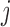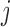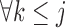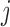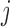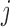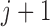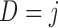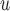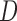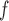SUMMARY
Cohort data are often incomplete because some subjects drop out of the study, and inverse probability weighting (IPW), multiple imputation (MI), and linear increments (LI) are methods that deal with such missing data. In cohort studies of ageing, missing data can arise from dropout or death. Methods that do not distinguish between these reasons for missingness typically provide inference about a hypothetical cohort where no one can die (immortal cohort). It has been suggested that inference about the cohort composed of those who are still alive at any time point (partly conditional inference) may be more meaningful. MI, LI, and IPW can all be adapted to provide partly conditional inference. In this article, we clarify and compare the assumptions required by these MI, LI, and IPW methods for partly conditional inference on continuous outcomes. We also propose augmented IPW estimators for making partly conditional inference. These are more efficient than IPW estimators and more robust to model misspecification. Our simulation studies show that the methods give approximately unbiased estimates of partly conditional estimands when their assumptions are met, but may be biased otherwise. We illustrate the application of the missing data methods using data from the ‘Origins of Variance in the Old–old’ Twin study.
Keywords: Discrete-time independent censoring, Dropout, Generalized estimating equation, Imputation, Longitudinal data, Missing at random, Partly conditional inference
1. Introduction
1.1. Motivation
Cohort studies involve measurements taken repeatedly over time, and studies with long follow-up often have missing data. A number of methods deal with missing data due to dropout in longitudinal studies (e.g. Schafer, 1997; Bang and Robins, 2005; Diggle and others, 2007, van Buuren and Groothuis-Oudshoorn, 2011), but not many describe how to handle missing data due to dropout and death (e.g. Kurland and Heagerty, 2005; Seaman and others, 2016).
We are motivated by the ‘Origins of Variance in the Old-Old’ (OCTO) Twin study. OCTO is a study of Swedish twins aged 80 years or older at recruitment, and it consists of five scheduled biennial visits. One continuous outcome measured in this study is peak expiratory function (henceforth, ‘lung function’) rate, which measures the maximal airflow at expiration, after maximal inspiration. In the older adults, lung function is associated with poor physical and cognitive health and mortality (Fragoso and others, 2007).
We assume a monotone missing data pattern, such as arises when subjects who drop out of the study do not return later. This is approximately true in the OCTO lung function data. In the OCTO study, 24% of the outcomes are missing due to death, and 27% are missing due to dropout, commonly because the subject had difficulty using the measuring instrument. Our goal is to estimate the expected lung function of survivors at each visit, and to understand how lung function is associated with smoking while subjects are alive. First, we describe estimands that may be of interest when outcomes are truncated by death.
1.2. Estimands
Suppose there are 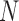 subjects in the study, and each subject has
subjects in the study, and each subject has 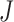 scheduled visits. Let
scheduled visits. Let 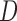 be the last visit before the subject dies, regardless of whether the subject actually attended the visit. If
be the last visit before the subject dies, regardless of whether the subject actually attended the visit. If 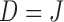 , the subject is still alive at the end of the study. Let
, the subject is still alive at the end of the study. Let 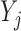 be the continuous outcome of interest at visit
be the continuous outcome of interest at visit 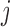 (
(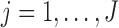 ).
).
Let 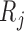 denote the corresponding response indicator, i.e.
denote the corresponding response indicator, i.e. 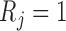 if
if 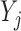 is observed, and
is observed, and 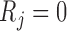 otherwise. Also let
otherwise. Also let 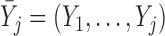 , and
, and 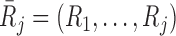 .
. 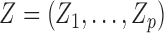 is a vector of
is a vector of 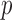 fully observed baseline covariates. We assume
fully observed baseline covariates. We assume 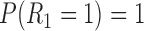 .
.
Three estimands of interest are unconditional, partly conditional, and fully conditional on death (Kurland and Heagerty, 2005). These are parameters in models for, respectively, 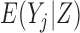 ,
, 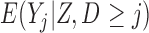 , and
, and 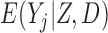 .
.
When missingness is due to death, it is important to distinguish between inference for a mortal cohort and an immortal cohort. Immortal cohort inference makes no distinction between missingness due to death and missingness due to dropout. Unconditional estimands describe associations in immortal cohorts because the definition of 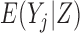 requires an implicit or explicit imputation of
requires an implicit or explicit imputation of 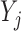 for those subjects who die before
for those subjects who die before 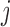 , thereby effectively creating a cohort that never dies (Dufouil and others, 2004). Since outcomes are undefined after death,
, thereby effectively creating a cohort that never dies (Dufouil and others, 2004). Since outcomes are undefined after death, 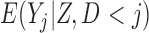 , and therefore
, and therefore 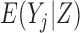 , is not meaningful. Hence, unconditional models (i.e. models for
, is not meaningful. Hence, unconditional models (i.e. models for 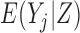 ) are generally inappropriate when there is a non-negligible amount of missing data due to death (Kurland and others, 2009). Linear Mixed-effects Models (LMM) provide immortal cohort inference unless
) are generally inappropriate when there is a non-negligible amount of missing data due to death (Kurland and others, 2009). Linear Mixed-effects Models (LMM) provide immortal cohort inference unless 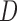 is included as a covariate or stratified on in the model.
is included as a covariate or stratified on in the model.
Partly conditional models (i.e. models for 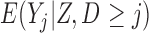 ) make inference about the expected outcome at a given time among survivors at that time point. Dufouil and others (2004) and Kurland and Heagerty (2005) favor partly conditional models. They argue that unconditional models would probably not be of interest unless the outcome and survival processes are independent. In this paper we focus on missing data methods that provide partly conditional inference.
) make inference about the expected outcome at a given time among survivors at that time point. Dufouil and others (2004) and Kurland and Heagerty (2005) favor partly conditional models. They argue that unconditional models would probably not be of interest unless the outcome and survival processes are independent. In this paper we focus on missing data methods that provide partly conditional inference.
Partly conditional models can be fitted using generalized estimating equations with independence working correlation matrix (IEE) when dropout among survivors at each time point is conditionally independent of the outcomes given 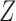 , but may give biased estimates otherwise. To weaken this assumption, Kurland and Heagerty (2005) described the inverse probability weighted (IPW) method using IEE to weight up observed subjects to represent subjects who are still alive but have dropped out. This requires the estimation of the probability of dropout among subjects who are alive, given earlier outcomes and covariates.
, but may give biased estimates otherwise. To weaken this assumption, Kurland and Heagerty (2005) described the inverse probability weighted (IPW) method using IEE to weight up observed subjects to represent subjects who are still alive but have dropped out. This requires the estimation of the probability of dropout among subjects who are alive, given earlier outcomes and covariates.
Multiple imputation (MI; Schafer, 1997) is a commonly used method for handling missing data. However, the literature on how to use MI to handle dropout and death is limited (Harel and others, 2007), and this literature does not address the assumptions under which MI gives valid partly conditional inference.
Diggle and others (2007) introduced linear increments (LI) as a further method for handling missing data. This method allows the underlying outcome to be measured with an error that is independent of the covariates and the underlying outcome process. The LI methods developed by Aalen and Gunnes (2010) allows for non-monotone missing data, but does not allow for independent measurement error (Seaman and others, 2016). Other work on LI includes Gran and others (2017) who use LI to make causal inference, and Hoff and others (2014) who provide software in R to implement the LI method. Building on the work by Aalen and Gunnes (2010) and Seaman and others (2016) provide the underlying assumptions under which LI is a valid method for making partly conditional inference.
We have several aims in this article. In Section 3, we define and compare the assumptions of MI, LI, and IPW for making partly conditional inference. In particular, we show that, when we do not stratify on 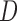 or include it as a covariate in the dropout or imputation model, the assumptions of MI and LI about the dropout and survival processes are different from those of IPW. In Section 4, we describe how to use MI, LI, and IPW when the imputation or dropout model stratifies on
or include it as a covariate in the dropout or imputation model, the assumptions of MI and LI about the dropout and survival processes are different from those of IPW. In Section 4, we describe how to use MI, LI, and IPW when the imputation or dropout model stratifies on 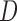 or includes it as a covariate. In Section 5, we illustrate graphically the assumptions described in Sections 3 and 4 using Directed Acyclic Graphs. In Section 6, we propose augmented IPW estimating equations for making partly conditional inference. These are attractive because they offer double protection against model misspecification. That is, they provide consistent estimations if the dropout or the imputation models are correctly specified. In Section 7, we provide simulation studies to compare bias and efficiency of the various missing data methods under various scenarios. Finally, in Section 8 we apply IPW, MI, and augmented IPW to data on lung function from the OCTO study. All proofs are in the supplementary material available at Biostatistics online (http://www.biostatistics.oxfordjournals.org).
or includes it as a covariate. In Section 5, we illustrate graphically the assumptions described in Sections 3 and 4 using Directed Acyclic Graphs. In Section 6, we propose augmented IPW estimating equations for making partly conditional inference. These are attractive because they offer double protection against model misspecification. That is, they provide consistent estimations if the dropout or the imputation models are correctly specified. In Section 7, we provide simulation studies to compare bias and efficiency of the various missing data methods under various scenarios. Finally, in Section 8 we apply IPW, MI, and augmented IPW to data on lung function from the OCTO study. All proofs are in the supplementary material available at Biostatistics online (http://www.biostatistics.oxfordjournals.org).
2. Motivating example
As a motivating example, we model the association of lung function with time, age at baseline (age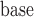 ), sex, education, smoking status, and the interactions between time and age
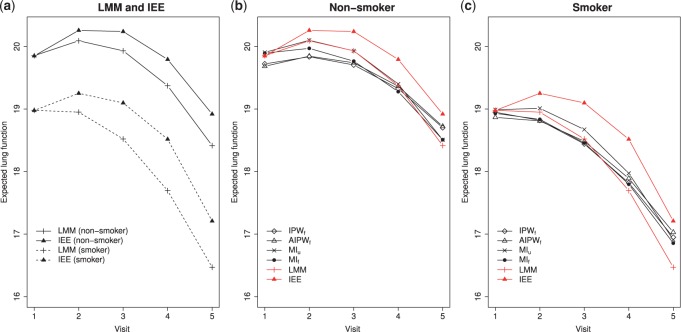
), sex, education, smoking status, and the interactions between time and age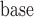 , sex, education, and smoking status in the OCTO study. Figure 1a shows the expected lung function of male smokers and non-smokers with average baseline age (83 years old) and average years of education (7 years). These means were estimated using both LMM and IEE. LMM do not distinguish between dropout and death, and IEE do not address dropout. In both smokers and non-smokers, the estimated means from LMM suggest a more rapid decline than do those from IEE. This is because unhealthier subjects (meaning those with poorer lung function) are more likely to die or drop out than are healthier subjects. Whereas IEE calculates the mean at each visit over only those subjects who have not yet dropped out or died (a healthier than average group), LMM calculates the mean over all subjects, implicitly imputing the (lower than average) missing lung functions. Moreover, because average lung function in smokers is lower than in non-smokers, there is more death in the smokers. Thus, the difference between the LMM and IEE estimates is greater for smokers than for non-smokers, with the result that LMM suggests smoking has a greater effect on rate of decline than does IEE. If dropout were independent of lung function, IEE would consistently estimate mean lung function conditional on still being alive. However, it is not. In the next two sections, we discuss various missing data methods that require weaker assumptions. A broad overview of these methods can be found in Table 1. Further discussion of this example can be found in Section 8.
, sex, education, and smoking status in the OCTO study. Figure 1a shows the expected lung function of male smokers and non-smokers with average baseline age (83 years old) and average years of education (7 years). These means were estimated using both LMM and IEE. LMM do not distinguish between dropout and death, and IEE do not address dropout. In both smokers and non-smokers, the estimated means from LMM suggest a more rapid decline than do those from IEE. This is because unhealthier subjects (meaning those with poorer lung function) are more likely to die or drop out than are healthier subjects. Whereas IEE calculates the mean at each visit over only those subjects who have not yet dropped out or died (a healthier than average group), LMM calculates the mean over all subjects, implicitly imputing the (lower than average) missing lung functions. Moreover, because average lung function in smokers is lower than in non-smokers, there is more death in the smokers. Thus, the difference between the LMM and IEE estimates is greater for smokers than for non-smokers, with the result that LMM suggests smoking has a greater effect on rate of decline than does IEE. If dropout were independent of lung function, IEE would consistently estimate mean lung function conditional on still being alive. However, it is not. In the next two sections, we discuss various missing data methods that require weaker assumptions. A broad overview of these methods can be found in Table 1. Further discussion of this example can be found in Section 8.
Fig. 1.
Expected lung function of men who were 83 years old at baseline with 7 years of education, stratified by smokers and non-smokers
Table 1.
Assumptions and general guidelines for making partly conditional inference
| Method | Assumption under which method is valid | Guidelines for use |
|---|---|---|
IPW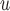
|
u-MAR (1) u-MAR is equivalent to mortal-cohort dDTIC and missingness-independent death (2) u-MAR is the discrete death-time version of MAR in Kurland and Heagerty (2005) |
|
IPW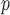
|
p-MAR Note: p-MAR is a weaker assumption than u-MAR |
|
IPW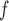
|
f-MAR (1) f-MAR is a weaker assumption than u-MAR (2) f-MAR is the discrete death-time version of MAR-S in Kurland and Heagerty (2005) |
|
MI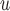 /LI /LI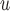
|
mortal-cohort dDTIC and independent death |
|
MI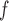 /LI /LI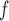
|
f-MAR |
|
AIPW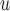
|
either i) u-MAR or ii) p-MAR, f-MAR and independent death |
|
AIPW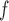
|
f-MAR |
|
3. Methods and assumptions
In this section, we describe how IPW, MI, and LI can be used to estimate the partly conditional estimand. We consider IPW, MI and LI where the dropout or imputation model do not stratify on 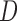 or include it as a covariate, but these situations are the focus of Section 4.
or include it as a covariate, but these situations are the focus of Section 4.
3.1. Conditions under which MI produces consistent parameter estimates
Joint multivariate normal MI (Schafer, 1997) is one of the most popular methods for imputing missing data. It is widely available in many general statistical packages and is particularly suitable for handling missing continuous outcome variables. As we point out later, MI is closely related to LI when data are monotone missing.
Let 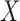 be a vector that includes
be a vector that includes 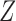 and possibly also fully observed variables that are predictive of
and possibly also fully observed variables that are predictive of 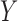 (so-called ‘auxiliary variables’). Suppose that
(so-called ‘auxiliary variables’). Suppose that
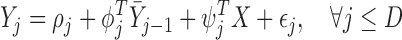 |
(3.1) |
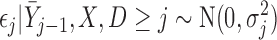 |
(3.2) |
In MI, (3.1) and (3.2) are assumed to hold and the data are assumed to be missing at random (MAR). If no distinction is made between outcomes missing due to death and those missing due to other reasons, then all missing outcomes are imputed. Consequently, inference obtained using the imputed data will be for a ‘supplemented’ outcome process that consists of the actual pre-death outcomes and additional hypothetical post-death outcomes (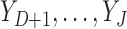 ). The pre-death outcomes are assumed to obey (3.1) and (3.2), and the hypothetical post-death outcomes are defined by
). The pre-death outcomes are assumed to obey (3.1) and (3.2), and the hypothetical post-death outcomes are defined by 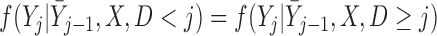 .
.
The joint distribution of  in this supplemented process is
in this supplemented process is 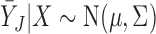 , where
, where  and
and 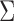 are functions of the
are functions of the 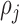 ,
, 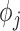 ,
, 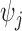 , and
, and 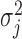 ’s. In MI, given a non-informative prior distribution for
’s. In MI, given a non-informative prior distribution for 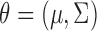 , missing outcomes in the supplemented process are drawn from their posterior predictive distribution. Conditional on
, missing outcomes in the supplemented process are drawn from their posterior predictive distribution. Conditional on 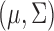 , if
, if 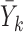 is observed but
is observed but 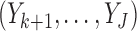 are missing,
are missing, 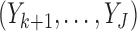 is sampled from the distribution
is sampled from the distribution 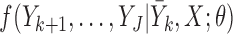 .
.
If a marginal model is fitted to all the imputed data including the post-death outcomes, an estimate of 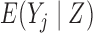 for the supplemented process is obtained. To fit a partly conditional model, it is necessary to delete the imputed post-death outcomes and retain only the pre-death outcomes. The following two conditions are then sufficient for consistent estimation of parameters in the partly-conditional model for
for the supplemented process is obtained. To fit a partly conditional model, it is necessary to delete the imputed post-death outcomes and retain only the pre-death outcomes. The following two conditions are then sufficient for consistent estimation of parameters in the partly-conditional model for 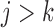 : (1)
: (1) 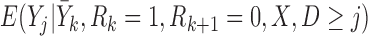 =
=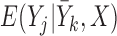 , and (2)
, and (2) 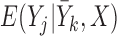 is consistently estimated. Suppose that condition (2) is true, and
is consistently estimated. Suppose that condition (2) is true, and 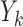 is the last observed outcome before visit
is the last observed outcome before visit 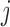 . Then the expected imputed value
. Then the expected imputed value 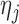 of
of 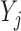 in a data set created by MI as
in a data set created by MI as 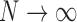 , is equal to
, is equal to 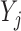 if
if 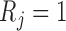 and
and 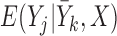 if
if 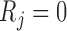 .
.
If condition (1) is satisfied, then as shown by Seaman and others (2016),  and this ensures that the parameters of the partly conditional model are consistently estimated. To estimate these parameters, we exclude post-death outcomes from the imputed data sets, and use IEE to analyze each.
and this ensures that the parameters of the partly conditional model are consistently estimated. To estimate these parameters, we exclude post-death outcomes from the imputed data sets, and use IEE to analyze each.
Condition (1) is satisfied if the conditional distribution of the missing outcomes in the supplemented process given the earlier observed outcomes and covariates is the same whether or not the missing outcomes are after death, i.e. if
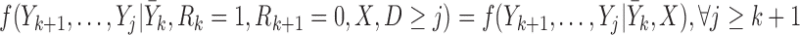 |
(3.3) |
Seaman and others (2016) proved that if (3.1) and (3.2) hold, and the two assumptions specified below (mortal-cohort dDTIC and independent death) are satisfied, then (3.3) (and hence condition (1)) holds. It can then be shown that these assumptions imply that the data on the supplemented process are MAR. This then implies that condition (2) holds.
Mortal-cohort discrete-time independent censoring in distribution (dDTIC) is defined by Seaman and others (2016) as
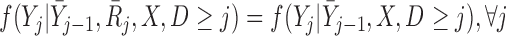 |
(3.4) |
which is equivalent to 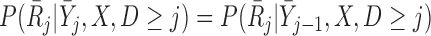 , i.e. conditional on survival up to visit
, i.e. conditional on survival up to visit 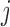 and outcomes prior to visit
and outcomes prior to visit 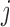 , the missingness history up and including visit
, the missingness history up and including visit 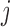 does not depend on the outcome at visit
does not depend on the outcome at visit 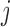 . Note that mortal cohort dDTIC looks similar to the classical MAR assumption conditional on subjects alive. However, in Appendix B of supplementary material available at Biostatistics online, we show that it is not the same.
. Note that mortal cohort dDTIC looks similar to the classical MAR assumption conditional on subjects alive. However, in Appendix B of supplementary material available at Biostatistics online, we show that it is not the same.
The independent death assumption is defined by Seaman and others (2016) as, 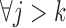 :
:
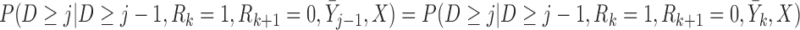 |
(3.5) |
This assumption says the probability of dying between visits  and
and 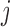 for people who attended visit
for people who attended visit 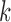 , but not visit
, but not visit 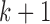 , could depend on the past observed outcomes, but not on any subsequent unobserved outcomes.
, could depend on the past observed outcomes, but not on any subsequent unobserved outcomes.
In summary, MI is a valid method for making partly conditional inference if (3.1) and (3.2) hold, mortal-cohort dDTIC and independent death are satisfied, and post-death outcomes are deleted before analyzing each imputed data set using IEE.
MI assumes that 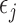 is normally distributed as described by (3.2). However, as shown in Theorem 4 of Seaman and others (2016), when data are monotone missing, this assumption is stronger than required for consistent estimation of the parameters in the partly conditional model. Instead, the following weaker condition is sufficient:
is normally distributed as described by (3.2). However, as shown in Theorem 4 of Seaman and others (2016), when data are monotone missing, this assumption is stronger than required for consistent estimation of the parameters in the partly conditional model. Instead, the following weaker condition is sufficient:
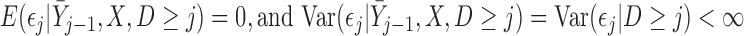 |
(3.6) |
Equation (3.2) is needed, though, for Rubin’s rules (Schafer, 1997) to give asymptotically unbiased estimation of the variance of the parameter estimators. Bootstrapping can be used to estimate this variance when (3.6) but not (3.2) holds.
3.2. A comparison with LI
For monotone missing data, LI imputation (Seaman and others, 2016) is asymptotically equivalent to MI with an infinite number of imputations. As with MI, the LI imputation method provides consistent parameter estimates of a model for 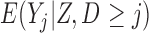 provided that (3.1) and (3.6) hold, mortal-cohort dDTIC and independent death hold, and post-death imputed outcomes are deleted before analyzing the imputed data using IEE (Seaman and others, 2016). Details about LI imputation are provided in Appendix A of supplementary material available at Biostatistics online.
provided that (3.1) and (3.6) hold, mortal-cohort dDTIC and independent death hold, and post-death imputed outcomes are deleted before analyzing the imputed data using IEE (Seaman and others, 2016). Details about LI imputation are provided in Appendix A of supplementary material available at Biostatistics online.
3.3. Conditions under which IPW produces consistent parameter estimates
Let 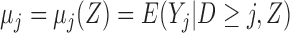 . A subject’s contribution to the IEE is
. A subject’s contribution to the IEE is
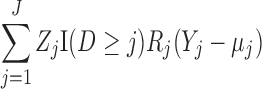 |
and the 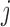 th element of the estimating equations is
th element of the estimating equations is 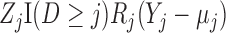 . In IPW, we multiply the
. In IPW, we multiply the 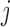 th element of the estimating equation by the inverse of
th element of the estimating equation by the inverse of 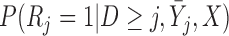 . Since the probability
. Since the probability 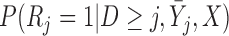 is unknown, it needs to be modeled. Let
is unknown, it needs to be modeled. Let  be a model for
be a model for 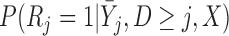 , with associated parameters
, with associated parameters  . The
. The  th element of the estimating equations becomes
th element of the estimating equations becomes
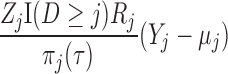 |
Dufouil and others (2004) provide non-algebraic descriptions of the assumptions of the IPW method. First, they say that they assume the “probability that [an outcome] is missing may depend on other observed parts of the response profile, but does not depend on unobserved [outcomes].” Second, they say that they assume the “mortality rates following dropout from the study are the same as the corresponding rates for subjects remaining in the study.” The first assumption is ambiguous because it appears to be describing MAR, which does not distinguish between subjects who are alive and dead, thereby making the assumption hard to interpret. The second assumption is ambiguous because it does not specify whether the mortality rates are conditional or unconditional on any of the past outcomes.
Kurland and Heagerty (2005) provide algebraic expressions for the assumptions of IPW and introduce three ways to model 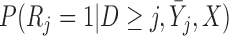 when the missingness depends on some of the past observed outcomes.
when the missingness depends on some of the past observed outcomes.
Neither Dufouil and others (2004) nor Kurland and Heagerty (2005) compare the assumptions of the various IPW methods or compare these methods with alternative methods. In this section and the next, we expand on their work by distinguishing the IPW assumptions and their corresponding estimators, and compare these assumptions to those of MI and LI imputation.
We distinguish between two MAR-type assumptions: unconditional MAR (u-MAR) and partly-conditional MAR (p-MAR). u-MAR is the assumption that
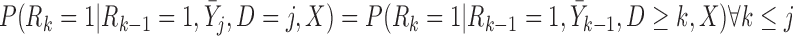 |
(3.7) |
which is equivalent to 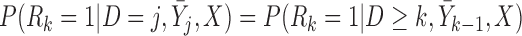 ,
, 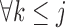 . Under u-MAR, the probability of observing the outcome at visit
. Under u-MAR, the probability of observing the outcome at visit 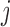 given survival up to that time is
given survival up to that time is
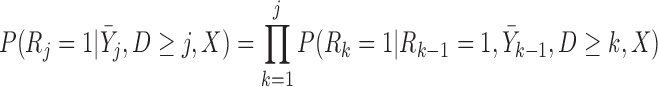 |
We can compare this assumption with the assumptions of MI and LI imputation. To do this, we define missingness-independent death as
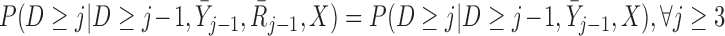 |
(3.8) |
In contrast to independent death, which says that the probability of dying between visits  and
and 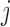 does not depend on the past missing outcomes given the past observed outcomes and the time of dropout, missingness-independent death says that the probability of dying between visits
does not depend on the past missing outcomes given the past observed outcomes and the time of dropout, missingness-independent death says that the probability of dying between visits  and
and 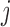 does not depend on the past missingness history given all of the past outcomes.
does not depend on the past missingness history given all of the past outcomes.
Theorem 1
u-MAR holds if and only if mortal-cohort dDTIC and missingness-independent death both hold.
p-MAR is the assumption that
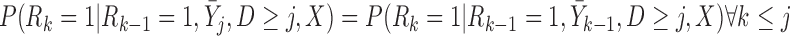 |
(3.9) |
or equivalently, 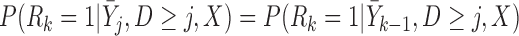 ,
, 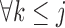 . Under p-MAR, the probability of observing the outcome at visit
. Under p-MAR, the probability of observing the outcome at visit 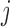 given survival up to that time is
given survival up to that time is
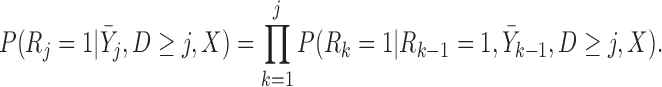 |
We might prefer to assume p-MAR rather than u-MAR because it is a weaker assumption than u-MAR. However, the IPW method based on the p-MAR assumption requires the survival statuses to be known up to the end of a study in order to fit models for 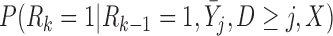 , whereas the IPW method based on the u-MAR assumption requires the survival statuses only to be known up to and including the time of dropout.
, whereas the IPW method based on the u-MAR assumption requires the survival statuses only to be known up to and including the time of dropout.
4. Methods fully conditional on death
4.1. IPW
If times of all deaths occurring prior to the end of the study are known, we could instead assume fully conditional MAR (f-MAR):
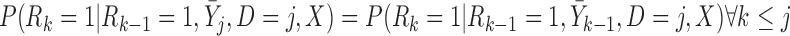 |
(4.1) |
or equivalently, 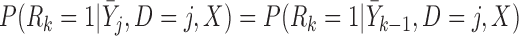 ,
, 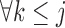 . Under f-MAR, the probability of observing the outcome at visit
. Under f-MAR, the probability of observing the outcome at visit 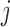 can be written as
can be written as
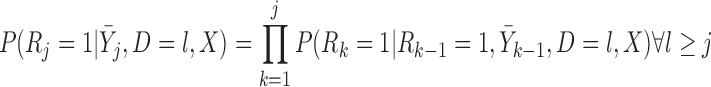 |
(4.2) |
u-MAR is a stronger assumption than p-MAR and f-MAR, so the IPW method that relies on the u-MAR assumption (henceforth called IPW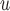 ) may be more efficient when this assumption is true than the IPW methods that rely on the p-MAR and f-MAR assumptions (henceforth called IPW
) may be more efficient when this assumption is true than the IPW methods that rely on the p-MAR and f-MAR assumptions (henceforth called IPW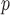 and IPW
and IPW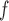 , respectively). Moreover, for IPW
, respectively). Moreover, for IPW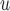 , survival statuses only need to be known up to and including the time of dropout, but for IPW
, survival statuses only need to be known up to and including the time of dropout, but for IPW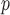 and IPW
and IPW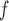 , survival statuses need to be known until the end of a study. Note that neither p-MAR nor f-MAR implies the other.
, survival statuses need to be known until the end of a study. Note that neither p-MAR nor f-MAR implies the other.
4.2. MI and LI
For MI and LI imputation, instead of assuming (3.1) and (3.2), suppose that for those subjects with 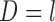 ,
,
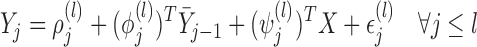 |
(4.3) |
with
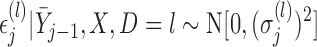 |
(4.4) |
The vector of outcomes 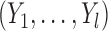 then has the joint distribution
then has the joint distribution 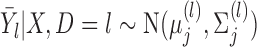 , where
, where 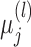 and
and 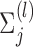 are functions of
are functions of 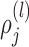 ,
, 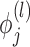 ,
, 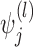 , and
, and 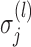 ’s. Under (4.3)–(4.4) and f-MAR, if visit
’s. Under (4.3)–(4.4) and f-MAR, if visit 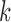 is the last visit attended before visit
is the last visit attended before visit 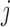 , then as
, then as 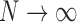 , the expected imputed value of
, the expected imputed value of 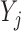 in a data set created by MI will be
in a data set created by MI will be 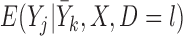 when
when 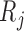 =0. As shown in Appendix H of supplementary material available at Biostatistics online, it is then valid to use the imputed data to estimate the partly conditional mean.
=0. As shown in Appendix H of supplementary material available at Biostatistics online, it is then valid to use the imputed data to estimate the partly conditional mean.
Henceforth, we will call MI and LI imputation based on (3.1) and (3.2) MI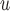 and LI
and LI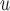 imputation, respectively. We will call MI and LI imputation based on (4.3) and (4.4) MI
imputation, respectively. We will call MI and LI imputation based on (4.3) and (4.4) MI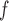 and LI
and LI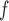 imputation, respectively.
imputation, respectively.
Instead of stratifying on 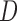 (as in (4.3) and (4.4)), we can include it as a covariate in the imputation model. Doing this may be a more feasible option when the sample size in at least some of the strata defined by
(as in (4.3) and (4.4)), we can include it as a covariate in the imputation model. Doing this may be a more feasible option when the sample size in at least some of the strata defined by 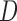 is small.
is small.
5. Directed Acyclic Graphs depicting scenarios for the missing data methods
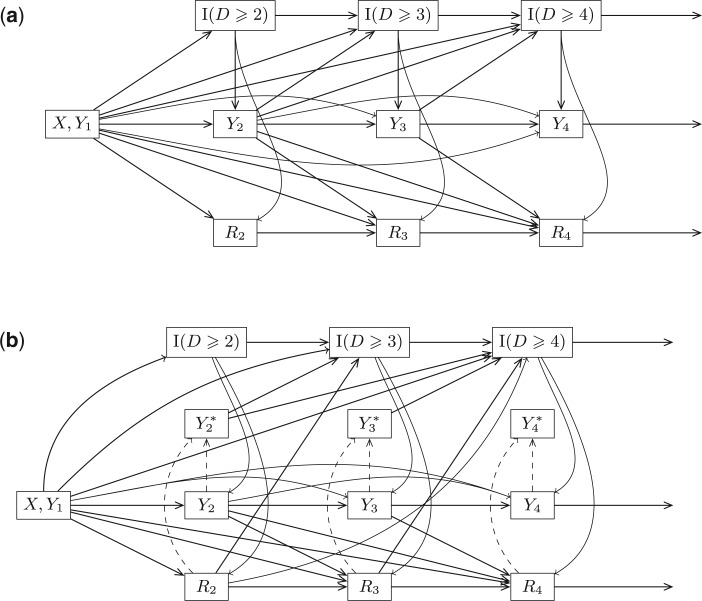
5.1. Graph 1: most complex scenario for u-MAR to hold
The Directed Acyclic Graphs in this section describe data generating mechanisms where the survival statuses, the longitudinal outcomes, and the dropout statuses are generated in a temporal order. For these graphs, we define 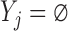 when
when 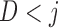 . Graph 1 (Figure 2a) shows the most complex scenario where u-MAR holds. It is the most complex scenario because adding a directed edge of the form
. Graph 1 (Figure 2a) shows the most complex scenario where u-MAR holds. It is the most complex scenario because adding a directed edge of the form 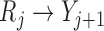 to Graph 1 would allow the mechanism to violate mortal-cohort dDTIC, and adding a directed edge
to Graph 1 would allow the mechanism to violate mortal-cohort dDTIC, and adding a directed edge 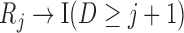 would allow it to violate missingness-independent death. When u-MAR holds, IPW
would allow it to violate missingness-independent death. When u-MAR holds, IPW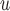 , IPW
, IPW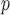 , and IPW
, and IPW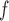 give consistent estimates, provided that the dropout models are correctly specified (Section 3.3).
give consistent estimates, provided that the dropout models are correctly specified (Section 3.3).
Fig. 2.
Directed Acyclic Graphs for scenarios 1 and 2. (a) Directed Acyclic Graph 1 for scenario 1 (Most complex scenario for u-MAR to hold). (b) Directed Acyclic Graph 2 for scenario 2 (Most complex scenario for mortal cohort dDTIC and independent death to hold). Dashed lines represent deterministic associations (e.g. 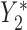 is determined by
is determined by 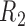 and
and 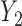 ).
).
Graph 1 allows the data generating mechanism to violate independent death, and so MI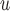 and LI
and LI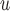 imputation may not provide consistent estimates under mechanisms described by Graph 1. However, u-MAR implies f-MAR. Consequently, MI
imputation may not provide consistent estimates under mechanisms described by Graph 1. However, u-MAR implies f-MAR. Consequently, MI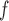 or LI
or LI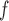 imputation would give consistent estimates, provided that the imputation models were correctly specified (Section 4.2).
imputation would give consistent estimates, provided that the imputation models were correctly specified (Section 4.2).
5.2. Graph 2: most complex scenario for mortal-cohort dDTIC and independent death to hold
Let 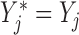 if
if 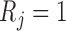 and
and 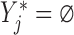 if
if 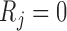 , and let
, and let 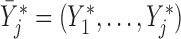 . So, if
. So, if 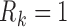 ,
, 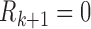 and
and 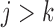 , then
, then 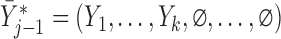 . Independent death can now be written as
. Independent death can now be written as
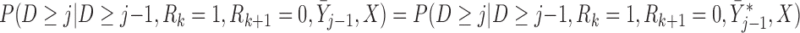 |
Introducing  allows us to draw a Directed Acyclic Graph that satisfies independent death. Graph 2 (Figure 2b) is the most complex scenario under which LI
allows us to draw a Directed Acyclic Graph that satisfies independent death. Graph 2 (Figure 2b) is the most complex scenario under which LI imputation and MI
imputation and MI provide consistent estimates. This is because adding a directed edge
provide consistent estimates. This is because adding a directed edge 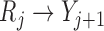 would allow violation of mortal-cohort dDTIC, and adding a directed edge
would allow violation of mortal-cohort dDTIC, and adding a directed edge 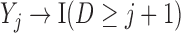 would allow violation of independent death.
would allow violation of independent death.
Graph 2 fails to satisfy missingness-independent death, and so, by Theorem 1, it fails to satisfy u-MAR. Hence, estimates from IPW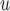 may be biased. Graph 2 also fails to satisfy p-MAR and f-MAR, because, for example, the directed edges
may be biased. Graph 2 also fails to satisfy p-MAR and f-MAR, because, for example, the directed edges 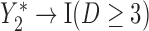 induce an association between
induce an association between 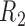 on
on 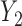 conditional on
conditional on 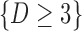 ,
, 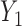 and
and 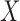 (as shown in Appendix C of supplementary material available at Biostatistics online). Therefore, estimates from IPW
(as shown in Appendix C of supplementary material available at Biostatistics online). Therefore, estimates from IPW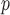 , IPW
, IPW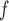 , MI
, MI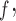 and LI
and LI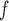 imputation may be biased.
imputation may be biased.
6. Augmented IPW
Robins, Rotnitzky, and colleagues introduced the augmented IPW (AIPW) method in a series of seminal papers including Robins and others (1995) and Scharfstein and others (1999). AIPW methods require specification of a model for the probability of dropout and an imputation model for the expectation of each missing outcome. The AIPW method is attractive because it provides consistent parameter estimates as long as one of these two models is correctly specified.
Bang and Robins (2005) and Seaman and Copas (2009) described AIPW to handle longitudinal data that are monotone missing due to dropout. We now adapt this method to handle missingness due to dropout and death. Like the IPW method for partly-conditional inference, this method requires an independence working correlation matrix.
Let 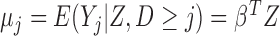 , where
, where 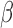 is a vector of parameters of interest, and assume that
is a vector of parameters of interest, and assume that 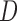 is included in
is included in 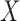 . For
. For 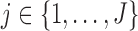 , let
, let 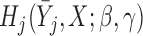 be a model for
be a model for 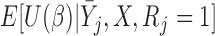 , where
, where 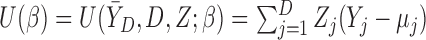 .
. 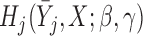 can be any function of
can be any function of 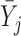 ,
, 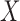 and parameters
and parameters 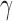 that obeys
that obeys 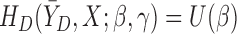 . Let
. Let 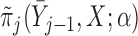 be a model for
be a model for 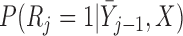 with parameters
with parameters 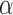 . Let
. Let 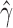 and
and 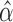 denote consistent estimators of
denote consistent estimators of 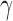 and
and  . A subject’s contribution to the AIPW estimating equations for
. A subject’s contribution to the AIPW estimating equations for  is,
is,
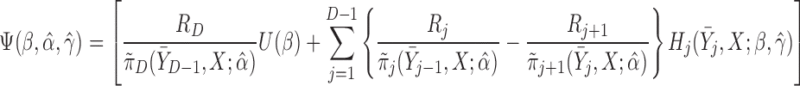 |
(6.1) |
The resulting estimators of  will be consistent if either
will be consistent if either 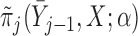 is correctly specified for all
is correctly specified for all  or
or 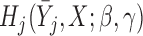 is correctly specified for all
is correctly specified for all  . If both models are correctly specified, this AIPW estimator is at least as efficient and usually more efficient than the IPW
. If both models are correctly specified, this AIPW estimator is at least as efficient and usually more efficient than the IPW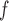 estimator.
estimator.
Theorem 2
If the data are f-MAR, the AIPW gives consistent estimations if either
or
is correctly specified. Moreover, consistent estimates of
are still obtained when the models for
and
omit the covariate
(or do not stratify on
) provided that either (1) u-MAR holds and the dropout model is correctly specified, or (2) f-MAR, p-MAR and independent death hold and
is correctly specified.
When models 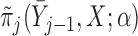 and
and 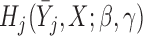 stratify on
stratify on  or include
or include  as a covariate, the AIPW method will be called AIPW
as a covariate, the AIPW method will be called AIPW . When
. When 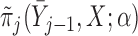 and
and 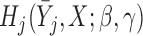 exclude
exclude 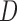 , it will be called AIPW
, it will be called AIPW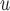 . Note that if the data generating mechanism satisfies u-MAR and independent death, then AIPW
. Note that if the data generating mechanism satisfies u-MAR and independent death, then AIPW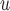 gives consistent estimations if either
gives consistent estimations if either 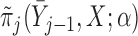 or
or 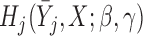 is correctly specified. We can use logistic regression to model dropout and Paik’s mean imputation (Paik, 1997) to estimate the conditional expectation of each missing outcome.
is correctly specified. We can use logistic regression to model dropout and Paik’s mean imputation (Paik, 1997) to estimate the conditional expectation of each missing outcome.
Under the data generating mechanism described by Directed Acyclic Graph 1, we expect AIPW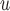 to yield consistent
to yield consistent  estimates provided that the dropout models are correctly specified, and AIPW
estimates provided that the dropout models are correctly specified, and AIPW to yield consistent
to yield consistent 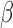 estimates provided that the dropout or the imputation models are correctly specified. However, under the data generating mechanism described by Directed Acyclic Graph 2, we expect AIPW
estimates provided that the dropout or the imputation models are correctly specified. However, under the data generating mechanism described by Directed Acyclic Graph 2, we expect AIPW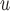 and AIPW
and AIPW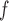 to yield inconsistent
to yield inconsistent 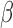 estimates in general.
estimates in general.
7. Simulation study
7.1. Methods
In the following simulation study, we compare the different assumptions underlying the missing data methods. The baseline covariate is generated and the parameters for the longitudinal, survival, and dropout models are chosen to mimic the data from the OCTO study.
We compare the bias, standardized bias and efficiency of the missing data methods. Standardized bias is 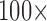 [(Average estimate
[(Average estimate True parameter)/empirical standard deviation of the parameter estimates]. It has been suggested that an absolute standardized bias of approximately 40% will be “practically significant” and will have a “noticeable adverse impact on efficiency, coverage, and error rates.” (Collins and others, 2001).
True parameter)/empirical standard deviation of the parameter estimates]. It has been suggested that an absolute standardized bias of approximately 40% will be “practically significant” and will have a “noticeable adverse impact on efficiency, coverage, and error rates.” (Collins and others, 2001).
Simulations 1 and 2 correspond to simplified versions (shown in Figure D.1 in the Appendix D of supplementary material available at Biostatistics online) of Directed Acyclic Graphs 1 and 2. In the simplified version of Graph 1, 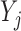 ,
, 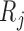 , and
, and 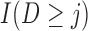 depend on
depend on 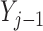 , but not on
, but not on 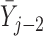 . In the simplified version of Graph 2,
. In the simplified version of Graph 2, 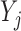 and
and 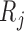 depend on
depend on 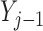 , but not on
, but not on 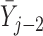 ;
; 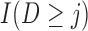 depends on the last observed outcome, but not on the ones before. In both of the simulations, data are generated in a sequential manner. For example, under simulation 1, data are generated by:
depends on the last observed outcome, but not on the ones before. In both of the simulations, data are generated in a sequential manner. For example, under simulation 1, data are generated by:
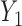 from
from 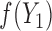 and
and 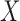 from
from 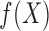
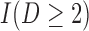 from
from 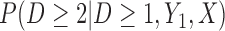
 from
from 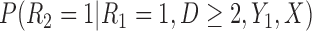 , and
, and 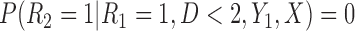
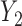 from
from 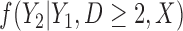 (
(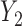 is not generated for those with
is not generated for those with 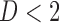 )
)
and so on. More details are given in Appendix D of supplementary material available at Biostatistics online. Approximately 24% of outcomes are missing due to death, and of those who are alive at each visit, approximately 27% are missing due to dropout. The analysis model we use for the simulations is
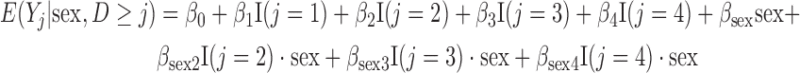 |
(7.1) |
For each simulation, we generate 1000 data sets, assume 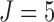 ,
, 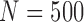 or 1000, and use 30 imputations in each MI procedure. The correct dropout and imputation models are used, with the exception of the AIPW
or 1000, and use 30 imputations in each MI procedure. The correct dropout and imputation models are used, with the exception of the AIPW method in simulation 1. In simulation 1, we show that AIPW
method in simulation 1. In simulation 1, we show that AIPW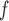 are doubly robust against model misspecification by using a misspecified dropout or a misspecified imputation model (omitting sex in one of the dropout and imputation models).
are doubly robust against model misspecification by using a misspecified dropout or a misspecified imputation model (omitting sex in one of the dropout and imputation models).
7.2. Results
In both simulation studies, IEE and complete case (subjects observed at all five visits) analysis overestimate the average outcomes in males and females at all visits. Table 2a and Table J.4 in Appendix J of supplementary material available at Biostatistics online, show, for 500 and 1000 subjects, the bias, standardized bias, and the empirical standard error for simulation 1. The biases from IPW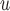 , IPW
, IPW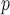 , IPW
, IPW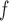 , AIPW
, AIPW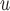 , AIPW
, AIPW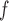 , LI
, LI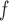 imputation, and MI
imputation, and MI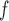 are negligible, but the biases from LI
are negligible, but the biases from LI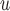 imputation and MI
imputation and MI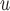 are bigger and/or practically significant. This is because, as explained in Section 5.1, Graph 1 fails to satisfy independent death.
are bigger and/or practically significant. This is because, as explained in Section 5.1, Graph 1 fails to satisfy independent death.
Table 2.
Results from simulation 1 and 2 (N=500). In the imputation and dropout models of method (1), 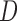 is modeled as a covariate, and in the imputation and dropout models of method (2),
is modeled as a covariate, and in the imputation and dropout models of method (2), 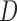 is stratified on.
is stratified on.
(a) Bias ( ), standardized bias (s-bias) and standard error ( ), standardized bias (s-bias) and standard error ( ) from simulation 1 ) from simulation 1 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parameter |
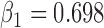
|
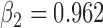
|
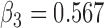
|
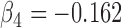
|
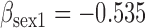
|
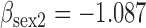
|
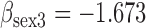
|
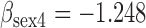
|
||||||||||||||||
| bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | |
CC
|
30.7 | 101.0 | 11.6 | 5.4 | 13.8 | 16.7 |
 14.0 14.0 |
 32.4 32.4 |
21.2 |
 64.1 64.1 |
 130.8 130.8 |
27.2 |
 8.1 8.1 |
 20.8 20.8 |
16.4 |
 8.0 8.0 |
 15.9 15.9 |
22.9 | 4.0 | 7.2 | 28.0 | 12.0 | 19.3 | 35.1 |
IEE
|
16.1 | 97.9 | 16.4 | 31.7 | 134.7 | 23.5 | 34.6 | 109.7 | 31.5 | 44.6 | 105.3 | 42.3 | 1.5 | 6.5 | 23.0 |
 5.2 5.2 |
 15.7 15.7 |
33.5 |
 6.7 6.7 |
 16.3 16.3 |
41.2 |
 4.5 4.5 |
 8.3 8.3 |
54.7 |
IPW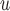 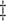
|
 0.3 0.3 |
 1.6 1.6 |
16.0 |
 0.7 0.7 |
 3.0 3.0 |
23.2 |
 0.8 0.8 |
 2.4 2.4 |
32.5 | 0.1 | 0.3 | 46.3 | 0.1 | 0.5 | 23.2 | 0.4 | 1.3 | 34.8 | 1.0 | 2.4 | 44.0 | 1.0 | 1.7 | 60.8 |
IPW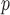 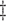
|
 0.3 0.3 |
 1.6 1.6 |
16.0 |
 0.8 0.8 |
 3.3 3.3 |
23.2 |
 0.9 0.9 |
 2.7 2.7 |
32.4 | 0.5 | 1.0 | 46.4 | 0.1 | 0.5 | 23.2 | 0.5 | 1.3 | 34.9 | 1.1 | 2.5 | 44.1 | 0.8 | 1.3 | 61.2 |
IPW (1) (1)
|
 0.3 0.3 |
 1.9 1.9 |
15.9 |
 0.8 0.8 |
 3.2 3.2 |
23.3 |
 0.8 0.8 |
 2.4 2.4 |
32.6 | 0.1 | 0.3 | 46.3 | 0.1 | 0.5 | 23.2 | 0.5 | 1.3 | 34.8 | 1.0 | 2.3 | 44.0 | 1.0 | 1.7 | 60.9 |
IPW (2) (2)
|
 0.4 0.4 |
 2.3 2.3 |
15.7 |
 1.0 1.0 |
 4.0 4.0 |
23.8 |
 0.6 0.6 |
 2.0 2.0 |
32.9 | 0.5 | 1.0 | 46.4 | 0.1 | 0.3 | 22.7 | 0.6 | 1.7 | 35.1 | 0.8 | 1.8 | 44.5 | 0.8 | 1.3 | 61.2 |
AIPW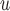 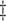
|
 0.2 0.2 |
 1.5 1.5 |
15.2 |
 1.1 1.1 |
 5.1 5.1 |
22.2 |
 1.3 1.3 |
 4.2 4.2 |
30.1 |
 1.3 1.3 |
 2.9 2.9 |
43.1 | 0.1 | 0.3 | 21.2 | 0.9 | 3.0 | 32.0 | 1.5 | 3.8 | 40.0 | 2.2 | 3.9 | 55.5 |
AIPW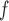 (1) (1)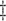
|
 0.3 0.3 |
 1.7 1.7 |
15.1 |
 1.3 1.3 |
 5.8 5.8 |
22.2 |
 1.4 1.4 |
 4.8 4.8 |
29.8 |
 0.9 0.9 |
 2.1 2.1 |
43.2 | 0.0 | 0.2 | 21.0 | 1.0 | 3.0 | 31.8 | 1.6 | 4.1 | 39.8 | 1.9 | 3.4 | 55.6 |
AIPW (2) (2)
|
 0.3 0.3 |
 2.0 2.0 |
15.1 |
 1.3 1.3 |
 5.8 5.8 |
22.6 |
 1.3 1.3 |
 4.2 4.2 |
30.2 |
 1.0 1.0 |
 2.3 2.3 |
43.2 | 0.0 | 0.0 | 21.0 | 0.9 | 2.7 | 32.2 | 1.4 | 3.6 | 40.4 | 2.0 | 3.6 | 55.7 |
LI 
|
 0.3 0.3 |
 1.6 1.6 |
15.3 |
 4.4 4.4 |
 20.2 20.2 |
21.8 |
 10.5 10.5 |
 36.5 36.5 |
28.7 |
 17.8 17.8 |
 43.9 43.9 |
40.5 | 0.1 | 0.5 | 21.2 | 1.1 | 3.5 | 31.0 | 4.8 | 12.6 | 37.9 | 7.0 | 13.6 | 51.4 |
LI (1) (1)
|
 0.4 0.4 |
 2.5 2.5 |
15.1 |
 2.3 2.3 |
 10.7 10.7 |
21.8 |
 1.0 1.0 |
 3.6 3.6 |
28.9 |
 1.6 1.6 |
 3.9 3.9 |
40.8 | 0.2 | 0.8 | 21.0 | 1.5 | 4.9 | 30.8 | 1.6 | 4.2 | 37.9 | 2.4 | 4.7 | 51.4 |
LI (2) (2)
|
 0.4 0.4 |
 2.9 2.9 |
15.2 |
 1.7 1.7 |
 7.8 7.8 |
22.0 |
 1.5 1.5 |
 5.1 5.1 |
29.7 |
 1.4 1.4 |
 3.4 3.4 |
41.4 | 0.2 | 0.8 | 21.1 | 1.4 | 4.6 | 31.2 | 1.8 | 4.5 | 39.0 | 2.4 | 4.6 | 52.4 |
MI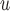 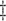
|
 0.4 0.4 |
 2.9 2.9 |
15.2 |
 5.3 5.3 |
 24.3 24.3 |
21.9 |
 12.0 12.0 |
 41.7 41.7 |
28.8 |
 19.7 19.7 |
 48.7 48.7 |
40.4 | 0.1 | 0.4 | 21.0 | 1.2 | 3.8 | 31.2 | 5.0 | 13.2 | 38.1 | 7.3 | 14.2 | 51.4 |
MI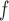 (1) (1)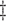
|
 0.6 0.6 |
 4.0 4.0 |
15.2 |
 1.7 1.7 |
 7.8 7.8 |
21.8 | 1.3 | 4.5 | 29.0 | 1.7 | 4.2 | 41.0 | 0.4 | 1.7 | 21.1 | 1.7 | 5.5 | 30.8 | 1.3 | 3.5 | 37.9 | 1.6 | 3.2 | 51.5 |
MI (2) (2)
|
 0.3 0.3 |
 2.2 2.2 |
15.2 |
 2.4 2.4 |
 10.8 10.8 |
22.1 |
 3.1 3.1 |
 10.4 10.4 |
29.8 |
 3.0 3.0 |
 7.3 7.3 |
41.4 | 0.2 | 0.9 | 21.1 | 1.6 | 5.1 | 31.4 | 1.9 | 4.7 | 39.1 | 2.6 | 4.9 | 52.6 |
 bias, standardized bias, and standard error for CC (complete case): bias, standardized bias, and standard error for CC (complete case): 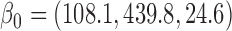 and and 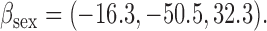
| ||||||||||||||||||||||||
 bias, standardized bias, and standard error for all other methods: bias, standardized bias, and standard error for all other methods: 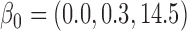 and and 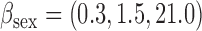 . . | ||||||||||||||||||||||||
(b) Bias ( ), standardized bias (s-bias) and standard error ( ), standardized bias (s-bias) and standard error ( ) from simulation 2 ) from simulation 2 | ||||||||||||||||||||||||
| Parameter |
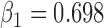
|
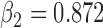
|
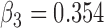
|
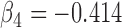
|
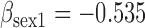
|
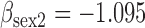
|
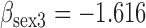
|
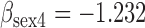
|
||||||||||||||||
| bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | bias | s-bias | SE | |
CC
|
25.7 | 92.0 | 11.6 | 7.2 | 20.5 | 16.7 | 0.9 | 2.2 | 21.2 |
 39.9 39.9 |
 88.9 88.9 |
27.2 |
 5.9 5.9 |
 16.2 16.2 |
16.4 |
 5.5 5.5 |
 12.1 12.1 |
22.9 |
 1.9 1.9 |
 3.7 3.7 |
28.0 | 6.8 | 11.9 | 35.1 |
| IEE¶ | 15.7 | 97.5 | 16.1 | 37.0 | 159.7 | 23.2 | 50.7 | 169.4 | 29.9 | 59.7 | 153.2 | 38.9 | 2.4 | 10.7 | 22.4 |
 4.1 4.1 |
 12.8 12.8 |
32.0 |
 13.6 13.6 |
 34.0 34.0 |
40.0 |
 9.7 9.7 |
 19.1 19.1 |
50.7 |
IPW ¶ ¶
|
 0.9 0.9 |
 5.5 5.5 |
15.6 | 3.1 | 13.0 | 23.6 | 13.3 | 42.2 | 31.5 | 11.5 | 28.1 | 41.0 | 1.1 | 5.0 | 22.7 | 2.0 | 6.0 | 32.9 |
 5.4 5.4 |
 12.7 12.7 |
42.4 |
 4.1 4.1 |
 7.4 7.4 |
55.9 |
IPW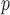 ¶ ¶
|
 0.9 0.9 |
 5.5 5.5 |
15.6 | 2.3 | 9.9 | 23.7 | 11.6 | 36.2 | 32.1 | 18.5 | 44.7 | 41.4 | 1.1 | 5.0 | 22.7 |
 0.2 0.2 |
 0.7 0.7 |
33.0 |
 10.4 10.4 |
 23.9 23.9 |
43.7 |
 14.3 14.3 |
 25.1 25.1 |
57.0 |
IPW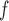 (1)¶ (1)¶
|
 1.3 1.3 |
 8.5 8.5 |
15.8 |
 17.4 17.4 |
 62.9 62.9 |
27.7 | 3.2 | 8.3 | 38.4 | 24.8 | 63.4 | 39.0 | 1.0 | 4.6 | 22.7 | 9.4 | 25.0 | 37.5 |
 12.1 12.1 |
 22.9 22.9 |
52.6 |
 10.0 10.0 |
 18.8 18.8 |
53.4 |
IPW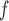 (2)¶ (2)¶
|
 3.6 3.6 |
 21.7 21.7 |
16.4 | 3.1 | 12.9 | 23.9 | 1.6 | 4.4 | 36.1 | 18.5 | 44.7 | 41.4 | 3.6 | 15.7 | 23.3 |
 7.3 7.3 |
 21.3 21.3 |
34.4 |
 8.0 8.0 |
 16.7 16.7 |
47.9 |
 14.3 14.3 |
 25.1 25.1 |
57.0 |
AIPW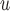 ¶ ¶
|
 0.8 0.8 |
 5.3 5.3 |
14.8 | 1.9 | 8.7 | 22.3 | 9.2 | 31.2 | 29.6 | 18.5 | 46.1 | 40.2 | 1.0 | 4.9 | 20.5 | 0.4 | 1.5 | 30.3 |
 5.0 5.0 |
 12.8 12.8 |
39.0 |
 12.2 12.2 |
 23.6 23.6 |
51.7 |
AIPW (1)¶ (1)¶
|
 1.9 1.9 |
 13.0 13.0 |
14.9 |
 2.3 2.3 |
 9.7 9.7 |
23.8 |
 3.7 3.7 |
 11.7 11.7 |
31.3 | 14.2 | 38.0 | 37.5 | 3.1 | 15.1 | 20.4 | 3.4 | 10.5 | 32.3 | 1.3 | 3.2 | 41.8 |
 8.2 8.2 |
 16.9 16.9 |
48.5 |
AIPW (2)¶ (2)¶
|
 4.7 4.7 |
 30.2 30.2 |
15.5 |
 1.5 1.5 |
 6.6 6.6 |
23.0 |
 3.2 3.2 |
 9.8 9.8 |
32.8 | 14.8 | 37.8 | 39.2 | 5.6 | 26.8 | 21.0 | 2.2 | 6.9 | 31.7 | 0.8 | 1.9 | 42.5 |
 8.9 8.9 |
 17.4 17.4 |
51.2 |
LI ¶ ¶
|
 0.8 0.8 |
 5.3 5.3 |
14.8 |
 1.0 1.0 |
 4.7 4.7 |
21.6 | 1.9 | 6.9 | 27.8 | 1.9 | 5.0 | 37.0 | 1.0 | 5.0 | 20.3 | 0.9 | 3.2 | 29.4 |
 1.8 1.8 |
 4.9 4.9 |
36.7 |
 3.2 3.2 |
 6.8 6.8 |
46.7 |
LI (1)¶ (1)¶
|
 2.0 2.0 |
 13.1 13.1 |
14.9 |
 2.6 2.6 |
 11.8 11.8 |
21.9 |
 3.5 3.5 |
 12.3 12.3 |
28.5 | 13.9 | 37.5 | 37.0 | 3.1 | 15.2 | 20.3 | 2.9 | 9.8 | 29.6 | 2.1 | 5.8 | 37.0 |
 7.2 7.2 |
 15.5 15.5 |
46.7 |
LI (2)¶ (2)¶
|
 4.6 4.6 |
 29.7 29.7 |
15.4 |
 2.1 2.1 |
 9.6 9.6 |
22.4 |
 4.1 4.1 |
 12.8 12.8 |
32.2 | 13.6 | 36.4 | 37.4 | 5.4 | 26.1 | 20.6 | 3.1 | 10.2 | 30.4 | 1.9 | 4.6 | 41.1 |
 7.3 7.3 |
 15.3 15.3 |
47.6 |
MI ¶ ¶
|
 1.0 1.0 |
 7.0 7.0 |
14.8 |
 1.8 1.8 |
 8.2 8.2 |
21.7 | 0.7 | 2.4 | 28.0 | 0.3 | 0.7 | 37.1 | 0.9 | 4.3 | 20.4 | 0.9 | 3.0 | 29.6 |
 1.8 1.8 |
 5.0 5.0 |
37.1 |
 3.2 3.2 |
 6.9 6.9 |
46.9 |
MI (1)¶ (1)¶
|
 2.3 2.3 |
 15.2 15.2 |
14.9 |
 1.8 1.8 |
 8.3 8.3 |
22.0 |
 1.4 1.4 |
 5.0 5.0 |
28.7 | 16.0 | 43.1 | 37.0 | 3.5 | 17.0 | 20.3 | 3.0 | 9.9 | 29.7 | 2.1 | 5.6 | 37.1 |
 7.3 7.3 |
 15.5 15.5 |
47.0 |
MI (2)¶ (2)¶
|
 4.7 4.7 |
 30.3 30.3 |
15.3 |
 2.4 2.4 |
 10.5 10.5 |
22.4 |
 4.4 4.4 |
 13.5 13.5 |
32.2 | 14.1 | 37.5 | 37.5 | 5.5 | 26.6 | 20.6 | 3.2 | 10.7 | 30.3 | 2.1 | 5.1 | 41.3 |
 7.2 7.2 |
 15.1 15.1 |
47.7 |
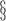 bias, standardized bias, and standard error for CC (complete case): bias, standardized bias, and standard error for CC (complete case): 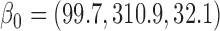 and and 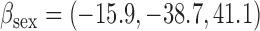 . . | ||||||||||||||||||||||||
¶bias, standardized bias, and standard error for all other methods: 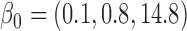 and and 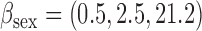 . . | ||||||||||||||||||||||||
Table J.1 in Appendix J of supplementary material available at Biostatistics online shows the double robust property of AIPW . When the dropout models are misspecified but the imputation models are correctly specified (and vice versa), the biases from AIPW
. When the dropout models are misspecified but the imputation models are correctly specified (and vice versa), the biases from AIPW are negligible. However, when both the dropout and the mean imputation models are misspecified, the biases are practically significant.
are negligible. However, when both the dropout and the mean imputation models are misspecified, the biases are practically significant.
Table 2b and Table J.5 in Appendix J of supplementary material available at Biostatistics online, show, for 500 and 1000 subjects, the bias, standardized bias, and the empirical standard error for simulation 2. The biases from LI imputation and MI
imputation and MI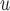 are negligible, but the biases from IPW
are negligible, but the biases from IPW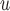 , IPW
, IPW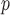 , IPW
, IPW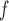 , AIPW
, AIPW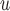 , AIPW
, AIPW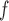 , LI
, LI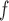 imputation, and MI
imputation, and MI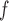 are bigger and/or practically significant. This is because, as explained in Section 5.2, Graph 2 fails to satisfy u-MAR, p-MAR, and f-MAR.
are bigger and/or practically significant. This is because, as explained in Section 5.2, Graph 2 fails to satisfy u-MAR, p-MAR, and f-MAR.
With respect to efficiency, the standard errors under the IPW methods are larger than those under MI and LI imputation (Table 2a). Moreover, the standard errors under AIPW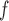 are equal to or less than equal to those under IPW
are equal to or less than equal to those under IPW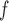 when both dropout and imputation models are correctly specified. The standard errors from IPW
when both dropout and imputation models are correctly specified. The standard errors from IPW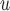 , IPW
, IPW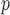 , and IPW
, and IPW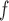 are not very different; the standard errors under IPW
are not very different; the standard errors under IPW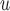 are slightly smaller than those under the other two IPW methods. We would expect to see a larger difference if the sample size were smaller, because unstable weights are more likely to be obtained when using IPW
are slightly smaller than those under the other two IPW methods. We would expect to see a larger difference if the sample size were smaller, because unstable weights are more likely to be obtained when using IPW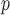 or IPW
or IPW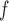 than when using IPW
than when using IPW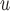 .
.
In Appendix E of supplementary material available at Biostatistics online, we show an additional simulation (simulation 3) to demonstrate bias from methods whose dropout and/or imputation models do not include 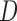 (i.e. IPW
(i.e. IPW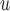 , IPW
, IPW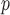 , AIPW
, AIPW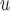 , and MI
, and MI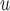 ) when data are f-MAR. We do not follow the sequential data generating mechanism in simulation 3. Instead,
) when data are f-MAR. We do not follow the sequential data generating mechanism in simulation 3. Instead, 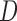 is generated first, so that it can be included in the models for the outcome and dropout process. Tables J.2 and J.3 in Appendix J of supplementary material available at Biostatistics online show the bias, standardized bias, and empirical standard error for 500 subjects for simulation 3. Biases from IPW
is generated first, so that it can be included in the models for the outcome and dropout process. Tables J.2 and J.3 in Appendix J of supplementary material available at Biostatistics online show the bias, standardized bias, and empirical standard error for 500 subjects for simulation 3. Biases from IPW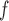 , AIPW
, AIPW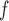 , and MI
, and MI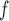 are negligible, but biases from IPW
are negligible, but biases from IPW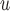 , IPW
, IPW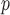 , AIPW
, AIPW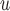 , and MI
, and MI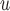 are practically significant.
are practically significant.
8. Application
In the OCTO study, for the subjects who had at least one lung function measurement, 22.5% of the outcomes are missing due to monotone dropout and 5.8% of the outcomes are intermittent missing. We ignored the 204 subjects with no data on lung function and forced the missingness pattern to be monotone by ignoring the relatively small number of outcomes observed after a subject’s first missing outcome. Data are then available on 437 subjects; 100 (22.9%) of whom have complete data up to visit 5.
To account for the slightly right skewness in the lung function variable (peak expiratory function), we took its square-root transform. The partly conditional model of interest is
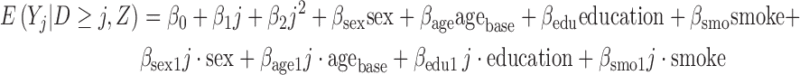 |
(8.1) |
where binary variables 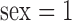 if subject is female, and
if subject is female, and 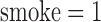 if a subject has ever smoked.
if a subject has ever smoked.
The dropout and imputation models at visit 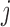 include sex, education, smoking status, past outcomes
include sex, education, smoking status, past outcomes 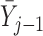 , and baseline age, Mini-Mental State Exam score, instrumental activities of daily living score, and health prevention score (a measure of the degree to which a subject’s health prevents him from doing things that he likes to do). Mini-Mental State Exam score, instrumental activities of daily living score, and health prevention score can tell us about a subject’s cognitive function, physical ableness, and overall health status, respectively. Exploratory analyses suggest that these auxiliary variables are all associated with both lung function and dropout.
, and baseline age, Mini-Mental State Exam score, instrumental activities of daily living score, and health prevention score (a measure of the degree to which a subject’s health prevents him from doing things that he likes to do). Mini-Mental State Exam score, instrumental activities of daily living score, and health prevention score can tell us about a subject’s cognitive function, physical ableness, and overall health status, respectively. Exploratory analyses suggest that these auxiliary variables are all associated with both lung function and dropout.
If we assume mortal-cohort dDTIC and independent death hold, then the probability of dying between two visits, given past observed lung function data, missingness history, and covariates such as age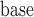 , does not depend on the past missing lung function data. But if we assume that the data were u-MAR, then probability of dying between two visits given covariates and all past lung function data does not depend on the missingness history. Under u-MAR, the probability of dropout at visit
, does not depend on the past missing lung function data. But if we assume that the data were u-MAR, then probability of dying between two visits given covariates and all past lung function data does not depend on the missingness history. Under u-MAR, the probability of dropout at visit 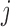 given past outcomes, covariates, and survival up to visit
given past outcomes, covariates, and survival up to visit 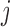 does not depend on lung function data after visit
does not depend on lung function data after visit  and
and 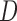 . But p-MAR allows this probability to depend on subjects who are alive at future visits, and f-MAR allow this probability to depend on
. But p-MAR allows this probability to depend on subjects who are alive at future visits, and f-MAR allow this probability to depend on 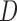 .
.
As a preliminary step in the data analysis we could test the u-MAR assumption if information on death is available by modeling the association between dropout and 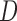 . In the OCTO study, we found strong associations between the probability of dropout and
. In the OCTO study, we found strong associations between the probability of dropout and 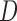 while controlling for past outcomes and covariates. Hence, we deduced that u-MAR was implausible in this example.
while controlling for past outcomes and covariates. Hence, we deduced that u-MAR was implausible in this example.
The parameters from the dropout models fitted for IPW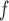 showed that dropout was associated with age
showed that dropout was associated with age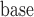 , lung function at last visit, and the auxiliary variables. The strongest predictor was lung function at the last visit. For example, for those who died between visits 3 and 4, the probabilities of dropout between visits 2 and 3 and between 3 and 4 were higher for subjects who had lower lung function at visits 1 and 2, respectively: the estimated log-odds ratios per unit increase in lung function were respectively
, lung function at last visit, and the auxiliary variables. The strongest predictor was lung function at the last visit. For example, for those who died between visits 3 and 4, the probabilities of dropout between visits 2 and 3 and between 3 and 4 were higher for subjects who had lower lung function at visits 1 and 2, respectively: the estimated log-odds ratios per unit increase in lung function were respectively  0.23 (
0.23 (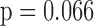 ) and
) and  0.75 (
0.75 (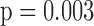 ).
).
Table 3 shows estimates from the complete case analysis (i.e. using only subjects with observed lung function at all five visits), LMM, IEE, IPW, AIPW, and MI. Standard errors were calculated by bootstrapping from the original data. Estimates from the complete case analysis are different from the other methods. Since subjects with better lung function are more likely to be observed than those with poorer lung function, the complete cases are not representative of all subjects who remain in the study at each visit. Hence, the complete case analysis is not valid for partly conditional inference, and we focus on the results from IPW, MI, and AIPW.
Table 3.
Parameter estimate (standard error) of OCTO data application
|
|
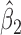
|
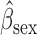
|
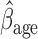
|
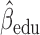
|
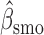
|
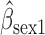
|
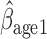
|
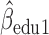
|
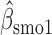
|
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | 20.326 (0.831)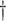
|
0.143 (0.108) |
 0.042 (0.009) 0.042 (0.009)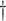
|
 2.502 (0.612) 2.502 (0.612)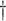
|
 0.108 (0.086) 0.108 (0.086) |
0.093 (0.085) |
 0.135 (0.556) 0.135 (0.556) |
 0.031 (0.070) 0.031 (0.070) |
0.008 (0.014) |
 0.004 (0.013) 0.004 (0.013) |
 0.132 (0.071) 0.132 (0.071) |
| LMM | 19.612 (0.446)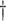
|
0.203 (0.093)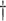
|
 0.050 (0.007) 0.050 (0.007)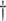
|
 3.084 (0.310) 3.084 (0.310)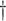
|
 0.233 (0.049) 0.233 (0.049)
|
0.235 (0.055)
|
 0.873 (0.309) 0.873 (0.309)
|
0.002 (0.055) | 0.012 (0.010) |
 0.007 (0.010) 0.007 (0.010) |
 0.134 (0.057) 0.134 (0.057)
|
| IEE | 19.657 (0.414)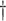
|
0.333 (0.111)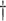
|
 0.053 (0.009) 0.053 (0.009)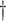
|
 3.085 (0.291) 3.085 (0.291)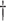
|
 0.238 (0.052) 0.238 (0.052)
|
0.228 (0.043)
|
 0.872 (0.291) 0.872 (0.291)
|
0.028 (0.070) | 0.022 (0.013) |
 0.022 (0.015) 0.022 (0.015) |
 0.067 (0.069) 0.067 (0.069) |
IPW
|
19.675 (0.438)
|
0.076 (0.149) |
 0.038 (0.013) 0.038 (0.013)
|
 3.018 (0.315) 3.018 (0.315)
|
 0.271 (0.064) 0.271 (0.064)
|
0.249 (0.052)
|
 0.932 (0.324) 0.932 (0.324)
|
0.071 (0.085) | 0.026 (0.015) |
 0.014 (0.019) 0.014 (0.019) |
 0.111 (0.088) 0.111 (0.088) |
IPW
|
19.618 (0.436)
|
0.156 (0.151) |
 0.043 (0.011) 0.043 (0.011)
|
 2.996 (0.308) 2.996 (0.308)
|
 0.272 (0.060) 0.272 (0.060)
|
0.250 (0.050)
|
 0.866 (0.315) 0.866 (0.315)
|
0.041 (0.089) | 0.030 (0.016) |
 0.019 (0.018) 0.019 (0.018) |
 0.120 (0.093) 0.120 (0.093) |
IPW
|
19.806 (0.442)
|
0.091 (0.147) |
 0.031 (0.012) 0.031 (0.012)
|
 3.073 (0.308) 3.073 (0.308)
|
 0.335 (0.078) 0.335 (0.078)
|
0.251 (0.048)
|
 0.772 (0.311) 0.772 (0.311)
|
0.042 (0.084) | 0.037 (0.015)
|
 0.024 (0.018) 0.024 (0.018) |
 0.123 (0.088) 0.123 (0.088) |
AIPW
|
19.560 (0.434)
|
0.218 (0.142) |
 0.048 (0.013) 0.048 (0.013)
|
 2.980 (0.297) 2.980 (0.297)
|
 0.246 (0.057) 0.246 (0.057)
|
0.236 (0.049)
|
 0.844 (0.298) 0.844 (0.298)
|
 0.032 (0.087) 0.032 (0.087) |
0.019 (0.022) |
 0.004 (0.016) 0.004 (0.016) |
 0.112 (0.087) 0.112 (0.087) |
AIPW
|
19.745 (0.460)
|
0.065 (0.157) |
 0.034 (0.016) 0.034 (0.016)
|
 3.026 (0.314) 3.026 (0.314)
|
 0.336 (0.091) 0.336 (0.091)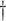
|
0.256 (0.049)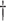
|
 0.817 (0.312) 0.817 (0.312)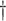
|
0.017 (0.079) | 0.036 (0.020) |
 0.012 (0.014) 0.012 (0.014) |
 0.110 (0.081) 0.110 (0.081) |
MI
|
19.722 (0.420)
|
0.172 (0.119) |
 0.045 (0.011) 0.045 (0.011)
|
 (0.293) (0.293)
|
0.000 (0.066) | 0.014 (0.014) |
 0.008 (0.013) 0.008 (0.013) |
 0.084 (0.066) 0.084 (0.066) |
|||
MI
|
19.750 (0.423)
|
0.079 (0.128) |
 0.035 (0.012) 0.035 (0.012)
|
 3.055 (0.301) 3.055 (0.301)
|
 0.253 (0.053) 0.253 (0.053)
|
0.231 (0.047)
|
 0.963 (0.299) 0.963 (0.299)
|
0.003 (0.064) | 0.016 (0.013) |
 0.007 (0.013) 0.007 (0.013) |
 0.086 (0.066) 0.086 (0.066) |
 Denote parameters that are statistically significant at the 5% level. CC, complete case analysis.
Denote parameters that are statistically significant at the 5% level. CC, complete case analysis.
In general, the rate of decline in lung function increases over time (MI :
: 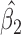 =
= 0.035,
0.035, 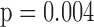 ), and this rate of decline is also estimated to be greater if we assume mortal-cohort dDTIC and independent death (MI
), and this rate of decline is also estimated to be greater if we assume mortal-cohort dDTIC and independent death (MI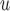 :
: 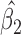 =
= 0.045,
0.045, 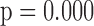 ). This is reflected in Figure 1 (b) and (c). The estimated rate of decline is also different between IPW
). This is reflected in Figure 1 (b) and (c). The estimated rate of decline is also different between IPW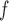 and IPW
and IPW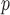 : IPW
: IPW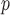 suggests a steeper rate of decline than IPW
suggests a steeper rate of decline than IPW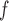 (IPW
(IPW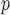 :
: 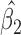 =
=  0.043 ,
0.043 , 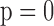 ; IPW
; IPW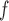 :
: 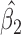 =
= 0.031 , p = 0.013).
0.031 , p = 0.013).
IPW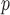 , IPW
, IPW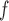 , and AIPW
, and AIPW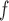 suggest the rate of decline in lung function among subjects who are older at recruitment is not as steep as that of younger subjects (IPW
suggest the rate of decline in lung function among subjects who are older at recruitment is not as steep as that of younger subjects (IPW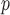 :
: 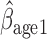 = 0.03, p = 0.057; IPW
= 0.03, p = 0.057; IPW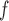 :
: 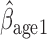 = 0.037, p = 0.017; AIPW
= 0.037, p = 0.017; AIPW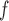 :
: 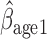 = 0.036, p=0.07), but this effect is smaller when determined by MI
= 0.036, p=0.07), but this effect is smaller when determined by MI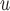 and MI
and MI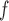 (MI
(MI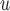 :
: 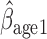 = 0.014, p = 0.301; MI
= 0.014, p = 0.301; MI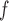 :
: 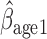 = 0.016, p=0.208).
= 0.016, p=0.208).
Overall conclusions about the effects of baseline covariates are, in general, consistent across the missing data methods (IPW, MI, and AIPW), which is reassuring. Using AIPW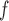 , we conclude that holding all other variables constant, (i) smokers have poorer lung function than non-smokers (
, we conclude that holding all other variables constant, (i) smokers have poorer lung function than non-smokers (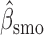 =
= 0.816, p=0.009), (ii) the older a person is at recruitment, the poorer their initial lung function is (
0.816, p=0.009), (ii) the older a person is at recruitment, the poorer their initial lung function is (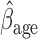 =
= 0.336,
0.336, 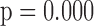 ); MI
); MI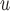 suggests a smaller effect than the other methods (
suggests a smaller effect than the other methods (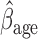 =
= 0.236,
0.236, 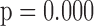 ), (iii) females have poorer baseline lung function than males (
), (iii) females have poorer baseline lung function than males (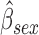 =
= 3.03,
3.03, 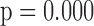 ), and (iv) the more education a person has, the better their initial lung function is (
), and (iv) the more education a person has, the better their initial lung function is (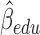 =0.256,
=0.256, 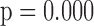 ).
).
9. Discussion
We described and compared the assumptions of IPW, LI imputation, MI, and AIPW for making partly conditional inference. IPW require mortal-cohort dDTIC and missingness-independent death to hold. MI
require mortal-cohort dDTIC and missingness-independent death to hold. MI and LI
and LI imputation require mortal-cohort dDTIC and independent death to hold. AIPW
imputation require mortal-cohort dDTIC and independent death to hold. AIPW requires either (i) u-MAR or (ii) f-MAR, p-MAR and independent death to hold. IPW
requires either (i) u-MAR or (ii) f-MAR, p-MAR and independent death to hold. IPW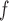 , LI
, LI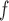 imputation, MI
imputation, MI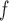 and AIPW
and AIPW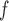 require f-MAR to hold.
require f-MAR to hold.
For data sets with a non-negligible number of deaths, the most appropriate method to handle dropout should be chosen on a case-by-case basis. As a guideline, IPW and AIPW may not be appropriate if the survival process depends on the dropout process, and LI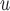 imputation or MI
imputation or MI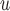 may not be appropriate if the survival process depends on the past missing longitudinal outcomes. Contrary to intuition, using imputation models that condition on D can sometimes induce bias that would not have been present if
may not be appropriate if the survival process depends on the past missing longitudinal outcomes. Contrary to intuition, using imputation models that condition on D can sometimes induce bias that would not have been present if 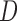 were not conditioned on. This is because, as explained in Section 5.2 and demonstrated in simulation 2, imputation methods that condition on
were not conditioned on. This is because, as explained in Section 5.2 and demonstrated in simulation 2, imputation methods that condition on 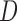 are valid when f-MAR is satisfied, but may be biased otherwise, whereas imputation methods that do not condition on
are valid when f-MAR is satisfied, but may be biased otherwise, whereas imputation methods that do not condition on 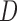 require other assumptions and so may still be valid. Hence, conditioning on time of death in the imputation model should not be done automatically. We suggest that it may be sensible to apply various methods to a data set as a simple form of sensitivity analysis to see if the conclusions change from one method to another.
require other assumptions and so may still be valid. Hence, conditioning on time of death in the imputation model should not be done automatically. We suggest that it may be sensible to apply various methods to a data set as a simple form of sensitivity analysis to see if the conclusions change from one method to another.
When there is dropout and death, Shardell and others (2015) provided an AIPW estimator to measure the causal effect of a time-varying exposure on a longitudinal outcome. The current paper provides AIPW estimating equations to obtain partly conditional means of longitudinal outcomes, and to study (possibly non-causal) associations between the outcomes and covariates among survivors. Our AIPW estimators are double robust to model misspecification. Moreover, when the dropout and imputation models are both correctly specified and the underlying assumptions are met, AIPW is at least as efficient as IPW.
IPW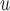 is a stronger assumption than both IPW
is a stronger assumption than both IPW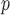 and IPW
and IPW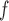 , but IPW
, but IPW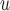 may be more efficient when u-MAR is true than IPW
may be more efficient when u-MAR is true than IPW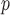 and IPW
and IPW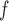 , which rely on p-MAR and f-MAR. We could assess the u-MAR assumption if information on death is available by modeling the association between dropout and
, which rely on p-MAR and f-MAR. We could assess the u-MAR assumption if information on death is available by modeling the association between dropout and 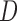 . Moreover, if we know certain auxiliary variables might make u-MAR more plausible, these variables should be included in the model for dropout.
. Moreover, if we know certain auxiliary variables might make u-MAR more plausible, these variables should be included in the model for dropout.
When the missing outcomes do not follow a multivariate normal distribution, the full conditional specification (van Buuren and Groothuis-Oudshoorn, 2011) is an alternative to the joint multivariate normal MI. Multivariate normal MI and full conditional specification, however, are equivalent for monotone missing continuous outcomes (Seaman and Hughes, 2016). Note that IPW methods can easily handle both continuous and non-continuous outcomes.
If possible, it is better to stratify on 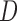 rather than to include
rather than to include 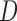 as a covariate in the dropout or imputation model. This is because stratifying on
as a covariate in the dropout or imputation model. This is because stratifying on 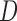 gives a richer dropout or imputation model that is less likely to be misspecified than just including
gives a richer dropout or imputation model that is less likely to be misspecified than just including 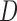 as a covariate. But stratification should be avoided if the sample sizes are small in some strata, since we might obtain unstable weights for IPW or obtain very imprecise estimates of the parameters of the imputation models.
as a covariate. But stratification should be avoided if the sample sizes are small in some strata, since we might obtain unstable weights for IPW or obtain very imprecise estimates of the parameters of the imputation models.
In this article, we have focussed on partly conditional estimation. Interpreting partly conditional estimands, unlike unconditional estimands, does not require defining post-death outcomes, e.g. (non-zero) lung functions in dead people. Dufouil and others (2004) argued for partly conditional models, saying that “immortal-cohort inference is generally inappropriate unless the longitudinal and survival processes are independent.” Note, however, this view is not held universally: Aalen and Gunnes (2010) argued that an unconditional model provides “a more fair comparison of treatments” when a treatment keeps subjects with poorer outcomes alive for longer. An alternative estimand when comparing two treatments is the survivor average causal effect. This is the effect of treatment on outcome in a group of subjects who would have survived regardless of treatment status. See, e.g. Hayden and others (2005), Egleston and others (2006), Yang and Small (2016), and references within for estimating this.
Finally, it is important to note that all of the methods in this article rely on the assumption that, conditional on survival at least to the current visit, the probability of observing an outcome at the current visit depends only on the past outcomes and not on the current outcome. However, we cannot rule out the possibility that dropout depends on the current outcome. In the future we will look at ways to assess the sensitivity of the results to a range of assumptions about the dependence of dropout on the current outcome.
10. Software
Software in the form of R code, together with a sample input data set and complete documentation is available on request from the corresponding author (lan.wen@mrc-bsu.cam.ac.uk).
Supplementary Material
Acknowledgements
The authors thank Professor Boo Johansson for the access to the OCTO data, as well as his comments about this research. Conflict of Interest: None declared.
Funding
Medical Research Council Grant (U105260558 to S.R.C.).
References
- Aalen, O. O. and Gunnes, N. (2010). A dynamic approach for reconstructing missing longitudinal data using the linear increments model. Biostatistics 11, 453–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang, H. and Robins, J. M. (2005). Doubly robust estimation in missing data and causal inference models. Biometrics 61, 962–973. [DOI] [PubMed] [Google Scholar]
- Collins, L. M., Schafer, J. L. and Kam, C. M. (2001). A comparison of inclusive and restrictive strategies in modern missing data procedures. Psychological Methods 6, 330–351. [PubMed] [Google Scholar]
- Diggle, P., Farewell, D. and Henderson, R. (2007). Analysis of longitudinal data with drop-out: objectives, assumptions and a proposal. Journal of the Royal Statistical Society Series C 56, 499–550. [Google Scholar]
- Dufouil, C., Brayne, C. and Clayton, D. (2004). Analysis of longitudinal studies with death and drop-out: a case study. Statistics in Medicine 23, 2215–2226. [DOI] [PubMed] [Google Scholar]
- Egleston, B. L., Scharfstein, D. O., Freeman, E. E. and West, S. K. (2006). Causal inference for non-mortality outcomes in the presence of death. Biostatistics 8, 526–545. [DOI] [PubMed] [Google Scholar]
- Fragoso, C. A., Gahbauer, E. A., Van Ness, P. H. and Gill, T. M. (2007). Reporting peak expiratory flow in older persons. Journal of Gerontology: Biological Sciences, 62, 1147–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gran, J. M., Hoff, R., Røysland, K., Ledergerber, B., Young, J. and Aalen, O. O. (2017). Estimating the treatment effect on the treated under time-dependent confounding in an application to the swiss hiv cohort study. Journal of the Royal Statistical Society: Series C. [Google Scholar]
- Harel, O., Hofer, S. M., Hoffman, L., Pedersen, N. L. and Johansson, B. (2007). Population inference with mortality and attrition in longitudinal studies on aging: a two-stage multiple imputation method. Experimental Aging Research 33, 187–203. [DOI] [PubMed] [Google Scholar]
- Hayden, D., Pauler, D. K. and Schoenfeld, D. (2005). An estimator for treatment comparisons among survivors in randomized trials. Biometrics 61, 305–310. [DOI] [PubMed] [Google Scholar]
- Hoff, R., Gran, J. M. and Farewell, D. (2014). Farewell’s linear increments model for missing data: The flim package. R Journal 6, 137–150. [Google Scholar]
- Kurland, B. F. and Heagerty, P. J. (2005). Directly parameterized regression conditioning on being alive: analysis of longitudinal data truncated by death. Biostatistics 6, 241–258. [DOI] [PubMed] [Google Scholar]
- Kurland, B. F., Johnson, L. L., Egleston, B. L. and Diehr, P. H. (2009). Longitudinal data with follow-up truncated by death: match the analysis method to research aims. Statistical Science 24, 211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik, M. C. (1997). The generalized estimating equation approach when data are not missing completely at random. Journal of the American Statistical Association 92, 1320–1329. [Google Scholar]
- Robins, J. M., Rotnitzky, A. and Zhao, L. P. (1995). Analysis of semiparametric regression models for repeated outcomes in the presence of missing data. Journal of the American Statistical Association 90, 106–121. [Google Scholar]
- Schafer, J. L. (1997). Analysis of Incomplete Multivariate Data, 1 edition.London: Chapman & Hall. [Google Scholar]
- Scharfstein, D. O., Rotnitzky, A. and Robins, J. M. (1999). Adjusting for nonignorable drop-out using semiparametric nonresponse models. Journal of the American Statistical Association 94, 1096–1120. [Google Scholar]
- Seaman, S. R. and Copas, A. (2009). Doubly robust generalized estimating equations for longitudinal data. Statistics in Medicine 28, 937–955. [DOI] [PubMed] [Google Scholar]
- Seaman, S. R., Farewell, D. and White, I. R. (2016). Linear increments with non-monotone missing data and measurement error. Scandinavian Journal of Statistics 43, 996–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman, S. R. and Hughes, R. A. (2016). Relative efficiency of joint-model and full-conditional-specification multiple imputation when conditional models are compatible: the general location model. Statistical Methods in Medical Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shardell, M., Hicks, G. E. and Ferrucci, L. (2015). Doubly robust estimation and causal inference in longitudinal studies with dropout and truncation by death. Biostatistics 16, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Buuren, S. and Groothuis-Oudshoorn, K. (2011). Mice: multivariate imputation by chained equations in r. Journal of Statistical Software 45. [Google Scholar]
- Yang, F. and Small, D. S. (2016). Using post-outcome measurement information in censoring-by-death problems. Journal of the Royal Statistical Society: Series B 78, 299–318. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.