Abstract
CENP-E is a kinesin-like protein that when depleted from mammalian kinetochores leads to mitotic arrest with a mixture of aligned and unaligned chromosomes. In the present study, we used immunofluorescence, video, and electron microscopy to demonstrate that depletion of CENP-E from kinetochores via antibody microinjection reduces kinetochore microtubule binding by 23% at aligned chromosomes, and severely reduces microtubule binding at unaligned chromosomes. Disruption of CENP-E function also reduces tension across the centromere, increases the incidence of spindle pole fragmentation, and results in monooriented chromosomes approaching abnormally close to the spindle pole. Nevertheless, chromosomes show typical patterns of congression, fast poleward motion, and oscillatory motions. Furthermore, kinetochores of aligned and unaligned chromosomes exhibit normal patterns of checkpoint protein localization. These data are explained by a model in which redundant mechanisms enable kinetochore microtubule binding and checkpoint monitoring in the absence of CENP-E at kinetochores, but where reduced microtubule-binding efficiency, exacerbated by poor positioning at the spindle poles, results in chronically monooriented chromosomes and mitotic arrest. Chromosome position within the spindle appears to be a critical determinant of CENP-E function at kinetochores.
INTRODUCTION
CENP-E is a kinesin-like protein that binds to kinetochores during mitosis (Yen et al., 1991,1992; Cooke et al., 1997; Yao et al., 1997). Depletion of CENP-E from HeLa cell kinetochores via antibody microinjection or expression of dominant negative mutants results in mitotic arrest with unaligned chromosomes (Schaar et al., 1997; Chan et al., 1998). Recent efforts to eliminate CENP-E expression by an antisense strategy yielded a similar phenotype (Yao et al., 2000). However, in all three approaches, many of the chromosomes do achieve metaphase alignment. Deconvolution immunofluorescence microscopy revealed that disruption of CENP-E functions also results in an increased incidence of spindle fragmentation and reduced tension across centromeres (Yao et al., 2000). These data have been interpreted to mean that CENP-E is required for stable attachment of kinetochore microtubules (kMts) and chromosome congression to the spindle equator (Schaar et al., 1997; Yao et al., 2000). The presence of bipolar aligned chromosomes in the absence of CENP-E function has been attributed to chromosomes located near the center of the forming spindle during early prometaphase. Such chromosomes would more readily achieve bipolar attachment because they are exposed to a greater number of microtubule (Mt) plus ends, and they would not have to undergo congression because they are already aligned.
Two CENP-E homologs have been discovered in Drosophila (Yucel et al., 2000). Live cell imaging demonstrated that P-element disruption or complete removal of one of the CENP-E homologs, CENP-meta, results in chromosomes that undergo congression but frequently fail to maintain a stable alignment at the metaphase plate. These data suggest that CENP-meta is required for maintaining equatorial alignment rather than achieving it. The contribution of CENP-ana to chromosome alignment remains unknown.
For mammalian cells, it is evident that CENP-E is required for full chromosome alignment and normal progression through mitosis. However, its role in any one of the component steps, such as initial monopolar attachment, bipolar attachment, congression, or stability of equatorial alignment, has yet to be established. In the current study, we use electron microscopy and live cell imaging to demonstrate that CENP-E has a redundant role in kMt attachment. In addition, depletion of CENP-E from the kinetochore results in reduced tension across the centromere, increased incidence of spindle pole fragmentation, and monooriented chromosomes located abnormally close to a spindle pole. The latter exhibit little or no oscillatory behavior and appear to be “stuck” at the poles. Disruption of CENP-E function does not reveal a demonstrable effect on the initial fast poleward motion, chromosome congression, oscillations, or stability of equatorial alignment. Kinetochores depleted of CENP-E also show normal patterns of recruiting and dissociating checkpoint proteins. These results indicate that CENP-E has critical roles in positioning monooriented chromosomes and affecting bioriented attachment that are only partially redundant with other kinetochore components.
MATERIALS AND METHODS
Cell Cultures and Injections
HeLa and CF-PAC cell monolayers were grown on glass coverslips at 37°C in DMEM and Iscou's Modified Eagle Medium, respectively. Both media were supplemented with fetal bovine serum and 5% CO2. Cells were synchronized by a double (HeLa) or single (CF-PAC) thymidine block and anti-CENP-E antibody (HX-1) was microinjected into cells ∼2 h after release from the G1/S boundary (Schaar et al., 1997). Injected cells were identified by their location within a scribed area and by coinjection with green fluorescent protein expression plasmid, Texas Red dextran, or Oregon Green dextran (Molecular Probes, Eugene, OR). Control cells were coinjected with nonimmune serum, at or above the concentration of HX-1.
Immunostaining and Light Microscopy
Cells on coverslips were fixed with 3.5% electron microscopy grade paraformaldehyde, permeabilized with 0.2% Triton X-100 in 1× KB buffer (10 mM Tris pH 7.5, 150 mM NaCl, 0.1% bovine serum albumin), and stained as described (Chan et al., 1998). Injected rabbit and rat antibodies were detected with Cy-5-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA). Endogenous CENP-E was detected with a mouse monoclonal antibody (mAb) 177 or rat polyclonal antibody that was generated and affinity-purified as described previously for rabbit HX-1 anti-CENP-E antibodies (Yen et al., 1991; Schaar et al., 1997). Rabbit anti-hBUB1 and rabbit anti-hBUBR1 were previously described (Chan et al., 1998; Jablonski et al., 1998). Rabbit anti-h MAD1 was generated against a GST-hMAD1 fusion protein containing the C-terminal 250 amino acids of hMAD1. The anti-hMAD1 antibodies were affinity-purified as described (Campbell et al., 2001). Mts were stained with mouse monoclonal anti-α-tubulin antibodies (Sigma, St. Louis, MO). The affinity-purified rabbit anti-XMAD2 antibodies were a kind gift from Dr. E. Salmon (University of North Carolina, Chapel Hill, NC). DNA was stained either with 4,6-diamidino-2-phenylindole (DAPI) or Hoechst. Stained cells were examined with the use of either a 40× or 100× PlanNeofluor objective mounted on a Nikon Microphot SA that was equipped with epifluorescence optics. Images were captured with a TEC-1 charge-coupled device camera (Dage-MTI) that was controlled with IP LabSpectrum version 3.1 (Scanalytics, Fairfax, VA) and contrast enhanced with Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA).
Electron Microscopy and Image Analysis
Cells were fixed with 0.5–1.0% glutaraldehyde in PHEM (60 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9) or phosphate-buffered saline either 16–18 h (prolonged M phase) or 8–9 h (M-phase peak) after G1/S release. Preextracted cells were treated with 0.5% Triton X-100 in PHEM buffer for 1 min then washed for 2 min in PHEM, before fixation. Coverslips were flat embedded in Epon and serial 80-nm-thick sections were cut, stained, and imaged at 5000× as previously described (McEwen et al., 1997; Rieder and Cassels, 1999). The Mt counts for individual kinetochores were determined in duplicate counting trials as described by McEwen et al. (1997). Counting variation was 1.3% for unextracted cells and 0.4% for extracted cells. Statistical computations and preparation of bar graphs was accomplished with the use of Microsoft Excel (Microsoft, Redmond, WA). Distances between sister kinetochores in the same serial section were determined from the separation of the average coordinates for sets of eight points taken along the outer plate of each sister kinetochore. Linearity of kinetochore outer plates was estimated as the R value for a straight line fit of each set of points. Minimum distances between centrioles and the nearest centromere were determined by taking sets of eight points along the closest approaching edges of each structure and finding the minimum pairwise point-to-point separation between the two sets of points.
Live Cell Imaging
CF-PAC cells were coinjected with HX-1 and Oregon Green dextran and later remounted into Rose chambers containing L-15 HEPES-buffered media. Approximately 7–8 h post thymidine release, the cells were transferred to a heated microscope stage (37°C) and screened for green fluorescent prophase cells. Cells were imaged on a Nikon Diaphot phase contrast microscope with the use of a 60× objective lens (1.4 numerical aperature) and 0.7 numerical aperature condenser (Savoian et al., 1999). Cells were illuminated with filtered and shuttered green (546 nm) light. Images were captured at 10–30-s intervals with a Paulteck charge-coupled device camera, with the use of the Image1 image acquisition package (Universal Imaging, West Chester, PA), and stored digitally on a PC.
RESULTS
Kinetochore Microtubule Binding in Anti-CENP-E–injected Cells
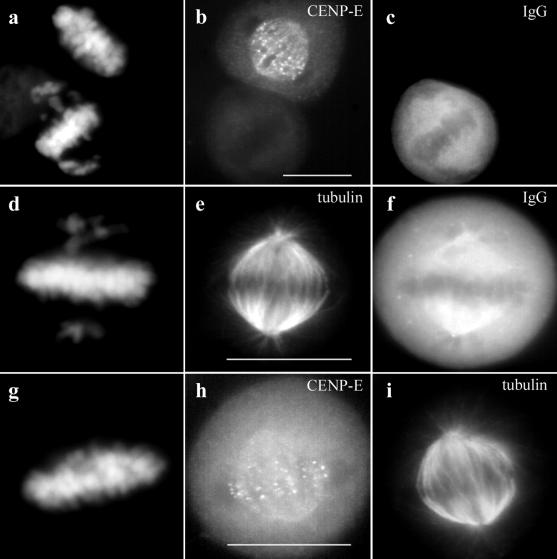
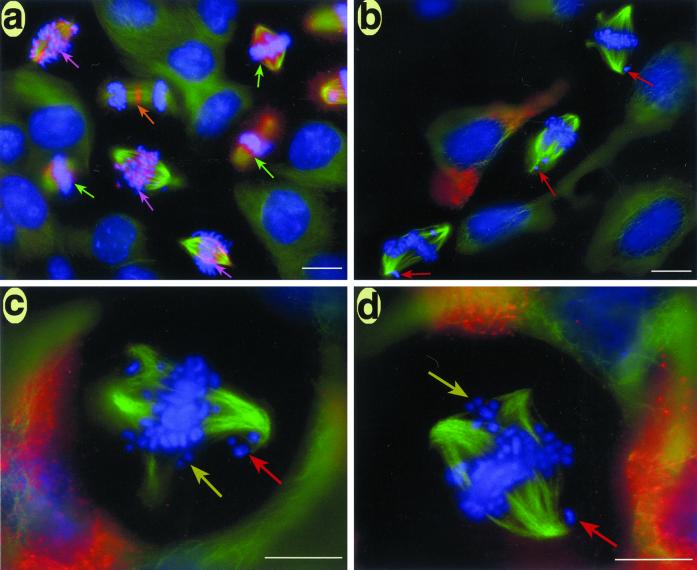
In agreement with previous studies, microinjection of CENP-E antibodies into interphase HeLa cells shortly after release from a G1/S block resulted in cell cycle arrest with misaligned chromosomes during the ensuing mitosis (Schaar et al., 1997). Nevertheless, in 60–70% of the cells we examined, the majority of the chromosomes were aligned at the spindle equator where they formed a robust metaphase plate (Figure 1, a and d). The unaligned chromosomes were generally located near the spindle poles so that DNA staining gives a distinct “cruciform” pattern as illustrated in Figure 1, a and d. Neither the aligned nor the unaligned chromosomes had detectable levels of CENP-E staining (Figure 1b).
Figure 1.
Depletion of CENP-E from kinetochores by microinjection of HX-1 CENP-E antibodies (rabbit). HeLa cells synchronized by double thymidine block were injected with HX-1 ∼2 h after release and fixed for indirect immunofluorescence at 12 h after release from the G1/S block. (a–c) An injected (bottom) and uninjected (top) cell side by side on the coverslip. (d–f) Single injected cell. (g–i) Uninjected metaphase control cell from the same coverslip stained for CENP-E and tubulin. The injected anti-CENP-E antibodies were detected with Cy-5-conjugated anti-rabbit antibodies (c and f). Localization of endogenous CENP-E was achieved with a rat polyclonal anti-CENP-E antibody (rHX-1) (b and h). rHX-1 staining was visualized with Cy2-conjugated anti-rat secondary antibodies. Chromosomes were stained with DAPI (a, d, and g). Mts were stained with a mouse monoclonal anti-tubulin antibody and visualized with Texas Red-conjugated anti-mouse secondary antibodies (e and i). Bar, 10 μm.
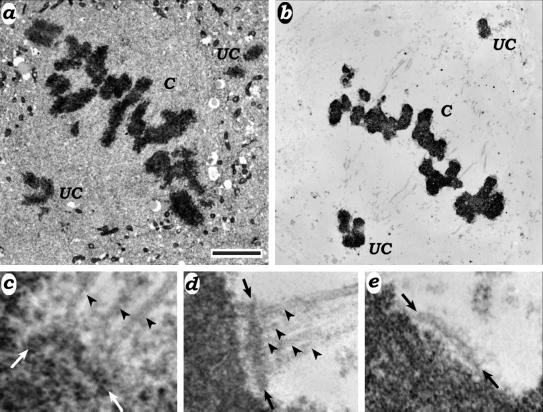
To assess the effect of removing CENP-E from the kinetochore on its ability to bind Mts, we used correlative light and electron microscopy to measure kMt binding on both aligned and unaligned chromosomes (Figure 2). Because kMts can be difficult to identify due to unextracted cytoplasm (Figure 2c), we also counted kMts in cells extracted with detergent in Mt-stabilizing buffer. For uninjected metaphase controls, we found an average of 16.9 kMts per kinetochore (Table 1A), regardless of whether the cells were unextracted, preextracted, or extracted while fixed (our unpublished data). These values were in excellent agreement with the value of 17.1 reported by Wendell et al. (1993) for metaphase HeLa cells. Anti-CENP-E–injected cells on the same coverslips had an average of 12.4 kMts per kinetochore on aligned chromosomes (Figure 3a and Table 1A). This was 74% of the kMt count found in controls and the difference was highly significant (p < 10−6). Nearly half of the kinetochores on uncongressed chromosomes were unattached, and on average attached kinetochores bound 1.9 Mts (range 1–5; Figure 3a and Table 1B). All uncongressed chromosomes where we could identify sister kinetochores (27 total) were monooriented, and the attached kinetochore faced the spindle pole.
Figure 2.
Electron micrographs of HeLa cells microinjected with HX-1. (a and b) Overview of unextracted (a) and detergent-extracted (b) cells. Congressed (C) indicates chromosomes that are aligned at the spindle equator, and uncongressed (UC) indicates chromosomes that are located near one of the poles. (c). Attached kinetochore from an unextracted cell. Arrows delineate the kinetochore, whereas arrowheads indicate kMts. (d and e) Attached (d) and unattached (e) kinetochores from extracted cells. Kinetochores and kMts are indicated as in c. Bar, 2.5 μm (a and b) and 0.30 μm (c–e).
Table 1.
Kinetochore microtubule binding in anti-CENP-E-injected HeLa cells
| A. Congressed chromosomes | |||
| M phase peaka (8–9h post G1/S) | Prolonged M phasea (16–18h post G1/S) | Uninjected control | |
|---|---|---|---|
| Average kMt binding | 12.5 | 14.9 | 16.9 |
| SD | 1.76 | 1.72 | 1.51 |
| Range | 8–17 | 11–23 | 13–22 |
| No. of attached kinetochores | 118 | 107 | 127 |
| No. of unattached kinetochores | 0 | 0 | 0 |
| B. Uncongressed chromosomes | ||
| M-phase peaka | Prolonged M phasea | |
|---|---|---|
| Average kMt bindingb | 1.9 | 3.7 |
| SD | 0.93 | 2.68 |
| Range | 1–5 | 1–15 |
| No. of attached kinetochores | 48 | 57 |
| No. of unattached kinetochores | 45 | 40 |
| No. of monooriented chromosomesc | 24 | 27 |
M-phase peak and prolonged M phase are synchronized HeLa cells fixed 8–9 and 16–18 h after release from G1/S block. The latter had presumably been blocked in M phase 4–10 h.
Does not include unattached kinetochores.
Chromosomes where an attached and unattached sister kinetochore were identified. No uncongressed bioriented chromosomes were found in cells with bipolar spindles.
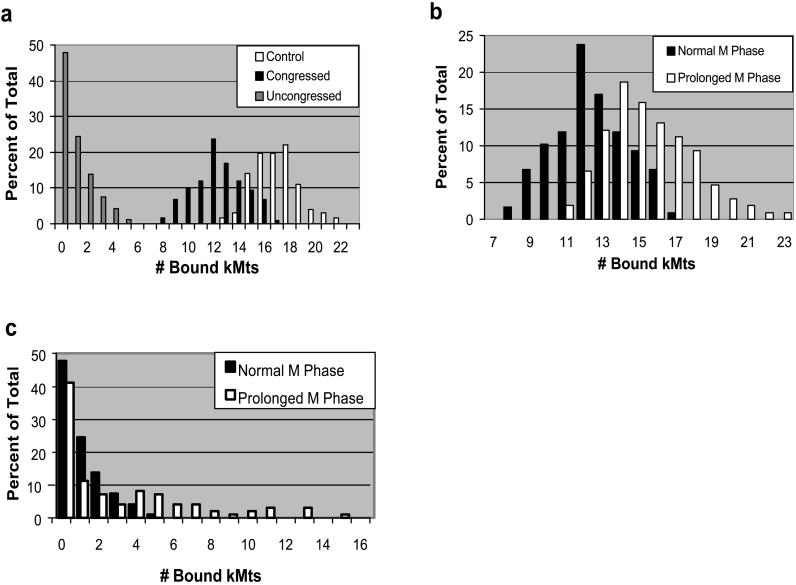
Figure 3.
Numbers of Mts binding to CENP-E–depleted kinetochores. (a) Synchronized HeLa cells were fixed 8–9 h after release from G1/S block and kMts counted via serial section electron microscopy. Congressed and uncongressed chromosomes are defined in Figure 2. Control were counts taken from chromosomes in uninjected metaphase cells on the same coverslips as the injected cells. Differences in kMt binding between congressed chromosomes from injected and control cells are highly significant (p < 10−6). (b) Comparison of kMt binding between congressed chromosomes from the M-phase peak and cells arrested in M phase for a prolonged period. Cells in prolonged M phase were fixed 16–18 h after release from G1/S block. Differences are highly significant (p < 10−5). (c) Comparison of kMt binding between uncongressed chromosomes from peak and prolonged M phase.
To assess the effect of a prolonged M-phase block, we also looked at anti-CENP-E–injected cells fixed 16–18 h after G1/S release. Because the major peak of mitotic cells occurs 8–12 h from the G1/S boundary, most mitotic cells fixed at 16–18 h had been arrested in M phase for 4–10 h (Schaar et al., 1997). Kinetochores on congressed chromosomes in these cells bound on average 14.9 Mts (Figure 3b and Table 1A). This is a significant increase from the 12.4 observed during the mitotic peak (p < 10−5) but still significantly lower than the 16.9 observed in uninjected control cells on the same coverslip (p = 1.4 × 10−5). The range of Mt binding was approximately the same as controls, indicating that under some circumstances CENP-E–depleted kinetochores are capable of binding a full complement of spindle Mts (Table 1A). As during the mitotic peak, nearly half of the kinetochores on uncongressed chromosomes were unattached, and all uncongressed chromosomes where we could identify sister kinetochores (24 total) were monooriented with the attached kinetochore facing the spindle pole. The attached kinetochores bound on average 3.8 Mts with a wider range than observed during the mitotic peak (Figure 3C and Table 1B). These results suggest that CENP-E–depleted kinetochores of both congressed and uncongressed chromosomes slowly continue to acquire kMts during a prolonged M-phase block.
We detected no gross morphological changes in kinetochores depleted of CENP-E (Figure 2). The corona on unbound kinetochores was clearly visible and similar to control unbound kinetochores (Figure 2e). Because detergent extraction and conventional fixation mask fine structure alterations (McEwen et al., 1998), we chose not to perform a more detailed analysis at this time.
Tension across Sister Kinetochores after CENP-E Depletion
We measured the separation of sister kinetochore outer plates as an indicator of the effect of anti-CENP-E microinjection on tension across the centromere (Waters et al., 1998). In addition, we estimated the flatness of each kinetochore outer plate by measuring how well points chosen along the outer plate fit to a straight line. Depletion of CENP-E from the kinetochore reduced the average separation between sister kinetochores from 1.56–0.79 μm, whereas nocodazole treatment reduced the separation to 0.67 μm (Table 2). Therefore, CENP-E–depleted kinetochores on congressed chromosomes only produce 13% of the Mt-induced centromere stretching found in untreated cells. In addition, the kinetochore outer plate was less distorted (had a greater degree of flatness) after anti-CENP-E injection or nocodazole treatment. Thus, anti-CENP-E injection dramatically reduces Mt-induced tension across the centromeres of congressed chromosomes.
Table 2.
Sister kinetochore separation in HeLa cells
| Uninjected control | Anti-CENP-E–injected | Nocodazole-treated | |
|---|---|---|---|
| Average sister kinetochore separation (μm)a | 1.56 | 0.79 | 0.67 |
| SD | 0.19 | 0.06 | 0.10 |
| Range | 1.14–2.10 | 0.65–1.02 | 0.40–0.85 |
| Kinetochore linearityb | 0.82 | 0.93 | 0.95 |
| No. measured | 36 | 57 | 23 |
Sister separation was determined from electron micrographs by choosing eight points on the outer plate of each sister kinetochore and measuring the distance between average of coordinates of each set of points.
R2 values of a least squares fit of the eight points chosen for each kinetochore. R2 values range from 0 (completely nonlinear) to 1 (perfect linear fit) and provide a measure of how straight or bent the kinetochore is.
Spindle Morphology and Position of Monooriented Chromosomes in Anti-CENP-E–injected Cells
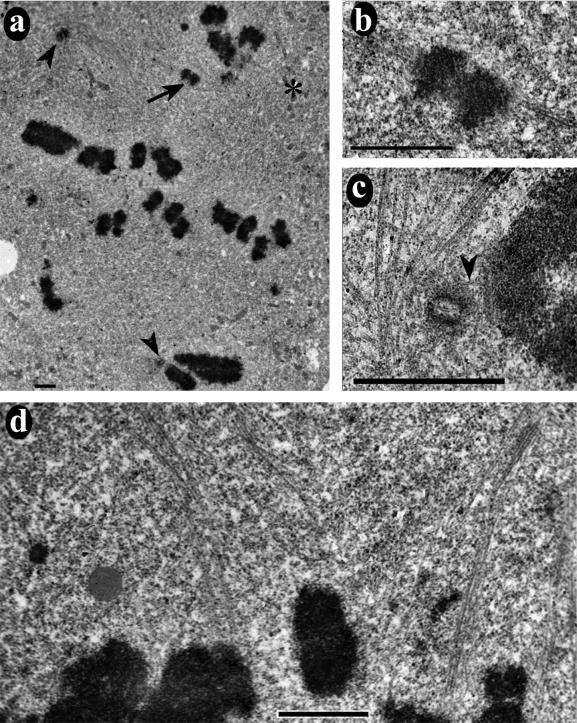
Previous studies have concluded that depleting CENP-E from the HeLa cell kinetochores inhibits metaphase alignment (Schaar et al., 1997; Yao et al., 2000). Here, we found that most anti-CENP-E–injected cells had a robust metaphase plate with only a few monooriented chromosomes. To reconcile our data with observations from previous studies, we used electron microscopy to examine antibody-injected cells that exhibited a more scattered distribution of chromosomes. Figure 4a shows a serial section containing centrioles from both primary spindle poles (arrowheads). The chromosome indicated by an arrow (see also Figure 4b) is bound to the upper primary pole and a secondary spindle (asterisk). This chromosome has in fact “congressed” to a position half way between the primary and secondary spindle poles. A neighboring serial section (Figure 4d) shows that even chromosomes located at the apparent spindle equator can be bound either to the primary or secondary pole.
Figure 4.
Anti-CENP-E injection causes an increased incidence in spindle distortion and chromosomes located abnormally close to the centriole. (a) Low-magnification electron micrograph showing an overview of an HX-1–injected HeLa cell. Centrioles at opposite spindle poles are indicated by arrowheads. The asterisk indicates the position of another spindle pole without a centriole. The chromosome indicated by the arrow is bioriented between the acentriolar pole and the upper centrosome. (b) Higher magnification view of the chromosome indicated in a, showing sister kinetochore fibers going to the centrosome and acentriolar spindle poles. (c) Higher magnification view of the lower centriole in a and the adjacent chromosome. This image was taken three serial sections away from the view in a. Separation between the centriole and kinetochore was only 0.16 μm (Table 3). A single Mt attached this kinetochore to the pole (arrowhead). (d) Upper half of the spindle shown in a at higher magnification and in a different serial section. It is evident that some of the chromosomes that appear to be located at the “metaphase plate” were actually attached to the acentriole pole, whereas others were attached to the centriole-containing pole. Bars, 1.5 μm.
Studies on HeLa cells disagree as to whether depleting CENP-E from kinetochores causes spindle fragmentation (Schaar et al., 1997; Yao et al., 2000). To resolve these differences, we examined CF-PAC cells, a human cell line that stays flat during mitosis (Gordon et al., 2001). As seen in Figure 5, a significant number of anti-CENP-E–injected CF-PAC cells exhibited multiple spindle poles with chromosomes aligned to the “minor” spindle poles (Figure 5, b–d, yellow arrows). The incidence of multiple spindle poles increased from 19% (8/42) for uninjected controls to nearly 50% (11/23) for injected cells on the same coverslips. Furthermore, even the bipolar injected cells often had poorly focused, swirl-shaped spindle poles (Figure 5, b–d), whereas multipolar control cells had well focused spindle poles (Figure 5a; our unpublished data). The swirling of the spindle fibers also was detected in injected HeLa cells via electron microscopy (Figure 4c).
Figure 5.
Immunofluorescence images of CF-PAC cells depleted of CENP-E via antibody injection. Synchronized CF-PAC cells were microinjected with HX-1 antibodies shortly after release from the G1/S block and fixed for straining 10–12 h later. To visualize endogenous CENP-E (red), cells were stained with rHX-1 (rat polyclonal) primary and Cy2-conjugated anti-rat secondary antibodies. Mts (green) were stained with a monoclonal primary and Alexa Green-conjugated secondary antibodies, whereas chromosomes (blue) were stained with Hoechst. (a) Uninjected control cells on the same coverslip as injected cells. CENP-E staining was prominent at the kinetochores in prometaphase cells (purple arrows), delocalized and less prominent at the kinetochore in out-of-focus metaphase cells (green arrows), and concentrated in the midbody in the late anaphase/teleophase cell (orange arrow). (b) Three injected cells showed no trace of CENP-E staining despite 6–8× exposure in the red channel (note staining in neighboring uninjected G2 cell). Injected cells showed an increased incidence of spindle pole fragmentation and accompanying spindle distortion. Monooriented chromosomes extremely close to one pole are indicated by red arrows. (c and d) Higher magnification view of injected cells with clear examples of bioriented chromosomes aligned to secondary poles (yellow arrows) and monooriented chromosomes stranded near a pole (red arrows). Bar, 20 μm (a and b) and 10 μm (c and d).
In addition to spindle pole fragmentation, we noticed that the chromosomes frequently approached unusually close to centrioles (Figure 4c). To quantify this effect, we measured the separation between each centriole we could identify and the closest centromere. For injected HeLa cells the average value was 0.69 μm (Table 3). In contrast, the average closest approach we observed in uninjected prometaphase HeLa cells was 3.6 μm. These results are corroborated by immunofluorescence images of injected CF-PAC cells that show a number of chromosomes unusually close to the spindle poles (Figure 5, b–d, red arrows). Thus, CENP-E depletion from the kinetochore results in a dramatic decrease in the minimum separation between centromeres and centrioles.
Table 3.
Closest approach of centromeres to a centriole in prometaphase HeLa cells
| Uninjected controlb | Anti-CENP-E– injected | |
|---|---|---|
| Average separation (μm)a | 3.6 | 0.69 |
| SD | 1.4 | 0.28 |
| Range | 2.1–7.1 | 0.16–1.27 |
| No. measured | 8 | 29 |
Measurements were made on electron micrographs between the centriole and the centromere of the nearest uncongressed chromosome for each pole where a centriole could be identified. Distances were determined from the minimum separation computed from all possible point pair combinations between sets of eight points chosen along the closest edges of both structures.
Control values were measured in uninjected cells in various stages of prometaphase.
Live Cell Imaging of Anti-CENP-E–injected CF-PAC Cells
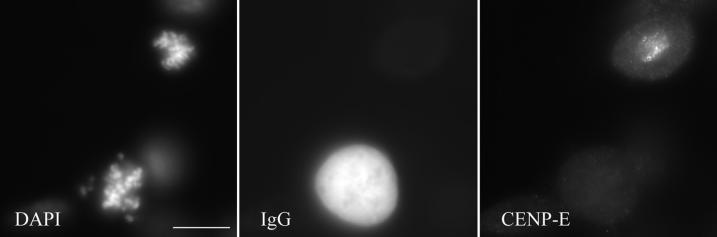
We used live cell imaging of CF-PAC cells to determine whether chromosomes in cells injected with CENP-E antibodies are able to undergo congression. In contrast to HeLa cells, CF-PAC cells are very amenable to video microscopy during mitosis due to their extreme flatness. To verify that antibody injection can depletes CENP-E from the CF-PAC cells, we first examined injected cells via immunofluorescence staining for the location of both the injected antibodies, and the mAb 177 CENP-E epitope. The latter is distinct from the HX-1 epitope of the injected antibody (Schaar et al., 1997). As seen in Figure 6, neither the injected antibodies nor mAb 177 stained kinetochores of injected cells. Thus, antibody injection can effectively deplete CENP-E from kinetochores in CF-PAC cells.
Figure 6.
Verification of CENP-E depletion from centromers of anti-CENP-E–injected CF-PAC cells. CF-PAC cells were injected with HX-1 as in Figure 5. (a) DAPI staining of an injected (lower middle) and uninjected (upper right) cell in prometaphase. (b) Injected antibodies are located via staining with Cy-5 anti-rabbit secondary antibody. Injected antibodies are located throughout the cytoplasm but not on the centromeres. (c) Visualization of endogenous CENP-E with the use of mouse monoclonal m 177 primary and Alexa Fluor 488 anti-mouse secondary antibodies. Note centromere staining is only detected in the uninjected cell. Bar, 20 μm.
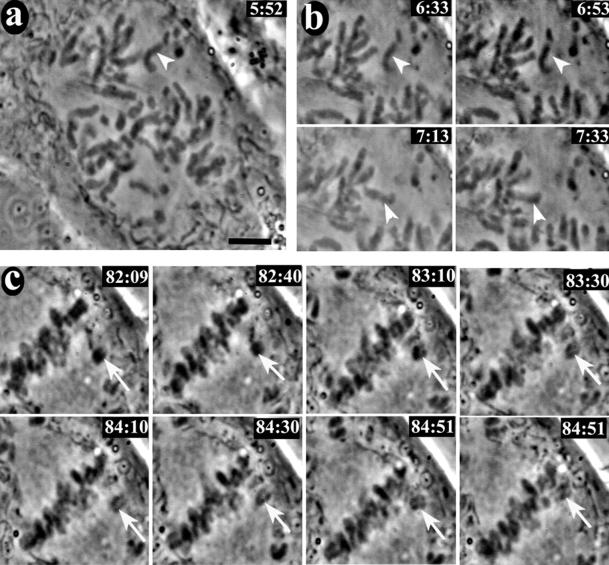
Figure 7 shows selected frames from the video sequence of one of four anti-CENP-E–injected CF-PAC cells filmed. All four cells were filmed for at least 2 h past nuclear envelope breakdown (NEB), at which time the cells were still arrested in prometaphase with a robust metaphase plate and monooriented chromosomes at the poles. In contrast, uninjected and nonimmune IgG-injected control cells all entered anaphase within 50 min of NEB (our unpublished data). Figure 7a shows a cell shortly after NEB. The indicated chromosome was about to undergo the classical fast poleward motion that accompanies initial monooriented attachment (Rieder and Alexander, 1990). Windowed frames from the video illustrating the fast poleward motion are presented in Figure 7b. A different set of windowed frames in Figure 7c shows a chromosome (arrow) undergoing normal congression to the spindle equator with a brief reversal of motion similar to that previously reported in newt lung (Skibbens et al., 1993) and PtK (Khodjakov and Rieder, 1996) cells. Congressed chromosomes at the metaphase plate and many of the monooriented chromosomes showed vigorous oscillations, but in contrast to what was reported for Drosophila (Yucel et al., 2000), we did not detect aligned chromosomes leaving the metaphase plate. Some of the monooriented chromosomes on the astral side of the pole did not appear to oscillate (see video). At least six clear examples of chromosome congression and four clear examples of fast poleward motion can be identified in this video. The other three cells filmed also showed numerous examples of congression, fast poleward movement, and oscillations.
Figure 7.
Video live cell imaging of a CF-PAC cell injected with HX-1 anti-CENP-E antibody. Synchronized CF-PAC cells were microinjected with antibody shortly after release from G1/S block and later remounted into Rose chambers. Coverslips were scanned for injected cells in prophase (injected cells were located via a scribe mark and coinjection with Oregon Green dextran). This cell was filmed for at total of 2 h past NEB with a 10-s filming rate (approximate time in minutes: seconds past NEB is located the upper right of each frame). (a) Early prometaphase. The unattached chromosome indicated by the arrowhead was about become monooriented and undergo fast poleward motion (see video 1 in online supplement). (b) Windowed frames showing the chromosome indicated in a undergoing fast motion to the upper spindle pole (see video 2 in online supplement). (c) Windowed frames illustrating congression. The congressing chromosome exhibited a typical transient reversal of motion before completing congression to the spindle equator (see video 3 in online supplement). Bar, 5.0 μm.
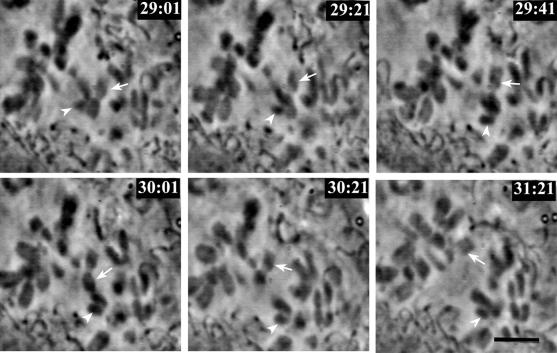
Figure 8 documents two chromosomes passing one another as one moved poleward and the other congressed to spindle equator. The chromosome indicated by the arrowhead started out as an unattached chromosome halfway between the pole and equator. As it became monooriented and traveled rapidly poleward, it brushed aside the arm of a newly bioriented chromosome (arrow) that was congressing to the spindle equator. This demonstrates that the chromosomes scattered between the pole and equator are not necessarily bioriented; they also can be unbound. Furthermore, examination of the full video sequence reveals that although some unattached chromosomes located in the center of the spindle became bioriented directly, without poleward migration, others under went fast poleward movement upon monooriented attachment and subsequently had to congress back to the spindle equator upon bioriented attachment.
Figure 8.
Unattached chromosomes can be located within the spindle. Selected windowed video frames showing two chromosomes sliding by one another. The chromosome indicated by the arrowhead obtained monooriented attachment and moved rapidly toward the spindle pole, moving past the chromosome indicated by the arrow. The latter obtained bioriented attachment at approximately the same time and congressed to the spindle equator (see video 4 in online supplement). Bar, 5.0 μm.
Status of Checkpoint Proteins at Kinetochores Depleted of CENP-E
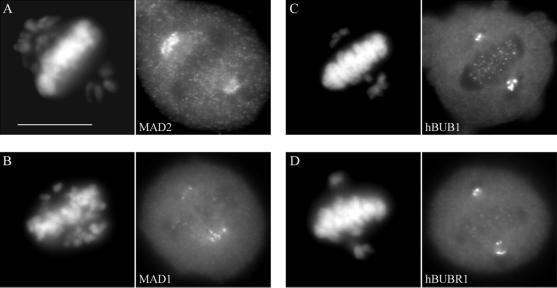
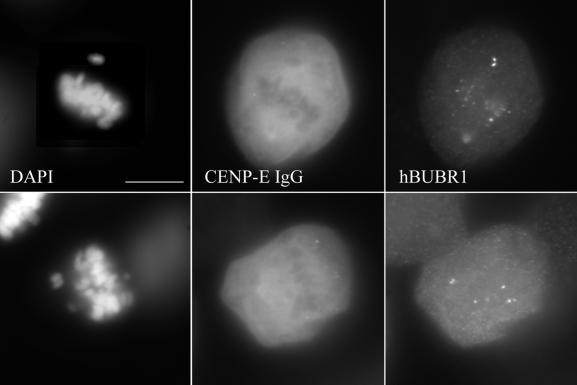
To determine how a 26% reduction in kMt binding and reduced kinetochore tension affect the checkpoint status at these kinetochores, mitotic HeLa cells were stained with antibodies to various checkpoint proteins. Figure 9a shows bright MAD2 staining at kinetochores of the monooriented chromosomes and only residual amounts of MAD2 on kinetochores of congressed chromosomes, in agreement with reports for uninjected metaphase cells (Chen et al., 1996; Li and Benezra, 1996). A similar staining pattern was found for MAD1 (Figure 9b), a checkpoint protein also known to be lost from kinetochores upon chromosome alignment (Chen et al., 1999; Campbell et al., 2001). The staining intensities of both hBUB1 and hBUBR1 were noticeably reduced at aligned kinetochores compared with the intensities at unaligned kinetochores (Figure 9, c and d), consistent with what has been reported previously for normal mitotic HeLa cells (Jablonski et al., 1998). A similar pattern of hBUBR1 staining was observed in CF-PAC cells (Figure 10), except that the more favorable imaging enables detection of bright staining on both sister kinetochores of monooriented chromosomes. Thus, the loss of CENP-E did not prevent kinetochores from recruiting checkpoint proteins and releasing them in response to spindle attachment and congression.
Figure 9.
Immunofluorescence staining of checkpoint proteins in anti-CENP-E–injected HeLa cells. Synchronized cells were injected with rat anti-CENP-E antibodies 2 h after release from the G1/S boundary and sampled 10 h later. (a–d) Cells were stained for hMAD2, hMAD1, hBUB1, and hBUBR1 with the use of appropriate rabbit antibodies and Texas Red-conjugated anti-rabbit secondary antibodies. Chromosomes and nuclei were stained with DAPI (left column). Injected cells exhibited the normal dissociation of checkpoint proteins from kinetochores of bioriented, aligned chromosomes. Bar, 10 μm.
Figure 10.
Localization of hBUBR1 in anti-CENP-E–injected CF-PAC cells. Chromosomes were stained with DAPI (a and d) and injected HX-1 antibodies with Cy-5 anti-rabbit secondary antibody (b and e). hBUBR1 was detected with rat anti-hBUBR1 primary and Alexa Fluor 488 anti-rat secondary antibodies (c and f). Note the bright hBUBR1 staining of both sister kinetochores on the monooriented chromosomes. As in Figure 6, centromeres of injected cells show no HX-1 staining. Bar, 20 μm.
DISCUSSION
CENP-E Is Not Required for Congression, Fast Poleward Motion, or Oscillation
Although previous studies all concluded that chromosome alignment requires CENP-E (Schaar et al., 1997; Wood et al., 1997; Yao et al., 2000), our time-lapse phase imaging of CF-PAC cells (Figures 7c and 8) clearly demonstrates that individual chromosomes in cells microinjected with anti-CENP-E antibodies are fully capable of undergoing congression that results in stable metaphase alignment. These data, along with the finding that all bioriented chromosomes are aligned either at the metaphase plate or to a secondary spindle pole (Table 1 and Figures 2, 4, and 5), clearly demonstrate that CENP-E is not required for congression. Despite this, unaligned chromosomes that were monooriented were always found along with aligned chromosomes. These results strongly suggest that the dependence of chromosome alignment on CENP-E is critically linked to the position of the chromosome within the spindle.
A potential caveat to this conclusion is that depletion via antibody injection is not equivalent to a genetic knockout. We show that anti-CENP-E injections deplete CENP-E staining from kinetochores to below levels detectable via immunofluorescence, even when the exposure time of the antibody–injected cells was 6–8 times longer than controls (Figure 5). Furthermore, neither the injected antibodies nor an antibody directed against a different CENP-E epitope (Figures 1, c and f, and 6, b and c) detected CENP-E at the kinetochore. Moreover, depletion of CENP-E from kinetochores via antibody injection has functional consequences on aligned chromosomes as exemplified by an 87% drop in Mt-induced stretching between sister kinetochores (Table 2), with remaining tension apparently contributed by other proteins (Gordon et al., 2001). Finally, the mixed phenotype of both congressed and uncongressed chromosomes is found regardless of whether CENP-E disruption is accomplished via antibody injection, transfection of mutants (Schaar et al., 1997), antisense RNA (Yao et al., 2000), or complete gene disruption in primary cells from mice (Putkey et al., 2000; data reported at the 40th annual meeting of the American Society for Cell Biology). Therefore, the weight of the combined evidence strongly supports the contention that we are observing the CENP-E null phenotype.
Previous conclusions that CENP-E is required for normal congression and chromosome alignment were largely based upon static images of fixed cells, or limited video imaging of cells that round up during mitosis (Schaar et al., 1997; Yao et al., 2000). However, the partial phenotype results in a range of chromosome alignment configurations when cells are depleted of CENP-E function. Although it is natural to emphasize cells with a large number of unaligned chromosomes as more strongly expressing the phenotype, these cells often show spindle pole fragmentation. This increased spindle pole fragmentation upon CENP-E depletion makes it potentially misleading to categorize the chromosomes scattered throughout the cell as being impaired in congression, because many of these chromosomes are actually aligned to a secondary pole (Figures 4 and 5). Furthermore, chromosomes located halfway between the spindle pole and equator are not necessarily bioriented (Figure 8). Therefore, the position of chromosomes in fixed cells is not sufficient to determine whether CENP-E is essential for chromosome alignment.
CENP-E Is Required for Reliable Spindle Attachment
The observation that CENP-E is not crucial for congression, fast poleward motion, or oscillations of bipolar chromosomes indicates there is functional redundancy. Our electron microscopy measurements demonstrate that CENP-E depletion reduces kMt binding on congressed chromosomes by 23% on average, even though these kinetochores appear to be capable of capturing a full complement of Mts given enough time (Figure 3 and Table 1). On the other hand, the chronic presence of monooriented chromosomes that fail to congress and exhibit very little Mt binding on attached kinetochores (Figure 3 and Table 1) shows that CENP-E must provide essential functions for these chromosomes.
It is unlikely that differences in chromosome behavior are due to variation in levels of CENP-E because, as noted above, antibody injection eliminates CENP-E staining at kinetochores and eliminates CENP-E's contribution to Mt-induced tension across the centromeres for both congressed and uncongressed chromosomes (Figures 1, 5, and 6 and Table 2; Yao et al., 2000). Furthermore, we do not observe intermediate levels of kMt binding or tension as expected for varying levels of residual CENP-E (Figure 3 and Table 2). A more likely explanation is the differences in environments of congressed and uncongressed chromosomes. Kinetochores on congressed chromosomes encounter relatively high numbers of Mt plus ends at the spindle equator (McIntosh and Landis, 1971; McEwen et al., 1997), which enables them to bind near normal numbers of Mts despite a loss of binding efficiency in the absence of CENP-E. In contrast, kinetochores on uncongressed chromosomes encounter few Mt plus ends, particularly from the opposite spindle pole (Nicklas and Ward, 1994), and reduced kMt binding efficiency results in significant delays to bioriented attachment. This situation is exacerbated for chromosomes approaching abnormally close to the spindle pole (Figures 4c and 5, b–d) where they exhibit little, if any, oscillatory behavior (video supplement 1). Without oscillations these chromosomes are unable to move to a location where they would have a better chance of capturing Mts emanating from the opposite spindle pole. Hence, they become chronically monooriented with a minimal probability of becoming bioriented.
The abnormally close approach of centromeres to the centriole when CENP-E is depleted from the kinetochore (Figures 4c and 5, b–d) is one of the striking findings of this study. This effect is probably not due to reduced polar ejection forces because antibody-injected cells have approximately normal numbers of spindle Mts (Figure 5) and astral Mts (our unpublished data), and we still see oscillations by many mono- and bioriented chromosomes. An alternative hypothesis is that CENP-E, a putative plus-end Mt motor (Wood et al., 1997), acts as a break to prevent fast poleward movement from carrying the kinetochore abnormally close to the spindle pole. Regardless of the mechanism, the loss of oscillations indicates that chromosomes close to the poles have been pulled into an area of few Mt plus ends and low polar ejection forces (Rieder et al., 1986). This is corroborated by the finding that these chromosomes are bound by only one or two kMts (Figure 4c and Table 1; our unpublished data).
Role of CENP-E in Maintaining Stability of Kinetochore Attachment and Tension across the Centromere
The kinetochore fiber is a dynamic entity with a constant turnover of Mts (Cassimeris et al., 1990; Zhai et al., 1995). Therefore, the number of Mts bound to any one kinetochore varies stochastically with time, and its average value is determined by the net sum of Mt capture and release. Depletion of CENP-E from the kinetochore reduces Mt binding by decreasing the capture rate, increasing the dissociation rate, or both. This in turn will reduce the efficiency with which monooriented chromosomes can form and maintain bipolar attachments. Nevertheless, functional redundancy enables a chromosome that is situated in an area of high Mt density to overcome the reduced efficiency of CENP-E–depleted kinetochores and establish bipolar attachments. This implies that simultaneous depletion of CENP-E function and a redundant component will produce a more pronounced phenotype where few, if any, chromosomes will achieve equatorial alignment. A potential example of this is with the use of CENP-E antibodies to disrupt in vitro chromosome motion driven by Mt disassembly (Lombillo et al., 1995). Because poleward motion in situ is not disrupted by injection of the same antibodies (Figure 7b and 8; our unpublished data), it is plausible that a CENP-E redundant component is lost from the kinetochore during chromosome isolation.
Even in situ, redundancy of CENP-E function is not absolute because in the absence of CENP-E at the kinetochore, unaligned chromosomes do not achieve significant kMt binding (Table 1) and very little tension is generated across the centromeres of aligned chromosomes (Table 2). The latter result is somewhat surprising because injected cells exhibit normal fast poleward motion, congression, and oscillations. Evidently, CENP-E generates 80–90% of the tension across the centromere independently of chromosome motion. Recent evidence suggests that 20–30% of the Mt-induced tension across the centromere is dependent upon the spindle poles acting as an anchor for kMts (Gordon et al., 2001), but it is still unclear how these different factors effecting tension interact.
In Drosophila, chromosomes readily form bioriented attachments and congress despite the loss of CENP-meta. Nevertheless, some chromosomes fall off the metaphase plate to resume a monopolar orientation (Yucel et al., 2000). We do not observe this instability of metaphase alignment in anti-CENP-E–injected CF-PAC cells, even though congressed chromosomes oscillate vigorously (video supplement 1). It is possible that this represents functional differences of CENP-E between evolutionarily distant species, but it is also possible that the differences arise from the fact that Drosophila kinetochores bind on average five Mts (Lin et al., 1981), compared with 17 for HeLa cells (Table 1A; Wendell et al., 1993). Therefore, a reduction in kMt binding in Drosophila due to the absence of CENP-meta, coupled with the normal stochastic variation in time, could easily result in some kinetochores losing all of their Mts and migrating to the attached spindle pole. In support of this hypothesis, sister chromatids of most unaligned chromosomes in mutant cells do not separate and segregate to opposite spindle poles at the onset of anaphase, indicating that one sister kinetochore was unbound (Yucel et al., 2000; note, the checkpoint is turned off in Drosophila embryos). Chromosomes that did segregate to opposite poles could have achieved bioriented attachment at, or just after, the start of anaphase because Drosophila kinetochores depleted of CENP-meta appear to achieve bioriented attachment more readily than do mammalian kinetochores depleted of CENP-E.
Role of CENP-E in Mitotic Checkpoint
Our data demonstrate that CENP-E depletion from the kinetochore does not affect the recruitment of normal amounts of MAD1, MAD2, hBUBR1, and hBUB1 to unattached kinetochores, or the release of these checkpoint proteins once chromosomes have become bioriented and congress to the spindle equator (Figures 9 and 10). Yao et al. (2000) reported similar results for MAD2 and hBUBR1 binding. These data indicate that in mammalian cells checkpoint proteins respond to Mt occupancy (Howell et al., 2000) or kinetochore tension (Nicklas, 1997) without requiring CENP-E. In contrast, the Xenopus extract system requires CENP-E for MAD2 localization at the kinetochore and checkpoint activation (Abrieu et al., 2000). The contradictory results between Xenopus extracts and mammalian cells are probably due to species-specific variations or differences between embryonic and somatic cells.
Waters et al. (1998, 1999) have postulated that MAD2 is recruited to kinetochores by low tension (via specific phosphorylation) and dissociates in response to kMt binding. Our electron microscopy data showed that kinetochores of congressed chromosomes in anti-CENP-E–injected cells had a 23% reduction in Mt occupancy and 87% reduction in centromere tension (Tables 1 and 2). Although we expected that reduced tension might recruit MAD2 and other checkpoint proteins to kinetochores, we detected low levels of hBUB1 and hBUBR1, and only residual amounts of MAD1 and MAD2, staining at the metaphase plate. This is similar to the case when taxol was used to relieve kinetochore tension without interfering with Mt occupancy (Waters et al., 1998). Therefore, our results indicate that 10–12 kMts are generally sufficient to promote dissociation of checkpoint proteins, even with minimal tension across the centromere. On the other hand, one or two kMts appears to be insufficient to dissociate checkpoint proteins because both kinetochores on uncongressed chromosomes in CF-PAC cells stain brightly for hBUBR1 (Figure 10). Hence, our data indicate that binding from three to nine Mts is required to dissociate checkpoint proteins from mammalian cell kinetochores.
Redundant Mechanisms for Spindle Attachment and Checkpoint Release
Although CENP-E is not a bona fide checkpoint protein in mammalian cells, it forms a stable complex with the checkpoint protein hBUBR1 (Chan et al., 1998, 1999; Yao et al., 2000). This has led Chan et al. (1999) to postulate that hBUBR1 monitors CENP-E–Mt interactions. If this postulate is correct then there should be a redundant mechanism for checkpoint release in response to kMt attachment because MAD1 and MAD2 dissociate normally from attached kinetochores in the absence of CENP-E (Figure 9; above discussion). One possibility, recently suggested by Chan et al. (2000), is that dynein-mediated kinetochore–Mt interactions are monitored by zeste white 10 (ZW10) and ROD (Rough Deal). ZW10 and ROD are required for dynein localization to the kinetochore (Starr et al., 1997) and checkpoint function in Drosophila and human cells (Basto et al., 2000; Chan et al., 2000; Savoian et al., 2000).
One possible explanation for these observations is that CENP-E and dynein form redundant pathways for kMt binding with the CENP-E pathway being monitored by hBUBR1, and the dynein pathway by ZW10/ROD. Both pathways require hBUB1 for localization to the kinetochore (Jablonski et al., 2000), and deactivation of either hBUBR1 or ZW10/ROD by respective kMt binding to CENP-E or dynein is sufficient to release the checkpoint. Thus, according to this model, neither CENP-E nor dynein is uniquely required for kMt attachment or checkpoint release, which explains why single depletion of either pathway has no affect on the recruitment or dissociation of MAD1 and MAD2 (Figure 9; Chan et al., 2000).
ACKNOWLEDGMENTS
We sincerely thank J. Hittle for antibody purifications and microinjections, R. Barnard for technical assistance, and Drs. M.S. Campbell and E.D. Salmon for anti-human MAD1 and MAD2 antibodies, respectively. We also thank Dr. A. Khodjakov and R. Cole for advice concerning video recording and image processing. Special thanks to Dr. D.A. Compton for suggesting the use of CF-PAC cells for videomicroscopy. Supported by National Science Foundation Grant MCB-9808879 (to B.F.M.), National Institutes of Health Grant RR-01219 to support the Wadsworth Center's Resource for Visualization of Biological Complexity as a National Biotech Resource, and Wadsworth Center's core facilities for light and electron microscopy. T.J.Y. was supported by National Institutes of Health Grant CA-75138, Core Grant CA-06927, the March of Dimes Foundation, and an Appropriation from the Commonwealth of Pennsylvania.
REFERENCES
- Abrieu A, Kahana JA, Wood KW, Cleveland DW. CENP-E as an essential component of mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Basto R, Gomes R, Karess RE. Rough Deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- Campbell MS, Chan GKT, Yen TJ. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. JCell Sci. 2001;114:953–963. doi: 10.1242/jcs.114.5.953. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Rieder CL, Rupp GR, Salmon ED. Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci. 1990;96:9–15. doi: 10.1242/jcs.96.1.9. [DOI] [PubMed] [Google Scholar]
- Chan GKT, Jablonski SA, Starr DA, Goldberg ML, Yen TJ. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Chan GKT, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at the kinetochore and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GKT, Schaar BT, Yen TJ. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J Cell Biol. 1998;143:49–63. doi: 10.1083/jcb.143.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Cooke CA, Schaar B, Yen T, Earnshaw WC. Localization of CENP-E in fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma. 1997;106:446–455. doi: 10.1007/s004120050266. [DOI] [PubMed] [Google Scholar]
- Gordon MB, Howard L, Compton DA. Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J Cell Biol. 2001;152:425–434. doi: 10.1083/jcb.152.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski SA, Chan GKT, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Jablonski SA, Hittle JC, Yen TJ. hBUB1 mediates assembly of hBUBR1, hMAD2, and hROD checkpoint proteins onto kinetochores. Mol Biol Cell. 2000;11:92a. [Google Scholar]
- Khodjakov A, Rieder CL. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J Cell Biol. 1996;135:315–327. doi: 10.1083/jcb.135.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMad2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Lin H-PP, Ault JG, Church K. Meiosis in Drosophila melanogaster. I. Chromosome identification and kinetochore microtubule number during the first and second meiotic divisions in males. Chromosoma. 1981;83:507–521. doi: 10.1007/BF00328276. [DOI] [PubMed] [Google Scholar]
- Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR. Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol. 1995;128:107–115. doi: 10.1083/jcb.128.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Hsieh C-E, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Landis SC. The distribution of spindle microtubules during mitosis in cultured human cells. J Cell Biol. 1971;49:468–497. doi: 10.1083/jcb.49.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey FR, Weaver BA, Cleveland DW. Determining the function of murine CENP-E in chromosome movement. Mol Biol Cell. 2000;11:96a. [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cassels G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol. 1986;103:581–591. doi: 10.1083/jcb.103.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Earnshaw W, Rieder CL. Cleavage furrows formed between centrosomes lacking an intervening spindle and chromosomes contain microtubule Bundles, INCENP, and CHO1 but not CENP-E. Mol Biol Cell. 1999;10:297–311. doi: 10.1091/mbc.10.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Goldberg ML, Rieder CL. The rate of poleward chromosome motion is attenuated in Drosaphila zw10 and rod mutants. Nat Cell Biol. 2000;2:948–952. doi: 10.1038/35046605. [DOI] [PubMed] [Google Scholar]
- Schaar BT, Chan GKT, Maddox P, Salmon ED, Yen TJ. CENP-E function at kinetochores is essential for chromosome alignment. J Cell Biol. 1997;139:1373–1382. doi: 10.1083/jcb.139.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Skeen VP, Salmon ED. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J Cell Biol. 1993;122:859–875. doi: 10.1083/jcb.122.4.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Li Z, Etmad-Moghadam B, Daw RK, Goldberg ML. Conservation of the centromere/kinetochore protein ZW10. J Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Gorbsky GJ, Salmon ED, Nicklas RB. Mad2 binding by phosphorylated kinetochores links error detection and checkpoint action in mitosis. Curr Biol. 1999;9:649–652. doi: 10.1016/s0960-9822(99)80287-5. [DOI] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends upon microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendell KL, Wilson L, Jordan MA. Mitotic block in HeLa cells by vinblastine: ultrastructural changes in kinetochore-microtubule attachment and in centrosomes. J Cell Sci. 1993;104:261–274. doi: 10.1242/jcs.104.2.261. [DOI] [PubMed] [Google Scholar]
- Wood KW, Sakowicz R, Goldstein LSB, Cleveland DW. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel human centrosome-associated protein required for progression from metaphase to anaphase. EMBO J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Li G, Schaar BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature. 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]
- Yucel JK, Marszalek JD, McIntosh JR, Goldstein LSB, Cleveland DW, Phlip AV. CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J Cell Biol. 2000;150:1–11. doi: 10.1083/jcb.150.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]