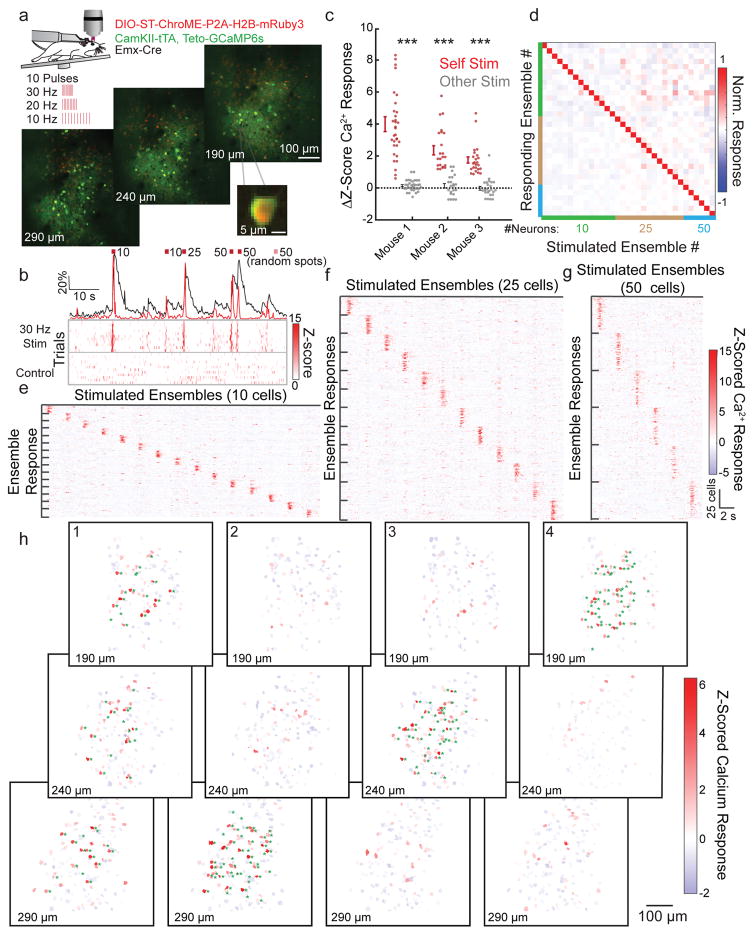
We next employed ST-ChroME to manipulate larger ensembles. When testing spatial resolution of ensemble stimulation in brain slices, we found that use of the ST-opsin, which increases stimulation resolution for one target31, was essential when stimulating groups of neurons with many holograms (Supplementary Fig. 19a–f), something not employed in previous manipulations of neural ensembles14,16,45. We tested multi-spot spatial resolution in vivo with cell attached recordings of ST-ChroME-expressing cells. The spiking PPSF was measured for each cell with holograms targeting 1–50 spots simultaneously throughout a large volume (400 × 400 × 200 μm). These experiments showed that 3D-SHOT stimulation in vivo remained spatially precise when targeting up to 50 locations simultaneously (Supplementary 19g–i).
To manipulate large groups of cells all-optically we prepared mice as in Figure 5, and selected 150 ST-ChroME-expressing neurons across three planes (Fig 7a). We randomly assigned them to unique neural ensembles containing overlapping sets of 10, 25, or 50 neurons and stimulated them with 10 pulses at 10, 20, or 30 Hz (Fig 7b, Supplementary Fig 20a, and Supplementary Movie 2). We did not stimulate more than 50 neurons simultaneously due to limitations in available laser power (4.1 Watts available from the objective resulting in approximately 0.13 mW/μm2, or 40 mW per target, accounting for losses from the imaging gate, but not for decreased diffraction efficiency of phase masks encoding 50 spots across the accessible volume). For control trials we either did not photostimulate the brain at all, or we directed the laser to 50 random spots (Supplementary Fig 20a). Neurons responded reliably to stimulation when targeted as a member of an ensemble, regardless of the identity of the other ensemble members. These neurons retained normal calcium dynamics when not being stimulated (Fig 7b, Supplementary Fig 20b). Stimulation of ensembles was selective and successful when targeting ensembles of different sizes, at different frequencies, and either within or across axial planes (Fig 7c–h, Supplementary Fig 20a–c).
Figure 7. Manipulating neural ensembles with high temporal and spatial precision.
a) Top, schematic of all-optical ensemble experiments as in (Fig 5). 33 ensembles of 10, 25, or 50 neurons are stimulated with 10 pulses at 10–30 Hz. Bottom, representative images of 3 plane FOV (550 × 550 × 100 μm), depth from pial surface noted. Inset, enlargement showing example calcium source expressing ST-ChroME.
b) A representative neuron stimulated as part of five different ensembles composed of varying numbers of cells. Top, normalized mean dF/F (black) or OASIS deconvolution (red) during stimulation. Bottom, raster plots showing z-scored deconvolved calcium activity from 10 stimulation trials (top) or control trials (bottom).
c) Summary data from experiments in three mice. Each point represents the mean change in z-scored calcium response of all ensemble members in response to stimulation of the target ensemble (red) or mean response to stimulation of other ensembles (gray). Ensembles significantly increased their fluorescence only when they were stimulated (*** indicates p<0.001. Mouse 1: n = 33 ensembles, p=3.5×10−10, Mouse 2: n = 22 ensembles, p=2.8×10−4, Mouse 3: n = 24 ensembles, p=5.5×10−4, paired two-sided t-test).
d) Normalized z-scored calcium response of the neurons that compose each stimulated ensemble upon stimulation of each ensemble. Color codes show the size of the ensembles (green: 10, brown: 25, blue: 50 neurons).
e–g) Responses of each neuron in each ensemble to each ensemble stimulation, grouped by ensemble identity and separated by size. Scalebars are shared between e–g. Data represent the mean z-scored deconvolved calcium response for each neuron.
h) Maps showing the mean response of all calcium sources to stimulation of four unique ensembles composed of 50 cells across 3 depths. Green asterisks indicate neurons that were targeted for stimulation. Note: ensembles can be distributed in 3D (ensemble 1) or confined to one depth (ensembles 2–4). Data calculated from 0–300 ms after stimulation.
During ensemble stimulation, we observed infrequent activation of neurons that were not holographically targeted. Cells located within the PPSF (0–11 μm distance from nearest target neuron) showed evidence of the expected facilitation from the stimulation laser (z-score 0.87±1.3 mean and s.d., Supplementary Figure 21a–c). However, just outside the PPSF, neurons were not modulated by the optogenetic stimulus (11.5–25 μm away from nearest target: z-score 0.07±0.86 mean and s.d). On average, neurons distal from the nearest target exhibited a small but significant suppression suggesting that optogenetic stimulation engaged cortical circuits for lateral inhibition46 (> 30 μm distal from the nearest target, z-score = −0.081±0.69 mean and s.d.). Similar effects were not observed in control trials (Supplementary Figure 21d). Non-target neurons exhibited high variability during both control and stimulation trials consistent with expected spontaneous activity in the awake, running mouse, whereas the variability of target neurons was strongly reduced by holographic stimulation (Supplementary Figure 21e–h).
Analysis of targeted neurons during interleaved control trials showed no evidence that repeated stimulation caused toxicity (Supplementary Figure 22a–c). To directly measure temperature changes induced by stimulation, we implanted a thermal probe into the cortex of anesthetized mice and repeated the stimulation protocol exactly as used above. This resulted in brain heating of 1.7–2.2° C over the course of an hour (Supplementary Figure 22d). We next addressed whether heat would constrain future all-optical experiments using full laser power at maximum rates. We found that one second of stimulation targeting 50 spots at full laser power (4.1W, 83% duty cycle) increased brain temperature by ~2° C (Supplementary Figure 22e). Continuous stimulation resulted in significant brain heating after one minute (6–8° C, Supplementary Figure 22f). These data define bounds for potentially acceptable levels of laser illumination.
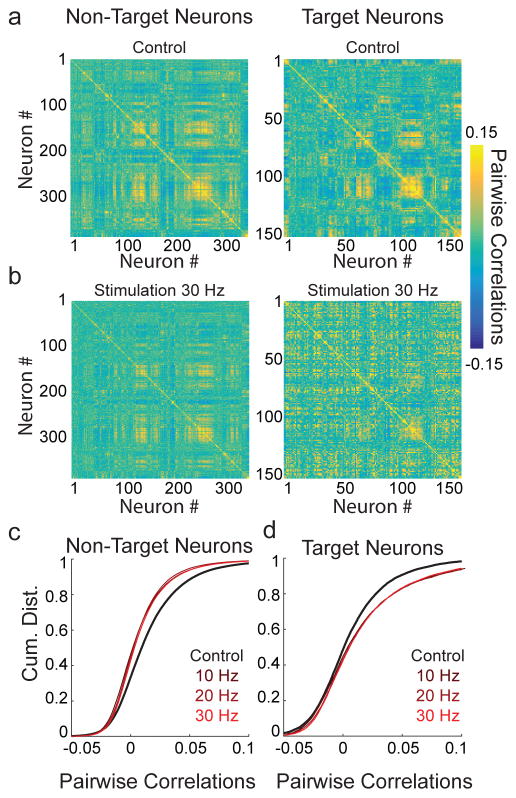
The ability to control the firing patterns of arbitrary neural ensembles raises the possibility of achieving optogenetic control over population activity47. Since sensory stimuli often decorrelate population activity48; we tested whether holographic ensemble stimulation could drive a change in population activity that mimicked a sensory stimulus. Towards this end, we computed the pairwise correlations during spontaneous activity of all neurons during control trials or during trials in which random ensembles were photostimulated. Ensemble photostimulation resulted in striking changes in the structure of population correlations (Fig 8a–b), and resulted in significant decorrelation of untargeted neurons during stimulation trials (Fig 8c). Conversely, targeted neurons exhibited the opposite effect, increasing their pairwise correlations on trials that they were stimulated (Fig 8d). These results show that high fidelity, temporally precise holographic activation of neural ensembles can provide the scale of experimental control need to directly manipulate previously inaccessible properties of neural networks such as correlational structure and shared variability with cellular resolution.
Figure 8. Altering population correlational structure with 2P ensemble stimulation.
a) Left, pairwise Pearson’s correlations for non-targeted neurons calculated based on firing during control trials (n = 365 neurons); right, pairwise correlations of target neurons during control trials (n = 150 neurons).
b) Left, pairwise correlations between non-target neurons or target neurons (right) during trials in which ensembles were stimulated at 30 Hz (10 × 5 ms pulses, see color bar on right).
c) Cumulative distributions of all pairwise correlations between non-target neurons during control trials (black) or during trials on which ensemble stimulation occurred at 10–30 Hz (red shaded). All stimulation conditions decorrelated population activity relative to control trials (p<0.01), but were not significantly different from each other (p>0.425, Friedman test with Tukey-Kramer correction for multiple comparisons). Cumulative distributions are from a representative experiment (Experiment performed in n = 3 mice. Stimulation vs. control trials, non-target cells: Mouse 1: p=0.007, Mouse 2: p=1.3×10−6, Mouse 3: p=0.004, Friedman test with Tukey-Kramer correction for multiple comparisons).
d) Cumulative distributions of all pairwise correlations between target neurons during control trials (black) or during trials in which ensembles were stimulated at 10–30 Hz (red shaded). All stimulation conditions increased correlations between target neurons relative to control trials (p<0.01), but were not significantly different from each other (p>0.186, Friedman test with Tukey-Kramer correction for multiple comparisons). Cumulative distributions are from a representative experiment (Experiment performed in n = 3 mice. Stimulation vs. control trials, target cells, Mouse 1: p=0.003, Mouse 2: p=5.4×10−3, Mouse 3: p=0.02, Friedman test with Tukey-Kramer correction for multiple comparisons).
Discussion
We developed an integrated experimental approach for multimodal control of neural ensemble activity with cellular resolution and millisecond precision in vivo. This new system achieves the simultaneous temporal precision, spatial resolution, reliability, and scale needed to generate or edit custom spatiotemporal activity patterns. It builds on previous 2P optogenetics manipulations of neural activity14–16, but offers the critical advances needed to achieve the faithful reproduction of naturalistic or artificial sequences of neural activity that could help parse temporal and spatial information in neural codes. The generation of ST-ChroME and IRES-ST-eGtACR1, the application and improvement of 3D-SHOT, and the integration and optimization of these systems with fast volumetric calcium imaging provides the increase in performance needed to address many fundamental, yet unanswered question in neuroscience.
Several in vivo mouse studies have previously employed 2P optogenetics with calcium imaging and/or electrophysiology to activate identified neurons13–16. Since these studies used C1V1T/T, a slow opsin exhibiting 2P photocurrents <500 pA12,13, they had comparatively poor control over the onset, timing, and, perhaps most importantly, the absolute number (and pattern) of evoked spikes over any given time window. Thus, spiral scanning of C1V1T/T-expressing neurons allows all-optical manipulation of neural ensembles (most recently in 3D45), but not the generation of spatiotemporally precise patterns of neural activity. However, recent work suggests that spiral scanning may be more power-efficient than scanless holography45, which should be considered when seeking to maximize the number of neurons simultaneously addressed if precise control of the underlying spike train is not required.
Although scanless 2P optogenetics holds the promise of eliciting short-latency, low jitter action potentials, it was initially limited by the strength of available opsins (photocurrents < 500 pA)19,49. However, two recent papers demonstrated sub-millisecond temporal resolution using scanless holographic optogenetics in brain slices24,25. One employed the ultra-fast opsin Chronos fused with GFP to demonstrate sub-millisecond control of firing at high rates in inhibitory interneurons24. This study reported average photocurrents of ~400 pA, agreeing with our observations that Chronos is not powerful enough to reliably elicit spiking in most L2/3 pyramidal neurons, which require larger photocurrents due to their low intrinsic excitability. Here, we extend these results by using ST-Chronos-mRuby2 to drive naturalistic spatiotemporal activity patterns in three genetically defined inhibitory neuron subtypes in vivo (Figure 4).
Another study presented soCoChR25, a combination of a novel soma-restriction sequence (‘so’ versus ‘ST’31,32) and CoChR, a previously characterized potent opsin with slow off kinetics28 (see Supplementary Fig 6). Direct comparison of the ‘so’ tag and the Kv2.1 tag in brain slices and in vivo are needed to address which tag is preferable. ST-ChroME exhibited larger 2P photocurrent amplitudes than those exhibited by soCoChR, suggesting that ChroME may be preferable for many applications.
Recently, multiple groups reported that opsins with slow kinetics can generate a temporally precise spike upon 2P stimulation25,45. We elicited spikes with sub-millisecond jitter in 1 of 11 ST-C1V1-expressing and 4 of 14 ST-ChrimsonR-expressing neurons, though the population average (8.6±2 and 12±5 ms respectively, Fig 1h) agreed with previous reports14. Our data indicate that opsins with slow decay constants are at an inherent disadvantage reproducing precise spike trains at physiological spike rates30. If more than one action potential per trial is required, faster opsins have a clear advantage. Almost every neuron expressing ST-ChroME exhibited sub-millisecond jitter, even in response to naturalistic Poisson stimulation in vivo.
We further optimized the extremely potent GtACR129 specifically for the purpose of multiphoton suppression, achieving fast, reliable, and potent silencing of neurons. Using IRES-ST-eGtACR1, we observe a significantly higher photocurrent than a previous report of 2P suppression13, which employed a different stimulation method and targeting sequences. Direct comparison of IRES-ST-eGtACR1 to ST-eArch3 yielded an 80-fold increase in photocurrent. Therefore, the approach presented here represents an important advance in optogenetic technology for the editing and synthesis of neural activity patterns that can be used to probe the fundamental logic of sensation, cognition, and behavior at the cellular scale.
Limiting optical cross talk between the read and write channels is a critical aspect of any all-optical approach. Our data show that we can minimize undesired activation of opsin-expressing neurons with the imaging laser. Since even red-shifted opsins absorb at the blue end of the two-photon spectrum, an alternative approach is to employ a blue shifted opsin and a red-shifted calcium indicator20. However, the lower efficacy of these red indicators compared to GCaMPs (at least at present), and the lack of high pulse energy lasers at 920 nm restricts the scale of this approach. Nevertheless, for applications seeking to minimize optical-cross talk, this color-flipped scheme may be preferable.
The utility of multiphoton optogenetics for biological applications depends on the number of neurons that can be photostimulated simultaneously and per unit time. The number of simultaneous targets is constrained by the available average power from the stimulation laser and the diffraction efficiency of the SLM phase mask. To estimate the maximum number of light-evoked spikes under various conditions, we built a model based on our empirical data and our hardware specifications. The model indicates that our system could produce thousands of light-evoked spikes in one second (Supplementary Fig. 23, and see methods), though detecting these responses using calcium imaging would be technically challenging. We note that laser-induced brain damage or heating will place an upper bound on the maximum duration and power of light that can be directed into brain tissue33, and may significantly constrain the maximum numbers of neurons that can be stimulated in practice.
This combination of temporally focused 3D holography with fast, potent actuators and suppressors and calcium imaging allows new experimental applications. This approach should enable experiments where specific statistical features of neural activity can be varied optogenetically to probe functional connectivity, perception, or behavior. For example, controlling the number, rate, and timing of action potentials written to specific ensembles will allow neuroscientists to test models of cortical dynamics by probing the boundary conditions for the initiation of ‘winner take all’ ensemble activity. Perhaps most intriguingly, our approach provides the tight stimulus control needed to rigorously test how parameterized manipulation of specific neurons can create or alter behavior. Various reports suggest that animals can engage behavioral responses based on the activity of a number of neurons compatible with our approach50. By recording neural activity and simultaneously writing or suppressing custom spatiotemporal sequences of neural activity, investigators can use this new technology to probe how specific, unique patterns of neural activity influence neural circuits and behavior.
Accession Codes (addgene)
| pCAG-ChroME-mRuby2-ST | 108902 |
| pAAV-CAG-DIO-ChroME-P2A-H2B-mRuby3 | 108912 |
| pCAG-H2B-mRuby3-IRES-eGtACR1-ST | 108960 |
| pAAV-CAG-DIO-NLS-H2B-mRuby3-IRES-eGtACR1-ST | 109048 |
Data Availability Statement
The datasets generated and analyzed in this study are available from the corresponding author on reasonable request. Additionally, the sequences for constructs created in this study will be made publicly available on Addgene.
Online Methods
All experiments were performed in accordance with the guidelines and regulations of the ACUC of the University of California, Berkeley. Protocol # AUP-2014010-6832. Every experiment was conducted in at least three mice unless otherwise stated.
Transgenic mice
The following mouse lines were used for this study: ICR (CRL:CD1), the PV-IRES-cre line (B6;129P2-Pvalbtm1(cre)Arbr/J ; JAX stock #008069), the SOM-IRES-cre line (JAX stock 013044), the VIP-IRES-cre line (JAX stock 010908), the Emx-IRES-cre line (JAX stock 005628), the Drd3-Cre line (JAX stock 002958) the teto-GCaMP6s line (JAX stock 024742), the CaMKII-tTA line (JAX stock 003010). Mice were housed in cohorts of five or fewer with a light:dark cycle of 12:12 hours, and were used for experimentation during their subjective night.
Plasmid construction and Mutagenesis
Chronos, eArch3.0 and iC++ were generated by gene synthesis (Chronos and eArch3.0 - Genewiz, South Plainfield, NJ and iC++ - Integrated DNA Technologies, Coralville, IA) and anion opsins PsuACR and GtACR1 were provided by Dr. John Spudich, (University of Texas Health Science Center, Houston, TX). C1V1T/T, ChR2, ChrimsonR, and eNpHR3.0 were obtained from Addgene. All opsins were fused to mRuby2 at their C-terminus either at a NotI site (Chronos and eArch3.0) or AgeI site (GtACR1, iC++, PsuACR, eNpHR3.0) and sub-cloned into the pCAGGS expression vector between KpnI and XhoI restriction sites by In-Fusion cloning (Clontech, Mountain View, CA). In order to target the opsins to the soma and proximal dendrites of neurons, the sequence encoding the proximal restriction and clustering domain of the Kv2.1 voltage-gated potassium channel consisting of amino acids 536–60031,32,51 (soma-targeting; ST) was codon optimized, synthesized (Integrated DNA Technologies, Coralville, IA) and inserted at the C-terminus of mRuby2 between BsrGI and XhoI restriction sites by In-Fusion cloning. To further enhance membrane trafficking, the endoplasmic reticulum (ER) export signal from the vertebrate inward rectifier potassium channel Kir and a neurite trafficking signal were appended to iC++ and the ER-export signal to GtACR1 and PsuACR. For the generation of a bicistronic pCAGGS vector encoding mRuby3 fused to histone H2B to promote its nuclear localization followed by GtACR1 under the control of an internal ribosome entry sequence (IRES), GtACR1 fused to the soma-targeting domain was subcloned downstream of an IRES. For GtACR1 AAV preparations, the H2B was replaced with a nuclear localization sequence (NLS) due to space constraints. Mutations in Chronos were introduced by overlap extension PCR and verified by DNA sequencing.
2P Holographic Microscope Setup
Our experimental setup (See Supplementary Figure 2) is a Movable Objective Microscope (MOM, Sutter instruments), modified with commercially available parts (listed in Supplementary Table 2). The microscope objective is mounted on a 3D mechanical stage (MP-285 controller, Sutter) enabling controlled mechanical displacement of the objective in 3D (We represent this mechanical displacement by the “true coordinates:” (x, y, z), expressed in microns). A polarizing beam-splitter (BS) merges the photostimulation and imaging laser paths with minimal losses.
Volumetric Photostimulation with two-photon holography
For photostimulation, we relied on femtosecond lasers (Coherent Monaco: 1040nm, 2MHz, 40W, or Amplitude Satsuma 1040 nm, 2MHz 20W on a separate identical setup). For conventional 3D Computer Generated Holography (CGH), the beam is expanded into a wider collimated Gaussian beam with a pair of lenses, L10, and L7, placed in a 4-f configuration, and illuminates the Spatial Light modulator (SLM). A telescope lens, L6, transforms the phase patterning in the pupil plane into a hologram with an accessible volume centered on the image plane. In the absence of a phase patterning, the SLM focuses all the incoming light into the 0 order, which is absorbed by a reverse pinhole filter. During operation, the 3D volume image is replicated by a set of relay lenses, L4, and L5, and aligned to maximally overlap with the accessible volume for two-photon imaging. All lenses in the optical path are separated by two focal distances on either side, so that the image plane (dashed green) and pupil planes (dashed red) alternate between lenses from the initial collimated laser beam to the focal plane under the microscope objective. A half wave plate adjusts the polarization of the laser to match the orientation of the liquid crystal on the SLM.
3D-SHOT implementation with a rotating diffuser for partially coherent 3D holography with temporal focusing
After alignment of the photostimulation path for conventional CGH is complete, we place two mirrors mounted on a pair of sliding stages to divert the beam on a separate path on either side of lens L10. The beam is diverted towards a blazed holographic diffraction grating. The incidence angle of the illumination and the orientation of the grating are adjusted so that the first diffracted order is reflected orthogonally to the grating surface and back into the optical path along the same optical axis. The outgoing beam is then demagnified five times with a pair of lenses L9, and L8. In the focal plane of the demagnified image (dashed green line), spectrally separated components interfere constructively after propagating along separate paths and reconstruct a custom temporally focused pattern (CTFP) matched to the dimensions of a neuron soma (here a disc of radius 10 μm). Let Δλ be the spectral bandwidth of the femtosecond laser, and 1/a the spatial frequency of the grating. The spectral separation d (on the SLM, Supplementary Figure 2b), is given by:
The fundamental principle of 3D-SHOT is to replicate identical copies of the CTFP anywhere in 3D with the SLM. Temporal focusing and 3D holography are made compatible by engineering the phase of the CTFP to simultaneously broaden the spatial footprint of all spectral components, and to ensure high diffraction efficiency by the SLM. Static phase masks introduce geometric secondary foci above or below the temporally focused plane27, so we rely on a rotating diffuser placed in the first virtual image plane to apply a rapidly varying randomized phase perturbation to CTFP. The characteristic angle of the Gaussian diffuser (θ =0.5°, 47–989, Edmund Optics) is chosen to simultaneously broaden the spatial footprint of all spectral components. The diffusion length, r, given by:
matches the dimensions of the SLM active area. The diffuser is glued on a brushless motor (from a modified 30 mm diameter computer hardware ventilation fan), and continuously rotates during operation.
Hologram Computation and Power Balancing
Variability in opsin expression as well as spatially dependent diffraction efficiency requires the ability to control the power distribution precisely in each target. To compensate for spatially dependent diffraction efficiency throughout the optical system, we proceed to a power calibration, as shown in Supplementary Figure 16g,h,i.
We computed holograms, each targeting one spot in a 3D grid pattern, and we used our substage camera system (Supplementary Figure 11a) to quantify the amount of two-photon absorption, I2, achieved by each hologram while supplying the same amount of laser power, I0, to the SLM. Experimental measurements of two photon absorption I2(x′i, y′i, z′i) (Supplementary Figure 16g), show how intensity degrades when targeting far away from the zero-order. To digitally compensate for this unwanted effect, we estimate losses with a 3D polynomial interpolation of the power calibration data (Supplementary Figure 16h). Interpolation error measurements (Supplementary Figure 16i) shows how our model fits experimental measurements within the operating volume, with the known exception of the blocked zero-order. Several methods have been developed for 3D hologram computation. Here, for precise control of the intensity distribution, we used either an iterative method: Global Gerchberg Saxton52, or two-photon optimized NOVO-CGH53 with a Euclidean cost function to maximize contrast in holograms where precise control of the power distribution is required, simultaneously in many targets. Relevant algorithms for 3D-alignment of 3D-SHOT with 3D imaging, and for digital power compensation with 3D polynomial interpolation are provided on our repository (https://github.com/adesniklab/3D-SHOT)
Synchronization of Stimulation Window to Imaging Frames (e.g. Laser Gate)
Since two-photon excitation by the photostimulation laser yields stimulation artifacts, we developed an electronic circuit to restrict the photostimulation laser to engage only as the resonant galvomirror reverses course, on either side of the imaging window. The electronic circuit is shown in Supplementary Fig 14a, with additional details on our repository (https://github.com/adesniklab/3D-SHOT).
Stroboscopic Imaging for SLM high speed performance testing
To characterize the illumination pattern during high speed phase transitions (using the Meadowlark 512L), we considered a test case with four holograms, each targeting several randomized points distributed throughout the volume. We employed stroboscopic imaging to measure the intensity during the phase transition at very high speeds by illuminating a fluorescent calibration slide at specific times during the cycle and with time-averaged imaging with a substage camera (See Supplementary Table 2). A repeating sequence was played at 300 Hz using the SLM built-in trigger. To observe holograms at any point of this cycle during SLM frame-to-frame transitions, the SLM trigger clock was synchronized with the laser controller to restrict illumination to a 1 ms pulse placed anywhere within the 13.3 ms duration of the cyclic sequence.
CHO cell recording
Chinese hamster ovary (CHO) cells were cultured and recorded as described27. One-photon photostimulation of cells was performed at 550nm for C1V1T/T, eNpHR3.0, eArch3.0 and PsuACR, 470nm for Chronos, ChroME, iC++ and GtACR1 and 630nm for ChrimsonR at a power of 0.55 mW using a Lumencor light engine (Lumencor). For activating opsins, currents were measured at a holding potential of −40 mV; for suppressing opsins, currents were measured at 0 mV. For two-photon spectra measurements in CHO cells, currents were evoked by rapidly raster scanning a diffraction limited spot from a Chameleon Ti-Saphirre Laser (Coherent) and average power was normalized across wavelengths by attenuating the laser beam using a pockels cell (Conoptics).
In Utero Electroporations
Electroporations were performed on pregnant CD1 (ICR) mice (E15, Charles River ca. SC:022). For each surgery, the mouse was initially anesthetized with 5% isoflurane and maintained with 2.5% isoflurane. The surgery was conducted on a heating pad, and warm sterile phosphate-buffered saline (PBS) was intermittently perfused over the pups throughout the procedure. A micropipette was used to inject ~2 μl of recombinant DNA at a concentration of 2 μg/μl and into the left ventricle of each neonate’s brain (typically DNA encoding opsins were doped with plasmids expressing GFP or mRuby3 at a concentration of 1:20 to facilitate screening for expression). Fast-green (Sigma-Aldrich) was used to visualize a successful injection. Following successful injection, platinum-plated 5mm Tweezertrodes (BTX Harvard Apparatus ca. 45–0489) were positioned along the frontal axis across the head of the neonate with the positive electrode of the tweezers positioned against the left side of the head. An Electro Square Porator (BTX Harvard Apparatus ca. 45-0052) was used to administer a train of 5 × 40 mV pulses with a 1s delay. After the procedure, the mouse was allowed to recover and come to term, and the delivered pups were allowed to develop normally.
Brain slice recording
Acute coronal slices were prepared and recorded from mice (ages p14–29) as described27. For measuring opsin kinetics, the photocurrent elicited by 0.5 or 1 second CGH stimulation (0.04 mW/μm2, 20 mW, disc r=12.5 μm) was measured in voltage clamp. The time to peak current was measured from average currents, and decay time constants were measured by fitting the traces from stimuli offset to 80% of baseline to a single exponential. In some neurons expressing ST-Chronos and ST-ChroME, the decay kinetics were better fit with a two-term exponential decay function of form I = aebt + cedt. The size of the primary decay tau (b) was unaffected by the two-term fit. This secondary decay Tau (d) averaged ~50 ms for Chronos and ~200 ms for ChroME, and the scale constant (c) for the secondary tau was maximally 0.3.
For current injection experiments in figure 1 and Supplementary figure 1, random white noise (mean 0 pA, range ± 60 pA) was generated on each sweep, and rheobase was determined by increasing current injections in a stepwise fashion (25 pA/step) until action potentials were recorded. This procedure was repeated for each stimulus duration tested. In supplementary figure 1, current injections were performed at the rheobase 5 ms or 1 second. White noise stimulus without additional current injection never resulted in action potentials.
For optogenetics experiments, mice were screened by a handheld 300 mW 594 nm laser and filtered goggles for expression after decapitation and before slicing. After slicing, recordings were made from the slices with strongest expression from the densest area as judged by handheld laser and filter googles. To be included in subsequent datasets neurons were required to pass an expression test: experiments were continued if the recorded neuron spiked in response to a brief one-photon stimulus (5 ms, 0.5 mW 490 nm Chronos and ChroME, and 5 ms 0.5 mW 510 nm for C1V1 T/T and ChrimsonR, for voltage clamp experiments this 1P test was performed in cell-attached mode before break-in). As some suppressing opsins had much smaller currents, cells were only excluded if no visible current was detected with a 250ms 10mW 490nm 1 photon pulse.
To determine a neuron’s optical rheobase, stimulus duration and power levels were increased in a step-wise fashion while recording in cell-attached or current-clamp configuration. We first increased laser power for a 5 ms stimulus by steps of ~25 mW average power to 0.4 mW/μm2 (200 mW, CGH disc radius = 12.5 μm) and then the stimulus duration was increased by 5 ms steps to 25 ms. If neurons still did not spike, power was increased in a step-wise fashion to a maximum of 0.8 mW/μm2 (400 mW, CGH disc radius = 12.5 μm). We defined a neuron’s optical rheobase to be the first observed stimulus combination (time and power) that elicited 100% spike probability while stimulating at 1–5 Hz. CGH stimulation at 1Hz to measure latency and jitter was performed at the optical rheobase. Neurons that did not spike using laser power 0.8 mW/μm2 (400 mW, disc r=12.5 μm) at stimulus durations < 30 ms were considered to not be spikeable using 2P holography. In practice, neurons that were not activated with 0.4 mW/μm2/10 ms pulses were rarely activated at higher powers.
Latency to spike was quantified as the time from the initiation of holographic stimulation until the membrane potential crossed 0 mV. Jitter is defined as the standard deviation for the spike latencies corresponding to a particular stimulus. For Poisson stimulation in pyramidal neurons, stimulation was conducted with a mean frequency of 5 Hz, and was performed at power levels approximately 25% higher than the optical rheobase. For Poisson stimulation, jitter was calculated as the standard deviation of all spike latencies across all instantaneous rates. To calculate the fidelity index, for each sweep, the pulsatile time indices of stimulation times and the spike times were each convolved with a Gaussian kernel with σ = 10 ms, and the fidelity index for that sweep was defined as the max normalized cross-correlation at lag 0–10 ms. Fidelity index reported for each cell is the mean score computed for each sweep.
For dual patch experiments using the fast SLM, pyramidal neurons expressing ST-ChroME virally (neonatally injected into Emx-Cre mice) were stimulated with a mean rate of 10 Hz. On each trial a Poisson train was generated for cell 1, and replicated with an offset of 3 ms with a flip of the SLM in between (exposure time: 2.5 ms with 0.5 ms to allow the SLM to flip frames). The time between spikes was calculated as the difference in spike times of cells A and B given a spike in cell A within the preceding 10 ms. Cells were excluded if a 2.5 ms stimulation time was unable to generate action potentials. To determine the onset of time of holographic suppression from current clamp recordings, the time of each action potential was binned into 1 ms bins. The observed bin counts were fit to an exponential decay from the mean firing rate to 0 spikes per bin starting at the time of light onset and assuming a Poisson noise distribution. The duration of suppression was defined as the mean time from suppression onset to the next detected spike after suppression ceased. For 2P imaging cross-talk experiments, neurons were placed in the center of the field of view in the focal plane of the natural 2P focus for volumetric imaging. 2P imaging was performed at the specified window size, speed, and power for one second sweeps after a one second baseline period.
Shaping 2P Stimulation Pulse and Survival Curve
For laser pulse shaping experiments, whole cell recordings were obtained and pulse features of CGH stimulation (radius = 12.5 μm) were varied on-line while holding the cell. The Satsuma HP 1040 nm stimulation laser is switchable between 2 and 40 MHz, allowing on-line testing of the response to multiple repetition rates. Peak power was controlled using the EOM to vary average power and the relationship between peak power and pulse energy was probed using the Satsuma HP laser’s onboard dispersion compensation that allowed us to control pulse-width online to test pulses of identical pulse energy with variable peak power. The stage positions necessary to chirp the laser pulses were determined before the experiment began using a Mini TPA Compact, Tuning-free Autocorrelator (Angewandte Physik & Elektronik GmbH).
To establish the maximum safe power levels useable before acute cellular damage, GCaMP6 positive cells (without opsin) were stimulated with increasing power densities until calcium responses (indicating cell death) or cavitation of the tissue was observed. We do not know the specific mechanisms of cellular damage, but only scored damage when a laser pulse resulted in an acute change in cellular fluorescence. Teto-GCaMP mice were headplated and windowed as if for in vivo imaging (see below), deeply anesthetized as in cell attached recordings (see below). Up to 800mW per target (3D-SHOT disc radius r=10μm) were used.
In Vivo Patch
Two-photon guided patch recordings were performed from adult mice (35 days old or older) as described27. For suppression experiments, whisker stimulation was achieved with an air puffer (PicoSpritzer II, General Value) directed towards the contralateral whisker pad. Six 50 ms puffs were applied before during and after each optogenetic stimulation, in general L2/3 neurons did not respond in a time locked manner but increased their overall firing rates during stimulation. Glass pipettes (2–5MΩ resistance) were filled with HEPES ACSF and Alexa 488 dye (100μM) for visualization. Cells were identified by the presence of mRuby2 or mRuby3 fluorescence imaged at 50–100 mW at 930–1000 nm. Data were acquired using a Multiclamp 700B Amplifier (Axon Instruments) and digitized at 20 kHz (National Instruments). Data was digitally band pass filtered 0.5–2.2 kHz for identification of spikes. All data were acquired using custom written MATLAB (Mathworks) software.
Cells were included for analysis if they were spontaneously active and those spikes were sufficiently larger than the noise (>5 Standard deviations of the noise). Furthermore, cells had to respond to a one photon LED stimulation (Sola SE, Lumencor), fire action potential(s) if they expressed an excitatory opsin or temporarily cease firing if they expressed a suppressing opsin (5 ms, 0.5 mW 490 nm Chronos and ChroME, 250 ms 10 mW 490 nm for GtACR).
To determine fraction spikeable neurons, cells that exhibited spontaneous activity and passed the 1P test were stimulated repetitively (1–5 Hz) with 5 ms light pulses of increasing laser power until they spiked to each laser pulse. Neurons that did not spike reliably with stimuli of 5 ms, 0.32 mW/μm2 (100 mW) 3D-SHOT disc r =10 μm were considered not spikeable. In practice, neurons that responded to holographic stimulation were activated with much less than maximum power. These laser powers and conditions were used for Poisson stimulus trains and tests of spatial resolution. Poisson stimuli were analyzed as in brain slices. Neurons were typically recorded at a depth of 75–250 μm below the pia surface. For 2P imaging cross-talk experiments, cells were centered in the field of view in the focal plane and 2P imaging was conducted at the specified window size, speed, and laser power for five seconds after five seconds of non-imaging. All experiments used 512×512 pixels for the imaging window.
In Vivo Temperature Measurements
To measure brain temperature during holographic photostimulation and imaging, animals were prepared and deeply anesthetized as above for in vivo cell attached patch, except they were additionally administered 2 mg/kg of dexamethasone as an anti-inflammatory agent. Once the craniotomy was completed, the dura was removed, and an 800 μm thick thermocouple coated in DiI was slowly lowered at 45° degree angle using a micromanipulator (Sutter MP285) until it was 300–500 μm beneath the pial surface. It was then secured in place with orthojet while the open craniotomy was protected by gelfoam and ACSF. The thermocouple was attached to a TC-324C temperature controller (Warner Instruments). The temperature to voltage conversion was calibrated in a series of water baths. The thermocouple was located under 2P illumination based on DiI signal, and the objective was placed over the thermocouple for the duration of stimulation. A single phase mask that targeted 50 spots was held static for the duration of the experiment, and duty cycling of the stimulation laser was performed using the EOM.
Histology
Mice were deeply anesthetized with ketamine/xylazine and transcardially perfused with phosphate-buffered saline (PBS) and 4% paraformaldehyde. Brains were post fixed for at least 2 h. Brains were embedded in 30% sucrose solution overnight, then frozen and 40 μm sections made on a microtome (American Optical Society). If the sections required immunohistochemistry, they were blocked for 1h at 4°C in blocking solution (0.6% Triton X-100, 0.2% Tween-20, 3% normal goat serum, and 3% BSA in PBS, all from fisher scientific), and then incubated in α-FLAG antibody at 1:1000 dilution (Sigma clone M2). Then sections were washed with PBS and 0.25% Triton X-100, before being incubated in secondary antibody Alexa Fluor 488 Mouse anti-mouse (1:1000). All sections were mounted on slides and sealed with Vectashield with DAPI (Vector Laboratories). Confocal images were acquired using an Olympus Fluoview system (Fv1000 Olympus Microscope) running the Fluoview software (Olympus), with 488 and 543nm lasers.
Viral Infection
For viral injection animals where anesthetized using 2% isoflurane on a heating pad and head fixed in a stereotactic apparatus (Kopf). After sterilizing the incision site, the skin was opened, and a small burr hole was drilled over S1 using a 0.24 mm drill bit (Busch) (3.5 mm lateral, 1.4 mm posterior to bregma). 200–600 nL of virus was injected using a micro syringe pump (Micro4) and a wiretrol II glass pipette (Drummond) at a rate of 25–50 nL/second at a depth of 150–300 μm below the pia surface. After the injection was complete, we waited five minutes before retracting the needle and closing the scalp using sutures. On some surgeries, the virus was injected directly after installation of a headplate (see below). For opsin injections, mice were used 2–6 weeks post injection. For GCaMP6 injections, mice were used for experimentation 1–2 weeks post injection. Viruses used were: AAV-syn-DIO-ST-Chronos-mRuby2 (UC Berkeley Vector core, titer: 4.8 ×1014), AAV-CAG-DIO-ST-ChroME-P2A-H2B-mRuby3 (Custom Order Penn Vector core, titer: 3.76×1012 – 7.53 ×1012), AAV-CAG-DIO-IRES-eGtACR1-NLS-mRuby3 (Custom Order Penn Vector Core, titer: 6.82e1014), AAV-syn-DIO-GCaMP6f (Penn Vector Core titer: 6.56×1012), AAV-Syn-Cre (titer: 1.4 ×1012). For some whole cell slice experiments using virus to test ST-Chronos, ST-ChroME, or IRES-ST-eGtACR1, viruses were introduced via neonatal injection as described27,54 into P2–3 littermate Emx-Cre or Drd3-Cre animals (p3, 3 injection site, 2 injections per site at 100 and 250 μm below the pial surface, 18.5 nL/injection).
In vivo two photon imaging and photostimulation
Three different combinations of opsin and GCaMP6s were used in this study for all-optical experiments. First, mice expressing GCaMP6s in excitatory neurons using CamKII-tTA crossed to tetO-GCaMP6s43 were co-injected with AAV-syn-Cre and AAV-CAG-DIO-ST-ChroME-P2A-H2B-mRuby3. Alternatively, triple transgenic animals expressing CamKII-tTA, teto-GCaMP6s, and Em×1-Cre were injected with AAV-CAG-DIO-ST-ChroME-P2A-H2B-mRuby3. For all-optical suppression experiments, mice expressing PV-Cre were co-injected with AAV DIO-Syn-GCaMP6f and AAV-CAG-DIO-IRES-eGtACR1-NLS-mRuby3. Mice were fitted with a custom stainless steel headplate described54. Mice were anesthetized with isoflurane (2%), and administered 2 mg/kg of dexamethasone as an anti-inflammatory and 0.05 mg/kg bupromorphine as an analgesic. The scalp was removed, the fascia retracted, and the skull lightly etched. Following application of Vetbond (3M) to the skull surface, a custom stainless steel headplate was fixed to the skull with two dental cements: Metabond (C&B) followed by Orthojet (Lang). After the dental cement dried, a 3 mm diameter craniotomy over the left primary somatosensory cortex was drilled, and residual bleeding stopped with repeated wet-dry cycles using sterile artificial cerebral spinal fluid, gauze, and Gelfoam (Pfizer). A window plug consisting of two 3mm diameter coverslips glued to the bottom of a single 5mm diameter coverslip (using Norland Optial Adhesive #71) was placed over the craniotomy and sealed permanently using Orthojet (Lang). Animals were allowed to recover in a heated recovery cage before being returned to their home cage. Two days after surgery, animals were habituated to head fixation under a freely moving circular treadmill for 2–7 days.
For calcium imaging, mice were head-fixed on a freely spinning running wheel under a Nixon 20x-magnification water immersion objective and imaged with a Sutter MOM two-photon resonant scanning microscope within a darkened box (see above for description of imaging setup). Volume acquisition occurred at 5.8–6.6 Hz for 550 × 550 μm fields of view with three Z-planes each separated by 50 μm. Imaging planes were 100–350 μm below the pia surface. 2P imaging was conducted at a wavelength of 930 nm with average powers at 50 mW. Neurons that co-expressed opsin tagged to a red fluorophore and GCaMP6 were identified based on average movies taken at 1000–1040 nm, and using custom Matlab software, regions of interest were circled and their centroids were used to compute holographic targets that were pre-loaded in sequence on the SLM. Custom Matlab digital acquisition software controlled the experiment by triggering Scanimage5 to acquire frames, the SLM to change hologram, and EOM to control laser power.
Two photon imaging data analysis
Motion correction, calcium source extraction, and deconvolution were performed using Suite2P as described55. Briefly, raw calcium movies were motion corrected using Suite2P with subpixel alignment = 2, and calcium sources extracted with key parameters Diameter = 12–16 μm and Signal Extraction = ‘raw’. Calcium sources were then manually examined and accepted or rejected based on their overlap with morphologically identifiable neurons. Neuropil subtracted Fluorescence vectors (F) or the OASIS deconvolution (S) were used for downstream analysis. Calcium signals were acquired continuously, and each cell’s fluorescence Z-scored or ΔF/Fo was calculated using Fo = the tenth percentile of Fluorescence observed over the entire experiment. If a trial had motion over threshold (5μm) during half or more of the stimulation period the entire trial was excluded, since the results of experiment may not be interpretable, but if motion occurred when not stimulating it was corrected post hoc and the data was included. Holographic targets were aligned to calcium sources by calculating the Euclidean distance between the centroids of all holographic targets and all calcium sources and finding the minimum. Very rarely, targets assigned were assigned to calcium sources with distance > 15 μm. These targets were assumed to have failed and were excluded from subsequence analysis.
We found that the OASIS deconvolution signals provided a good estimate for the first derivate of the calcium signal, since local peaks in the S vector aligned with the frame on which stimulation occurred better than ΔF/Fo. Since deconvolved calcium signals decay much faster than fluorescent signals this ameliorates analysis problems where slowly decaying fluorescence from a recently stimulated neuron may be attributed to holographic stimulation of the next cell in a sequence. Therefore, most subsequent analysis for ST-ChroME expressing neurons was performed based on the S vector.
In GtACR1 experiments with GCaMP6f in PV neurons, reduction of calcium activity was most apparent when analyzing Z-scored fluorescence responses, likely due to the relatively high firing rates of PV neurons making true baseline F0 hard to determine. Each cell’s fluorescence was Z-scored on data from the entire experiment, and then had its baseline (determined during a non-stimulation period) subtracted to remove state dependent variability. In single-neuron suppression experiments neurons were stimulated one by one at a rate of 0.5Hz for 1s at 0.16mW/μm2 (50mW per Neuron). As the fluorescence response to self-stimulation lasted more than 2s the trial immediately following self-stimulation was excluded from analysis. Similarly, small ensemble stimulation was performed at 0.5Hz for 1s but at 0.08mW/μm2 (25mW per Neuron) but for 4 neurons simultaneously or 100mW in the field of view. In both cases cells that fell within the region of optical artifact were excluded from subsequent analysis, but were not considered a failed stimulation. The laser gate was used in every all optical experiment. Small ensemble responses were calculated as the average of every recorded member of the 4 stimulated cells, in the case that a member of the ensemble was in the optical artifact that cells response would be ignored, and the remaining cell responses would comprise the ensemble response. The neurons that make up each ensemble were selected randomly using matlab ‘randperm’ from a list of identified neurons.
For analysis of single-neuron stimulation experiments the S vector was z-scored on a trial-wise basis. For optical rheobase experiments, neurons were stimulated one-by-one at a rate of 2 Hz with 10 × 5 ms pulses at 30 Hz with varying laser intensities (corrected for varying diffraction efficiency of holograms targeting areas across the accessible volume). To determine the power needed to activate neurons, the relationship between stimulation power and the mean z-scored S vectors fit with a smoothing interpolant and the power to reach 80% of saturation was reported. To determine the single-neuron response matrix, neurons were sequentially stimulated with 10 × 5 ms pulses at 30 Hz and the mean z-scored S vector for each neuron was averaged for two frames (~300 ms) after stimulation of each target.
For ensemble stimulation experiments, the S vector was z-scored on a trial-wise basis as before, but with a small modification. Since stimulated neurons were not stimulated once per trial, but instead were repetitively stimulated over the course of every trial as part of many different ensembles, we evaluated the response of each neuron to each unique stimulation by discarding frames in which it was directly stimulated as part of a different ensemble. This analysis essentially treats each 73 second sweep as a series of concatenated 2 second single trials, but preserves the baseline information obtained during non-stimulation periods of the long sweep. Responses to ensemble stimulation of 10 × 5 ms pulses at 10, 20, or 30 Hz are shown as the mean z-scored S vector for each stimulus. Maps showing the response of all neurons to ensemble stimuli show the mean z-scored S vector for all neurons for two frames (~300 ms) after the marked ensemble was stimulated.
For correlational analysis, for each pair of neurons, Pearson’s ρ was calculated for each trial type from the raw S vector corresponding to those trials (i.e., one correlation coefficient was calculated from the concatenation of all trials of a given type, corresponding to 10–13 trials of length 73 seconds, or ~12–16 minutes of imaging data). Differences in the distributions of correlation coefficients across trial types were assessed for significance by the Friedman test with the Tukey-Kramer correction for multiple comparisons.
Modeling the speed and scale of photoactivation
A description of the model used to calculate the maximum number of light-evoked spikes per second (relating to supplemental figure 23) is available online in our repository (https://github.com/adesniklab/3D-SHOT).
Statistics
All analyses were performed MATLAB (Mathworks). The analyses performed were: paired t test, Mann-Whitney U-test, Wilcoxon Signed Rank Test, Fisher Exact Test, Friedman’s test, Kruskal-Wallis Test. All tests were two-sided unless noted otherwise. For parametric tests, data distribution was assumed to be normal but was not formally tested. No statistical methods were used to pre-determine sample sizes, but we collected sample sizes that were similar to or exceeded those reported in previous publications13,14,25. Data collection and analysis was not performed blind to the conditions of the experiments. Randomization was used in all applicable experiments (for experiments with multiple trial types, the order of trials was randomized). Animals and data points were only excluded based on criteria described above.
Code Availability Statement
Code for alignment of 3D holography with 3D imaging, holographic control, hologram computation, and analysis will be hosted online upon publication.
Supplementary Material
Acknowledgments
We thank Marla Feller, Alex Naka, and Jennifer Brown for critical feedback on the manuscript and discussions. We thank Christopher Baker and Maclean Bolton for soma-targeted ChR2 AAVs. We thank Mei Li and the UC Berkeley Vision Science Core, Gene Delivery Module for preparation of AAVs (Supported by NIH Core Grant P30 EY003176). We deeply appreciate the efforts of Desiree Chu, Chris Douglas and Rich Hakim for important technical assistance. We thank David Taylor for help with mouse work and histology. H.A. is a New York Stem Cell Foundation-Robertson Investigator. This work was supported by The New York Stem Cell Foundation, and by grants from the Arnold and Mabel Beckman Foundation, NINDS grant DP2NS087725-01, the McKnight Foundation, NINDS award F32NS095690-01 to A.M., the Simon’s Foundation Collaboration for the Global Brain award 415569 to I.O., and a fellowship from the David and Lucille Packard Foundation to L.W. This research was developed with funding from the Defense Advanced Research Projects Agency (DARPA), Contract No. N660011-17-C-4015.
Footnotes
Author contributions.
A.M., I.O., N.P. and H.A conceived the project and built the system.
A.M. designed and performed all experiments involving excitatory opsins.
I.O. designed and performed all experiments involving inhibitory opsins.
N.P. designed and assembled the light paths and wrote custom holography software
S.S. performed cloning, mutagenesis, cell culture, and 1P recordings in CHO cells.
A.M., I.O., N.P., and E.L. wrote code and developed software for experimental control.
K.C. performed histology.
S.B. performed modeling of the Chronos pore region.
L.W. contributed expertise on holographic design.
A.M., I.O., N.P., and H.A. wrote the manuscript.
Competing Interest Statement
The authors declare that they have no competing interests.
References
- 1.London M, Roth A, Beeren L, Häusser M, Latham PE. Sensitivity to perturbations in vivo implies high noise and suggests rate coding in cortex. Nature. 2010;466:123–127. doi: 10.1038/nature09086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gollisch T, Meister M. Rapid neural coding in the retina with relative spike latencies. Science. 2008;319:1108–1111. doi: 10.1126/science.1149639. [DOI] [PubMed] [Google Scholar]
- 3.Histed MH, Maunsell JHR. Cortical neural populations can guide behavior by integrating inputs linearly, independent of synchrony. Proc Natl Acad Sci U S A. 2014;111:E178–87. doi: 10.1073/pnas.1318750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruno RM, Sakmann B. Cortex is driven by weak but synchronously active thalamocortical synapses. Science. 2006;312:1622–1627. doi: 10.1126/science.1124593. [DOI] [PubMed] [Google Scholar]
- 5.Harris KD, Mrsic-Flogel TD. Cortical connectivity and sensory coding. Nature. 2013;503:51–8. doi: 10.1038/nature12654. [DOI] [PubMed] [Google Scholar]
- 6.Panzeri S, Harvey CD, Piasini E, Latham PE, Fellin T. Cracking the neural code for sensory perception by combining statistics, intervention and behavior. Neuron. 2017;93:1–30. doi: 10.1016/j.neuron.2016.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jepson LH, et al. High-fidelity reproduction of spatiotemporal visual signals for retinal prosthesis. Neuron. 2014;83:87–92. doi: 10.1016/j.neuron.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clancy KB, Schnepel P, Rao AT, Feldman DE. Structure of a Single Whisker Representation in Layer 2 of Mouse Somatosensory Cortex. J Neurosci. 2015;35:3946–3958. doi: 10.1523/JNEUROSCI.3887-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohki K, Chung S, Ch’ng YH, Kara P, Reid RC. Functional imaging with cellular resolution reveals precise micro-architecture in visual cortex. Nature. 2005;433:597–603. doi: 10.1038/nature03274. [DOI] [PubMed] [Google Scholar]
- 10.Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci U S A. 2009;106:15025–15030. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaziri A, Emiliani V. Reshaping the optical dimension in optogenetics. Curr Opin Neurobiol. 2012;22:128–137. doi: 10.1016/j.conb.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Packer AM, et al. Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods. 2012;9:1202–5. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash R, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Packer AM, Russell LE, Dalgleish HWP, Häusser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods. 2015;12:140–146. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci. 2014;17:1816–1824. doi: 10.1038/nn.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrillo-reid L, Yang W, Bando Y, Peterka DS, Yuste R. Imprinting and recalling cortical ensembles. Science (80− ) 2016;353:691–694. doi: 10.1126/science.aaf7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emiliani V, Cohen AE, Deisseroth K, Ha M. All-Optical Interrogation of Neural Circuits. J Neurosci. 2015;35:13917–13926. doi: 10.1523/JNEUROSCI.2916-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.dal Maschio M, Donovan JC, Helmbrecht TO, Baier H. Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging. Neuron. 2017;94:774–789.e5. doi: 10.1016/j.neuron.2017.04.034. [DOI] [PubMed] [Google Scholar]
- 19.Papagiakoumou E, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat Methods. 2010;7:848–854. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forli A, et al. Two-Photon Bidirectional Control and Imaging of Neuronal Excitability with High Spatial Resolution In Vivo. Cell Rep. 2018;22:3087–3098. doi: 10.1016/j.celrep.2018.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolenko V, et al. SLM Microscopy: Scanless Two-Photon Imaging and Photostimulation with Spatial Light Modulators. Front Neural Circuits. 2008;2:1–15. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papagiakoumou E, de Sars V, Oron D, Emiliani V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt Express. 2008;16:22039–22047. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- 23.Emiliani V, et al. Wave front engineering for microscopy of living cells. Opt Express. 2005;13:1395–1405. doi: 10.1364/opex.13.001395. [DOI] [PubMed] [Google Scholar]
- 24.Ronzitti E, Conti R, Zampini V, Tanese D, Foust AJ, Klapoetke N, Boyden ES, Papagiakoumou EEV. Sub-millisecond optogenetic control of neuronal firing by two-photon holographic photoactivation of Chronos. J Neurosci. 2017:1246–17. doi: 10.1101/062182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shemesh OA, et al. Temporally precise single-cell-resolution optogenetics. Nat Neurosci. 2017;20:1796–1806. doi: 10.1038/s41593-017-0018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez O, et al. Three-dimensional spatiotemporal focusing of holographic patterns. Nat Commun. 2016;7:1–10. doi: 10.1038/ncomms11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pégard NC, et al. Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT) Nat Commun. 2017;8:1–14. doi: 10.1038/s41467-017-01031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–46. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science (80− ) 2015;349:647–50. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahmadian Y, Packer AM, Yuste R, Paninski L. Designing optimal stimuli to control neuronal spike timing. J Neurophysiol. 2011;106:1038–1053. doi: 10.1152/jn.00427.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker CA, Elyada YM, Parra A, Bolton MML. Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. Elife. 2016;5:1–15. doi: 10.7554/eLife.14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu C, Ivanova E, Zhang Y, Pan Z. rAAV-Mediated Subcellular Targeting of Optogenetic Tools in Retinal Ganglion Cells In Vivo. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0066332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podgorski K, et al. Brain heating induced by near infrared lasers during multi-photon microscopy. bioRxiv. 2016;116 doi: 10.1152/jn.00275.2016. jn.00275.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato HE, et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature. 2012;482:369–74. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gradinaru V, et al. Molecular and Cellular Approaches for Diversifying and Extending Optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuong AS, et al. Noninvasive optical inhibition with a red-shifted microbial rhodopsin. Nat Neurosci. 2014;17:1123–9. doi: 10.1038/nn.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Govorunova EG, Sineshchekov OA, Spudich JL. Proteomonas sulcata ACR1: A Fast Anion Channelrhodopsin. Photochem Photobiol. 2016;92:257–263. doi: 10.1111/php.12558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berndt A, et al. Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity. Proc Natl Acad Sci U S A. 2016;113:822–829. doi: 10.1073/pnas.1523341113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markram H, et al. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 41.Taniguchi H, et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hippenmeyer S, et al. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:0878–0890. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wekselblatt JB, Flister ED, Piscopo DM, Niell CM. Large-scale imaging of cortical dynamics during sensory perception and behavior. J Neurophysiol. 2016;115:2852–2866. doi: 10.1152/jn.01056.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grewe BF, Voigt FF, van ‘t Hoff M, Helmchen F. Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed Opt Express. 2011;2:2035–2046. doi: 10.1364/BOE.2.002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W, Carrillo-Reid L, Bando Y, Peterka DS, Yuste R. Simultaneous two-photon optogenetics and imaging of cortical circuits in three dimensions. Elife. 2018;7 doi: 10.7554/eLife.32671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adesnik H, Scanziani M. Lateral competition for cortical space by layer-specific horizontal circuits. Nature. 2010;464:1155–1160. doi: 10.1038/nature08935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris KD, Thiele A. Cortical state and attention. 2011;12 doi: 10.1038/nrn3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Churchland AK, et al. Article Variance as a Signature of Neural Computations during Decision Making. Neuron. 2010;69:818–831. doi: 10.1016/j.neuron.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci U S A. 2010;107:11981–6. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huber D, et al. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lim ST, Antonucci DE, Scannevin RH, Trimmer JS. A Novel Targeting Signal for Proximal Clustering of the Kv2.1 K+ Channel in Hippocampal Neurons. Neuron. 2000;25:385–397. doi: 10.1016/s0896-6273(00)80902-2. [DOI] [PubMed] [Google Scholar]
- 52.Gerchberg RW, Saxton WO. A Practical Algorithm for the Determination of Phase from Image and Diffraction Plane Pictures. Optik (Stuttg) 1972;35:237–246. [Google Scholar]
- 53.Zhang J, Pégard N, Zhong J, Waller L. 3D computer generated holograms by nonconvex optimization. Optica. 2017;2017:2016–2018. [Google Scholar]
- 54.Pluta S, et al. A direct translaminar inhibitory circuit tunes cortical output. Nat Neurosci. 2015;18:1631–1640. doi: 10.1038/nn.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pachitariu M, et al. Suite2p: beyond 10, 000 neurons with standard two-photon microscopy. bioRxiv. 2016:1–14. doi: 10.1101/061507. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in this study are available from the corresponding author on reasonable request. Additionally, the sequences for constructs created in this study will be made publicly available on Addgene.