Abstract
Quantitative investigations into functional properties of purified ion channel proteins using standard electrophysiological methods is challenging, in particular for the determination of average ion channel behavior following rapid changes in experimental conditions (e.g., ligand concentration). Here we describe a method for determining the functional activity of liposome-reconstituted K+ channels using a stopped-flow fluorometric ion flux assay. Channel activity is quantified by measuring the rate of fluorescence decrease of a liposome-encapsulated fluorophore, specifically quenched by thallium ions entering the liposomes via open channels. This method is well suited for studying the lipid bilayer dependence of channel activity, the activation and desensitization kinetics of ligand-dependent K+ channels, and channel modulation by channel agonists, blockers or other antagonists.
Keywords: Ion channel function, Stopped-flow assay, Liposomal ion flux assay, ANTS quenching, Thallium
1 Introduction
Ion channel activity has most commonly been studied in a cellular context using standardized electrophysiological measurements [1,2]. However, techniques such as patch clamp electrophysiology are technically challenging for purified and lipid-reconstituted ion channel proteins [3,4]. Such purified systems are not only suitable for structural characterization; they also offer the opportunity to study intrinsic channel properties and lipid-dependent changes in channel function, thereby providing for further studies into fundamental biophysical mechanisms underlying channel function. Electrophysiology, using proteoliposomes fusing into artificial lipid bilayers (black lipid membranes), is one solution, but is mostly applicable for systems already characterized in bulk and is difficult to use for non-equilibrium studies, such detection of the instantaneous channel response to fast ligand application. Here we describe a quantitative method to study ligand-gated ion channel activity and kinetics using a spectrofluorometric ion flux assay performed with a stopped-flow instrument. In the protocol below, we describe the channel reconstitution and ion flux assay steps we routinely use to characterize ligand-dependent channel kinetics. Similar procedures can be adapted for the study of channel blockade, the dependence of lipid composition, the influence of temperature, the channel response to agonists and antagonists, etc.
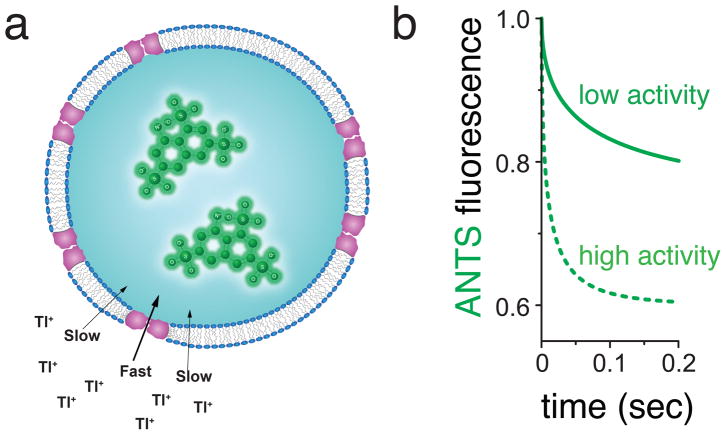
In this flux assay, open K+ channels reconstituted into large unilamellar vesicles (LUVs) allow the diffusive flux of external thallium ions (Tl+) into the liposomes, which then quench an encapsulated 8-Aminonaphthalene-1,3,6-Trisulfonic acid (ANTS) fluorophore (Figure 1a). The flux is rapid because K+ channels are permeable to Tl+ ions [2] (see Note 1). As Tl+ enters the liposomes, a charge-compensating exchange occurs, resulting in K+ exiting the liposomes. If the assay conditions favor high channel activity, the Tl+ influx rate will be high and the fluorescence quenching fast. If conditions favor less channel activity, the Tl+ flux will be comparably slower, as will the fluorescence quenching rate (Figure 1b). This assay format was first used for acetylcholine receptors [5,6] and fragmented sarcoplasmic reticulum membrane preparations [7,8]. A variant, based on Tl+ selective fluorescent indicators, later was employed and commercialized for cell-based flux assays [9]. The method also has been adapted for use in gramicidin channel assays [10–12], and more recently was employed using updated methods for biophysical studies on purified, reconstituted K+ channels [13–15]. Thus, although we here describe the assay as it was used for K+ selective channels, the assay will work equally well for any cation-selective ion channel that is permeable to Tl+.
Fig. 1.
General principle underlying the detection of K+ channel activity using Tl+ flux measurements. (a) K+ channels are reconstituted into liposomes with ANTS fluorophore inside, and rapidly mixed with Tl+-containing solutions using a stopped-flow mixing device. When channels are open, external Tl+ enter and exchange for K+ exiting the liposomes. The increasing internal Tl+ concentration creates a time-dependent quenching of the ANTS fluorescence inside the liposomes. (b) Higher K+ channel activity leads to faster ANTS fluorescence quenching rate than lower K+ channel activity (dotted line compared to solid line).
The first step in studying the function of a purified ion channel is to obtain the channel protein free from a cellular environment. In principle, the protein could derive from an in vitro translation system or extraction and purification from native biological specimens, but in practice this is most often accomplished using cultures of heterologous expression systems. The protocol we describe here typically requires several hundred micrograms of purified protein, a quantity achievable for numerous prokaryotic and eukaryotic ion channels. The protein expression and purification methodologies are not explicitly given here, as many reviews and books are available [16–19].
Once pure ion channel protein is available, it needs to be reconstituted into liposomes for the stopped-flow assay. Numerous methods for membrane protein reconstitution have been described [20]; we used SM-2 Bio-Beads to remove the detergent from mixtures of solubilized, purified protein and synthetic lipids, as this is an efficient way for making protein-reconstituted liposomes (proteoliposomes). The liposomes are next extruded through a 100 nm-pore-size membrane in order to obtain the LUVs. Since the liposomes are formed in the presence of ANTS, the fluorophore initially is both inside and outside the LUVs at equal concentrations. The liposome preparation is finished by passing the sample through a gravity-driven desalting column that exchanges the extra-liposomal solution with ANTS-free buffer, leaving the fluorophore only inside the liposomes. The sample is now ready to be used in channel activity assays [13].
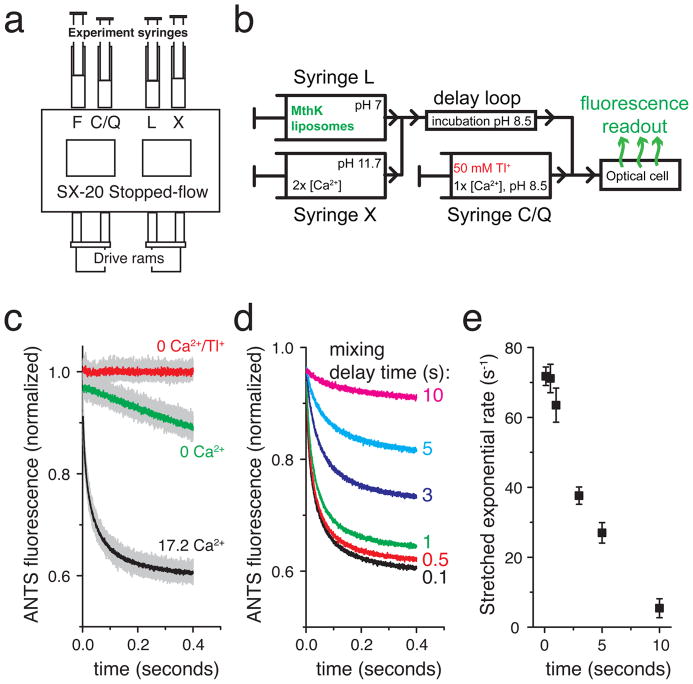
The assay is performed using a sequential mixing stopped flow device, in which the channel is activated in the first mixing reaction and channel activity is assayed in the second reaction. This multi-step mixing is crucial for accurate tracking of kinetic processes, and it allows for varying the time between the application of a particular assay condition such as ligand application (sample mixing #1, syringes L + X, Figure 2a–b) and testing the channel activity by application of Tl+ ions (sample mixing #2, syringe Q, Figure 2a–b). To accomplish this, we have used the SX20 instrument from Applied Photophysics, due to the high-performance specifications of this instrument, including low mixing dead time (<2 ms) and short minimum delay (~10 ms) between sample mixing #1 and #2. To illustrate the method explicitly, consider an experiment in which a Ca2+-activated K+ channel (in this case the MthK channel [21–24]) has been reconstituted into proteoliposomes. If the liposomes are mixed first with a Ca2+ solution (sample mixing #1, at 17.2 mM Ca2+ final concentration), Ca2+ will bind to the channels and activate them during incubation in the delay loop (Figure 2a–b). If this incubation is short (~100 ms), the channels are fully activated when sample mixing #2 occurs, resulting in a relatively fast flux rate (black, Figure 2c) compared with the slow TlNO3 leak across liposome membranes (green, Figure 2c). If the experiment is repeated using a longer incubation in the delay loop (up to 10 s), the fluorescence rate displays a gradual decrease over time, indicating that the MthK activity is decreasing; the channels desensitize in the presence of sustained Ca2+ (Figure 2d–e).
Fig. 2.
Kinetic measurements of channel activity are performed using a sequential-mixing stopped-flow spectrofluorometer. (a) Cartoon of the sample handling chamber of the SX20 stopped-flow device (Applied Photophysics). Liposomes in syringe L are mixed with experimental test conditions in syringe X. Following a user-defined reaction delay time, the L+X mixture is further mixed with the contents of syringe C/Q; C denotes a non-quenching control experiment and Q denotes a Tl+-containing quenching experiment. (b) Schematic flow diagram for the sequential mixing steps performed inside the stopped-flow apparatus for an experiment testing the Ca2+ activation of the MthK K+ channel. (c–e) Example data obtained with the MthK Ca2+-activated K+ channel. (c) Raw data from repeated mixing reactions (light gray data) with mean signal (dark data overlay) using 100 ms delay between mixing 1 and 2. Red: 0 Ca2+ in syringe X and 0 Tl+ in syringe C, Green: 0 Ca2+ in syringe X and 50 mM Tl+ in syringe Q, Black: 34.4 mM Ca2+ in syringe X and 50 mM Tl+ in syringe Q. (d) Mean ANTS fluorescence quenching time courses from the experiment in b with the mixing delay time varied between 0.1 and 10 seconds, illustrating the loss of MthK activity upon sustained exposure to Ca2+. (e) Mean stretched exponential rates from fitting individual mixing repeats from c–d to Equation 1 and calculating the rates using Equation 2. The error bars are standard deviations from the repeated mixing reactions as in c. Experimental replicates on independent samples should also be acquired.
To obtain quantitative estimates of flux rates from the raw fluorescence quench time courses (Figure 2c) we need a measure of the Tl+ flux into the liposomes immediately following solution mixing, i.e. just after the instrument mixing dead time of ~1.5 ms. This could be done by determining the slope of the fluorescence change at time t = 2 ms. However, due to the inevitable noise in the fluorescence signal, such a measurement is not always practical. Instead, a fit to the initial 100 ms of fluorescence data is used to describe the time course of fluorescence quenching. Due to the heterogeneity in liposomal volume and number of channels incorporated in the membrane, the fluorescence time course cannot be described by a single exponential decay (see Note 2), and we used a stretched exponential function instead [25] (Equation 1).
| (Equation 1) |
F(t) is the normalized fluorescence as a function of time t, F(0) the initial fluorescence, F(∞) the final steady-state fluorescence, τo a parameter with units of time, and β a parameter that depends on the dispersity of the proteoliposome preparation and describes the deviation from a single exponential decay. The fluorescence decay rate, and thus Tl+ influx rate following sample mixing (Equation 2), is evaluated at 2 ms (see Note 3):
| (Equation 2) |
The stretched exponential function was extensively tested and validated for this purpose during the development of the stopped-flow method for gramicidin activity assays [10] and has worked equally well in the analysis of Tl+ flux through K+ channels [13–15]. Given the large number of experiments typically performed, including repeated mixing reactions for a specific sample and experimental condition, multiple experimental conditions of interest, and ultimately, multiple sample preparations to confirm experimental reproducibility, it is convenient to develop a semi-automated analysis procedure using a software package such as Matlab® (Mathworks). However, analysis can similarly be implemented using any robust numerical data fitting routine.
2 Materials
Care should be taken when handling and disposing thallium reagents as they are toxic.
2.1 Preparations for K+ channel reconstitution
1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), in chloroform (Avanti Polar Lipids)
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (POPG), in chloroform (Avanti)
N2 gas tank, installed next to a fume hood
50 mL round-bottom flasks
SM-2 BioBeads (Bio-Rad)
Reconstitution buffer (Buffer R) (see Note 4): 100 mM KNO3, 10 mM HEPES pH 7.0
2.2 K+ channel reconstitution
Assay buffer (Buffer A): 140 mM KNO3, 10 mM HEPES pH 7.0
CHAPS detergent
ANTS (8-Aminonaphthalene-1,3,6-Trisulfonic acid, disodium salt, ThermoFisher), prepare a pH-adjusted 75 mM stock solution in reconstitution buffer (Buffer R)
15 mL glass test tubes
Mini-extruder with 100 nm polycarbonate membranes (Avanti)
PD-10 columns (GE Healthcare)
2.3 Tl+ flux assay
SX20 stopped-flow fluorescence spectrometer (Applied Photophysics) or similar with sequential push capability
Reaction buffer (Buffer X): Buffer A + 2x-concentrated ligands/blockers. For example, if the channel requires 100 μM Ca2+ to fully activate, use Buffer A + 200 μM Ca2+ to mix 1:1 with liposomes and fully activate the channel (see Note 5).
Control buffer (Buffer C): Buffer A + 1x-concentrated ligands/blockers. For example, if the channel requires 100 μM Ca2+ to fully activate, use Buffer A + 100 μM Ca2+ to mix 1:1 with the previous solution mixture of liposomes with Buffer X (see Note 5).
Quenching buffer (Buffer Q): 94 mM KNO3, 50 mM TlNO3, 10 mM HEPES pH 7.0 + 1x-concentrated ligands/blockers. For example, if the channel requires 100 μM Ca2+ to fully activate, use 100 μM Ca2+, as in Buffer C (see Note 5).
3 Methods
All steps are performed at room temperature. Glassware should be thoroughly cleaned, rinsed and dried. Make two independent samples with the procedure below. The first sample contains the protein to be studied and the second sample is a protein-free control.
3.1 Preparations for K+ channel reconstitution into liposomes
The day before ion channel reconstitution, mix 11.25 mg of DOPC and 3.75 mg of POPG (15 mg total of 3:1 ratio of DOPC and POPG from chloroform stock solutions) in a 50 mL round-bottom flask for a single batch of liposomes, either made with or without protein (see Note 6).
Evaporate chloroform using a stream of N2 gas inside a fume hood, rotating the flask by hand so that a thin, opaque film of lipid covers the bottom and sides of the flask.
Store the lipid-film containing flask in a vacuum desiccator overnight to eliminate trace chloroform.
Degas 5 g of SM-2 BioBeads by combining with 5 mL of reconstitution buffer (Buffer R) in a small vacuum flask and apply vacuum for two hours (see Note 7).
Sonicate the 50% BioBeads slurry for a couple of minutes in a bath sonicator to ensure the removal of trapped air. Most of the BioBeads should not float (see Note 7).
Remove the 5 mL of Buffer R from the BioBeads slurry carefully by pipetting and replace with a fresh 5 mL of Buffer R. Perform additional washes if the slurry buffer does not appear clear. Store the BioBeads at 4 °C for up to a few weeks.
Perform the expression and purification steps required to obtain a high-quality ion channel protein sample for the following day’s liposome reconstitution.
3.2 K+ channel reconstitution into liposomes
Finalize preparation of K+ channel sample. This could be an overnight-dialysis or gel purification step using reconstitution buffer (buffer R) or another compatible buffer (see Note 8).
Rehydrate lipid film in round-bottom flask by the addition of 1 mL Buffer R and 0.5 mL ANTS fluorophore stock solution.
Using a bath sonicator (see Note 9), sonicate the lipids into solution by the incremental addition CHAPS detergent to a final concentration of 35 mM (32 mg CHAPS).
Transfer lipid-detergent mixture to a glass test tube (~15 mL) with screw cap.
Add purified K+ channel to the lipid solution at a ratio of 20–30 μg protein per mg of lipid (see Note 10), mix and incubate for 30 minutes.
Add an additional 0.5 mL ANTS fluorophore stock solution and 2 mL 50% Bio-Beads slurry. The liposome batch is formed in ~3 mL final volume of Buffer R (see Note 11).
Incubate for 2 hours on a tube rocker. The solution will become turbid as detergent adsorbs to the Bio-Beads and liposomes are formed.
Sonicate the newly formed liposomes for 20 seconds in a bath sonicator.
Remove the Bio-Beads from the sample and convert the liposomes into large unilamellar vesicles (LUVs) by passing the sample through a 100 nm-pore-size polycarbonate membrane 42 times using an Avanti mini-extruder.
3.3 Transfer ANTS-filled liposomes to ANTS-free buffer
Equilibrate two PD-10 desalting columns each with 25 mL assay buffer (Buffer A) for a single 3 mL batch of liposomes. Allow the buffer to completely enter the column.
Add 2 mL of buffer A to the 3 mL liposomes sample and mix, bringing the sample volume up to 5 mL total.
Load 2.5 mL of sample onto each equilibrated PD-10 column. Allow the sample to completely enter the column.
Elute the liposomes by pipetting 3 mL buffer A onto each PD-10 column and combine the 3 mL eluates in a standard plastic conical tube. The liposomes solution should now be a turbid, colorless solution and the yellow ANTS should be visibly trapped inside the PD-10 columns. The 6 mL liposomal eluate has ANTS fluorophore inside but not outside the liposomes.
The concentrated, buffer-exchanged liposomes can be stored at 12 °C for several days without loss of intra-vesicular ANTS (see Note 12).
3.4 Stopped-flow Tl+ flux assay
Dilute the liposomes from a single preparation to 30 mL in assay buffer – buffer A (see Note 6).
Prepare the SX20 (or similar) stopped flow spectrofluorometer by turning on the system, any temperature control systems, light sources, and setting the excitation wavelength for the ANTS fluorophore (~350 nm) (see Note 13). Ensure that you are using a detection system/emission filter that can record ANTS fluorescence (~520 nm peak). Wash the fluid handling system thoroughly with water. Set the software to record 1 sec of data using 5000 recorded points and set the delay time between sequential sample mixing events to a desired value (Figure 1A, liposomes in syringe L are first mixed in a 1:1 volume ratio with the reaction buffer (Buffer X) in syringe X and incubated in the delay loop for the specified delay time. At the end of the delay time, the mixture L + X is further mixed with the Control/Quench buffer contained in syringe C/Q, again using a 1:1 volume ratio by concurrent loading of the optical cell with the contents of the delay loop (by the flushing action of syringe F) and those of syringe C/Q. Upon mixing L + X with C/Q the fluorescence time course data is recorded.
Begin by loading the fluorescent liposomes into syringe L and Assay buffer (Buffer A) into syringes X, C, and F. Ensure that the solutions are equilibrated to the experimental temperature (including room temperature). Wash in the experimental solutions to clear the system of water and filling the detection cell with the final experimental dilution of ANTS-containing liposomes (for the SX20 this requires about four repeated injections). Set the detector gain so that this maximum unquenched fluorescence signal is of appropriate magnitude for data acquisition instrument.
Wash the fluid handling system with water until the fluorescence reading has returned to the water-only baseline value to prepare for the actual experiment. For the SX20, this takes ten to twelve drives/injections from all four syringes to fully wash the system with water.
The experiment consists of 8 repeated control dual-mixing reactions (Tl+-free, Buffer C) and 11 repeated quenching measurements (Tl+ measurements, Buffer Q). Load sufficient proteoliposomes into drive syringe L for 19 measurements; sufficient Reaction buffer (Buffer X) in drive syringe X for 19 measurements; sufficient Flush buffer (Assay Buffer A) in drive syringe F for 19 measurements; sufficient Control buffer (Buffer C) for 8 control fluorescence measurements and, once the control traces have been acquired, sufficient Quenching buffer (Buffer Q) for 11 fluorescence quench measurements into drive syringe Q.
Acquire the control fluorescence signals using the SX20 control software to collect 8 repeated mixing reactions with the desired delay time. For the SX20 spectrofluorometer, the first 4–5 mixing reactions are necessary to completely wash out the water (or other solution) in the lines and measuring cell, and should not be used for analysis. The remaining traces are used for analysis and determination of the control fluorescence.
Switch the content of drive syringe C/Q to Buffer Q, and acquire the fluorescence quench measurements by using the software to collect 11 repeated mixing reactions. Again, discard the first 4–5 mixing reactions, which are necessary to wash out the control solutions. Following these, assuming that the K+ channels are active, you should see a time-dependent quenching of the ANTS fluorescence as Tl+ flows into the proteoliposomes through the active channels (exchanging with K+ inside the liposomes), as in Figure 1A.
Repeat steps 4–7 for all experimental conditions of interest. For example, perform titrations of activating ligands and/or channel blockers in Buffers C/Q and X. Alternatively, use a specific set of conditions and vary the delay time between consecutive series of mixing reactions to determine how the channel activity changes as a function of incubation time (see Figure 2) with the Reaction buffer (Buffer X).
Examine the data and discard any traces with obvious mixing artifacts. Normalize the fluorescence quench data to the control fluorescence, and fit a stretched exponential function (Equation 1) to the first 100 ms of the fluorescence quench trace for each experimental repeat. Use these fitted values of τ0 and β to calculate the relative fluorescence quench (Tl+ influx) rates at 2 ms (Equation 2), and use the flux rates from all experimental conditions to analyze ligand dose-response curves and/or ligand kinetics.
Acknowledgments
This work was supported by NIH grants R01 GM088352 (to CMN) and R01 GM021342 (to OSA), NIH fellowship F32GM087865 (to DJP), and a Kellen Fellowship (RR).
Footnotes
Tl+ can also be used for non-selective cation channels. The same fundamental assay principle can also be applied to anion channels, typically using a fluorophore such as N-(ethoxycarbonylmethyl)-6-methoxyquinolinium (MQAE), which can be quenched by chloride ions [26].
More precisely, if the liposomes were of uniform size, with the same number of channels per liposome, and the channels were in a perfect steady-state condition (conditions which never occur in practice), the internal [Tl+] concentration would increase according to a single exponential and the fluorescence would follow a simple Stern-Volmer fluorescence ratio according to that [Tl+]int time course [10].
The rate of the stretched exponential function cannot be extrapolated back to 0 ms, as the rate (according to Equation 1) goes to infinity as time goes to zero. This non-physical property of the stretched exponential can be side-stepped by using a “modified stretched exponential” function that derives from a simple transformation along the time axis [25]. Generally, this approach has not been necessary in the analysis of these experiments, but has been used to compare “modified” stretched exponential rates at 0 ms in specific cases [27,15].
All assay solutions employ NO3− anion because the chloride salt of thallium has limited water solubility.
We prepare our liposomes using pH 7.0 buffer. If, however, a flux experiment is done at another pH, it is necessary for the reaction buffer (Buffer X) to be a ‘pH-changing’ solution and the pH of Buffer X must be set so that a 1:1 volume-ratio mixture with pH 7.0 generates the desired experimental pH. Upon the second mixing with either the control buffer (Buffer C) or the quenching buffer (Buffer Q), the pH should be maintained at the desired pH and thus Buffer C and Buffer Q are ‘pH-maintaining’ buffers and are pH-adjusted to the experimental pH value. This procedure can be scaled-up or scaled-down as needed. Care should be taken to maintain the specified ratio of components and volumes used with PD-10 desalting columns. The final concentration of liposomes is diluted 10-fold from the initial liposome formation step. As written, 15 mg of lipid produce a 3 mL liposome suspension, which is diluted to 30 mL for the assays.
This procedure can be scaled-up or scaled-down as needed. Care should be taken to maintain the specified ratio of components and volumes used with PD-10 desalting columns. The final concentration of liposomes is diluted 10-fold from the initial liposome formation step. As written, 15 mg of lipid produce a 3 mL liposome suspension, which is diluted to 30 mL for the assays.
Initially washing the Bio-Beads with methanol followed by multiple washes with water before buffer can also be used.
We have always used gel filtration as the last step, performed immediately before the reconstitution procedure. If the purified protein can be concentrated to a fairly high concentration, and the buffer thus significantly diluted during reconstitution, a buffer containing chloride salts or other components may be used. If there is difficulty obtaining high-concentration protein, an effort should be made to ensure that the buffer is compatible with the experiment (ideally, identical to the assay buffer) and volumes and concentrations of the lipid components can be adjusted accordingly.
Do not allow the lipid-detergent solution get too hot. Both time and sonication will produce a clear solution, but this may take up to 60 minutes.
The protein amount to be added is dependent on the identity of the protein and sample quality. Don’t forget to make an identical protein-free control sample.
Dry BioBeads may also be added at the ration of 1 g BioBeads to 35 mg CHAPS. In this case addition of 0.5 mL ANTS to correct for the extra volume when adding BioBeads in 0.5% suspension is not necessary.
The liposomes are cooled (12 °C), but remain above the liquid-gel phase transition temperature to keep the ANTS from leaking out. The 12 °C incubator is most conveniently placed close to the stopped-flow device so that liposome samples can be taken out and warmed to room temperature immediately prior to use in the assay.
A 360 nm LED light source (Applied Photophysics) may also be used, which provides higher signal intensity and greater stability.
References
- 1.Aidley DJ. The physiology of excitable cells. 4. Cambridge University Press; Cambridge, UK ; New York, NY, USA: 1998. [Google Scholar]
- 2.Hille B. Ion channels of excitable membranes. 3. Sinauer; Sunderland, Mass: 2001. [Google Scholar]
- 3.Martinac B, Saimi Y, Kung C. Ion channels in microbes. Physiol Rev. 2008;88(4):1449–1490. doi: 10.1152/physrev.00005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordero-Morales JF, Cuello LG, Zhao Y, Jogini V, Cortes DM, Roux B, Perozo E. Molecular determinants of gating at the potassium-channel selectivity filter. Nature structural & molecular biology. 2006;13(4):311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
- 5.Moore HP, Raftery MA. Direct spectroscopic studies of cation translocation by Torpedo acetylcholine receptor on a time scale of physiological relevance. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(8):4509–4513. doi: 10.1073/pnas.77.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu WC, Moore HP, Raftery MA. Quantitation of cation transport by reconstituted membrane vesicles containing purified acetylcholine receptor. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(2):775–779. doi: 10.1073/pnas.78.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia AM, Miller C. Channel-mediated monovalent cation fluxes in isolated sarcoplasmic reticulum vesicles. The Journal of general physiology. 1984;83(6):819–839. doi: 10.1085/jgp.83.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia AM. Determination of ion permeability by fluorescence quenching. Methods Enzymol. 1992;207:501–510. doi: 10.1016/0076-6879(92)07035-m. [DOI] [PubMed] [Google Scholar]
- 9.Weaver CD, Harden D, Dworetzky SI, Robertson B, Knox RJ. A thallium-sensitive, fluorescence-based assay for detecting and characterizing potassium channel modulators in mammalian cells. J Biomol Screen. 2004;9(8):671–677. doi: 10.1177/1087057104268749. [DOI] [PubMed] [Google Scholar]
- 10.Ingolfsson HI, Andersen OS. Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay and drug development technologies. 2010;8(4):427–436. doi: 10.1089/adt.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingolfsson HI, Andersen OS. Alcohol’s effects on lipid bilayer properties. Biophysical journal. 2011;101(4):847–855. doi: 10.1016/j.bpj.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingolfsson HI, Sanford RL, Kapoor R, Andersen OS. Gramicidin-based fluorescence assay; for determining small molecules potential for modifying lipid bilayer properties. Journal of visualized experiments : JoVE. 2010;(44) doi: 10.3791/2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusinova R, Kim DM, Nimigean CM, Andersen OS. Regulation of ion channel function by the host lipid bilayer examined by a stopped-flow spectrofluorometric assay. Biophysical journal. 2014;106(5):1070–1078. doi: 10.1016/j.bpj.2014.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCoy JG, Rusinova R, Kim DM, Kowal J, Banerjee S, Jaramillo Cartagena A, Thompson AN, Kolmakova-Partensky L, Stahlberg H, Andersen OS, Nimigean CM. A KcsA/MloK1 chimeric ion channel has lipid-dependent ligand-binding energetics. The Journal of biological chemistry. 2014;289(14):9535–9546. doi: 10.1074/jbc.M113.543389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Posson DJ, Rusinova R, Andersen OS, Nimigean CM. Calcium ions open a selectivity filter gate during activation of the MthK potassium channel. Nature communications. 2015;6:8342. doi: 10.1038/ncomms9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Shaffer PL, Huang X, Rose PE. Rapid screening of membrane protein expression in transiently transfected insect cells. Protein expression and purification. 2013;88(1):134–142. doi: 10.1016/j.pep.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Schlegel S, Lofblom J, Lee C, Hjelm A, Klepsch M, Strous M, Drew D, Slotboom DJ, de Gier JW. Optimizing membrane protein overexpression in the Escherichia coli strain Lemo21(DE3) Journal of molecular biology. 2012;423(4):648–659. doi: 10.1016/j.jmb.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 18.Hays FA, Roe-Zurz Z, Stroud RM. Overexpression and purification of integral membrane proteins in yeast. Methods Enzymol. 2010;470:695–707. doi: 10.1016/S0076-6879(10)70029-X. [DOI] [PubMed] [Google Scholar]
- 19.Drew D, Newstead S, Sonoda Y, Kim H, von Heijne G, Iwata S. GFP-based optimization scheme for the overexpression and purification of eukaryotic membrane proteins in Saccharomyces cerevisiae. Nature protocols. 2008;3(5):784–798. doi: 10.1038/nprot.2008.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 21.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417(6888):515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 22.Zadek B, Nimigean CM. Calcium-dependent gating of MthK, a prokaryotic potassium channel. The Journal of general physiology. 2006;127(6):673–685. doi: 10.1085/jgp.200609534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Berke I, Chen L, Jiang Y. Gating and inward rectifying properties of the MthK K+ channel with and without the gating ring. The Journal of general physiology. 2007;129(2):109–120. doi: 10.1085/jgp.200609655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pau VP, Abarca-Heidemann K, Rothberg BS. Allosteric mechanism of Ca2+ activation and H+-inhibited gating of the MthK K+ channel. The Journal of general physiology. 2010;135(5):509–526. doi: 10.1085/jgp.200910387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berberan-Santos MN, Bodunov EN, Valeur B. Mathematical functions for the analysis of luminescence decays with underlying distributions 1. Kohlrausch decay function (stretched exponential) Chem Phys. 2005;315(1–2):171–182. doi: 10.1016/J.Chemphys.2005.04.006. [DOI] [Google Scholar]
- 26.Verkman AS. Development and biological applications of chloride-sensitive fluorescent indicators. Am J Physiol. 1990;259(3 Pt 1):C375–388. doi: 10.1152/ajpcell.1990.259.3.C375. [DOI] [PubMed] [Google Scholar]
- 27.Alejo JL, Blanchard SC, Andersen OS. Small-molecule photostabilizing agents are modifiers of lipid bilayer properties. Biophysical journal. 2013;104(11):2410–2418. doi: 10.1016/j.bpj.2013.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]