Abstract
Dysregulation of genetic pathways during human germ cell development leads to infertility. Here, we analyzed bona fide human primordial germ cells (hPGCs) to probe the developmental genetics of human germ cell specification and differentiation. We examined distribution of OCT4 occupancy in hPGCs relative to human embryonic stem cells (hESCs). We demonstrate that development, from pluripotent stem cells to germ cells, is driven by switching partners with OCT4 from SOX2 to PAX5 and PRDM1. Gain- and loss-of-function studies revealed that PAX5 encodes a critical regulator of hPGC development. Moreover, analysis of epistasis indicates that PAX5 acts upstream of OCT4 and PRDM1. The PAX5-OCT4-PRDM1 proteins form a core transcriptional network that activates germline and represses somatic programs during human germ cell differentiation. These findings illustrate the power of combined genome editing, cell differentiation and engraftment for probing human developmental genetics that have historically been difficult to study.
Introduction
Substantial research has centered on identification and characterization of genes that are required for specification, maintenance and differentiation of the mammalian primordial germ cells (PGCs) that ultimately give rise to the sperm and eggs required to perpetuate life1–13. In mice, several transcription factors have been identified that are required for in vitro specification and induction of the earliest stages of germ cells; these germ cells are ultimately able to mature and fulfill the greatest test of germ cell identity, the ability to produce live offspring10,14–18. However, the transcriptional network of human primordial germ cells (hPGCs) differs substantially from that of mice, making it difficult to translate knowledge directly to humans11. For example, hPGCs express lineage specifier genes that are not expressed in mouse PGCs, including SOX1719.
Although hPGCs are committed to the germ cell lineage, they share expression profiles of several pluripotency genes with human embryonic stem cells (hESCs), including OCT4 (also known as POU5F1); however, other key pluripotency genes, such as SOX2, are not expressed in hPGCs11,20,21. How the network of pluripotency genes, that encodes transcription factors, functions differently in hESCs and hPGCs, is a fundamental question in the field of human germ cell developmental genetics that has remained unaddressed. Thus, in this study, we elucidated genetic mechanisms that underlie development of hPGCs by identifying transcription factors that might function in a network to mediate hPGC specification and differentiation. We focused on OCT4, an essential gene that encodes a transcription factor that is expressed in both hESCs and hPGCs where it is required to maintain cell identity in both cell types22–24. We developed methods to map the genome-wide binding of OCT4 protein in highly heterogeneous tissue samples and identified OCT4 protein partners in hPGCs. We then used gain- and loss-of-function gene analyses to probe the function of PAX5, a member of the paired box (PAX) family, and discovered that it encodes a critical component of a genetic switch that is required for the transition from pluripotent stem cells to differentiation of hPGCs.
Results
Global redistribution of OCT4 occupancy in the transition from hPSCs to hPGCs
To probe the role of OCT4 in hPGCs, we performed OCT4 Chromatin Immunoprecipitation Sequencing (ChIP-seq) analysis on germ cells from second trimester human fetal testis, a developmental stage when hPGCs have colonized the testis and are in the process of expanding to approximately 1–2M total cells, but have not differentiated to spermatogonia or spermatocytes6. We note that OCT4-positive cells are only present in the seminiferous tubules of the testis and not within the interstitial spaces (Fig. 1a, Supplementary Fig. 1a). Immunostaining data also indicated that OCT4-positive cells are a subpopulation of cKIT-positive cells and do not express the DDX4 gene, which is an evolutionarily-conserved germ cell marker of later stages of development (post-PGC; Supplementary Fig. 1b, c). However, since only 1% of the cells in the human fetal testis are OCT4-positive hPGCs (Fig. 1a, Supplementary Fig. 1a), and conventional ChIP protocols require a large number of homogenous cells, we adapted protocols from carrier ChIP25 and tissue ChIP26 to detect binding specificity of individual transcription factors within a heterogeneous cell mixture. We validated our protocol using a heterogeneous control mixture of 10,000 OCT4-positive hESCs mixed with 990,000 OCT4-negative fibroblast cells to model composition of fetal testis (Supplementary Fig. 1d). We compared these data to that generated by conventional ChIP on a pure population of 1 million hESCs by quantitative PCR (Supplementary Fig. 1e) and ChIP-seq and found the result from mixed-ChIP highly correlates that from conventional ChIP (Supplementary Fig. 1f–h). Thus, our methods are reliable for generation of binding data from a heterogeneous mixture of cells when coupled with highly-specific antibodies.
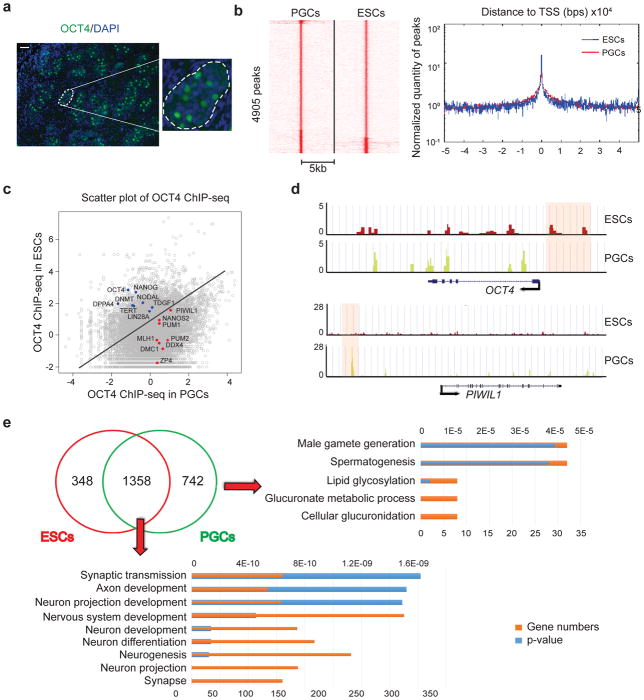
Figure 1. Global redistribution of OCT4 binding in PGCs compared with ESCs.
(a) Cross-section of a human fetal testis (22 weeks) with immunostaining for OCT4. Enlarged panel on the right represents the region enclosed within the white dashed lines of the left panel. Scale bar represents 50 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (b) Left panel: Heatmap visualization of OCT4 ChIP-seq data, depicting all binding events centered on the peak region within a 5kb window around the peak. Right panel: Distribution and peak heights of OCT4 peaks around the transcription start site (TSS). Peak heights are reported in reads per million (RPM). (c) Scatterplot comparing OCT4 binding in PGCs and ESCs. Selected genes known to be associated with pluripotency are highlighted in blue, and those associated with germline are highlighted in red. (d) Genome browser representation of ChIP-seq tracks for OCT4 in ESCs (red) and PGCs (yellow) at the OCT4 and PIWIL1 loci. Regions that were bound by OCT4 exclusively in ESCs or PGCs are highlighted by pink shaded boxes. ChIP-seq were independently repeated twice with similar results. (e) Venn diagram of unique and shared genes bound by OCT4 in ESCs and PGCs. Gene ontology analysis are shown in the right and bottom. Analysis were performed twice with similar results based on two independent ChIP-seq data.
We then applied the mixed-ChIP protocol to human fetal testis and generated a global binding profile for OCT4 in bona fide hPGCs. Two biological replicates were used and demonstrated gene overlap >90% (Supplementary Fig. 1h). Although the enrichment profile of OCT4 around the transcription start sites (TSS) was similar in both hPGCs and hESCs (Fig. 1b), there was a substantial redistribution of OCT4 binding, characterized by reduced binding near pluripotency-related genes (e.g. OCT4, NANOG, LIN28A) and an enrichment of binding near germ cell-related genes (e.g. PIWIL1, DDX4, NANOS2) in hPGCs relative to hESCs (Fig. 1c, d). Furthermore, Gene Ontology (GO) analysis of genes bound by OCT4 only in hPGCs revealed that genes were enriched in GO terms that include male gamete generation and spermatogenesis, while genes bound by OCT4 in both hESCs and hPGCs were enriched in neuronal development, potentially suggesting that OCT4 may repress ectoderm differentiation in both cell types (Fig. 1e). Together, our data demonstrated that the cell fate change from pluripotent hESCs to bona fide germline hPGCs is associated with global reorganization of OCT4 occupancy.
OCT4 switches partners to PAX5 and PRDM1 in hPGCs
We next set out to determine whether the genomic redistribution of OCT4 could be due to alternative OCT4 binding partners in hPGCs relative to hESCs. Immunostaining and RNA expression data showed that the expression of the SOX2, OTX2 and ZIC2, genes which encode well-characterized functional partners of OCT4 in hESCs27,28,29,30, are significantly downregulated in hPGCs (Supplementary Fig. 2a, b)31. This indicated that OCT4 might require different partners in hPGCs than in hESCs. To screen for potential OCT4-interacting transcription factors in hPGCs, we performed de novo sequence motif searches using OCT4-bound sequences exclusively in hPGCs and discovered motifs that are similar to consensus motifs for the transcription factors PAX5 (Paired Box Homeotic Gene 5) and PRDM1 (Positive Regulatory Domain I-Binding Factor 1) (Fig. 2a). Immunostaining demonstrated extensive co-expression of PAX5 and PRDM1 in OCT4-positive cells (Fig. 2b). Quantitation indicates that approximately 60% of the OCT4 +cells are also positive for PAX5 and PRDM1. RNA expression analysis also showed significant induction of PAX5 and PRDM1 in hPGCs compared to hESCs (Supplementary Fig. 2b)31. This suggested that PAX5 and PRDM1 might co-occupy genomic loci with OCT4 as functional complexes in a germ cell specific transcriptional network.
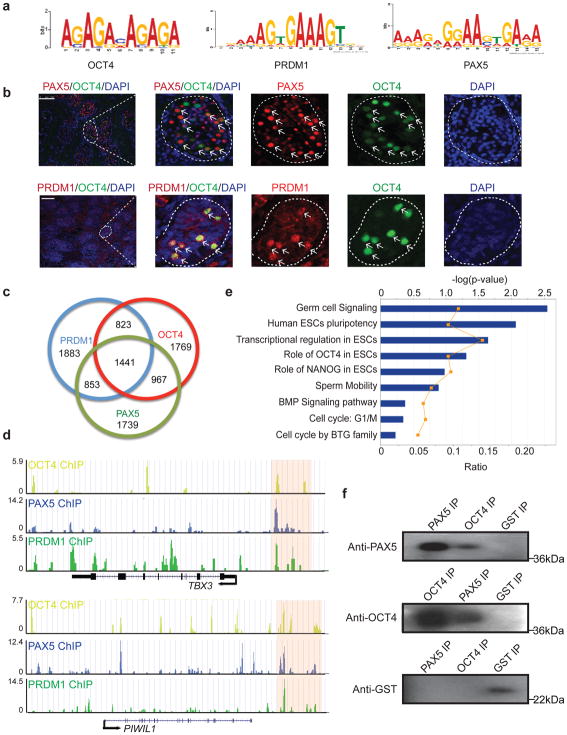
Figure 2. Co-occurrence of OCT4 with PAX5 and PRDM1 in PGCs.
(a) The position weight matrix of an enriched motif found in OCT4 ChIP-seq data from PGCs. The motif resembles the binding motifs for PRDM1 and PAX5. (b) Cross-section of a human fetal testis (22 weeks) with immunostaining for PAX5 (red), OCT4 (green) and DAPI stained nuclei (blue) (upper panel); PRDM1 (red), OCT4 (green) and DAPI stained nuclei (blue) (lower panel). Enlarged panels on the right represent the region enclosed within the white dashed lines of the left panel. White arrows indicate co-localization of PAX5 and OCT4 or PRDM1 and OCT4. Scale bars represent 100 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (c) Venn diagram of unique and shared genes bound by OCT4, PRDM1 and PAX5 in PGCs. The number of genes bound exclusively by each transcription factor or co-bound by multiple transcription factors are labelled. (d) Genome browser representation of ChIP-seq tracks for OCT4 (yellow), PAX5 (blue) and PRDM1 (green) at the TBX3 and PIWIL1 loci. Regions that are bound collectively by OCT4, PAX5 and PRDM1 in PGCs are highlighted by pink shaded boxes. ChIP-seq were independently repeated twice with similar results. (e) Gene ontology analysis of co-bound genes. Analysis were performed twice with similar results. (f) GST-pull down assay performed using OCT4 and PAX5 recombinant proteins. Pull-down was repeated three times with similar results. Unprocessed scans of western blot analysis are available in Supplementary Fig. 8.
To determine the binding profiles of PAX5 and PRDM1 and probe potential association with OCT4 in hPGCs, we performed ChIP-seq analysis for both PAX5 and PRDM1 (Supplementary Fig. 2c). Among the top 5000 genes bound by each transcription factor, 1441 genes, including germ cell specific genes (eg., DDX4, DAZL and PIWIL1), were collectively bound by all three transcription factors (Fig. 2c, d). These results suggested extensive co-occupancy of PAX5, PRDM1 and OCT4 in hPGCs. Annotation analysis revealed that these co-bound genes are enriched for GO terms of germ cell signaling, hESC pluripotency and bone morphogenetic proteins (BMP) signaling pathways (Fig. 2e). In addition, we observed that recombinant OCT4-GST fusion protein can immunoprecipitate recombinant PAX5 protein in vitro (Fig. 2f), indicating that PAX5 may interact directly with OCT4 protein in hPGCs. We did not detect direct protein interactions between recombinant OCT4 and PRDM1 proteins. However, PRDM1 protein was pulled down by PAX5 and vice versa (Supplementary Fig. 2d), indicating that OCT4, PAX5 and PRDM1 may assemble as a protein complex through both direct and indirect interactions. In summary, our observations suggested that OCT4, partnering with PAX5 and PRDM1 proteins, might constitute an extensive and unique transcription network in hPGCs.
Overexpression of PAX5 and PRDM1 induces human germ cell differentiation
Although PRDM1 is a well-studied, key regulator of PGC development in both mouse and human15,19,32, the discovery of PRDM1 as a binding partner of OCT4 has never been shown. Additionally, although PAX5 is most commonly known for its role in development of the blood system33, a potential role for PAX5 in PGC development or specification has not been reported. To investigate the roles of both genes in hPGCs, we examined whether overexpression (OE) of PAX5 and PRDM1 is capable of directing hESCs to the germ cell lineage in an in vitro differentiation system. We overexpressed PAX5 or PRDM1 by approximately 10–15 times above the endogenous expression levels (Supplementary Fig. 3a), to a level comparable to that of bona fide hPGCs (Supplementary Fig. 2b). Forced expression of either PAX5 or PRDM1 does not alter the identity of ESCs in routine hESC maintenance (Supplementary Fig. 3b, c). Next, we differentiated PAX5 OE and PRDM1 OE hESCs by subjecting the cells to BMPs for 7 days as previously described5,34. The expression of germ cell marker genes, including DAZL, DDX4, DPPA3 and NANOS3, is upregulated significantly in OE cells compared with non-OE control cells (Fig. 3a, Supplementary Fig. 3d). PAX5 also demonstrated binding on several later stage germ cells genes and activated their expression during differentiation, such as SYCP1 and SYCP3 (Fig. 3a, Supplementary Fig. 3e). Interestingly, co-expression of PAX5 and PRDM1 showed no obvious additive effects relative to single factor OE (Supplementary Fig. 3f). Immunostaining reveals DDX4 signal in PAX5 OE cells, but not control cells (Fig. 3b). We also used a hESC-DDX4-mOrange knock-in reporter cell line to better characterize the effects of in vitro differentiation. We observed a significantly higher percentage of DDX4-mOrange+ cells in the PAX5 OE cells by FACS (fluorescence-activated cell sorting) relative to control cells (Fig. 3c). These data demonstrated that induced expression of PAX5 and PRDM1 in hESCs strongly promotes differentiation of germ cells in vitro and prompted us to explore whether these cells may further mature if placed in the somatic niche via xenotransplantation.
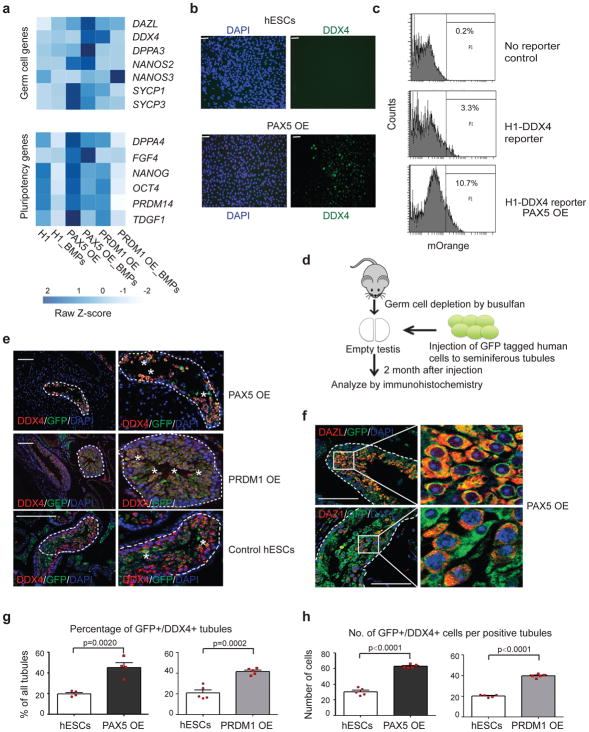
Figure 3. Overexpression PAX5 and PRDM1 enhance germ cell potential of ESCs.
(a) Heatmap of FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values for genes associated with germline (top) and pluripotency (bottom). _BMPs: differentiated by BMPs; OE: overexpression. (b) Immunostaining of differentiated cells from hESCs and PAX5 OE cells for DDX4 and DAPI. Scale bars represent 50 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (c) Gating strategy to sort mOrange+ cells from the H1 hESC line after differentiation by BMPs. (d) Schematic experimental design of xenotransplantion. Transplantations were performed by independently injecting GFP tagged human cells directly into seminiferous tubules of busulfan-treated mouse testes that were depleted of endogenous germ cells. Testis xenografts were analyzed by immunohistochemistry 2 months after injection. (e) Immunohistochemical analysis of testis xenografts derived from PAX5 OE, PRDM1 OE and control H1 hESCs. In all panels, dashed white lines indicate the outer edges of spermatogonial tubules and enlarged view are shown on the right. White asterisks represent GFP+/DDX4+ donor cells near the basement membrane. Scale bars represent 50 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (f) Immunostaining of testis xenografts derived from PAX5 OE H1 hESCs for later stage PGC markers DAZL and DAZ1. Enlarged panel on the right represents the region enclosed within the white rectangles of the left panel. Scale bars represent 50 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (g) Percentage of tubules positive for GFP+/DDX4+ cells were calculated across multiple cross-sections (relative to total number of tubules). Data are represented as mean ± SD of n= 4 independent replicates. P-values were calculated by two-tailed Student’s t-test. (h) For each positive tubule, the ratio of GFP+/DDX4+ cells per tubule was determined. Data are represented as mean ± SD of n=5 independent replicates. P-values were calculated by two-tailed Student’s t-test. Source data for g and h are in Supplementary Table 2.
To investigate differentiation potential in vivo, we used a previously-developed xenotransplantation platform2–4. Briefly, we transplanted GFP-tagged human cells into busulfan-treated immunodeficient mice, which are depleted of endogenous germ cells (Fig. 3d, Supplementary Fig. 4a). Two months post-transplantation, we analyzed the testes and observed significant human germ cell engraftment in the tubules for OE cells, as indicated by the presence of GFP+ cells that co-expressed DDX4 (Fig. 3e). More importantly, PAX5 OE cells differentiated to a later stage germ cell fate that expressed mature germ cell markers, such as DAZL and DAZ1 (Fig. 3f). To quantify germ cell potential, we counted GFP+/DDX4+ human germ cells and tubules across entire cross-sections. We then calculated the percentage of positive tubules and determined the number of GFP+/DDX4+ cells in each positively stained tubule. Both values were significantly higher in PAX5 OE and PRDM1 OE cells (Fig. 3g, h). Consistent with in vitro differentiation results, PAX5 OE and PRDM1 OE promoted germ cell differentiation of hESCs in the mouse seminiferous tubule; however, no additive effects have been detected compared to single OE cells (Supplementary Fig. 4b–d). These data provide strong evidence that overexpression of PAX5 and PRDM1 is able to greatly promote differentiation potential of hESCs towards germ cell lineage in vitro and in vivo.
Knockout of PAX5 or PRDM1 reduces germ cell potential of hESCs
We further examined the role of PAX5 in germ cell differentiation by loss-of-function studies. We generated a PAX5 knockout (KO) hESC line via use of CRISPR/Cas9-based genome editing (Supplementary Fig. 5a, b). PAX5 KO was confirmed by immunofluorescence and Western blot analysis upon induced neuronal differentiation (Supplementary Fig. S5c, d)35,36. Moreover, a significant reduction of germ cell gene expression was observed after in vitro differentiation (Fig. 4a) and DDX4 was not detected (Fig. 4b) further indicating a severe reduction of germ cell differentiation in vitro. We then examined whether germ cells could be differentiated and maintained from these cell lines via in vivo xenotransplantation. As noted, PAX5 KO cells gave rise to significantly fewer positive tubules with human germ cells and fewer GFP+/DDX4+ cells in the positive tubules; most of the DDX4+ cells were mouse germ cells regenerated after treatment (Fig. 4c). There was a >3-fold reduction of positive tubules and >5-fold reduction of GFP+/DDX4+ cells in the positive tubules (Fig. 4d, e) indicating that genetic knockout of PAX5 greatly reduced germ cell differentiation from hESCs.
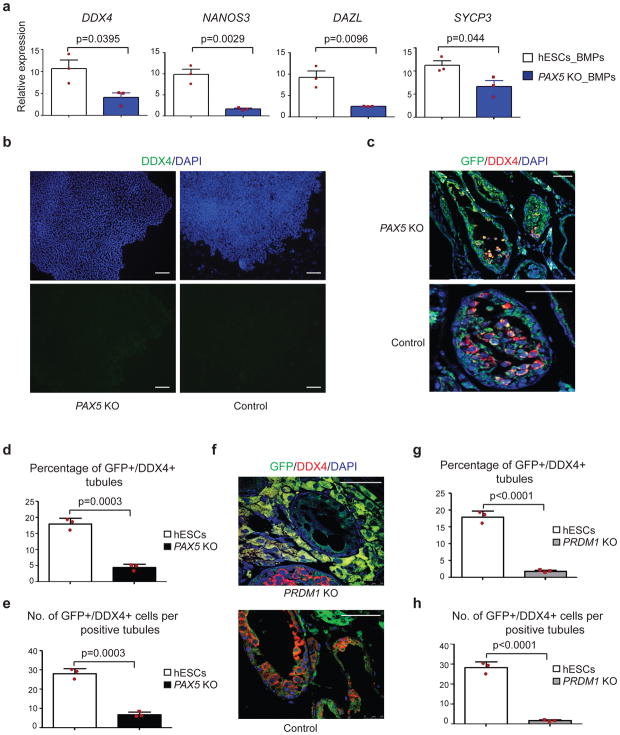
Figure 4. Knock out of PAX5 or PRDM1 reduce germ cell potential of ESCs.
(a) RT-qPCR analysis of control and PAX5 KO H1 hESCs after BMPs-induced differentiation. Abbreviations: _BMPs represents cells were differentiated by BMPs; KO represents knockout. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (b) Immunostaining of differentiated cells for DDX4 (green) and DAPI stained nuclei (blue). Scale bars represent 100 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (c, f) Immunohistochemical analysis of testis xenografts derived from PAX5 KO (c) and PRDM1 KO (f) and control H1 hESCs. All images are merged from DDX4 (red), GFP (green)and DAPI-stained nuclei. Scale bars represent 50 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (d, g) Percentage of tubules positive for GFP+/DDX4+ cells were calculated across multiple cross-sections (relative to total number of tubules) for PAX5 KO (d) and PRDM1 KO (g) H1 hESCs. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (e, h) For each positive tubule, the ratio of GFP+/DDX4+ cells per tubule was determined for PAX5 KO (e) and PRDM1 KO (h) H1 hESCs. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. Source data for a, d, e, g and h are in Supplementary Table 2.
Previous studies revealed that PRDM1-knockout hESCs fail to develop into PGCs in vitro19. To determine if these cells were also compromised in terms of germ cell differentiation in vivo, we used previously-reported PRDM1 KO hESCs19 to test ability to differentiate to germ cells in murine xenotransplants. We observed that PRDM1 KO hESCs were severely deficient in germ cell differentiation in vivo (Fig. 4f) with the majority of tubules devoid of any human germ cell engraftment. Only a small number of tubules were observed with sparse GFP+/DDX4+ cells. Counts of the engraftment revealed a >10-fold reduction in formation of human germ cells in the mouse tubules (Fig. 4g, h), resulting in a more severe defect in germ cell potential compared to PAX5 KO cells.
Epistasis of PAX5, OCT4 and PRDM1 in hPGCs: PAX5 acts upstream of OCT4 and PRDM1
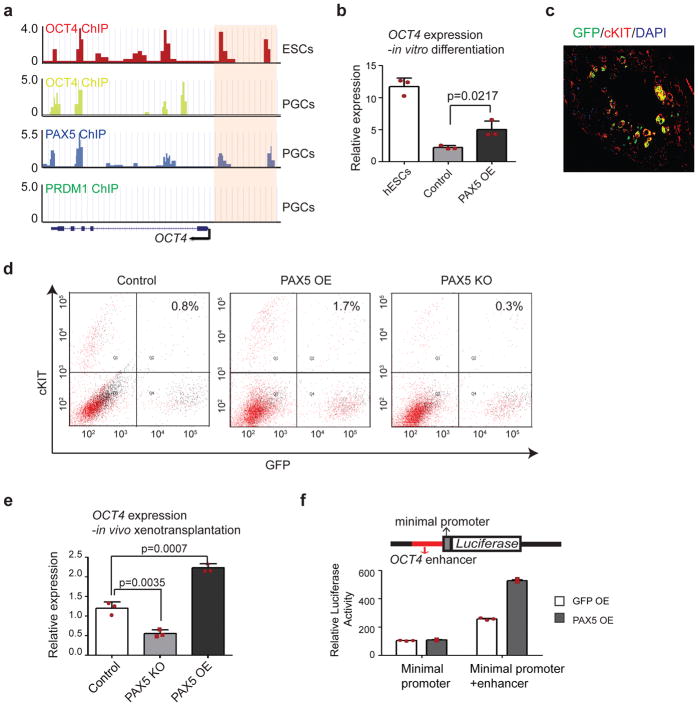
To explore the molecular mechanism of PAX5 function in hPGCs, we re-analyzed our ChIP-seq data and observed that the enhancers of OCT4, which are bound by OCT4 itself in hESCs, are bound by PAX5 in hPGCs (Fig. 5a). Thus, we hypothesized that one role of PAX5 is to regulate and maintain OCT4 expression, as germ cell differentiation proceeds and requires expression of OCT4 at moderate levels37. We tested this hypothesis, first, by examining OCT4 expression during in vitro differentiation. Following BMP-induced differentiation, OCT4 expression in PAX5 OE cells was substantially elevated relative to differentiated controls (Fig. 5b). However, due to the developmental limitation of current in vitro differentiation, genes essential for later stage germ cells (eg., DDX4, DAZL), including PAX5, cannot be induced to the functional level (Supplementary Fig. 6a, b) 19,31,38. Thus, PAX5 KO cells only exhibited a minor decrease in OCT4 expression, relative to control differentiated cells, by all three protocols (Supplementary Fig. 6c). To overcome this limitation, we sorted hPGCs that were formed in vivo in mouse seminiferous tubules via use of GFP and cKIT and analyzed effects of PAX5 KO in vivo (Fig. 5c, d). Note that the niche of mouse seminiferous tubules provides a superior differentiation environment for hESCs to develop to more mature hPGCs that express genes essential for later stage germ cells, including DDX4 (Fig. 3e, f). In these in vivo derived hPGCs, there was significant downregulation of OCT4 expression in cells formed by PAX5 KO cells, and as also expected, significant upregulation of OCT4 expression in cells formed by PAX5 OE cells, compared to cells formed by control hESCs (Fig. 5e).
Figure 5. PAX5 acts upstream of OCT4.
(a) Genome browser representation of ChIP-seq tracks at the OCT4 locus. Enhancer regions are highlighted by pink shaded boxes. ChIP-seq were independently repeated twice with similar results. (b) OCT4 expression in hESCs and in control and PAX5 OE cells during in vitro differentiation. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (c) Immunostaining of GFP and CKIT in mouse testis xenografts. Scale bar represents 50 μm. Immunostaining experiments were independently repeated a minimum of three times with similar results. (d) Flow cytometry analysis for GFP and CKIT of mouse testis xenografted. (e) RT-qPCR analysis of OCT4 expression in hPGCs formed in the mouse seminiferous tubules by PAX5 OE, PAX5 KO and control hESCs. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (f) Reporter constructs used to assay for testing OCT4 enhancer activity are shown. Genomic fragment bound by PAX5 and OCT4 (in red) was inserted upstream of a luciferase gene driven by minimal promoter. Y-axis represents the fold enrichment of luciferase activity. Data are represented as mean ± SD of n=3 independent replicates. Source data for b, e and f are in Supplementary Table 2.
To further determine whether PAX5 regulates OCT4 expression by regulating the enhancer of OCT4, a luciferase reporter assay was performed in 293T cells. As expected, PAX5 OE caused a significant increase in luciferase activity, suggesting that PAX5 could activate OCT4 expression through its enhancer (Fig. 5f). We also identified the binding motif of PAX5 in the region of the OCT4 enhancer sequences (Supplementary Fig. 7a–c). Results indicated that mutation of PAX5 binding motif abolished the induction effect of PAX5 protein (Supplementary Fig. 7d).
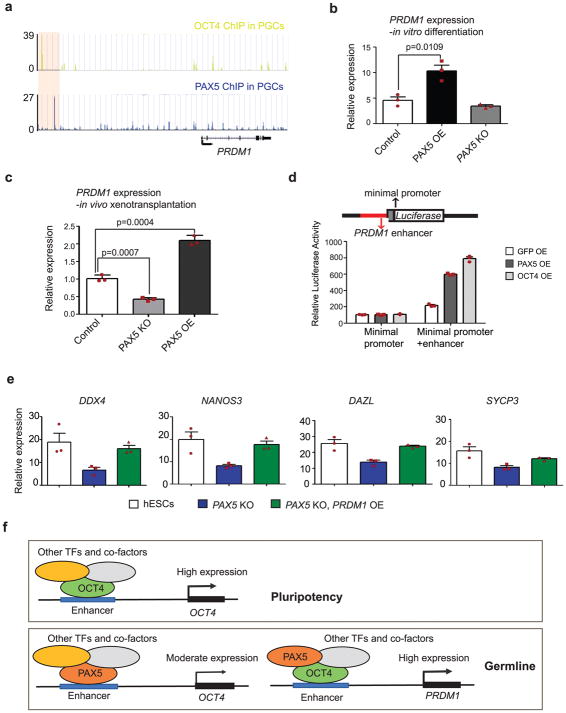
Further analysis of ChIP-seq data indicated that PAX5 and OCT4 bind to PRDM1 enhancers with high intensity (Fig. 6a), suggesting that PAX5 and OCT4 might act upstream of PRDM1 to regulate its expression. We found an increase of PRDM1 in PAX5 OE formed hPGCs and a significant decrease in PAX5 KO formed hPGCs (Fig. 6c). Luciferase reporter assay in 293T cells showed that either PAX5 or OCT4 was able to significantly increase luciferase activity driven by PRDM1 enhancer (Fig. 6d). These results indicated that PAX5 and OCT4 could act on the PRDM1 enhancer as activators to induce PRDM1 expression, potentially during germline differentiation. Mutation of the PAX5 binding motif in PRDM1 enhancer region abolished the induction effects of PAX5 protein (Supplementary Fig. 7e–h). Since PRDM1 is a critical gene for germ cell specification and it could be downstream of PAX5 and OCT4, we then overexpressed PRDM1 in PAX5 KO cells to test whether PRDM1 could rescue the defect of PAX5 KO cells. Indeed, we observed that OE of PRDM1 restores germ cell potential of PAX5 KO cells (Fig. 6e).
Figure 6. PAX5 and OCT4 acts upstream of PRDM1.
(a) Genome browser representation of ChIP-seq tracks at the PRDM1 locus. Enhancer regions bound by OCT4 and PAX5 in PGCs are highlighted by pink shaded boxes. ChIP-seq were independently repeated twice with similar results. (b) PRDM1 expression in control, PAX5 OE and PAX5 KO cells during in vitro differentiation. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (c) RT-qPCR analysis of PRDM1 expression in hPGCs formed in the mouse seminiferous tubules by PAX5 OE, PAX5 KO and control hESCs. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (d) Reporter constructs used to assay for testing PRDM1 enhancer activity are shown. Genomic fragment bound by PAX5 and OCT4 (in red) was inserted upstream of a luciferase gene driven by minimal promoter. Y-axis represents the fold enrichment of luciferase activity. Data are represented as mean ± SD of n=3 independent replicates. (e) RT-qPCR analysis of the expression of genes associated with germline. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (f) Model for gene regulation in pluripotency and germline: In pluripotent stem cells, OCT4, together with other transcription factors and cofactors, binds to its own enhancer to activate and maintain its high expression. While differentiation towards germline, PAX5 replaces OCT4 and binds to the enhancer of OCT4 to maintain a moderate expression of OCT4; In the meantime, PAX5 and OCT4 bind to the enhancer of PRDM1 and activate its expression to initiate the germ cell program. TFs: transcription factors. Source data for b, c, d and e are in Supplementary Table 2.
Taken together, our data shed light on the epistasis of these three transcription factors during differentiation from hESCs to hPGCs (Fig. 6f): In pluripotent stem cells, OCT4 interacts with SOX2 and other cofactors, and binds to its own enhancer to activate and maintain high expression. In contrast, with differentiation to germ cells, PAX5 replaces OCT4, recognizes its own binding motif and binds to the enhancer of OCT4 to maintain a moderate expression of OCT4. Concurrently, PAX5 and OCT4 may bind to the enhancer of PRDM1 and activate its expression to initiate the germ cell program.
Molecular model of PAX5-OCT4-PRDM1 network
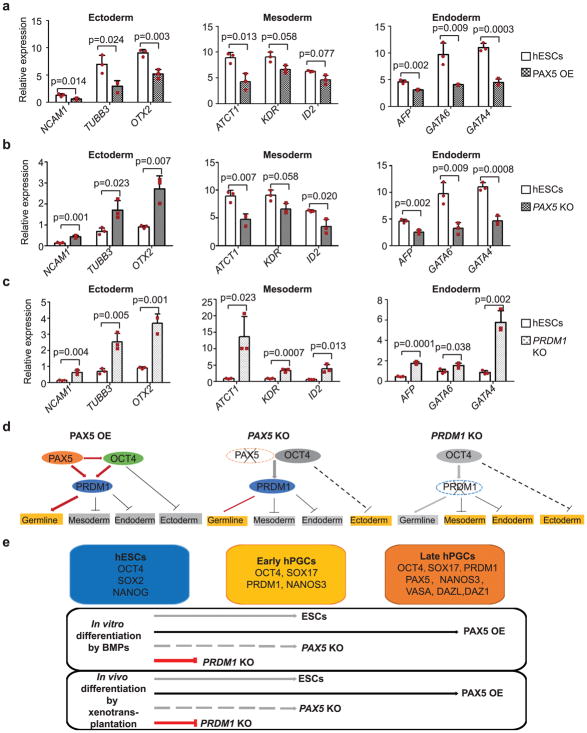
OCT4 is known to repress ectoderm formation from hESCs39 and during germ cell differentiation (Fig. 1e). In addition, PRDM1 has been shown to suppress endoderm and other somatic genes during gem cell specification19. Thus, we wondered whether PAX5, together with OCT4 and PRDM1, might function globally during germ cell differentiation. We examined expression of somatic genes during in vitro differentiation (Supplementary Table 1). We detected downregulation of somatic genes belonging to the three primary germ layers in PAX5 OE cells (Fig. 7a), while in PAX5 KO cells we observed a significant upregulation of expression of ectodermal genes (Fig. 7b). This suggests that PAX5 could repress ectoderm and that the repression might be mediated through the ability of PAX5 to activate/maintain OCT4 expression. Moreover, consistent with previous studies on PRDM119, we observed significant upregulation of somatic genes in all three germ layers during differentiation of PRDM1 KO cells (Fig. 7c), confirming the role of this gene in suppression of differentiation of somatic lineages during human germ cell development.
Figure 7. Role of PAX5 and PRDM1 in hPGC specification in vitro.
(a–c) RT-qPCR analysis of gene expression in all three germ layers in H1 hESCs, PAX5 OE cells (a) and H1 hESCs, PAX5 KO cells (b) and H1 hESCs, PRDM1 KO cells (c) after BMP induced differentiation. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (d) Proposed molecular model for transcriptional network centered by PAX5, OCT4 and PRDM1 in hPGCs. Upon induced germ cell differentiation with BMPs, OCT4 expression is reduced to moderate levels and maintained in partnership with PAX5. To efficiently induce germline programs, OCT4 represses ectodermal genes and at the same time, together with PAX5, activates PRDM1 to repress mesodermal and endodermal genes. In PAX5 KO cells, OCT4 expression has decreased to levels so low that the expression of ectodermal genes has not been suppressed effectively. Thus, the efficiency of induction of germ cells is low in PAX5 KO cells and lower in PRDM1 KO cells: due to low expression of OCT4 and loss of PRDM1 function, genes in all somatic lineages are upregulated and germ cell programs fail to be activated. (e) Summary of data establishing roles of PAX5 and PRDM1 in hPGC specification in vitro and in vivo. The identity of hESCs is maintained by core transcriptional network centered by OCT4, SOX2 and NANOG. Induced by BMP signals in vitro or in vivo by xenotransplantation, hESCs start to differentiate to early hPGCs, which express early germ cell markers, such as OCT4, SOX17, PRDM1 and NANOS3 (Grey line with arrowhead); Overexpression of PAX5 is able to enhance the efficiency to early hPGCs and promote early hPGCs to the later stage, which express mature germ cell markers, such as DDX4, DAZL and DAZ1(Black line with arrowhead). Loss of PAX5 significantly reduces germ cell potential of hESCs (Grey dotted line with arrowhead), while loss of PRDM1 leads to failure of hPGC specification (red line with an end bar). Source data for a–c are in Supplementary Table 2.
Based on our data, we propose a molecular model of human germ cell development (Fig. 7d). Upon external signaling via factors such as the BMPs, germ cell differentiation is induced under both in vitro or in vivo conditions and OCT4 expression is reduced to a moderate level that is at least in part, maintained by partnership with PAX5. To efficiently induce and maintain germline programs, OCT4 represses ectodermal genes and at the same time, together with PAX5, activates PRDM1 to repress mesodermal and endodermal genes. In PAX5 KO cells, OCT4 expression was so low that the expression of ectodermal genes was not suppressed effectively. Thus, the efficiency of induction of germ cells is very low in PAX5 KO cells. A more severe case is observed in PRDM1 KO cells with almost complete loss of expression of OCT4 and PRDM1, genes in all somatic lineages are upregulated and the germ cell programs fail to be activated.
Discussion
This work prompts a molecular model for germ cell development (Fig. 7e). In hESCs, OCT4 partners with pluripotent master regulators, including SOX2, to form the core transcriptional network that governs self-renewal and pluripotency. Induced by BMP signals in vitro and in vivo, hESCs differentiate into early hPGCs with low efficiency. When expression of PAX5 is induced in hESCs, differentiation into more mature human germ cells, with significantly higher efficiency, results. These later stage differentiated cells closely resemble late hPGCs or gonocytes and express genes, such as DDX4, c-KIT and DAZL, which mark mature human germ cells. Conversely, loss of function of PAX5 results in lower efficiency of germ cell differentiation and loss of function of PRDM1 results in complete failure of germ cell specification. Thus, this study provides evidence that human cell fate determination, at the juncture of pluripotency and somatic and germ line differentiation, may be the result of a balance of forces: Co-expression of pluripotency genes (eg., OCT4, NANOG) simultaneously with lineage specifiers (eg., SOX17, PAX5, PRDM1) distinguishes hPGCs from all other human cells and from mouse PGCs. To maintain cell identity, hPGCs require a precise regulation/balance of pluripotency and lineage specifiers to move forward from pluripotent stem cell while repressing somatic lineage development and activating germ cell programs. The PAX5-OCT4-PRDM1 axis that has been identified, along with the transcription factor genome-wide binding profiles, define the identity of bona fide hPGCs at stages beyond those commonly reported in vitro. The results of these studies may shed light on genetic requirements for human germ cell differentiation, enable more faithful and efficient production of human germ cells in vitro and contribute to knowledge and models of human germ cell pathologies.
Methods
hESC culture and differentiation
H1 hESCs (WiCell) were maintained feeder-free on Matrigel (BD Biosciences)-coated plates as previously described 1–5. All cultures were grown at 37 °C with 5% CO2 in mTeSR1 medium (STEMCELL Technologies). To differentiate, cells were seeded overnight at a density of 200,000 per well in 6 well plate and medium was replaced the next day with differentiation media (knockout DMEM supplemented with 20% fetal bovine serum, 1 mM l-glutamine, 0.1 mM nonessential amino acids, 0.1 mM β-mercaptoethanol and 50 ng ml−1 recombinant human BMP4, BMP7 and BMP8b (R&D systems) as previously described5,34, 40. Differentiation medium was changed every other day for 7 days.
Xenotransplantation assay
Human cell lines were transplanted into the testes of busulfan-treated, immune-deficient nude mice (NCr nu/nu; Taconic) as previously described1–4,41. Immune-deficient nude mice were treated with a single dose of busulfan (40 mg/kg; Sigma-Aldrich) at 5–6 weeks of age to eliminate endogenous spermatogenesis. Xenotransplantation was then performed 5–6 weeks after busulfan treatment by injecting 7–8 ul cell suspensions (1.5–3million cells/testis) containing 10% trypan blue (Invitrogen) into the seminiferous tubules of each recipient testes via cannulation of the efferent ducts. At 8 weeks after transplantation, recipient mouse testes were harvested for histology and immunohistochemical analyses. Procedures were evaluated and approved by the University of Pittsburgh and Montana State University Institutional Animal Care and Use Committee (IACUC); all procedures were compliant with all relevant ethical regulations regarding animal research.
ChIP on human fetal testis
Second trimester human fetal testes were staged and procured from Advanced Bioscience Resources (ABR Inc, CA). The chromatin immunoprecipitation (ChIP) protocol for human fetal testis (22 weeks) was adapted from published protocols25, 26 and optimized as follows. Note that the protocol for tissue procurement and use was approved by the Institutional Review Board of Montana State University (RR-P031014-EX); all procedures were compliant with all relevant ethical regulations regarding human research with unidentifiable banked tissue. The consent was obtained from all the participants. Human fetal testis samples (~25mg for each ChIP) were placed in a 100mm dish on ice and finely minced using a clean scalpel. Minced tissue was then transferred into a 15ml conical tube. 1ml PBS with protease inhibitor cocktail (PIC) (Roche) was added to the tissue. To crosslink, tissue was fixed in 1.5% formaldehyde and rocked at room temperature for 20 min, quenched with 1 vol 250 mM glycine (room temperature, 5 min), and rinsed with chilled TBSE buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA) twice. Crosslinked tissue was resuspended in PBS with PIC (1ml 0.1%SDS buffer for every 100ul nuclear pellet) and transferred to a Dounce homogenizer. Tissue pieces were then disaggregated into single cell suspension with 20–25 strokes. Cell suspension was transferred to a 15ml conical tube and centrifuged at 3,000 rpm in a bench top centrifuge for 5 min at 4°C. Cells were lysed with 1% SDS Lysis Buffer (on ice, 5 min) and then centrifuged (2,000 rpm, 10 min). Supernatant was removed as described above and samples were resuspended in 0.1% SDS buffer with PIC. Samples were then sonicated with glass beads for 12 times (30 s pulses with 30 s break interval) using the Bioruptor water bath sonicator (Diagenode). Chromatin extracts were then precleared with Dynal Magnetic Beads (Invitrogen) (4°C, 1 hr) followed by centrifugation (2,000 rpm, 30 min). Supernatant (precleared chromatin) was immunoprecipitated overnight with Dynal Magnetic Beads coupled with anti-OCT4 (sc-8628, Santa Cruz Biotechnology), anti-PAX5 (sc-1974, Santa Cruz Biotechnology), anti-PRDM1 (C14A4, Cell Signaling Technology) antibodies. On the next day, Beads were washed by 0.1% SDS buffer for three times followed by a wash with TE. Chromatin was eluted by incubating beads in TE supplemented with 1 % SDS at 65 °C for 30 min. Afterwards, chromatin underwent reverse crosslinking with pronase at 42°C for 2 hrs and 67°C for 6 hrs and DNA was purified using phenol-chloroform extraction. ChIP DNA was subjected to 15 cycles of amplification with the SeqPlex DNA Amplification (Sigma). Real-time qPCR was performed (ABI PRISM 7900 Sequence Detection System) and relative occupancy values were determined by the immunoprecipitation efficiency (amount of immunoprecipitated DNA relative to input). Illumina HiSeq 2 × 60 bp paired end reads were used for sequencing. [Isn’t this redundant with the statement above?]
Immunofluorescence analysis for cell culture
Cells were fixed with 4% paraformaldehyde at room temperature for 20 minutes, permeabilized with 0.3% Triton X-100 in PBS (PBST) for 10 minutes, and blocked for 45 minutes at room temperature in PBST containing 5% normal donkey serum (Jackson ImmunoResearch Laboratories). Primary antibodies were diluted at 1:200 in blocking solution and incubated at 4 °C overnight. The primary antibodies used in this study are: DDX4 (R&D #AF2030), OCT4 (Santa Cruz; sc-8628), PAX5 (Santa Cruz; sc-1974), PRDM1 (Cell Signaling #9115) and C-KIT (DAKO; A4502). Appropriate Alexa Fluor 488, 594 or 647-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were diluted at 1: 300 in PBS containing 0.1% bovine serum albumin (BSA) and incubated at room temperature for 1 hour. One μg/mL DAPI was used for nuclear staining. Images shown are representative of at least three independent experiments.
RNA isolation, library preparation and sequencing analysis
Total RNA was extracted using Acruturus PicoPure RNA Isolation Kit (Life Technologies). RNA quality was determined with Bioanalyzer 2100 (Agilent). Sequencing libraries were constructed by SMARTer universal low input RNA Kit (Clonetech) according to the manufacturer’s instructions. DNA library samples were submitted to the Stanford Genomics Facility and 100-base paired-end high throughput sequencing was performed. All sequenced libraries were mapped to the human genome using TopHat and Cufflink 42, 43 with default parameter setup. Differential expression was analyzed using StrandNGS (AvadisNGS).
Gene expression analysis by qPCR
Total RNA for qPCR was extracted from cells using the RNeasy Plus Mini Kit (Qiagen) and 1ug RNA was reverse transcribed using the SuperScript® III First-Strand Synthesis System. Quantitative PCR was done in triplicate using the Power SYBR® Green PCR Master Mix (both from Life Technologies) with the data normalized to housekeeping genes. The primer sequences were listed in Supplementary Table 1. Data shown are representative of at least three independent experiments.
Immunohistochemistry of recipient mouse testes
Paraformaldehyde (PFA) fixed mouse testes were sectioned by AML laboratories (Baltimore, MD) by paraffin embedding and conducting serial cross-sectioning every 5 mm. Deparafinization was conducted by two consecutive 10min. xylenes (Sigma-Aldrich) treatments followed by rehydration in 100%, 100%, 90%, 80% and 70% ethanol treatments followed by a 10-minute wash in tap water. Antigen retrieval was conducted by boiling slides for 30 minutes in 0.01 M Sodium Citrate (pH 6.0; Sigma-Aldrich), cooling slides for 30 minutes, followed by a 10 minute wash in PBS. Blocking and permeabilization was conducted by addition of 10% normal Donkey serum (Jackson Immunoreearch) with 0.1% Triton X (Sigma-Aldrich) in PBS for 1 hour, followed by incubation with the following primary antibodies diluted in 1% blocking solution overnight at 4°C in a humidified chamber. Antibodies used were: DDX4 (R&D #AF2030), GFP (Abcam; ab13970), OCT4 (Santa Cruz; sc-8628) and DAZL (Novus; NB100-2437). Slides were washed with PBST, followed by an hour incubation with fluorescently labeled secondary antibodies raised in Donkey, followed by additional washes with PBST. The primary antibodies were used at 1:200 and secondary antibodies were used at 1:300. All samples were mounted with ProLong Gold Anti-fade mounting media containing DAPI (Life Technologies). Samples were then imaged using a confocal microscope (Zeiss). Quantification of GFP/DDX4 double positive staining was determined manually from multiple sections taken from 2–3 different depths within the testis (technical replicates) and from multiple clonal replicates for each transplanted cell line (biological replicates).
Motif analysis
Motif analysis was performed using MEME-ChIP suite with default parameters. The novel motif for OCT4 in hPGCs were searched in TRANSFAC and JASPER databases to find transcription factors with similar consensus sequences 44,45.
CRISPR design and PAX5-KO derivation
CRISPR guide RNAs (gRNAs) were designed using the online CRISPR design tool from the Massachusetts Institute of Technology (http://crispr.mit.edu/). Candidate gRNAs with the highest score were chosen for each genomic region. Oligonucleotides for these gRNAs were synthesized and cloned into plasmid pX459 (Addgene 48139) carrying both Cas9 and gRNA expression cassettes, with one modification of the original plasmid in which the Cas9 promoter was replaced by the EEF1A1 promoter. The cutting efficiency of each gRNA construct was validated by transfecting HEK293T cells and sequencing the target regions in the genome. CRISPR pairs were nucleofected into H1 hESCs and plated as single cells. Single cells were clonally expanded and isolated for PCR to test for successful PAX5 deletion and sequencing.
Construction of overexpression constructs by lentiviral vectors
The sequences of PAX5 or PRDM1 were assembled with Gibson Assembly Cloning technology (NEB). Briefly, gBlocks (gene fragments) were synthesized by Integrated DNA Technologies (IDT), and individual gBlocks were assembled to one gene transcript with Gibson Assembly technology followed by an amplification reaction with Phusion DNA polymerase according to the manufacturer’s instructions. Amplified genes were ligated into the pENTR/D-TOPO vector (Life Technologies) for the Multisite Gateway system. Clones were transformed into One-Shot Competent Escherichia coli, DNA was purified and sequenced, and positive clones were used for a recombination reaction with the Gateway destination vector (pcDNA-DEST40). Subsequent transformation into One-Shot Competent E. coli, followed by DNA purification and sequencing for verification of correct cloning, resulted in overexpression vectors for PAX5 and PRDM1.
Luciferase assay for enhancer activity
Enhancer sequences were generated by PCR of human genomic DNA discovered for gene POU5F1 and PRDM1 and then cloned into pGL4.28(luc2CP/minP/Hygro) (#E8461, Promega) with restricted enzymes KpnI and BmtI (NEB). The minimal promoter pGL4.28 is used as negative control. Luciferase activity was measured using dual-luciferase reporter assay system (Promega) as described in the manufacturer’s Manu. Gene Ontology analysis. Gene Ontology (GO) enrichment was done using GREAT analysis, with default parameters 46.
FACS and flow cytometry
Cells were dissociated in 0.25% trypsin--EDTA (Gibco BRL) at 37°C for 5 min and collected by centrifugation at 200g in an Eppendorf 5702 R centrifuge. Then the cells were passed through the 70uM strainers (BD Biosciences) to make sure they were digested as single cells before they were subject to the flow cytometry. Mouse testes cells were dissociated with 0.25% Trypsin–EDTA for 30 minutes at 37 °C. Dissociated cells were incubated in 1% BSA in PBS containing primary antibodies (CKIT (A4502; DAKO)) on ice for 20 minutes. Cells were then analyzed for mOrange expression or CKIT/GFP using the BD FACSAriaII cell sorter. Analysis was performed using LSRII (Becton Dickinson) and FlowJo software (Tree Star).
GST pulldown assay
The recombinant protein, OCT4 (Novus; H00005460-P01) was bound to glutathione-sepharose beads (Amersham) and incubated with recombinant PAX5 (Novus; H00005079-P01) protein overnight at 4 °C. Beads were washed 6 times with cell lysis buffer. The eluents were analyzed by Western blot. The blots shown are representative of at least three independent experiments.
Differentiation to neuronal progenitor cells from hESCs
Differentiation of hESCs to neuronal progenitor cells was carried out as previously described 47, 48. hESCs were dissociated into single cells with accutase, depleted of MEF feeders by incubating on gelatin-treated culture dish for 30 minutes, and then plated onto matrigel-coated dish at the density of 36000 cells/cm2 in mTeSR1 (Stemcell Technologies) in the presence of 2 uM thiazovivin (Santa Cruz Biotechnology). Differentiation was started 48 hours later with KSR differentiation medium supplemented with different combinations of small molecules6. Diluted fibronectin stock solution (with cold (4 °C) PBS to 2 μg/ml), was added to 1 ml to 1 well of 6-well plates, and plates were incubated at 37 °C overnight. After 20 days of differentiation, cells were replated to the poly-L-ornithine/laminin/fibronectin plates and cultured with B27 differentiation medium.
Statistics and reproducibility
No statistical methods were used to predetermine samples or outcomes. For the xenotransplantation studies, animals were randomly allocated into groups receiving various cell line injections. All statistical analyses were conducted using GraphPad Prism (version 5). Two tailed Student’s t-test were used when data met criteria for parametric analysis (normal distribution or equal variances). P-value is shown in the figures and the number of biological replicates for each experiment is indicated in the figure legends. Experiments were repeated independently at least three times.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data Availability
All sequencing data that support the findings of this study have been deposited in NCBI’s GEO under accession number GSE100639. Previously published sequencing data that were re-analyzed here are available under accession codes GSE60138, GSE39821 and GSE67259. Source data for Fig. 3g, h; 4a, d, e, g, h; 5d, e, g, h, I; 6a-c and Supplementary Fig. 2b; 3a, d, f; 4c-d; 6a, b; 7a, c, d, e, g, h is provided in Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgments
This work was supported by P50 HD 068158 to RRP (Project I). The authors declare that they have no conflicts of interest.
Footnotes
Author contributions
The study was conceived and designed by F.F. and R.R.P.; F.F. performed most experiments (including ChIP-seq, immunohistochemistry, RNA-seq, protein pull-down, luciferase reporter assay, gene expression profiling) and analyzed the data. X.N. performed bioinformatic analyses for ChIP-seq and luciferase reporter assay, flow cytometry and gene expression analysis for the xenotransplantation. B.A generated the PAX5 knockout hESC lines and performed part of the immunohistochemistry in xenotransplantation samples. Z.W. performed the initial bioinformatics analysis for ChIP-seq data. M.S. and K.E.O. conducted the xenotransplantation. C.C.C and A. M performed RNA-seq analysis. J.C. constructed the H1-DDX4 reporter. R.W. and B.W. designed and constructed the PAX5 knockout plasmids. A.M.S. and N.I. provided the PRDM1 knockout hESC line and protocol. The manuscript was written by F.F., and R.R.P. with input from the other authors.
References
- 1.Ramathal C, et al. DDX3Y gene rescue of a Y chromosome AZFa deletion restores germ cell formation and transcriptional programs. Sci Rep. 2015;5:15041. doi: 10.1038/srep15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez A, Chiang H, Sukhwani M, Orwig K, Reijo Pera RA. Human germ cell formation in xenotransplants of induced pluripotent stem cells carrying X chromosome aneuploidies. Sci Rep. 2014;4:6432. doi: 10.1038/srep06432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramathal C, et al. Fate of iPSCs derived from azoospermic and fertile men following xenotransplantation to seminiferous tubules. Cell Rep. 2014;7:1284–1297. doi: 10.1016/j.celrep.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durruthy JD, et al. Fate of induced pluripotent stem cells following transplantation to murine seminiferous tubules. Hum Mol Genet. 2014;23:3071–3084. doi: 10.1093/hmg/ddu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kee K, Angeles V, Flores M, Nguyen H, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salto Mamsen L, Lutterodt MC, Andersen EW, Byskov AG, Yding Andersen C. Germ cell numbers in human embryonic and fetal gonads during the first two trimesters of pregnancy: analysis of six published studies. Hum Reprod. 2011;26:2140–2145. doi: 10.1093/humrep/der149. [DOI] [PubMed] [Google Scholar]
- 7.Clark AT, et al. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 8.Reijo Pera RA, Alagappan RK, Patrizio P, Page DC. Severe oligospermia resulting from deletions of the Azoospermia Factor gene on the Y chromosome. Lancet. 1996;347:1290–1293. doi: 10.1016/s0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 9.Irie N, Tang WW, Azim Surani M. Germ cell specification and pluripotency in mammals: a perspective from early embryogenesis. Reprod Med Biol. 2014;13:203–215. doi: 10.1007/s12522-014-0184-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnusdottir E, et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nature Cell Biol. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nature Rev Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 12.Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 13.Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol. 2012;4(11):a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakaki F, et al. Induction of mouse germ-cell fate by transcription factors in vitro. Nature. 2013;501:222–226. doi: 10.1038/nature12417. [DOI] [PubMed] [Google Scholar]
- 15.Ohinata Y, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- 16.Weber S, et al. Critical function of AP-2 gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod. 2010;82:214–223. doi: 10.1095/biolreprod.109.078717. [DOI] [PubMed] [Google Scholar]
- 17.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nature Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, et al. Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell. 2016;18:330–340. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Irie N, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang WW, et al. A Unique gene regulatory network resets the human germline epigenome for development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:858–873. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Kehler J, et al. Oct4 is required for primordial germ cell survival. EMBO Rep. 2004;5:1078–1083. doi: 10.1038/sj.embor.7400279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 24.Scholer HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nature Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- 26.Cotney JL, Noonan JP. Chromatin immunoprecipitation with fixed animal tissues and preparation for high-throughput sequencing. Cold Spring Harb Protoc. 2015;2015:191–199. doi: 10.1101/pdb.prot084848. [DOI] [PubMed] [Google Scholar]
- 27.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pardo M, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gkountela S, et al. The ontogeny of cKIT(+) human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nature Cell Biol. 2012;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent SD, et al. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- 33.Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 34.Kee K, Reijo Pera RA. Human germ cell lineage differentiation from embryonic stem cells. Cold Spring Harb Protoc. 2008 doi: 10.1101/pdb.prot5048. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Xia N, Reijo Pera RA. Directed dopaminergic neuron differentiation from human pluripotent stem cells. J Vis Exp. 2014:51737. doi: 10.3791/51737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrier AL, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeom YI, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 38.Sasaki K, et al. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Oron E, Nelson B, Razis S, Ivanova N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell. 2012;10:440–454. doi: 10.1016/j.stem.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 40.Kee K, Gonsalves JM, Clark AT, Reijo Pera RA. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006;15:831–837. doi: 10.1089/scd.2006.15.831. [DOI] [PubMed] [Google Scholar]
- 41.Hermann BP, et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandelin A, et al. Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics. 2004;5:99. doi: 10.1186/1471-2164-5-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang P, Xia N, Reijo Pera RA. Directed dopaminergic neuron differentiation from human pluripotent stem cells. J Vis Exp. 2014:51737. doi: 10.3791/51737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perrier AL, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci U S A. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data that support the findings of this study have been deposited in NCBI’s GEO under accession number GSE100639. Previously published sequencing data that were re-analyzed here are available under accession codes GSE60138, GSE39821 and GSE67259. Source data for Fig. 3g, h; 4a, d, e, g, h; 5d, e, g, h, I; 6a-c and Supplementary Fig. 2b; 3a, d, f; 4c-d; 6a, b; 7a, c, d, e, g, h is provided in Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.