Abstract
N6-Methyladenosine (m6A) represents the most prevalent internal modification in messenger and long noncoding RNAs. There has been a surge of interest toward understanding the biological significance of m6A modification. In this chapter, we describe the methods for biochemically studying the recently uncovered m6A methyltransferases (METTL3 and METTL14) and demethylases (FTO and ALKBH5). How to express these proteins, perform their biochemistry reactions against various RNA probes, and characterize the methylation and demethylation activity will be discussed.
1. INTRODUCTION
Discovered four decades ago, N6-methyladenosine (m6A) is the most abundant internal modification in polyadenylated messenger RNA (mRNA) and long noncoding RNA (lncRNA) in eukaryotes (Desrosiers, Friderici, & Rottman, 1974; Fu, Dominissini, Rechavi, & He, 2014). It is conserved in eukaryotes that range from yeast (Agarwala, Blitzblau, Hochwagen, & Fink, 2012; Bodi, Button, Grierson, & Fray, 2010; Clancy, Shambaugh, Timpte, & Bokar, 2002), plants (Zhong et al., 2008), flies (Hongay & Orr-Weaver, 2011) to mammals (Narayan & Rottman, 1988; Schibler, Kelley, & Perry, 1977; Wei, Gershowitz, & Moss, 1976), as well as in virus with a nuclear phase (Beemon & Keith, 1977; Krug, Morgan, & Shatkin, 1976). The modification site of m6A was demonstrated to be confined in a consensus motif of Pu[G>A] m6AC[U>A>C] (Pu=purine). Its abundance was estimated to be 0.1–0.4% of that of adenines (that is, ~3–5 m6A sites per mRNA) in mammals (Harper, Miceli, Roberts, & Manley, 1990; Horowitz, Horowitz, Nilsen, Munns, & Rottman, 1984; Narayan & Rottman, 1988) and ~0.25% in meiotic Saccharomyces cerevisiae (Bodi et al., 2010). In higher eukaryotes, the m6A modification plays critical roles because knockdown or knockout of the m6A methyltransferase has been shown to result in development arrest (Hongay & Orr-Weaver, 2011; Zhong et al., 2008), cell apoptosis (Bokar, 2005), and inhibition of stem cell differentiation (Batista et al., 2014; Geula et al., 2015). The discovery of an m6A demethylase FTO (fat mass and obesity-associated protein) in 2011 (Jia et al., 2011) sparked extensive research interest in the roles of RNA modifications in gene expression regulation. The field has been expanding rapidly with the first transcriptome-wide maps of m6A-modified mRNA obtained (Dominissini et al., 2012; Meyer et al., 2012), and the identification of N6-adenosine methyltransferases (writers) (Liu et al., 2014), m6A demethylase (erasers) (Zheng et al., 2013), and m6A-binding proteins (readers) (Dominissini et al., 2012; Wang et al., 2014). These emerging new results began to define cellular pathways involving the m6A modification and indicate possible functions in mRNA splicing, mRNA transport, mRNA stability, translation, immune tolerance, and stress response. In this chapter, we summarize methods for the preparation of human nuclear RNA m6A methyltransferases and demethylases as well as methods for biochemical characterization of their catalytic activity.
2. EXPRESSION OF HUMAN NUCLEAR RNA m6A METHYLTRANSFERASE CORE COMPLEX METTL3–METTL14–WTAP IN INSECT CELL EXPRESSION SYSTEM
2.1 Subcloning of METTL3, METTL14, and WTAP
The recombinant proteins METTL3 (GenBank Accession No. NP_062826.2), METTL14 (GenBank Accession No. NP_066012.1), and WTAP (GenBank Accession No. NP_001257460.1) could not be expressed in a bacteria system. We successfully expressed these proteins in insect cells with different N-terminal tags (Flag, His6, and GST) by using a Bac-to-Bac baculovirus expression system (Invitrogen). Their corresponding cDNAs (Open Biosystems) were cloned into a pFastBac™ dual expression vector (Invitrogen) with choices of suitable restriction enzymes. Meanwhile, two genes can be simultaneously cloned into one expression vector in order to maintain the stoichiometry of the encoded proteins if they form a heterodimeric complex. This strategy works out very well for the expression of the METTL3–METTL14 complex (Liu et al., 2014).
2.2 Generate Recombinant Bacmids
Thaw the MAX Efficiency® DH10Bac™ competent cells on ice.
Dispense 50 µL of the cells into 15-mL round-bottom polypropylene tubes.
Add 1 µL (around 500 ng) of specific gene-containing pFastBac™ plasmid into the cells. Mix well by tapping the side of the tube.
Incubate on ice for 30 min.
Apply heat shock by partially immersing the tube in a 42 °C water bath for 45 s.
Chill on ice for 2 min.
Add SOC medium up to 1 mL.
Shake at 37 °C with medium agitation at 225 rpm for 4 h.
Plate 100 µL of a 1/100 diluted culture onto Bac-to-Bac plates (1 L SOB agar for plates contains: 10 g Agar, 20 g tryptone, 5 g yeast extract, 2 mL 5 M NaCl, 2.5 mL 1 M KCl, 10 mL 1 M MgCl2, 10mL 1 M MgSO4, and remaining water for filling up to 1 L for autoclave. The resultant casting Bac-to-Bac plate contains 50 µg/mL Kanamycin, 7 µg/mL Gentamicin, 10 µg/mL Tetracycline, 100 µg/mL Blue-gal, and 40 µg/mL IPTG).
Incubate the plates for 24–48 h in darkness at 37 °C. Colonies are very small and blue colonies may not be discernible before 24 h incubation.
Inoculate a single, isolated white colony from the Bac-to-Bac plate into 2 mL LB medium containing 50 µg/mL Kanamycin, 7 µg/mL Gentamicin, and 10 µg/mL Tetracycline. Grow at 37 °C for 24 h shaking at 250 rpm.
Transfer 1.5 mL culture into a mcf tube.
Spin down and remove the supernatant.
Resuspend cells in 0.3 mL of cold P1.
Lyse cells by adding 0.3 mL of RT P2. Incubate at RT for 5 min.
Neutralize by adding 0.3 mL of P3. Incubate on ice for 5–10 min.
Centrifuge at 14,000 rpm for 10 min.
Prepare 1.5 mL tubes with 0.8 mL isopropanol.
Transfer the supernatant to 0.8 mL isopropanol.
Centrifuge at 14,000 rpm for 15 min.
Add 0.5 mL of 70% EtOH to the pellet. Invert the tube several times.
Centrifuge at 14,000 rpm for 5 min.
Remove as much of the supernatant as possible. Air-dry the bacmid pellet at RT for 5–10 min.
Redissolve the pellet in 40 µL TE (100 mM Tris, pH 8.0; 10 mM EDTA pH 8.0).
2.3 Produce Recombinant Baculovirus P0, P1, and P2 in SF9 Cell Line
Seed SF9 insect cells by splitting confluent 15-cm plate with 1:12 ratio into 6-well tissue culture plate.
Let sit for at least 1 h.
- Make the following mixes A and B in mcf tubes.
- A: 12 µL Bacmid+100 µL Grace’s medium without supplement.
- B: 8 µL Cellfectin II+100 µL Grace’s medium without supplement.
Combine A and B (mix C).
Incubate at RT for 15–30 min.
Add 780 µL Grace’s medium without supplement to mix C.
Wash cells in the 6-well plate with 2 mL Grace’s medium without supplement.
Overlay mix C and incubate at 27 °C for 5 h.
Change medium to Grace’s medium with supplement.
Harvest virus from ~2 mL supernatant at 72 h posttransfection and label it P0 virus.
Make one 15-cm plate of SF9 cell culture ready when the P0 virus is obtained.
Drain the medium from the prepared 15-cm plate of SF9 cell with 1 mL left and add 1 mL P0 virus into the plate. Gently shake every 20 min 3×, and then add 20 mL supplemented medium to the plate and let it grow for 72 h.
Harvest the P1 virus from the supernatant by centrifugation at 1000 rpm for 10 min. Store at 4 °C.
Culture SF9 cells in as many plates as planned. Perform the same procedure as above (P0–P1) in order to amplify the baculoviral stock by adding 1 mL P1 virus to each plate for infection. Harvest the P2 virus from the supernatant by centrifugation after 72 h.
2.4 Expression and Purification of Proteins
Infect SF9 cells with P2 baculovirus stock in order to express recombinant proteins. To each 15-cm culture plate, 1 mL P2 virus and 20 mL of fresh medium were added followed by incubation at 27 °C for 48 h.
Collect cells from 50 plates by using culture medium to flush cells away from the dish bottom and centrifuging at 5000 rpm for 10 min at 4 °C. Wash cells (~10–13 g pellet) with the cell lysis buffer containing 500 mM NaCl, 10 mM Tris pH 7.4, 5% glycerol, and 1 mM DTT. Then, centrifuge and resuspend cells in 40 mL of cell lysis buffer. In this step, cell pellets can be frozen in liquid nitrogen and stored at −80 °C for at least 3 months. All purification steps should be performed on ice or at 4 °C. Here, we show the purification procedure for Flag-tagged proteins.
Add 1× protease cocktail inhibitor (Nacalai) to the cell suspension.
Break cells by sonication for 15 min with 30% amplitude, 15 s on, and 15 s off.
Centrifuge the cell lysate at 37,000 rpm for 40 min at 4 °C.
Add 5 mL of anti-Flag M2 affinity gel (Sigma) to the glass Econo-Column and prewash it with at least 50 mL of cell lysis buffer using gravity.
Load the supernatant of the cell lysate to the equilibrated anti-Flag M2 affinity beads. Resuspend and incubate at 4 °C for 2 h using a rotating mixer.
Let the cell lysate flow through using gravity.
Wash the beads with 5 × 50 mL of cell lysis buffer containing 1 mM DTT, 1 mM PMSF, 1 µg/mL leupeptin, 1 µg/mL pepstatin, and 2 µg/mL aprotinin using gravity.
Elute protein from the beads using 3× Flag peptide (Sigma). Add 6 mL 0.2 mg/mL 3× Flag peptide in cell lysis buffer to the beads and incubate for 30 min with a rotating mixer. Collect the eluate by gravity.
Repeat step 10 using another 4 mL 0.2 mg/mL 3× Flag peptide for the second elution.
Concentrate the eluate to 2 mL and then apply it to the 120 mL Superdex-200 column with gel filtration running buffer containing 150 mM NaCl, 10 mM Tris pH 7.4, and 1 mM DTT.
Collect the peak fraction and concentrate the protein with an Amicon Ultra-4 filter (Millipore) to around 5–10 mg/mL. Run SDS-PAGE in order to characterize the purity of the protein. See Fig. 1 for gel filtration traces and SDS-PAGE images.
Add glycerol to the purified protein to 30% (vol/vol) and aliquot. Freeze the aliquots in liquid nitrogen and store at −80 °C.
Figure 1.
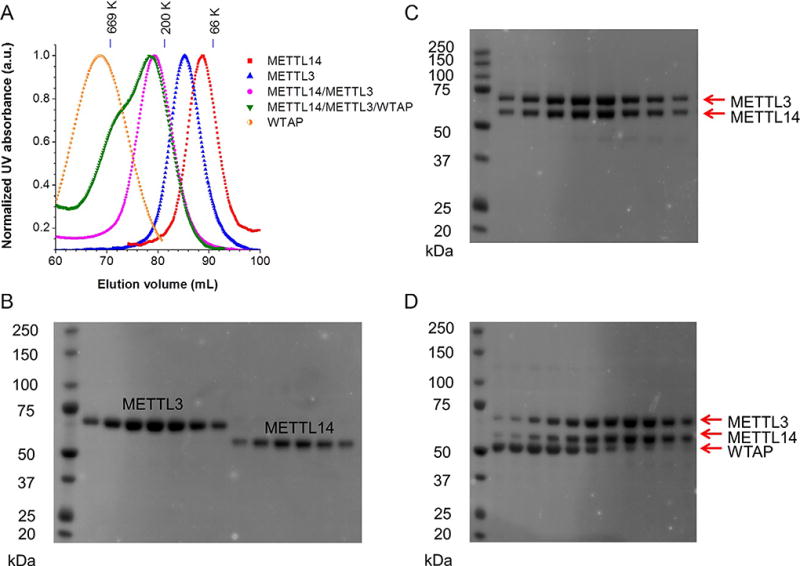
(A) Gel filtration traces of individual Flag-tagged METTL3, METTL14, and WTAP; coexpressed Flag-METTL14 and His6-METTL3; and mixed Flag-METTL14, Flag-METTL3, and Flag-WTAP with equal molar amounts. All proteins were expressed in insect cells and purified by Flag antibody immunoprecipitation. Markers: 669 kDa (thyroglobulin, bovine), 200 kDa (β-amylase from sweet potato), and 66 kDa (BSA). (B–D) The SDS-PAGE images illustrate purified proteins from gel filtration fractions shown in panel A. The panels B–D correspond to METTL3 and METTL14, METTL3–METTL14 complex, and mixed METTL3/METTL14/WTAP, respectively. Figure (A) is adapted from figure 1B in Liu et al. (2014), with permission from Nature Publishing Group.
3. EXPRESSION OF HUMAN NUCLEAR RNA m6A DEMETHYLASES FTO AND ALKBH5
3.1 Subcloning and Expression of FTO and ALKBH5 in Bacteria
The human FTO gene (GenBank Accession No. NP_001073901.1) was subcloned into pET28a vector (Novagen) with choices of suitable restriction enzymes to generate a His6-tagged fusion protein. The human ALKBH5 (GenBank Accession No. NP_060228.3) with deletion of the amino-terminal 66 amino acids was subcloned into a pMCSG19 vector with a His6-tag by ligation-independent cloning strategy (Donnelly et al., 2006). Both FTO and ALKBH5 were successfully expressed in the BL21 (DE3) Escherichia coli strain and purified in a soluble form (Jia et al., 2011; Zheng et al., 2013).
4. BIOCHEMICAL CHARACTERIZATION OF THE CATALYTIC ACTIVITY OF THE m6A METHYLTRANSFERASES AND DEMETHYLASES
4.1 Methylation Reactions of RNA Probes
A typical 50 µL reaction mixture contains the following components: 0.15 nmol RNA probe, 0.15 nmol each recombinant protein (single METTL3, METTL14, WTAP, or their combinations with a molar ratio of 0.15 nmol/0.15 nmol for two components, 0.8 mM d3-SAM, 80 mM KCl, 1.5 mM MgCl2, 0.2 U/µL RNasin, 10 mM DTT, 4% glycerol, and 15 mM HEPES at pH 7.9). The reaction occurs at 16 °C overnight. Prior to the reaction, RNA probes were annealed using a program of: (i) 90 °C for 3 min and (ii) −2 °C/cycle for 40 cycles within 30 min. The reaction was incubated at 16 °C for 12 h or overnight. The resultant RNA was recovered by phenol/chloroform (low pH) extraction followed by ethanol precipitation.
4.2 Demethylation Reactions of RNA/DNA Probes
Here, an example of FTO demethylation reaction is shown. The demethylation assay was performed in 100 µL of the reaction mixture containing 1 nmol RNA/DNA probe with m6A, FTO, 283 µM of (NH4)2Fe(SO4)2·6H2O, 300 µM of α-KG, 2 mM of l-ascorbic acid, 50 µg/mL of BSA, and 50 mM of HEPES buffer (pH 7.0). The reaction was incubated for ~3 h at room temperature or overnight at 16 °C and quenched by addition of 5 mM EDTA followed by heating for 5 min at 95 °C. The resultant RNA/DNA was recovered by phenol/chloroform (low pH) extraction followed by ethanol precipitation. Alternatively, the whole resultant reaction mixture could be directly subjected to digestion into free nucleosides for further quantification of the reaction yield.
4.3 LC–MS/MS Characterization of m6A Methylation Activity
The LC–MS/MS method is very useful in order to characterize in vitro m6A methylation reaction yield and the m6A content in mRNA. Here, we include an example of the quantification of m6A methyltransferase activity. Typically, 200–300 ng of recovered RNA from biochemistry reaction is digested by nuclease P1 (1–2 U, Wako USA) in 30 µL of buffer containing 25 mM NaCl and 2.5 mM ZnCl2 at 42 °C for 2 h, followed by additions of NH4HCO3 (1 M, 3 µL) and alkaline phosphatase (0.5–1 U, Sigma) and incubation at 37 °C for 2 h (Fig. 2). The sample is then filtered (0.22 µm pore size, 4 mm diameter, Millipore), and 5 µL of the solution is injected into LC–MS/MS. The nucleosides are separated by reverse phase ultra-performance liquid chromatography on a C18 column with online mass spectrometry detection using Agilent 6460 QQQ triple-quadrupole LC mass spectrometer in positive electrospray ionization mode. The nucleosides are quantified by using the nucleoside to base ion mass transitions of 285–153 (d3-m6A) and 284–152 (G). The amount of reacted probe can be calculated through the newly formed d3-m6A, while G serves as an internal control with which the total amount of RNA probe can be calculated.
Figure 2.
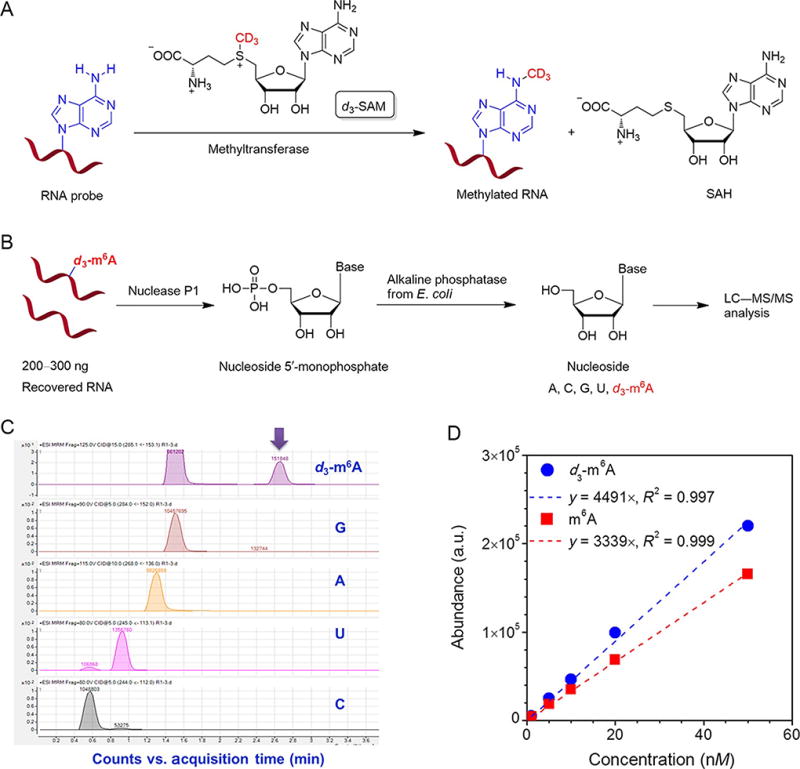
Typical procedure for quantification of m6A methylation reaction yield by LC–MS/MS. (A) Schematic illustration of m6A biochemistry reaction. (B) A flowchart of digestion of RNA for quantitative LC–MS/MS analysis. (C) On a LC–MS/MS system, the amount of each nucleoside was quantified by its integration area in the corresponding chromatogram. (D) The m6A and d3-m6A standard calibration curves were obtained from five standard samples with different amounts of pure m6A and d3-m6A, respectively. For in vitro biochemistry reaction, the d3-m6A calibration curve is applied to calculate the methylation reaction yield, while an m6A calibration curve could be used to determine the m6A content from cellular mRNA or other RNAs. Figure (D) is adapted from supplementary figure 2B in Liu et al. (2014), with permission from Nature Publishing Group.
4.4 HPLC Characterization of m6A Demethylation Activity
The recovered RNA/DNA from the demethylation reactions were digested by nuclease P1 and alkaline phosphatase. The reaction condition is the same as the one indicated for the characterization of m6A methylation activity. Digested free nucleosides were analyzed on a HPLC system equipped with an Agilent Eclipse XDB-C18 analysis column (150×4.6 mm) eluted with buffer A (25 mM NaH2PO4) and buffer B (acetonitrile) with a flow rate of 1 mL/min at room temperature. The detection wavelength was set at 266 nm. The demethylation yield can be estimated from the integration area of the m6A peak. For more accurate characterization of demethylation yield, QQQ-LC–MS/MS can be used as described above.
4.5 Methyltransferase Activity and Selectivity Toward Different RNA Probes
Biochemical tests of the methyltransferase activity were performed for the individual recombinant protein or their combinations toward various synthetic RNA probes with or without the consensus sequence GGACU and the stem-loop secondary structure. The d3-SAM (S-(5′-adenosyl)-l-methionine-d3) with a deuterium-substituted methyl group was utilized as the cofactor to avoid potential m6A contamination by RNA already present in the recombinant protein during the purification process and for accurate mass spectrometry quantification.
The RNA substrate was incubated with the recombinant protein at pH 7.9 and 16 °C overnight, followed by complete digestion to a single nucleoside by nuclease P1 and alkaline phosphatase, and subsequently analyzed by LC–MS/MS (Fig. 2). We used the molar ratio of the product d3-m6A to each probe to quantify the methylation efficiency. Results have been reported previously (Liu et al., 2014). We revealed that WTAP showed no methyltransferase activity with all probes tested, which is consistent with its lack of an active methyltransferase domain. Both METTL3 and METTL14 exhibited methyltransferase activity with METTL14 showing much higher in vitro activity (close to 10-fold with several probes) than METTL3. A noticeable synergistic effect was observed in that the combinations of METTL3/METTL14 dramatically enhanced the methyltransferase activity. This observation strongly implies that METTL3 and METTL14 interact with each other to enhance methyltransferase activity.
Recent m6A profiling results revealed that most m6A modifications in mammalian mRNAs occur in a consensus sequence of (Pu[G>A]m6AC[U>A>C]). We examined potential sequence preference of METTL3 and METTL14 and found that they preferentially methylate substrates containing the consensus sequence, but exhibited no obvious preference to the stem-loop structure for m6A deposition. These results support the notion that these methyltransferases have certain sequence specificity but show less structural preference to RNA substrates.
4.6 Demethylase Activity and Selectivity Toward Different RNA/DNA Probes
Several m6A-containing single-stranded RNAs (ssRNAs) and DNAs (ssDNAs) were synthesized for the characterization of the FTO demethylation activity (Fig. 3). These probes were incubated with 0.2 mol equivalent of FTO for 3 h at room temperature. Afterward, the probes were digested into free nucleosides, which were further separated on a C18 column by HPLC. The HPLC trace profile showed that m6A in ssRNA or ssDNA was completely converted to adenosine upon FTO treatment. The m6A-containing double-stranded RNA (dsRNA) and DNA (dsDNA) were also tested and only negligible activity was observed under the same reaction conditions. When the FTO amount was increased to 1 mol equivalent at 16 °C overnight, demethylation yields of 24% and 40% for dsRNA and dsDNA were obtained, respectively (Fig. 3).
Figure 3.
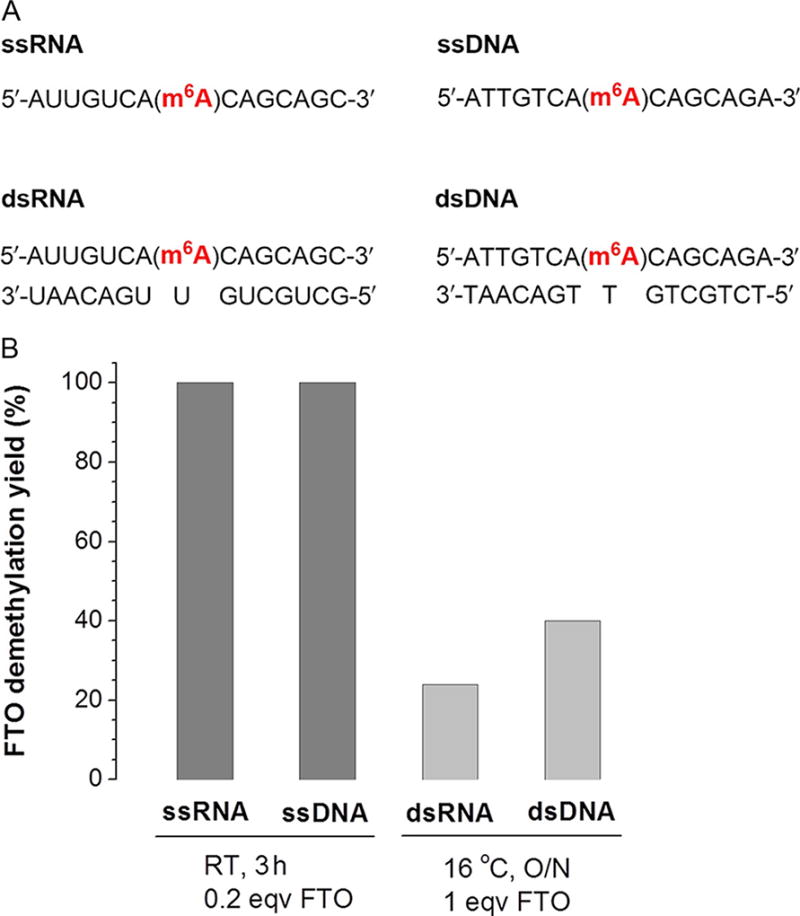
Examples showing demethylation activity of FTO against various RNA/DNA probes. (A) Sequences of RNA/DNA probes. (B) Comparison of the demethylation activity and selectivity among the probes. The demethylation yields were calculated by HPLC method (Jia et al., 2011).
Similar to FTO, ALKBH5 also catalyzes the demethylation of m6A in ssRNA. As shown in Fig. 4, the probe ssRNA-1 could be completely demethylated by an equal equivalent of ALKBH5 at 16 °C after overnight incubation. When the reaction was performed at room temperature for 2.5 h, the demethylation yield decreased to around 40%. Similar activity for ssDNA was observed. The strong preference of ALKBH5 for single-stranded substrates was shown by an almost unnoticeable demethylation activity toward m6A in dsRNA (Fig. 4B). Most m6A modifications in mammalian mRNA exist in a consensus sequence of (Pu[G/A]m6AC[U/A/C]). Three ssRNAs (ssRNA1–3) with the currently known consensus sequences of (GGm6ACU)/(CAm6ACA) and a random sequence of (CUm6AUU) were synthesized and tested for demethylation activity (Fig. 4). While ALKBH5 showed similar activity toward the two consensus sequences (~40%), a reduced demethylation yield of ~20% was observed toward the nonconsensus sequence under the same conditions, indicating that ALKBH5 may possess a sequence preference in demethylation.
Figure 4.
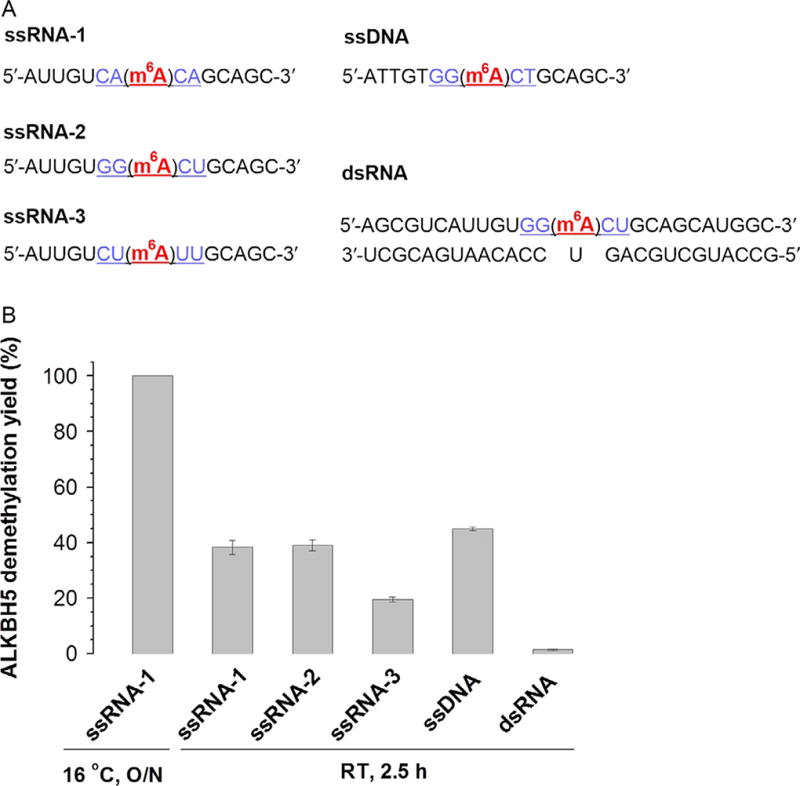
Examples showing demethylation activity of ALKBH5 against various RNA/DNA probes. (A) Sequences of RNA/DNA probes. (B) Comparison of the demethylation activity and selectivity among the probes. Error bars indicate ±SEM (n=3). Figure (B) is adapted from supplementary figure 1F and 1G in Zheng et al. (2013), with permission from Elsevier.
5. CONCLUSIONS
In summary, we have described the preparation of human nuclear RNA m6A methyltransferases (METTL3 and METTL14) and demethylases (FTO and ALKBH5) as well as procedures to biochemically characterize their catalytic activity. The introduced characterization method could serve as a general strategy for future investigations of other RNA or DNA modification enzymes.
Acknowledgments
This work was supported by National Institutes of Health (GM071440 to C.H.). S.F. Reichard contributed editing.
References
- Agarwala SD, Blitzblau HG, Hochwagen A, Fink GR. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genetics. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, et al. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemon K, Keith J. Localization of N6-methyladenosine in rous-sarcoma virus genome. Journal of Molecular Biology. 1977;113:165–179. doi: 10.1016/0022-2836(77)90047-x. [DOI] [PubMed] [Google Scholar]
- Bodi Z, Button JD, Grierson D, Fray RG. Yeast targets for mRNA methylation. Nucleic Acids Research. 2010;38:5327–5335. doi: 10.1093/nar/gkq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. In: Grosjean H, editor. Fine-tuning of RNA functions by modification and editing. Vol. 12. Berlin: Springer-Verlag; 2005. pp. 141–177. [Google Scholar]
- Clancy MJ, Shambaugh ME, Timpte CS, Bokar JA. Induction of sporulation in Saccharomyces cerevisiae leads to the formation of N6-methyladenosine in mRNA: A potential mechanism for the activity of the IME4 gene. Nucleic Acids Research. 2002;30:4509–4518. doi: 10.1093/nar/gkf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proceedings of the National Academy of Sciences of the United States of America. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, et al. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expression and Purification. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nature Reviews Genetics. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, et al. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. http://dx.doi.org/10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Harper JE, Miceli SM, Roberts RJ, Manley JL. Sequence specificity of the human messenger-RNA N6-adenosine methylase in vitro. Nucleic Acids Research. 1990;18:5735–5741. doi: 10.1093/nar/18.19.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongay CF, Orr-Weaver TL. Drosophila Inducer of MEiosis 4 (IME4) is required for Notch signaling during oogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:14855–14860. doi: 10.1073/pnas.1111577108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz S, Horowitz A, Nilsen TW, Munns TW, Rottman FM. Mapping of N6-methyladenosine residues in bovine prolactin messenger-RNA. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:5667–5671. doi: 10.1073/pnas.81.18.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nature Chemical Biology. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug RM, Morgan MA, Shatkin AJ. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. Journal of Virology. 1976;20:45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nature Chemical Biology. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan P, Rottman FM. An in vitro system for accurate methylation of internal adenosine residues in messenger-RNA. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- Schibler U, Kelley DE, Perry RP. Comparison of methylated sequences in messenger-RNA and heterogeneous nuclear-RNA from mouse L-cells. Journal of Molecular Biology. 1977;115:695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, Moss B. 5′-Terminal and internal methylated nucleotide-sequences in HeLa-cell messenger-RNA. Biochemistry. 1976;15:397–401. doi: 10.1021/bi00647a024. [DOI] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Molecular Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]


