Abstract
N -glycans are normally involved in crucial physiological and disease processes by interactions with glycan binding proteins. So far structurally defined N-glycans are good candidates for glycan binding study. Herein, a class of homogeneous asymmetric N-glycans was synthesized by coupling glycan-oxazoline and N-glycans using EndoM N175Q catalyzed quick glycan extension. Branch-biased binding and spacial inhibition caused by bulky group on the other branch of N-glycan were observed in glycan protein interactions involving lectins and these glycans by glycan microarray study. These new compounds are valuable for functional glycomic studies to better understand new functions of glycans and glycan-binding proteins.
Introduction
N-linked glycans are the sugar chains bonded to a peptide and a protein through the side chain of asparagine residue, which is a predominant protein modification in eukaryotes, also widely exists in archaea, but is very rare in eubacteria.1–3 N-glycans are involved in many crucial biological functions such as protein folding, degradation,4–7 cell differentiation, cell adhesion, host-pathogen interaction and cancer metastasis.8–10 In nature, N-glycans possess an inherited complexity and diversity. In mammalian cells, N-glycans have numerous structures which usually contain different glycan epitopes (sugar residues linked to each other in a specific sequence) in different branches.11 Since glycan epitopes are usually the binding partners of specific antibodies or glycan-binding proteins (e.g. lectins), the study of binding pattern of a glycan epitope in an asymmetric N-glycan will deepen our understanding of the roles of context in glycan-binding events involving asymmetric N-glycans.
Owing to the structural microheterogeneity, it is difficult to acquire homogeneous and structurally defined N-glycans from natural source. Although synthetic efforts on symmetric N-glycans are prominent,12–18 unfortunately, due to structural complexity, strategies for chemically, chemoenzymatically or enzymatically synthesis of asymmetrically branched N-glycans are very limited. By chemical synthesis of core structure and enzymatic extension of terminal epitopes, a group of asymmetric single- or bi-antennary N-glycans was synthesized with terminal glycan epitopes reported by our gourp.19 With the aid of orthogonal protection groups, Boons group achieved synthesis of tri-antennary N-glycans with distinctive appendages to different branches.11 Later, they further developed the strategy by incorporation of unnatural glycosyl linkages to fulfil selective installation of distinct appendages at different branching points, and generating N-glycans with more complex structures.20 Recently, with the aid of a series of glycosidases and glycosyltransferases, our group demonstrated completely enzymatic preparation of asymmetric N-glycans from naturally isolated N-glycans, which can circumvent the dependence on chemical preparation of N-glycan core structures.21 Based on the limited accessibility of asymmetrically branched N-glycans, glycan-binding studies involving asymmetric N-glycans11 are rare, although glycan microarray studies has burst over the past two decades.22–23 EndoM is an endo-β-N-acetylglucosaminidases (ENGases) from Mucor hiemali that can hydrolyze the N,N′-diacetylchitobiose moiety in the core pentasacharide of mammalian N-glycans. The mutant EndoM N175Q, which has a decreased hydrolytic activity, can transfer natural N-glycans or oxazoline N-glycans to any peptide or protein with a GlcNAc residue to form a β1–4-glycosidic linkage, hence becoming a useful tool in the synthesis of homogeneous glycopeptides and glycoproteins.24–26 Lai-Xi Wang and co-workers found that ENGase-based glycosynthases can synthesize a class of symmetric N-glycan clusters with isolated glycan as donor and acceptor.27 Based on the above approaches, herein, by one-step transglycosylation, we alternatively developed a strategy to produce a new class of homogeneous and structurally well-defined asymmetric single- or bi-antennary N-glycans in which one branch is terminated by a tetrasaccharide motif (Man2GlcNAc2) in the core pentasaccharide of mammalian N-glycans, and investigated the influence of context on the binding to terminal glycan epitopes by glycan microarray.
Results and discussion
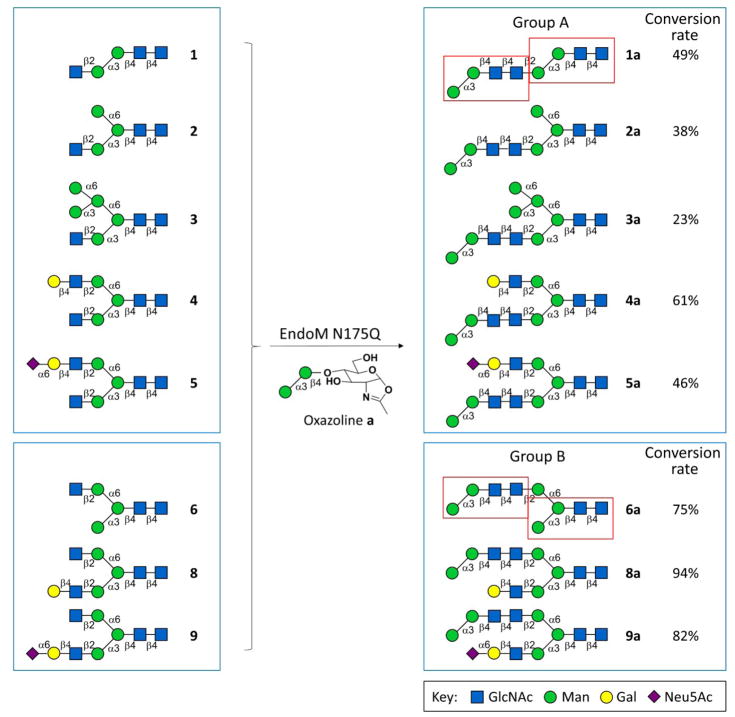
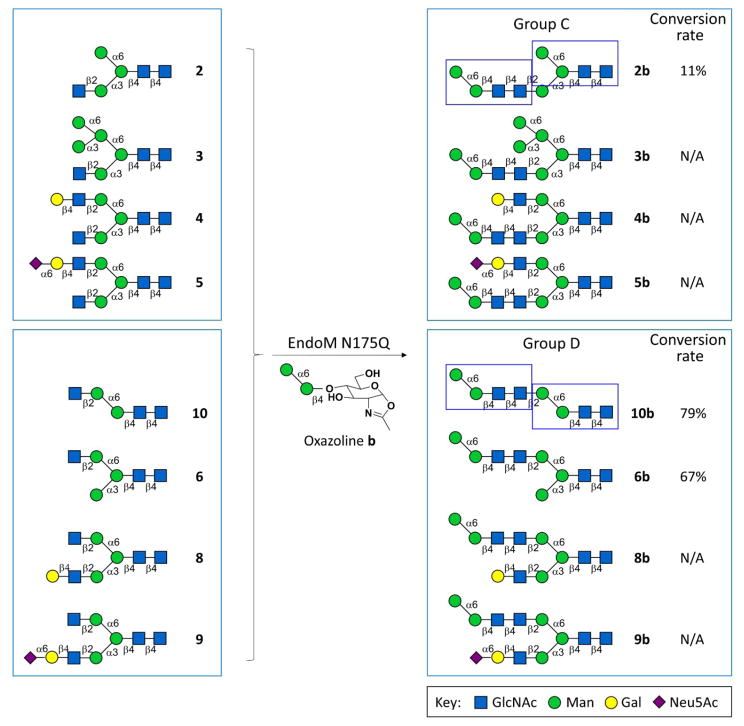
The class of asymmetric N-glycans we synthesized in this study contains core glycan motifs at both core position and terminal position (red rectangles in Groups A and B in Scheme 1, blue rectangles in Groups C and D in Scheme 2), and may serve as a good candidate for deep exploration of the function of core glycan and terminal glycan motif. The synthesis of the asymmetric N-glycans were performed (Scheme 1–3, Groups A–D) by using two N-glycan oxazolines (a and b) that were chemically synthesized as donors (Supplementary Information V), and ten asymmetric N-glycans with only one free terminal β1–2GlcNAc residue (1–10) that were chemoenzymatically synthesized as acceptor (Supplementary Information V). Group A or B glycans contain double α1–3Man branch core glycan motifs (i.e. core pentasaccharide minus α1–6Man), and the terminal α1–3Man branch core glycan motif located in α1–3Man branch or α1–6Man branch, respectively. Similarly, Group C or D glycans contain double α1–6Man branch core glycan motifs (i.e. core pentasaccharide minus α1–3Man) and the terminal α1–6Man branch core glycan motif located in α1–3Man branch or α1–6Man branch, respectively.
Scheme 1.
Groups A and B glycans were synthesized by EndoM N175Q with α1–3Man branch glycan oxazoline as donor
Scheme 2.
Groups C and D glycans were synthesized by EndoM N175Q with α1–6Man branch glycan oxazoline as donor (N/A: No product detected, hence conversion rate is not applicable.)
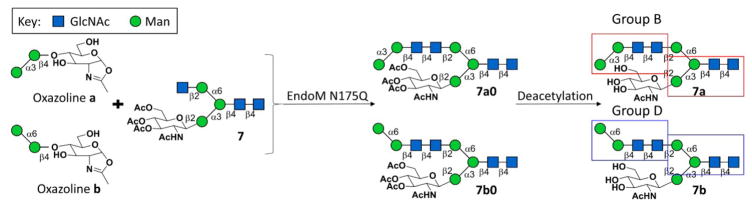
Scheme 3.
Selective transglycosylation achieved by using selectively acetylated glycan 7 as acceptor.
Glycan oxazolines (50 nM) and asymmetric N-glycans (1–10, 10 nM) were incubated with EndoM N175Q (50 μg) in 50 mM PBS (pH 6.5) at 30°C for 1 h. The reaction mixtures were analyzed by HPLC-HILIC-ELSD, and the conversion rate were calculated according to the peak area ratio of product peak over the sum of product peak and remaining acceptor peak. Then, the glycan products were subsequently purified under the monitor of A210nm according to the retention time from the analytical run, as previously reported19,28,29(Supplementary Information III). The collected substance was lyophilized and analyzed by MS (Supplementary Information VIII).
For compounds 1–6 and 8–10, which contain only one terminal β1–2GlcNAc residue, the corresponding asymmetric N-glycans were prepared simply by one-step transglycosylation and subsequent HPLC purification. However, to achieve selective transglycosylation on N-glycans with two terminal β1–2GlcNAc branches, acceptor 7 was partially protected by peracetylation of GlcNAc residue on the α1,3Man branch for the synthesis of asymmetric N-glycans. After 7a0 and 7b0 were synthesized by EndoM N175Q, deacetylation was performed to provide 7a and 7b (Scheme 3, Supplementary Information V), which belong to Group B and Group D, respectively. Since the peak of 7 was overlapped with peak of impurity in HPLC chromatograph, conversion rate was unable to calculate for compound 7a0 or 7b0.
The substrate specificity of EndoM N175Q is reflected by the conversion rate of transglycosylation reactions. Generally speaking, the conversion rate was higher for transglycosylation to α1–6Man branch than to α1–3Man branch (Group B vs. Group A, Group D vs. Group C, respectively), regardless of which donor was used (Schemes 1–2). However, when b was donor, out of 9 structures we designed, only 4 glycans were synthesized. The failure of producing the corresponding xb glycans can be a result of space hindrance caused by the appendage on the non-acceptor branch (i.e. the branch without terminal β1–2GlcNAc residue).
However, the length of the non-acceptor branch is not the only factor that determines the conversion rate of this transglycosylation reaction, which can be illustrated by the highest conversion rates for 4a and 8a comparing to those for other glycans in each group, respectively (Scheme 1).
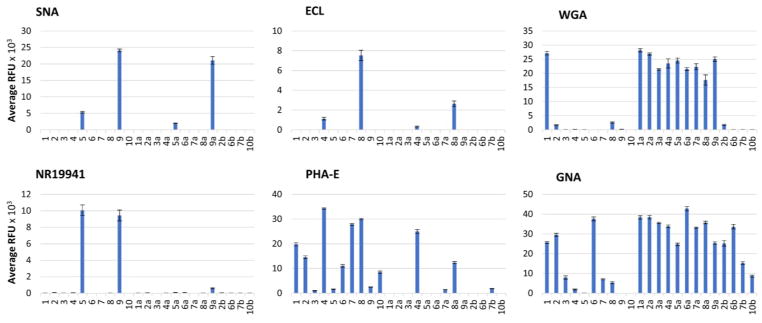
To better understand the interactions between glycan and proteins, we labelled all asymmetric N-glycans (both acceptors and products for comparison) with AEAB (N-(aminoethyl)-2-amino benzamide, Supplementary Information VI), generated customized glycan arrays via Z Biotech (Aurora, CO, USA; the slide is available from Z biotech and Chemily LLC.; Supplementary Information VII) and examined the recognition of 6 plant lectins (SNA, ECL, PHA-E, GNA, WGA, and ConA) as well as 2 hemoagglutinin proteins (HAs) of 2009 H1N1 influenza A viruses to the asymmetric N-glycans [NR19941 (HA of A/New York/18/2009 H1N1) and NR42486 (HA of A/Czech Republic/32/2011 H1N1; A/Czech Republic/32/2011 (H1N1) is an influenza A virus isolated from the seasonal outbreaks of 2009 H1N1 virus and the viruses have been adapted to the human population for two influenza seasons); BEI Resources Repository] (Supplementary Information VII). The results are illustrated as histograms in Figure 1 and Figure S1.
Fig. 1.
Glycan microarray analyses. Binding profiles with five plant lectins (including Sambucus nigra lectin (SNA), Erythrina cristagalli lectin (ECL), Phaseolus vulgaris erythroagglutinin (PHA-E), wheat germ agglutinin (WGA) and Galanthus nivalis lectin (GNA)) and one viral lectins (hemoagglutinin (HA) NR19941 from human influenza viruses) were examined.
SNA30 and HAs of 2009 H1N1 influenza A viruses31 are known to bind Siaα2–6Gal. It is interesting that results from our experiments suggested that they exhibited different preferences to the two branches in the asymmetric N-glycans. Specifically, SNA preferred to bind α1–3Man branch (9>5), while both HAs showed higher binding to α1–6Man branch (5>9). Nevertheless, the binding of all three proteins to their favorite branch was hampered slightly (for SNA, 5a<5, 9a<9) or significantly (for HAs, almost no detectable binding for 5a and 9a) when the other branch was extended by the transglycosylation reaction, demonstrating the extended glycans in the other antennary have impaired glycan epitope binding between glycans and SNA or HA proteins of the 2009 H1N1 virus.
ECL can bind Galβ1–4GlcNAc,32 and in this study, it demonstrated preferential binding to the epitope in α1–3Man branch (8>4). PHA-E, another lectin that recognizes outer Gal,27 favors the Gal located in α1–6Man branch (4>8). When the other branch was elongated by transglycosylation, the binding to Gal of both lectins decreased (8a<8, 4a<4). PHA-E also binds terminal GlcNAc,27 but preferred GlcNAc in α1–3Man branch (1>10, 2>6). Likewise, bulky groups on the other branch (branched Man3 or sialylated LacNAc) hindered the binding of PHA-E to GlcNAc regardless of which branch this GlcNAc is in (3, 5, and 9).
WGA, which is reported to bind chitin oligomers (GlcNAcn),27 in this study liked GlcNAc in α1–3Man branch (1>10), and displayed substantial increases in binding for most glycans (2a–9a vs. 2–9) when the glycans were extended with α1–3Man branch core glycan motif, no matter the extension located in α1–3Man branch or α1–6Man branch. In contrast, elongation with α1–6Man branch core glycan motif did not show this effect (2b vs. 2), indicating that WGA favors binding to GlcNAc oligomers linked with α1–3Man branch. In addition, no binding to Siaα2–6Gal was detected further confirmed that WGA only binds to Sia in Siaα2–3Gal, consistent with our previous observation.29
GNA is a lectin that binds high-mannose glycans especially Manα1–3Man epitope.27 Before transglycosylation, it exhibited higher binding to glycans exposing only Manα1–3Man at the terminal than those exposing only Manα1–6Man (2 vs. 6) or exposing both Manα1–3Man and Manα1–6Man (3 vs. 6). The binding also decreased as the branches extended, due to the masking effect of the extension on Man in glycan core. After transglycosylation, the glycans containing terminal Manα1–3Man epitope displayed higher binding than the original glycans (xa vs. x) and than those containing terminal Manα1–6Man epitope (xa vs. xb).
Conclusions
In summary, we have synthesized a class of asymmetric N-glycans by a facile transglycosylation approach, which can form structurally defined asymmetric N-glycans by easily assembly of glycan oxazoline and N-glycan acceptor, and applied them for the binding study of lectins to glycan epitopes locating in different branches. Also, donor and acceptor substrate specificity of EndoM N175Q was investigated. The binding study revealed that lectins selectively bind to their recognizable epitopes on different branches in asymmetric N-glycans. Meanwhile, the elongation of glycans on the other antenna is influential to the binding pattern of glycan terminal epitope. In addition, these new compounds may serve as valuable tools for functional glycomic studies, which may function as specific inhibitors in a given biological or pathological process.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (U01GM0116263 to P. G. Wang and Lei Li; R01AI116744 to X.F. Wan), National Science Foundation of China (31200605 to W. Guan). Z. Wu is grateful to Provost’s Dissertation Fellowship from Georgia State University. We would also like to acknowledge the proteins provided from NIH BEI resources repository.
Footnotes
Electronic Supplementary Information (ESI) available: Materials and enzymes, general methods for EndoM N175Q treatment, general methods for HPLC analysis and purification of N-glycans, general methods for mass spectrometry analyses, synthesis of acceptor N-glycans, N-glycan oxazolines (as donor) and deacetylation of asymmetric N-glycan products, labelling of N-glycans with AEAB, general methods for glycan microarray, HPLC analyses of N-glycan synthesizing mixtures and MS data of purified N-glycans, NMR spectrum and data of purified N-glycan. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Varki A. Glycobiology. 1993;3:97. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dwek RA. Chem Rev. 1996;96:683. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 3.Helenius A, Aebi M. Science. 2001;291:2364. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 4.Helenius A, Aebi M. Annu Rev Biochem. 2004;73:1019. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 5.Hebert DN, Molinari M. Physiol Rev. 2007;87:1377. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 6.Molinari M. Nat Chem Biol. 2007;3:313. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- 7.Stolz A, Wolf DH. Biochim Biophys Acta. 2010;1803:694. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Zhao YY, Takahashi M, Gu JG, Miyoshi E, Matsumoto A, Kitazume S, Taniguchi N. Cancer Sci. 2008;99:1304. doi: 10.1111/j.1349-7006.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamblin DP, Scanlan EM, Davis BG. Chem Rev. 2009;109:131. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 10.Sperandio M, Gleissner CA, Ley K. Immunol Rev. 2009;230:97. doi: 10.1111/j.1600-065X.2009.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Chinoy ZS, Ambre SG, Peng W, McBride R, de Vries RP, Glushka J, Paulson JC, Boons GJ. Science. 2013;341:379. doi: 10.1126/science.1236231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unverzagt C. Angew Chem Int Ed. 1996;35:2350. [Google Scholar]
- 13.Ratner DM, Swanson ER, Seeberger PH. Org Lett. 2003;5:4717. doi: 10.1021/ol035887t. [DOI] [PubMed] [Google Scholar]
- 14.Hanashima S, Manabe S, Ito Y. Angew Chem Int Ed. 2005;44:4218. doi: 10.1002/anie.200500777. [DOI] [PubMed] [Google Scholar]
- 15.Eller S, Schuberth R, Gundel G, Seifert J, Unverzagt C. Angew Chem Int Ed. 2007;46:4173. doi: 10.1002/anie.200604788. [DOI] [PubMed] [Google Scholar]
- 16.Serna S, Yan S, Martin-Lomas M, Wilson IBH, Reichardt NC. J Am Chem Soc. 2011;133:16495. doi: 10.1021/ja205392z. [DOI] [PubMed] [Google Scholar]
- 17.Walczak MA, Hayashida J, Danishefsky SJ. J Am Chem Soc. 2013;135:4700. doi: 10.1021/ja401385v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maki Y, Okamoto R, Izumi M, Murase T, Kajihara Y. J Am Chem Soc. 2016;138:3461. doi: 10.1021/jacs.5b13098. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Liu Y, Ma C, Qu J, Calderon AD, Wu B, Wei N, Wang X, Guo Y, Xiao Z, Song J, Sugiarto G, Li Y, Yu H, Chen X, Wang PG. Chem Sci. 2015;6:5652. doi: 10.1039/c5sc02025e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gagarinov IA, Li T, Toraño JS, Caval T, Srivastava AD, Kruijtzer JA, Heck AJ, Boons GJ. J Am Chem Soc. 2017;139:1011. doi: 10.1021/jacs.6b12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calderon AD, Zhou J, Guan W, Wu Z, Guo Y, Bai J, Li Q, Wang PG. J Fang and L Li Org Biomol Chem. 2017;15:7258. doi: 10.1039/c7ob01765k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyelaran O, Gildersleeve JC. Curr Opin Chem Biol. 2009;13:406. doi: 10.1016/j.cbpa.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rillahan CD, Paulson JC. Annu Rev Biochem. 2011;80:797. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto K. J Biosci Bioeng. 2001;92:493. doi: 10.1263/jbb.92.493. [DOI] [PubMed] [Google Scholar]
- 25.Wang LX. Carbohydr Res. 2008;343:1509. doi: 10.1016/j.carres.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umekawa M, Li C, Higashiyama T, Huang W, Ashida H, Yamamoto K, Wang LX. J Biol Chem. 2010;285:511. doi: 10.1074/jbc.M109.059832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Wang D, Yamada M, Wang LX. J Am Chem Soc. 2009;131:17963. doi: 10.1021/ja9078539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Z, Guo Y, Liu Y, Li L, Zhang Q, Wen L, Wang X, Kondengaden SM, Wu Z, Zhou J, Cao X, Li X, Ma C, Wang PG. J Org Chem. 2016;81:5851. doi: 10.1021/acs.joc.6b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z, Liu Y, Ma C, Li L, Bai J, Byrd-Leotis L, Lasanajak Y, Guo Y, Wen L, Zhu H, Song J, Li Y, Steinhauer DA, Smith DF, Zhao B, Chen X, Guan W, Wang PG. Org Biomol Chem. 2016;14:11106. doi: 10.1039/c6ob01982j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. J Biol Chem. 1987;262:1596. [PubMed] [Google Scholar]
- 31.Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. J Virol. 2012;86:982. doi: 10.1128/JVI.06322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itakura Y, Nakamura-Tsuruta S, Kominami J, Sharon N, Kasai K, Hirabayashi J. J Biochem. 2007;142:459. doi: 10.1093/jb/mvm153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.