Abstract
Abstract
Meroterpenoids are hybrid natural products that partially originate from the terpenoid pathway. Ganoderma meroterpenoids (GMs) are a type of meroterpenoids containing a 1,2,4-trisubstituted phenyl and a polyunsaturated terpenoid part. Over last 5 years, great efforts have been made to conduct phytochemistry research on the genus Ganoderma, which have led to the isolation and identification of a number of GMs. These newly reported GMs showed diverse structures and a wide range of biological activities. This review gives an overview of new GMs from genus Ganoderma and their biological activities and biosynthetic pathway, focusing on the period from 2013 until 2018.
Graphical Abstract
Keywords: Ganoderma, Ganoderma meroterpenoids, New structures, Biological activities
Introduction
Ganoderma is a ganodermataceae (basidiomycete) white rot fungus, normally growing on woody plants and wood logs [1], and is used for medicinal purposes in China, Japan, and South Korea (Chinese Higher Fungi: 18 volumes). It was first recorded in the Shennong’s Classic of Meteria Medica, and classified as an upper-grade medicine in medical books [2]. About 78 species of Ganoderma are recorded in Chinese Higher Fungi, of which, G. lucidum and G. sinense, were found to be edible and medicinally-beneficial fungi, and were registered in Chinese Pharmacopoeia (2010 and 2015 edition). However, other species, such as G. capense, G. cochlear, and G. tsuage, also play an important part in traditional folk medicines. In addition, pharmacological studies have also involved the extract and chemical constituents of other species [3–5]. Until now, the chemical constituents and biological activities of 22 species of Ganoderma have been studied.
Ganoderma is rich in novel “mycochemicals”, including polysaccharide, triterpenoids, steroids, fatty acids, etc. Although polysaccharide is found to be one of the main bioactive constituents, its high molecular weight and complex structure limits its use in the drug market. Meanwhile, the small molecular constituents have played a significant role over the last 200 years in treating and preventing diseases, and are continuing to serve as important leads in modern drug discovery [6–11].
Since the discovery of ganomycins A and B [12], more than 100 aromatic meroterpenoids, derived by a hybrid of shikimic acid and mevalonic acid biogenetical pathway, were isolated from the genus Ganoderma (Ganodermataceae) [13]. Ganoderma meroterpenoids (GMs) have attracted increasing attention because they showed diverse structural skeletons and series of bioactivities, such as NO production inhibitory [14], anti-oxidant [15, 16], anti-allergic [17, 18], anti-fibrotic [19], anti-Acetyl cholinesterase (AChE) [20], cytotoxic [21], antimicrobial [12], and aldose reductase inhibitory activities [22]. As a result, chemists have synthesized polycyclic meroterpenoids by employing many steps [23–26].
Herein, we review the structure, bioactivities, and biosynthesis pathways of GMs from Ganoderma species to lay the foundation for the further research and provide the important sources for the development of lead compounds.
Biosynthetic Pathway of GMs
The prenylation of aromatic compounds plays an important role in the natural product research because it not only gives rise to an astounding diversity of small molecular constituents in plants, fungi and bacteria, but also enhances the bioactivities and bioavailabilities of these compounds [27]. Aromatic prenyltransferase is the key enzyme for the prenylation of aromatic compounds. Meroterpenoids including ubiquinone, plastoquinone, menadione, vitamin E, prenylflavonoids, shikonin and prenylated alkaloids, are formed under prenyltransferase [28]. The analysis of the genome showed that abundant carbohydrate-active enzymes and ligninolytic enzymes were present in the G. lucidum genome [29]. All the meroterpenoids from Ganoderma consist of a 1,2,4-trisubstituted phenyl group and a polyunstaturated terpenoid parts, suggesting that lignin was degraded to phenyl group by the liginolytic enzymes of Ganoderma, and the terpenoid parts were further assembled under prenyltransferase.
Chemical Structures and Bioactivities of GMs
A class of GMs, which had a 1,2,4-trisubtituted phenyl group connecting with C10 or C15 polyunsaturated side chain or polycyclic substructure, widely distributed in genus Ganoderma. According to the difference in their terpenoid parts, these GMs can be divided into three types.
Chain-Contained GMs
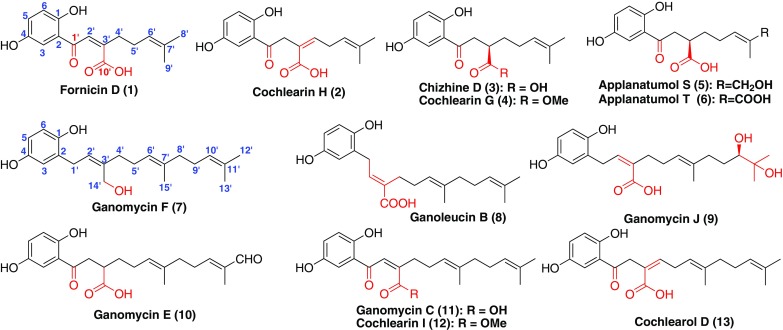
Due to the presence of double bonds in terpenoid part, the redox reaction can take place in allylic position (Fig. 1, Table 1). Thus, compounds 1–6, and 9–13 had a ketone carbonyl at C-1′ and a carboxyl or methyl ester at C-10′ or C-14′ [15, 16, 30–35]. Among them, compounds 2 and 13 existed positional isomerization of olefinic bond because of the shift of the double bond at C-2′ and C-3′ [30, 35], whereas, the reduction of the ∆2′,3′ in chizhine D (3), cochlearin G (4), applanatumols S, T (5, 6) and ganomycin E (9) was occurred [30, 31, 34]. The C-14′ of ganomycin F (7) was connected to a hydroxyl group [16]. The of ganoleucin B (8) was isomerized to cis under conditions of enzyme or light [33]. The of ganomycin J (9) was oxidized to two hydroxyls. Fornicin D (1), cochlearins H, G, I (2, 4, 12) and ganomycin C (11) isolated from Ganoderma cochlear, as well as ganomycins F and E (7 and 10) gained from G. capense, showed significant anti-oxidant activities [15, 16, 30]. Compound 3 was isolated from G. lucidum and displayed weak renoprotective effect [31]. The biological assay of applanatumols S and T (5, 6) from G. applanatum [32], and ganoleucin B (8) from G. leucocontextum didn’t show inhibitory activities against COX-1, COX-2, HMG-CoA reductase and α-glucosidase, respectively [33]. However, ganomycin J (9) from G. lucidum showed strong inhibitory activity against HMG-CoA reductase with an IC50 value of 30.3 μM [34].
Fig. 1.
Structures of GMs with a 10-carbon or 15-carbon chain
Table 1.
Name, source and their bioactivities of chain-containing GMs
| Number | Name | Bioactivity | Source | Reference |
|---|---|---|---|---|
| 1 | Fornicin D | Antioxidant activity | G. cochlear | [15] |
| 2 | Cochlearin H | Antioxidant activity | G. cochlear | [30] |
| 3 | Chizhine D | Renoprotective effect | G. lucidum | [31] |
| 4 | Cochlearin G | Antioxidant activity | G. cochlear | [30] |
| 5 | Applanatumol S | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 6 | Applanatumol T | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 7 | Ganomycin F | Antioxidant activity | G. capense | [15] |
| 8 | Ganoleucin B | Inhibitory activities against HMG-CoA reductase and α-glucosidase | G. leucocontextum | [33] |
| 9 | Ganomycin J | Inhibitory activity against HMGs reductase (IC50: 30 μM), aldose reductase and α-glucosidase | G. lucidum | [34] |
| 10 | Ganomycin E | DPPH radical scavenging activity | G. capense | [16] |
| 11 | Ganomycin C | Antioxidant activity | G. cochlear | [15] |
| 12 | Cochlearin I | DPPH radical scavenging | G. cochlear | [30] |
| 13 | Cochlearol D | G. cochlear | [35] | |
| 14 | (+)-Applanatumol U | Inhibitory activity against COX-1 and COX-2 | G. applanatum | [32] |
| 15 | (+)-Chizhine E | Renoprotective effects | G. lucidum | [31] |
| 16 | (+)-Lucidulactone B | G. lucidum | [36] | |
| 17 | (+)-Zizhine A | Renoprotective effects | G. sinense | [37] |
| 18 | (+)-Ganoleucin C | Inhibition against HMG-CoA reductase and a-glucosidase | G. leucocontextum | [33] |
| 19 | (+)-Chizhine F | Renoprotective effects | G. lucidum | [31] |
| 20 | (+)-Zizhine B | Renoprotective effects | G. sinense | [37] |
| 21 | (+)-Zizhine C | Renoprotective effects | G. sinense | [37] |
| 22 | (+)-Zizhine D | Renoprotective effects | G. sinense | [37] |
| 23 | (+)-Zizhine E | Renoprotective effects | G. sinense | [37] |
| 24 | (+)-Zizhine F | Renoprotective effects | G. sinense | [37] |
| 25 | (+)-Fornicin E | Renoprotective effects | G. cochlear | [16] |
| 26 | Chizhine A | Renoprotective effects | G. lucidum | [31] |
| 27 | Chizhine B | Renoprotective effects | G. lucidum | [31] |
| 28 | Chizhine C | Renoprotective effects | G. lucidum | [31] |
| 29 | (+)-Cochlearin B | Antioxidant activity | G. cochlear | [30] |
| 30 | (±)-Cochlearin D | Antioxidant activity | G. cochlear | [30] |
| 31 | (+)-Lingzhine E | Neural stem cell proliferation | G. lucidum | [38] |
| 32 | (+)-Applanatumol P | Inhibitory activity against COX-1 and COX-2 | G. applanatum | [32] |
| 33 | (+)-Applanatumol Q | Inhibitory activity against COX-1 and COX-2 | G. applanatum | [32] |
| 34 | (+)-Applanatumol R | Inhibitory activity against COX-1 and COX-2 | G. applanatum | [32] |
| 35 | (±)-Ganocapensin A | Inhibitory activity against COX-1 and COX-2 | G. capense | [16] |
| 36 | Ganocapensin B | Antioxidant activity | G. capense | [16] |
| 37 | (±)-Cochlearin E | Antioxidant activity | G. cochlear | [30] |
| 38 | Cochelarin F | Antioxidant activity | G. cochlear | [30] |
| 39 | Applanatumol Z1 | Inhibitory activity against COX-1 and COX-2 | G. applanatum | [32] |
| 40 | Cochlearol C | G. cochlear | [33] |
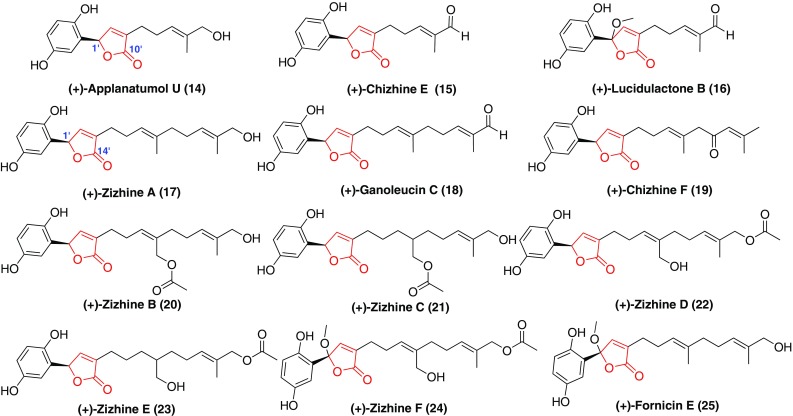
An α,β-unsaturated γ-lactone fraction can be formed through a nucleophilic reaction from the carboxyl at C-10′ or C-14′ to the ketone carbonyl at C-1′ (Fig. 2, Table 1). Cao et al [37] investigated the fruiting bodies of G. sinense and a series of GMs with an α,β-unsaturated γ-lactone fraction, namely (+)-zizhines A–F (17, 20–24), were isolated. All the compounds were evaluated for their inhibition on extracellular matrix component (fibronectin) generation by using TGF-β1 induced rat kidney tubular epithelial cells. However, all of them didn’t show any inhibitory activities. (±)-Chizhine E and F (15, 19) and (±)-lucidulactone (16) were isolated from G. lucidum and the individual enantiomers of compounds 15 and 19 significantly inhibit monocyte chemotactic protein 1 (MCP-1) and fibronectin production in a dose-dependent manner [31, 36]. Fornicin E (25) obtained from G. capense also was a pair of enantiomers, which showed stronger DPPH scavenging activity than vitamin E (positive control) [16]. (±)-Applanatumol U (14) was identified from G. applanatum and showed no inhibition against COX-1 and COX-2 [32].
Fig. 2.
Structures of GMs with a γ-lactone
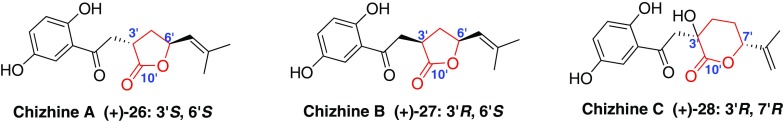
Three pairs of enatiomers (Fig. 3, Table 1), (±)-chizhines A–C (26–28) possessing a (6′ → 10′)-γ-lactone ring and a (7′ → 10′)-δ-lactone ring, respectively, were isolated from the fruiting bodies of G. lucidum. These compounds showed weak renoprotective effects [31].
Fig. 3.
Structures of GMs with a (6′ → 10′)- or (7′ → 10′)-lactone
With the help of oxidases, the ether ring was present in many GMs (Fig. 4, Table 1). For example, compounds 29–35 had different ether ring in the terpenoid part, whereas, the ether rings in compounds 36–40 were formed through a cyclization between the hydroxyl at C-1 and the hydroxyls of the terpenoid part. Compounds 29, 30, and 35–38 displayed significant antioxidant activities in the DPPH scavenging assay [16, 30]. Among them, (±)-cochlearin D (30) and (+)-30 exhibited weak inhibitory effects for the proliferation of hepatic stellate cells (HSCs) induced by transforming growth factor-β1 (TGF-β1) [30]. Except for above compounds, the rest of compounds didn’t show renoprotective activities [32, 33].
Fig. 4.
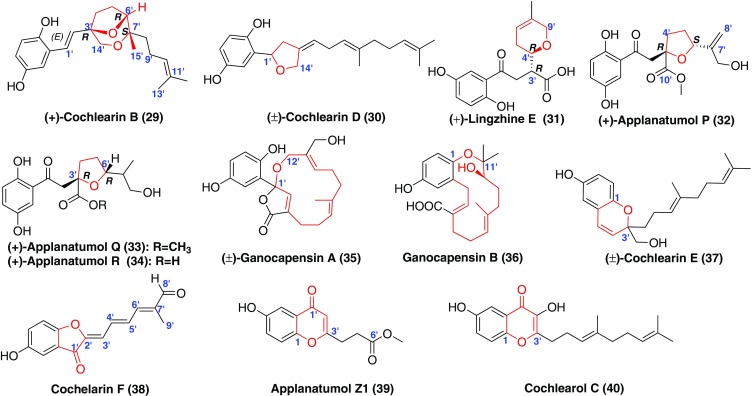
Structures of GMs with an ether ring
Polycyclic GMs
Because of the presence of polyunsaturated terpenoid part, free radical reaction can be occurred in GMs under the conditions of enzyme and light, which led to the formation of polycyclic structures (Table 2).
Table 2.
Name, source and bioactivities of polycyclic GMs
| Number | Name | Bioactivity | Source | References |
|---|---|---|---|---|
| 41 | Applanatumol V | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 42 | Applanatumol W | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 43 | Applanatumol X | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 44 | Applanatumol Y | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 45 | Applanatumol Z | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 46 | Applanatumol Z2 | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 47 | Applanatumol K | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 48 | Applanatumol L | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 49 | Applanatumol M | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 50 | Applanatumol N | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 51 | Applanatumol O | Inhibitory activities against COX-1 and COX-2 | G. applanatum | [32] |
| 52 | Chizhiol A | Inhibitory activities against COX-1 and COX-2 | G. lucidum | [39] |
| 53 | Ganotheaecoloid L | Inhibitory activities against COX-1 and COX-2 | G. theaecolum | [40] |
| 54 | (+)-Ganotheaecoloid M | Inhibitory activities against COX-1 and COX-2 | G. theaecolum | [40] |
| 55 | (−)-Ganotheaecoloid N | Inhibitory activities against COX-1 and COX-2 | G. theaecoloum | [40] |
| 56 | Petchiene A | Inhibitory activities against COX-1 and COX-2 | G. petchii | [41] |
| 57 | Lingzhine C | Promote proliferation of neural stem cells (NSCs) | G. lucidum | [38] |
| 58 | (±)-Lingzhine B | Inhibit NSC proliferation | G. lucidum | [38] |
| 59 | (−)-Ganotheaecoloid A | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 60 | (−)-Ganotheaecoloid B | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 61 | Ganotheaecoloid C | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 62 | Ganotheaecoloid D | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 63 | Ganotheaecoloid E | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 64 | (−)-Ganotheaecoloid F | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 65 | Ganotheaecoloid G | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 66 | Ganotheaecoloid H | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 67 | Ganotheaecoloid I | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 68 | (+)-Ganotheaecoloid J | COX-2 inhibitory activity (IC50: 9.96 μM) | G. theaecolum | [40] |
| 69 | Ganotheaecoloid K | Inhibitory activities against COX-2 | G. theaecolum | [40] |
| 70 | (+)-Cochlearin A | Antioxidant activity | G. cochlear | [30] |
| 71 | Spiroapplanatumine A | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 72 | Spiroapplanatumine C | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 73 | Spiroapplanatumine E | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 74 | Spiroapplanatumine G | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 75 | Spiroapplanatumine I | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 76 | Spiroapplanatumine B | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 77 | Spiroapplanatumine D | Inhibitory activities against JAK3 kinase (IC50: 7.0 ± 3.2 μM) | G. applanatum | [42] |
| 78 | Spiroapplanatumine F | Inhibitory activities against JAK3 kinase (IC50: 34.8 ± 21.1 μM) | G. applanatum | [42] |
| 79 | Spiroapplanatumine H | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 80 | Spiroapplanatumine J | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 81 | Spiroapplanatumine K | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 82 | Spiroapplanatumine L | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 83 | Spiroapplanatumine M | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 84 | (+)-Spiroapplanatumine N | Inhibitory activity against JAK3 kinase | G. applanatum | [42] |
| 85 | Spiroapplanatumine O | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 86 | (−)-Spiroapplanatumine N | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 87 | Spiroapplanatumine P | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 88 | Spiroapplanatumine Q | Inhibitory activities against JAK3 kinase | G. applanatum | [42] |
| 89 | (+)- Spirolingzhine A | Protective effects for NSC | G. lucidum | [38] |
| 90 | (+)-Spirolingzhine B | Protective effects for NSC | G. lucidum | [38] |
| 91 | (+)-Spirolingzhine C | Protective effects for NSC | G. lucidum | [38] |
| 92 | Spirolingzhine D | Protective effects for NSC | G. lucidum | [38] |
| 93 | (±)-Ganoderin A | Antioxidant activity | G. cochlear | [15] |
| 94 | Applanatumol H | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 95 | Applanatumol I | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 96 | Applanatumol J | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 97 | Applanatumol D | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 98 | Applanatumol E | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 99 | Applanatumol J | Inhibitory activities against COX-1, COX-2 | G. applanatum | [32] |
| 100 | Applanatumol F | Inhibitory activities against J COX-1, COX-2 | G. applanatum | [32] |
| 101 | Lingzhilactone A | Renoprotective effect | G. lucidum | [43] |
| 102 | Lingzhilactone B | Renoprotective effect | G. lucidum | [43] |
| 103 | Lingzhilactone C | Renoprotective effect | G. lucidum | [43] |
| 104 | Applanatumol Z3 | Inhibitory activities against JAK3 kinase | G. applanatum | [32] |
| 105 | Applanatumol Z4 | Inhibitory activities against JAK3 and DDR1 kinases | G. applanatum | [32] |
| 106 | Applanatumol C | Inhibitory activities against JAK3 and DDR1 kinases | G. applanatum | [32] |
| 107 | (−)-Lingzhiol | Renoprotective effect | G. lucidum | [44] |
| 108 | (±)-Ganocochlearin A | Antioxidant activity | G. cochlear | [15] |
| 109 | (±)-Ganocochlearin B | Antioxidant activity | G. cochlear | [15] |
| 110 | (±)-Ganocochlearin C | Antioxidant activity | G. cochlear | [15] |
| 111 | (±)-Ganocochlearin D | Antioxidant activity | G. cochlear | [15] |
| 112 | Lingzhine D | Anti-BuChE activity | G. lucidum | [38] |
| 113 | (±)-Ganocin A | Anti-BuChE activity | G. cochlear | [45] |
| 114 | (±)-Ganocin B | Anti-BuChE activity | G. cochlear | [45] |
| 115 | (±)-Ganocin C | Anti-BuChE activity | G. cochlear | [45] |
| 116 | (±)-Ganocin D | Anti-BuChE activity | G. cochlear | [45] |
| 117 | Cochlearol A | Renoprotective effect | G. cochlear | [46] |
| 118 | Cochlearol B | Renoprotective effect | G. cochlear | [46] |
| 119 | Applanatumol A | Anti-renal fibrosis | G. applanatum | [18] |
| 120 | (±)-Applanatumol B | Anti-renal fibrosis | G. applanatum | [18] |
Compounds 41–58 (Fig. 5) were derived from the biogenetic precusor fornicin D (1), of which compounds 41–46 had a five-membered carbon ring in the terpenoid part through the connection between C-2′ and C-6′ [32]; wheares, compounds 47–57 possessed a six-membered carbon ring by a linkage between C-3′ and C-9′ [32, 38–41]. The presence of a seven-membered carbon ring in compound 58 was formed due to the carbon bond at C-2′ and C-9′ [38]. The inhibitory activities against COX-1 and COX-2 of compounds 41–56 were evaluated and they didn’t show obvious inhibition [32, 39–41]. Compound 57 was found to promote proliferation of neural stem cells (NSCs) [38]. However, compound 58 can inhibit NSC proliferation compared with a DMSO control [38].
Fig. 5.
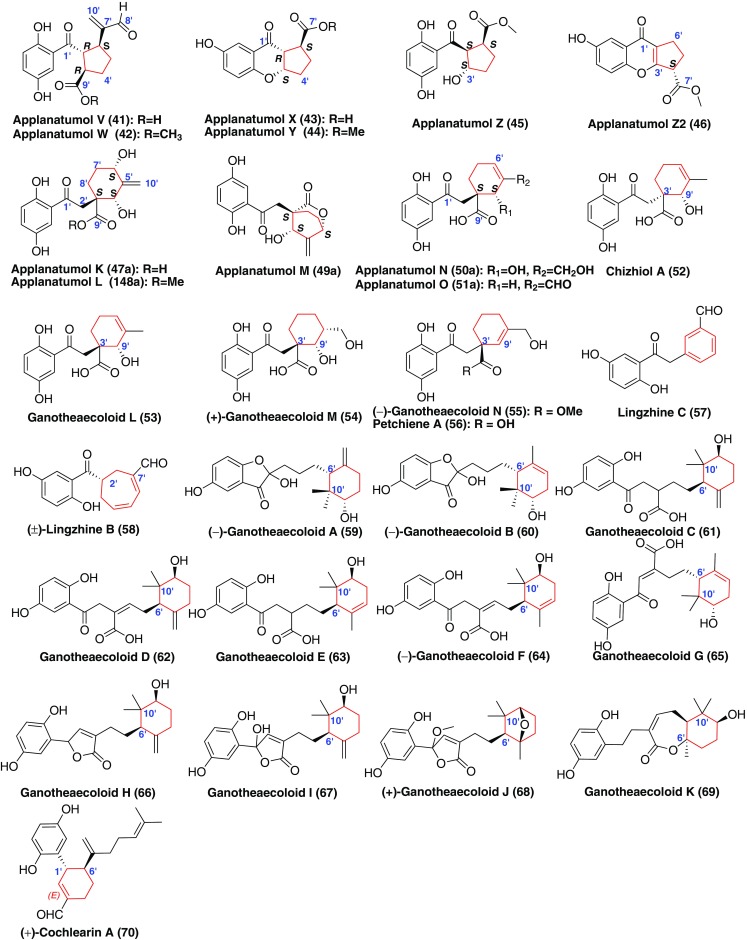
Structures of GMs with a five-membered or six-membered carbon ring
When ganomycin C (11) was the biosynthetic precusor, compounds 59–70 (Fig. 5) were formed through the cyclization between C-6′ and C-10′ [30, 40]. Biological activity of all the GMs against COX-2 was evaluated in vitro, only ganotheaecoloid J (68) was found to have COX-2 inhibitory activity with an IC50 value of 9.96 μM [40]. Cochlearin A (70) showing DPPH scavenging activity had a cyclohexane fraction, which was formed by C-1′ binding with C-6′ [30].
Furthermore, compounds bearing seven-membered carbon ring or five-membered carbon ring were as the precursor, the formation of an ether bond between C-1 and C-2′ resulted in the occurrence of sipro ring. For instance, compounds 71–80 (Fig. 6) contained a 6/5/7 ring system [42] and compounds 81–92 (Fig. 6) possessed a 6/5/5 ring system [38, 42]. Biological evaluation disclosed that compounds 77 and 78 inhibited JAK3 kinase with IC50 values of 7.0 ± 3.2 and 34.8 ± 21.1 μM, respectively [42]. The most potent member of this series, (−)-spirolingzhine A (89), was shown to affect NSC cell cycle progression using the 5-bromo-2-deoxyuridine (BrdU) incorporation assay [38].
Fig. 6.
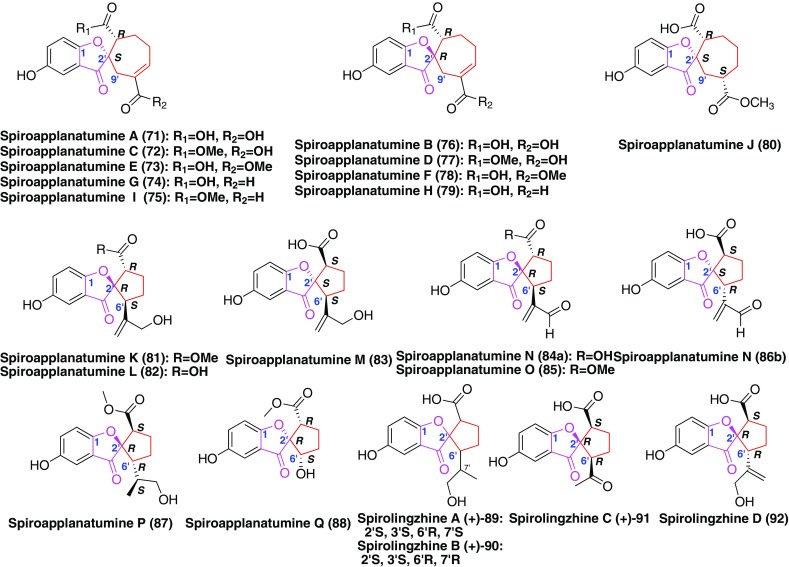
Structures of GMs with spiro ring
A series of bridge-ring compounds were formed through the free radical reactions. The structures of compounds 93–105 (Fig. 7) had a five-membered carbon ring fraction fusing with a γ-lactone ring [15, 32, 43]. Among them, ganoderin A (93) disclosed significant antioxdiant activities [15]. In the bioassay, compounds 94–100 didn’t exhibit inhibition aganist COX-1 and COX-2 [32]. The in vitro and in vivo results suggested that lingzhilactone B (102) could protect against renal injuries by increasing the activities of antioxidants and inhibiting inflammation [43]. The inhibition of Smad3 phosphorylation suggested that this substance displays in vivo antifibrotic activity by a mechanism that is dependent on disruption of Smad3. Applanatumol C (106) and linzhiol (107) beared an unusual 5/5/6/6 ring systerm characteristic of sharing a C-3′–C-7′ axis (Fig. 7) [32, 44]. The mirror of compound 106 was found to have COX-2 inhibitory effect with IC50 value of 25.5 mM [32]. (+)-Lingzhiol (107) and (−)-lingzhiol (107) could selectively inhibit the phosphorylation of Smad3 in TGF-β1-induced rat renal proximal tubular cells and activate Nrf2/Keap1 in mesangial cells under diabetic conditions [44].
Fig. 7.
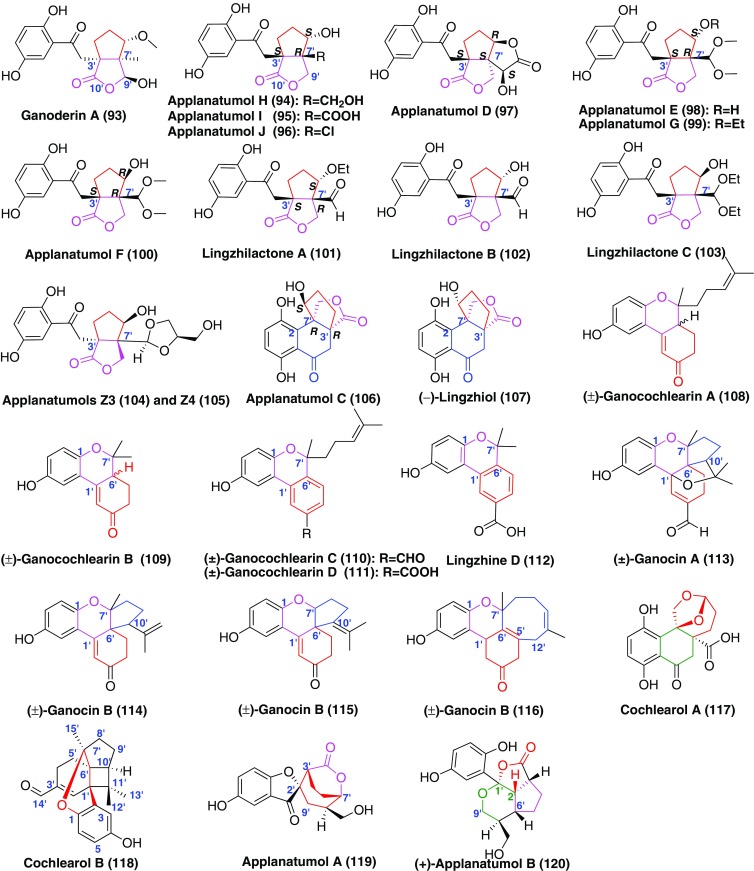
Structures of GMs with bridge ring
Cochlearin A (70) was as the biogenetic precursor for compounds 108–112 (Fig. 7) with an additional ether bond (C-1–C-7′) [15, 38]. The further cyclization led to the formation of ganoderins A–C (113–115) (Fig. 7) possessing a spiro[4,5]decane ring system, along with ganocin D (116) (Fig. 7) with an eight-membered ring [45]. Similarly, compounds 108–112 showed comparable antioxidant effects compared to the positive control (Vitamin E) [15, 45], while compounds 113–116 (Fig. 7) displayed anti-BuChE activities [45]. Cochlearol A (117) was a new normeroterpenoid containing a naturally unusual dioxaspiro[4.5]decane motif [46]. Compound 118 (Fig. 7) was a novel meroterpenoid possessing respective 4/5/6/6/6 polycyclic ring systems [46]. Meanwhile, biological studies showed that (−)-118 was a strong inhibitor of pSmads, exhibiting renoprotective activities in TGF-β1 induced rat renal proximal tubular cells [46]. Applanatumols A (120) and B [(±)-121] (Fig. 7) possessed a novel spiro[benzofuran-2,2′-biocyclo[3.2.2]nonane] ring system and a naturally unusual dioxacyclopenta[cd]inden motif, respectively [18]. Both of them didn’t show inhibitory activities against renal fibrosis in rat proximal tubular epithelial cells [18].
Dimeric GMs
Except for the intramolecular cyclization, the intermolecular cyclization was present in GMs, which resulted in the formation of dimeric GMs (Fig. 8, Table 3). (+)- and (−)-siensilactam A (121) was a novel hybrid metabolites possessing a unique 2H-pyrrolo[2,1-b][1,3]oxaz-in-6(7H)-one ring system [47]. (−)-121 was found to be a Smad3 phosphorylation inhibitor in TGF-β1 induced human renal proximal tubular cells [47]. (±)-Ganoapplanin (122) feartured an unprecedented dioxaspirocyclic skeleton, which was constructed from a 2,4-dihydroxy benzoic acid and a bridge-ring compound 102 [48]. Biological studies showed that (±)-122 and its enantomers exhibited different inhibitory activities on T-tpye voltage-gated calcium channels [48]. Applanatumin A (123) possessed a new hexacyclic skeleton containing spro[benzofuran-2,1′-cyclopentane] motif [17]. The analysis of its sturcture showed that it consisted of two meroterpenoid parts, sproapplanatumine N (84) and applanatumol S (5), which were connected by a key Diels-Alder reaction. In TGF-β1-induced human renal proximal tubular cells, applanatumin A (123) diclosed potent antifibrotic activity [17]. Cochlearoids A–E (124–128) containing a unique methanobenzo[c]oxocino[2,3,4-ij]-isochromene scafflod were also constructed by two meroterpenoids [49]. Among them, (+)-124, and (−)-126 significantly inhibited Cav3.1 TTCC and showed noticeable selectivity against Cav1.2, Cav2.1, Cav2.2 and Kv11.1 (hERG) channels [49]. The combination of two chian-contained GMs formed (+)-ganodilactone (129), cochlearoids F and G (130 and 131) [50, 51]. Similarly, when 2,4-dihydroy benzoic acid was linked with chain-contained GMs by the same method as ganoapplanin (124), compounds 132–135 were taken place. (±)-, (+)-, and (−)-ganodilactone (129) showed pancreatic lipase inhibitory activities and exhibited the IC50 values as 27.3, 4.0, and 2.5 μM, respectively [50]. In addition, other compounds were tested for their renoprotective activity against fibronectin inhibition in human proximal tubular epithelial cells (HKC-8). Compounds 130–133 and 135 exhibited potent inhibitory activity on fibronectin overproduction in TGF-β1-induced HKC-8 cells [51].
Fig. 8.
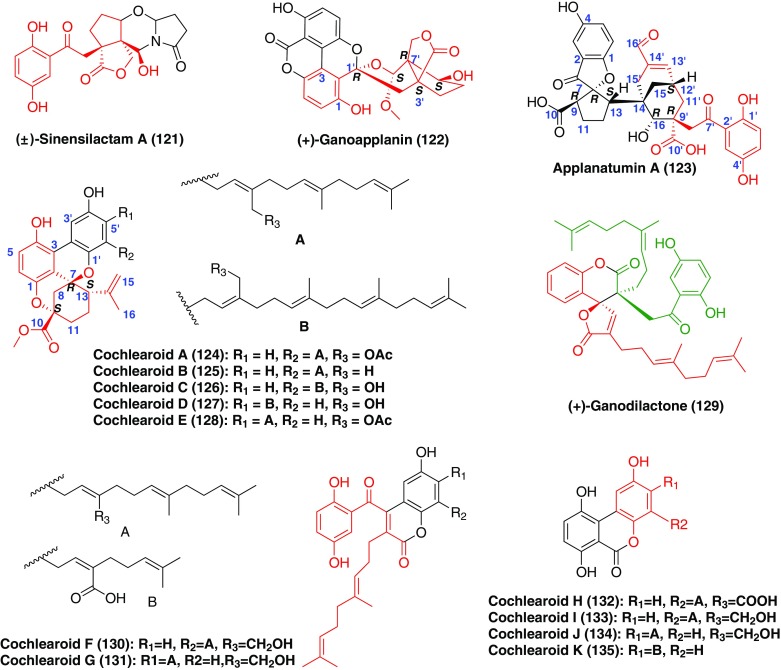
Structures of dimeric GMs
Table 3.
Name, source and bioactivities of dimeric GMs
| Number | Name | Bioactivity | Source | Reference |
|---|---|---|---|---|
| 121 | (−)-Sinensilactam A | Renoprotective activity | G. sinense | [47] |
| 122 | (+)-Ganoapplanin | Inhibitory activities on T-tpye voltage-gated calcium channels | G. applanatum | [48] |
| 123 | Applanatumin A | Antifibrotic activity | G. applanatum | [17] |
| 124 | (−)-Cochlearoid A | Inhibitory activities on T-tpye voltage-gated calcium channels | G. cochlear | [49] |
| 125 | (−)-Cochlearoid B | Inhibitory activities on T-tpye voltage-gated calcium channels | G. cochlear | [49] |
| 126 | (−)-Cochlearoid C | Inhibitory activities on T-tpye voltage-gated calcium channels | G. cochlear | [49] |
| 127 | (−)-Cochlearoid D | Inhibitory activities on T-tpye voltage-gated calcium channels | G. cochlear | [49] |
| 128 | (−)-Cochlearoid E | Inhibitory activities on T-tpye voltage-gated calcium channels | G. cochlear | [49] |
| 129 | (+)-Ganodilactone | Inhibitory activity against pancreatic lipase | G. leucocontextum | [50] |
| 130 | Cochlearoid F | Renoprotective effect | G. cochlear | [51] |
| 131 | Cochlearoid G | Renoprotective effect | G. cochlear | [51] |
| 132 | Cochlearoid H | Renoprotective effect | G. cochlear | [51] |
| 133 | Cochlearoid I | Renoprotective effect | G. cochlear | [51] |
| 134 | Cochlearoid J | Renoprotective effect | G. cochlear | [51] |
| 135 | Cochlearoid K | Renoprotective effect | G. cochlear | [51] |
Conclusion
In this review, we summarized the chemical structures and biological activities of 135 GMs in the last five years. Although the first GMs have been isolated in 2000, until recent years GMs were studied in-depth. Moreover, except for G. lucidum and G. sinense registered in Chinese Pharmacopoeia (2010 and 2015 edition), GMs were widely present in many other Ganoderma species, such as G. appalantum, G. capense, G. cochlear, and G. petchii. Above information indicated that GMs could play an important role in explaining the efficacy of Ganoderma. Thus, more bioactive studies should be carried out in the future for finding and developing lead compounds.
Furthermore, GMs possessed multiple prenyl groups or complex ring systems, which provided plentiful molecular model for various biological activities. However, we found that the majority of GMs showed racemic nature, which had impact on their bioactivites. Therefore, it is need to be separated using chiral HPLC method or be stereoselectively synthsized.
Addtionally, the formation of racemic GMs also attracted us attention. Analysis of these polycyclic GMs showed that their polycyclic structures are formed based on the polyunsaturated terpenoid fraction. Studies found that the cyclizations, such as cationic cyclization and radical cyclization, are the key factor to generate racemes. And these reactions can be taken place under conditions of acid, light and heating. However, the reactions in the plants mostly involved in enzyme system, which led to the generation of stereoselective compounds. Thus, we deduced that these polycyclic GMs with racemic nature may be formed for defending high temperature, strong light and diseases.
In all, the efforts to discover novel GMs with interesting biological activity and intriguing strutures from Ganoderma species have long been a hot topic in natural products chemistry. Meanwhile, novel GMs will serve as an abundant resource for synthetic chemists.
Acknowledgements
The research work was financially supported by the National Natural Science Foundation of China (No. 21702209 and No. 81172940), as well as Foundation of State Key Laboratory of Phytochemistry and Plant Resources in West China (P2010-ZZ14).
Compliance with Ethical Standards
Conflict of interest
All authors declare no conflict of interest.
References
- 1.Upadhyay M, Shrivastava B, Jain A, Kidwai M, Kumar S, Gomes J, Goswami D, Panda A, Kuhad R. Ann. Microbiol. 2014;64:839–846. doi: 10.1007/s13213-013-0723-9. [DOI] [Google Scholar]
- 2.Li Y, Zhu Z, Yao W, Chen R. Med. Plant. 2012;3:75–81. [Google Scholar]
- 3.Lee S, Shim SH, Kim JS, Shin KH, Kang SS. Biol. Pharm. Bull. 2005;28:1103–1105. doi: 10.1248/bpb.28.1103. [DOI] [PubMed] [Google Scholar]
- 4.Ngai PHK, Ng TB. Biochem. Biophys. Res. Commun. 2004;314:988–993. doi: 10.1016/j.bbrc.2003.12.196. [DOI] [PubMed] [Google Scholar]
- 5.Gurunathan S, Raman J, Malek SNA, John PA, Vikineswary S. Int. J. Nanomed. 2013;8:4399–4413. doi: 10.2147/IJN.S51881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cragg GM, Newman DJ, Snader KM. J. Nat. Prod. 1997;60:52–60. doi: 10.1021/np9604893. [DOI] [PubMed] [Google Scholar]
- 7.Newman DJ, Cragg GM, Snader KM. Nat. Prod. Rep. 2000;17:215–234. doi: 10.1039/a902202c. [DOI] [PubMed] [Google Scholar]
- 8.Newman DJ, Cragg GM, Snader KM. J. Nat. Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 9.Butler MS. J. Nat. Prod. 2004;67:2141–2153. doi: 10.1021/np040106y. [DOI] [PubMed] [Google Scholar]
- 10.Paterson I, Anderson EA. Science. 2005;310:451–453. doi: 10.1126/science.1116364. [DOI] [PubMed] [Google Scholar]
- 11.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461–477. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 12.Mothana RAA, Jansen R, Julich WD, Lindequist U. J. Nat. Prod. 2000;63:416–418. doi: 10.1021/np990381y. [DOI] [PubMed] [Google Scholar]
- 13.Chen XQ, Chen LX, Li SP, Zhao J. Phytochem. Lett. 2017;22:214–218. doi: 10.1016/j.phytol.2017.10.015. [DOI] [Google Scholar]
- 14.Wang M, Wang F, Xu F. Bioorg. Med. Chem. Lett. 2016;26:3342–3345. doi: 10.1016/j.bmcl.2016.05.037. [DOI] [PubMed] [Google Scholar]
- 15.Peng XR, Liu JQ, Wang CF, Han ZH, Shu Y, Li XY, Zhou L, Qiu MH. Food Chem. 2015;171:251–257. doi: 10.1016/j.foodchem.2014.08.127. [DOI] [PubMed] [Google Scholar]
- 16.Peng XR, Li L, Wang X, Zhu GL, Zhou L, Qiu MH. Fitoterapia. 2016;111:18–23. doi: 10.1016/j.fitote.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Luo Q, Di L, Dai WF, Lu Q, Yan YM, Yang ZL, Li RT, Cheng YX. Org. Lett. 2015;17:1110–1113. doi: 10.1021/ol503610b. [DOI] [PubMed] [Google Scholar]
- 18.Luo Q, Di L, Yang XH, Cheng YX. RSC Adv. 2016;6:45963–45967. doi: 10.1039/C6RA05148K. [DOI] [Google Scholar]
- 19.Huang SZ, Cheng BH, Ma QY, Wang Q, Kong FD, Dai HF, Qiu SQ, Zheng PY, Liu ZQ, Zhao YX. RSC. Adv. 2016;6:21139–21147. doi: 10.1039/C6RA01466F. [DOI] [Google Scholar]
- 20.Peng XR, Liu JQ, Han ZH, Yuan XX, Luo HR, Qiu MH. Food Chem. 2013;141:920–926. doi: 10.1016/j.foodchem.2013.03.071. [DOI] [PubMed] [Google Scholar]
- 21.Niu XM, Li SH, Sun HD, Che CT. J. Nat. Prod. 2006;69:1364–1365. doi: 10.1021/np060218k. [DOI] [PubMed] [Google Scholar]
- 22.Zhang JJ, Ma K, Chen HY, Wang K, Xiong WP, Bao L, Liu HW. J. Antibiot. 2017;70:915–917. doi: 10.1038/ja.2017.57. [DOI] [PubMed] [Google Scholar]
- 23.Gautam KS, Birman VB. Org. Lett. 2016;18:1499–1501. doi: 10.1021/acs.orglett.5b03212. [DOI] [PubMed] [Google Scholar]
- 24.Li XY, Liu XY, Jiao XZ, Yang HG, Yao YY, Xie P. Org. Lett. 2016;18:1944–1946. doi: 10.1021/acs.orglett.6b00542. [DOI] [PubMed] [Google Scholar]
- 25.Chen D, Liu HM, Li MM, Yan YM, Xu WD, Li XN, Cheng YX. Chem. Commun. 2015;51:14594–14596. doi: 10.1039/C5CC05680B. [DOI] [PubMed] [Google Scholar]
- 26.Liu HX, Hou LQ, Yang B, Yuan YF, Zhang WM, Xu ZF, Qiu SX, Tan HB. Org. Lett. 2017;19:4786–4789. doi: 10.1021/acs.orglett.7b02159. [DOI] [PubMed] [Google Scholar]
- 27.Liu X, Chen RD, Xie D, Li JH, Wang RS, Dai JG. Yaoxue Xuebao. 2013;48:161–169. [PubMed] [Google Scholar]
- 28.Gao J, Zeng Y, Lu S. Zhiwu Xuebao (Beijing, China) 2010;45:751–759. [Google Scholar]
- 29.Liu D, Gong J, Dai W, Kang X, Huang Z, Zhang HM, Liu W, Liu L, Ma J, Xia Z, Chen Y, Chen Y, Wang D, Ni P, Guo AY, Xiong X. PLoS ONE. 2012;7:e36146. doi: 10.1371/journal.pone.0036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng XR, Wang X, Chen L, Yang H, Lei L, Lu SY, Zhou L, Qiu MH. Fitoterapia. 2018 [Google Scholar]
- 31.Luo Q, Wang XL, Di L, Yan YM, Lu Q, Yang XH, Hu DB, Cheng YX. Tetrahedron. 2015;71:840–845. doi: 10.1016/j.tet.2014.12.052. [DOI] [Google Scholar]
- 32.Luo Q, Yang XH, Yang ZL, Tu ZC, Cheng YX. Tetrahedron. 2016;30:4564–4574. doi: 10.1016/j.tet.2016.06.019. [DOI] [Google Scholar]
- 33.Wang K, Bao L, Ma K, Zhang JJ, Chen BS, Han JJ, Ren JW, Luo HJ, Liu HW. Eur. J. Med. Chem. 2017;127:1035–1046. doi: 10.1016/j.ejmech.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 34.Dou M, Li RT, Chen YX. Chin. Herb. Med. 2016;8:85–88. doi: 10.1016/S1674-6384(16)60013-8. [DOI] [Google Scholar]
- 35.Chen BS, Tian J, Zhang JJ, Wang K, Liu L, Yang B, Bao L, Liu HW. Fitoterapia. 2017;120:6–16. doi: 10.1016/j.fitote.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Wang XF, Yan YM, Wang XL, Ma XJ, Fu XY, Cheng YX. J. Asian Nat. Prod. Res. 2015;17:329–332. doi: 10.1080/10286020.2014.960858. [DOI] [PubMed] [Google Scholar]
- 37.Cao WW, Luo Q, Chen YX, Wang SM. Fitoterapia. 2016;110:110–115. doi: 10.1016/j.fitote.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Yan YM, Wang XL, Luo Q, Jiang LP, Yang CP, Hou B, Zuo ZL, Chen YB, Cheng YX. Phytochemistry. 2015;114:155–162. doi: 10.1016/j.phytochem.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Zhou FJ, Wang XL, Wang SM, Cheng YX. Nat. Prod. Res. Dev. 2015;27:22–25. [Google Scholar]
- 40.Luo Q, Tu ZC, Yang ZL, Cheng YX. Fitoterapia. 2018;125:273–280. doi: 10.1016/j.fitote.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Gao QL, Guo PX, Luo Q, Yan H, Cheng YX. Nat. Prod. Commun. 2015;10:2019–2022. [PubMed] [Google Scholar]
- 42.Luo Q, Wei XY, Yang J, Luo JF, Liang R, Tu ZC, Cheng YX. J. Nat. Prod. 2017;80:61–70. doi: 10.1021/acs.jnatprod.6b00431. [DOI] [PubMed] [Google Scholar]
- 43.Yan YM, Wang XL, Zhou LL, Zhou FJ, Li R, Tian Y, Zuo ZL, Fang P, Chung ACK, Hou FF, Cheng YX. J. Ethnopharm. 2015;176:385–393. doi: 10.1016/j.jep.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 44.Yan YM, Ai J, Zhou LL, Chung ACK, Li R, Nie J, Fang P, Wang XL, Luo J, Hu Q, Hou FF, Cheng YX. Org. Lett. 2013;15:5488–5491. doi: 10.1021/ol4026364. [DOI] [PubMed] [Google Scholar]
- 45.Peng XR, Liu JQ, Wan LS, Li XN, Yan YY, Qiu MH. Org. Lett. 2014;16:5262–5265. doi: 10.1021/ol5023189. [DOI] [PubMed] [Google Scholar]
- 46.Dou M, Di L, Zhou LL, Yan YM, Wang XL, Zhou FJ, Yang ZL, Li RT, Hou FF, Cheng YX. Org. Lett. 2014;16:6064–6067. doi: 10.1021/ol502806j. [DOI] [PubMed] [Google Scholar]
- 47.Luo Q, Tian L, Di L, Yan YM, Wei XY, Wang XF, Cheng YX. Org. Lett. 2015;17:1565–1568. doi: 10.1021/acs.orglett.5b00448. [DOI] [PubMed] [Google Scholar]
- 48.Li L, Li H, Peng XR, Hou B, Yu MY, Dong JR, Li XN, Zhou L, Yang J, Qiu MH. Org. Lett. 2016;18:6078–6081. doi: 10.1021/acs.orglett.6b03064. [DOI] [PubMed] [Google Scholar]
- 49.Zhou FJ, Nian Y, Yan YM, Gong Y, Luo Q, Zhang Y, Hou B, Zuo ZL, Wang SM, Jiang HH, Yang J, Cheng YX. Org. Lett. 2015;17:3082–3085. doi: 10.1021/acs.orglett.5b01353. [DOI] [PubMed] [Google Scholar]
- 50.Chen HP, Zhao ZZ, Zhang Y, Bai X, Zhang L, Liu JK. RSC Adv. 2016;6:64469–64473. doi: 10.1039/C6RA10638B. [DOI] [Google Scholar]
- 51.Wang XL, Zhou FJ, Dou M, Yan YM, Wang SM, Di L, Cheng YX. Bioorg. Med. Chem. Lett. 2016;26:5507–5512. doi: 10.1016/j.bmcl.2016.10.011. [DOI] [PubMed] [Google Scholar]