Abstract
Amphibian embryos provide a powerful system to study early cell fate determination because their eggs are externally fertilised, large, and easy to manipulate. Ultraviolet (UV) or lithium chloride (LiCl) treatment are classic embryonic manipulations frequently used to perturb specification of the dorso-ventral (DV) axis by affecting the stability of the maternal Wnt mediator β-catenin. Such treatments result in the formation of so-called ventralised or dorsalised embryos. Although these phenotypes have been well described with respect to their morphology and some aspects of gene expression, their whole transcriptomes have never been systematically characterised and compared. Here we show that at the early gastrula stage UV-treated embryos are transcriptionally more closely related to untreated embryos than to LiCl-treated embryos. Transcriptional comparisons with dissected ventral and dorsal regions of unperturbed gastrula embryos indicate that UV and LiCl treatments indeed enrich for ventral and dorsal cells, respectively. However, these treatments also affect the balance of neural induction in the ectodermal germ layer, with LiCl stimulating pro-neural BMP inhibition and UV preferentially generating epidermis because of elevated BMP levels. Thus the transcriptomes of UV- and LiCl-treated embryos can best be described as ventro-epidermalised and dorso-neuralised. These descriptions notwithstanding, our profiling reveals several hitherto uncharacterized genes with differential expression along the DV axis. At least one of these genes, a RNF220-like ubiquitin ligase, is activated dorsally by β-catenin. Our analysis of UV/LiCl-mediated axis perturbation will enhance the mechanistic understanding of DV axis determination in vertebrates.
Keywords: Xenopus, UV, Lithium, Wnt, β-catenin, Dorso-ventral, Neural induction, Cell fate, Transcriptome, RNF220
Highlights
-
•
We perturb DV axis specification in Xenopus tropicalis by UV or LiCl treatment.
-
•
Transcriptome reveals that UV-treated embryos are more similar to untreated embryos.
-
•
Comparison with ventral and dorsal cells confirms the effects of LiCl and UV in DV axis perturbation.
-
•
We identify a RNF220-like ubiquitin ligase as a dorsal marker regulated by β-catenin.
1. Introduction
Amphibian eggs have an animal–vegetal axis that is defined by the differential deposition of various proteins and RNA transcripts during oogenesis. This initial radial symmetry is broken immediately after fertilisation along the prospective dorso-ventral (DV) axis. At the sperm entry point (SEP) the paternal centriole initiates microtubule polymerisation which enables a 30° rotation of the cortical layer relative to the cytoplasmic core (Elinson, 1975; Elinson and Manes, 1978; Kao and Elinson, 1988; Manes and Elinson, 1980; Vincent et al., 1986). This causes the so-called grey crescent to appear opposite the SEP, which marks the prospective dorsal side of the embryo.
The disruption of microtubule polymerisation provides a powerful approach to investigate DV axis determination (Kao and Elinson, 1988). The method of choice has frequently been the ultraviolet (UV) irradiation of the vegetal pole immediately after fertilisation. Without intact vegetal-cortical microtubules, various components of the canonical Wnt signalling pathway such as Dishevelled (Dsh), Wnt11 and GSK3-binding protein (GBP) fail to be transferred from the vegetal pole to the prospective dorsal side (Miller et al., 1999, Tao et al., 2005, Weaver et al., 2003). Consequently, the Wnt mediator β-catenin remains unstable and does not accumulate in nuclei, where it normally triggers zygotic gene expression and so regulates DV patterning (Larabell et al., 1997).
In contrast to UV irradiation, lithium chloride (LiCl) is frequently used to promote dorsal specification. LiCl stabilises β-catenin by inhibiting GSK3β, which is ubiquitously expressed and belongs to the destruction complex of the canonical Wnt pathway (Klein and Melton, 1996, Stambolic et al., 1996). The phenotypic effects of LiCl treatment on amphibian embryos depend on dosage and stage of administration (Hall, 1942, Pasteels, 1945). Addition of LiCl to 0.3 M to the medium of embryos at the 32-cell stage generates truncated tadpoles with enlarged dorso-anterior structures (Kao et al., 1986). By the 32-cell stage, the activity of canonical Wnt is normally restricted to the dorsal side of the embryo due to Dsh-mediated GSK3β inhibition; widespread LiCl-mediated inhibition of GSK3β causes ventral cells to accumulate nuclear β-catenin and thus to adopt a dorsal fate.
The degree of ventralisation and dorsalisation can be classified morphologically according to the dorso-anterior index (DAI), which ranges from 0 to 10 (Kao and Elinson, 1988): Hyperdorsalised embryos are scored as 10; normal embryos as 5; and hyperventralised embryos resembling a belly piece with no dorso-anterior structures as 0.
Here we use UV and LiCl treatment to study DV axis determination on a transcriptome-wide scale. The genomes of the allotetraploid X. laevis and the diploid X. tropicalis have been sequenced (Hellsten et al., 2010, Session et al., 2016), and this has allowed thorough analyses of whole transcriptomes in time (Collart et al., 2014, Owens et al., 2016) and space (Blitz et al., 2017, De Domenico et al., 2015; Ding et al., 2017a; Popov et al., 2017). Our comparison with the transcriptomes of dorsal- and ventral embryonic regions confirms that UV or LiCl treatments confer ventral or dorsal cell fate specification, respectively. However, these treatments also affect the balance of neural induction in the ectodermal germ layer, prompting us to characterise UV- and LiCl-treated embryos as ventro-epidermalised and dorso-neuralised. This revised characterisation of UV and LiCl phenotypes notwithstanding, we have identified some hitherto uncharacterized genes with differential expression along the DV axis. We show that a gene coding for a novel RN220-like ubiquitin ligase is directly activated by β-catenin in dorsal cells.
2. Results
2.1. UV and LiCl treatments perturb DV axis formation in X. tropicalis
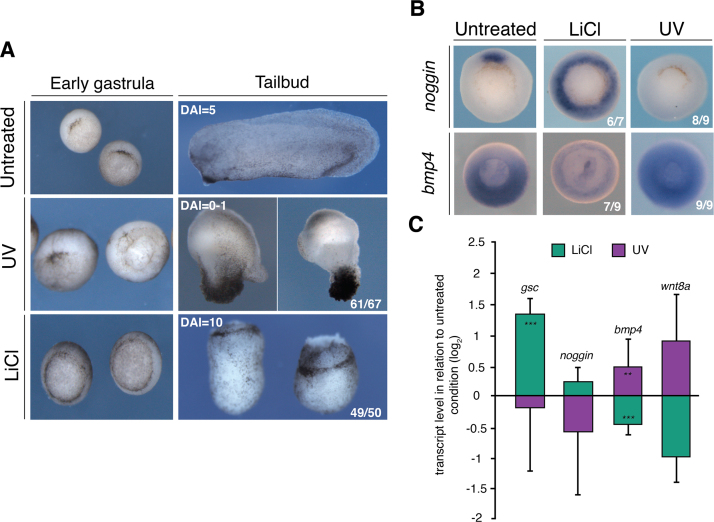
The conditions required to ventralise or dorsalise X. tropicalis embryos were determined empirically starting with those that had been optimised for X. laevis (Sive et al., 1999). Hyperventralisation was achieved by irradiating the vegetal pole with shortwave UV for 2 min at a range of 2 cm from the light source within 30 min after fertilisation. This treatment often caused the blastopore lip to develop abnormally: the initial appearance of the blastopore lip was delayed, and it formed circumferentially rather than on the dorsal side. By the tailbud stage, these embryos appeared as amorphic tissue lumps known as belly pieces (DAI = 0–1) (Fig. 1A). Dorsalisation was most effective when 32-cell embryos were incubated in 0.3 M LiCl for 5 min. Hyperdorsalisation caused blastopore lip formation to be initiated on time but circumferentially (Fig. 1A). At the tailbud stage, hyperdorsalised embryos consisted of radially symmetric, extended anterior structures such as a circular cement gland at the expense of any posterior structures such as trunk and tail (DAI = 10).
Fig. 1.
Optimised LiCl and UV treatments ofX. tropicalisembryos perturb their DV axis determination in line with the original experiments inX. laevis. (A-C) DV perturbations in X. tropicalis embryos caused by LiCl or UV treatments. (A) Phenotypical morphologies. (B) WMISH of dorsal (noggin) and ventral (bmp4) markers in mid-gastrula embryos. (C) RT-qPCR of dorsal (gsc and noggin) and ventral (bmp4 and wnt8a) markers in mid-gastrula embryos. Error bars, standard error of the mean (SEM) of biological triplicates. Student's two-tailed t-test: *, p < 0.1; **, p < 0.05; and ***, p < 0.01. DAI, dorso-anterior index.
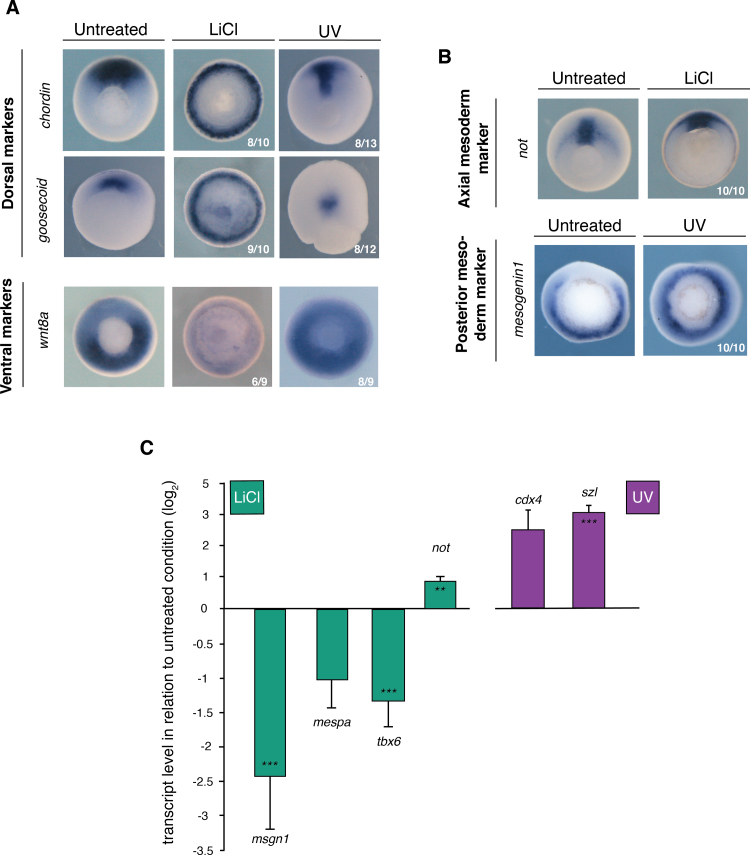
LiCl treatment had a high penetrance with 95–100% (e.g., 49 out of 50) of embryos being hyperdorsalised, while UV irradiation caused 85–95% (e.g., 61 out of 67) of the embryos being hyperventralised. In addition to the DAI score, the treatment efficacies were validated by analysing the expression levels of known dorsal (gsc, chordin, noggin and not) and ventral (wnt8a, bmp4 and msgn1) markers (Fig. 1B,C and Fig. S1). Expression of dorsal markers expanded ventrally in LiCl-treated embryos while being substantially reduced under UV treatment. Conversely, the expression levels of ventral markers increased in UV-treated embryos and decreased in LiCl-treated embryos.
2.2. Transcriptomics of UV or LiCl treatment
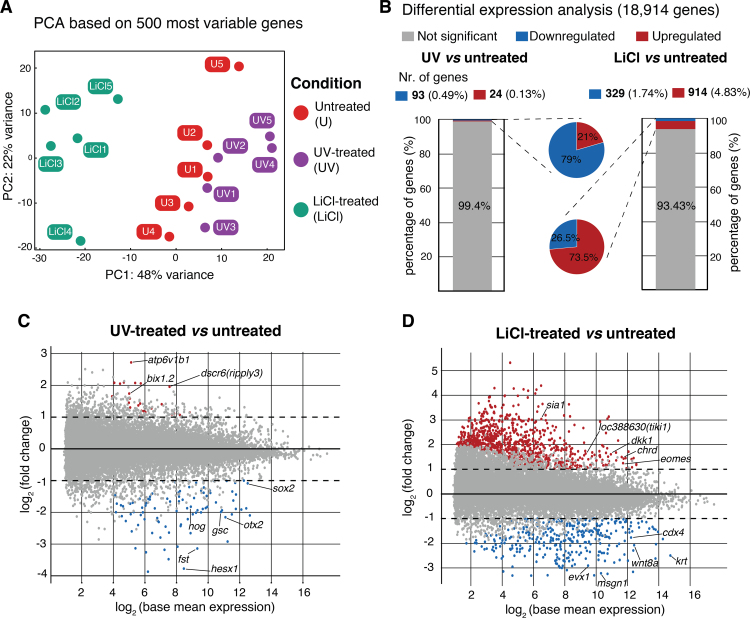
While the morphological and some transcriptional effects of UV or LiCl treatment have been well studied, there has been no systematic comparison of the transcriptomes of such embryos. Therefore, poly-A enriched transcriptomes of untreated, LiCl- and UV-treated embryos were analysed for differential expression and enrichment of gene function. For each condition, mid-gastrula embryos (stage 11–11.5) from five independent experiments were profiled, so as to provide statistical power to allow for transcript level variability between clutches and differing efficacies of UV or LiCl treatment. For each biological replicate, some sibling embryos were maintained to the tailbud stage, and only clutches that showed high phenotype penetrance (≥85%) were subjected to total RNA extraction, poly(A) RNA-Seq library generation and deep paired-end sequencing. Results of aligning sequencing reads to gene models v7.2 of the X. tropicalis genome assembly v7.1 are summarised in Table S1.
Overall, principal component analysis (PCA) suggested that the majority of observed variance reflected treatment-induced changes, while the second principal component revealed some variance that separated biological replicates (Fig. 2A). The transcriptome of LiCl-treated embryos was significantly different from that of untreated embryos, and it was slightly more different from that of UV-treated embryos. By contrast, UV treatment did not induce any large-scale shifts of gene expression by the mid-gastrula stage (Fig. 2A). Pairwise comparison with the normal transcriptome corroborated these relationships: LiCl-treated embryos had ten times more significant (at least two-fold change at FDR ≤ 1%) gene expression changes (~ 6.6%) than did UV-treated embryos (~ 0.6%) (Fig. 2B-D). Of the genes whose expression were affected in LiCl-treated embryos, almost 75% were upregulated (914 out of 1243 genes), while genes that were affected by UV treatment were predominantly downregulated (93 out of 117 genes) (Fig. 2B-D).
Fig. 2.
UV-treated embryos are transcriptionally more closely related to unperturbed embryos than LiCl-treated embryos. (A) Principal component analysis (PCA) of five biological replicates for each condition. (B-D) Identification of affected genes: transcriptional fold change ≥ 2 with FDR ≤ 1%. (C,D) MA plots of transcriptional fold changes against base mean expressions between indicated conditions.
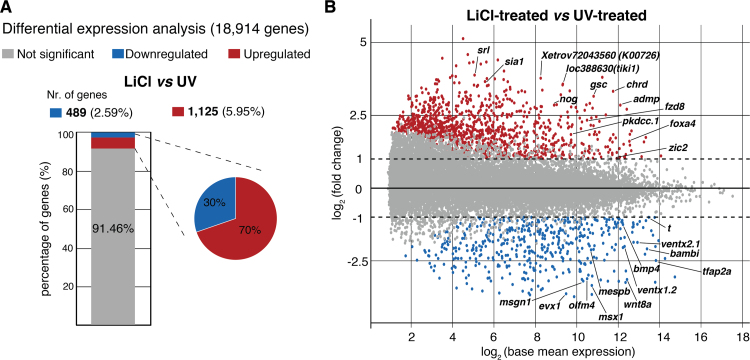
UV and LiCl have opposing effects on development, so to identify genes that are differentially expressed along the DV axis we made a direct comparison of gene expression following the two treatments. About 8.6% of the genes analysed showed significantly different transcript levels (at least two-fold change at FDR ≤ 1%), between UV and LiCl, 70% of which were elevated upon LiCl treatment (Fig. 3A; Table S2 and S3). Among the genes with the highest upregulation in LiCl-treated embryos were early dorsal fate determinants expressed in the Spemann organiser: the homeobox genes siamois 1 (sia1) (Lemaire et al., 1995) and goosecoid (gsc) (Laurent et al., 1997); BMP antagonists noggin (nog) (Kessler et al., 1997) and chordin (chrd) (Piccolo et al., 1999); canonical Wnt regulators frizzled 8 (fzd8) (Itoh et al., 1998) and loc3888630 (tiki1) (Zhang et al., 2012); and the secreted tyrosine kinase pkdcc.1 implicated in morphogenetic movements and planar cell polarity (Ding et al., 2016; Popov et al., 2017; Vitorino et al., 2015). Similar transcriptional stimulation was observed for later markers of dorsal axial and paraxial cell specification, including the neural transcription factor zic2, the muscle-specific calcium ion regulator sarcalumenin (srl), and the chordamesoderm and notochord marker foxa4 (Fig. 3B).
Fig. 3.
Sensitive transcriptomics of DV axis determination. (A) Differential expression analysis between LiCl- and UV-treated embryos. (B) MA plots of transcriptional fold changes against base mean expressions between indicated conditions. Identification of affected genes: transcriptional fold change ≥ 2 with FDR ≤ 1%.
By contrast, among the 485 genes significantly upregulated in UV-treated embryos were markers of ventro-posterior and non-neural ectoderm development. The former included both ligands (bmp4 and wnt8a) and tissue-specific transcription factors (evx1, msx1, mespb, msgn1, ventx1.1 and ventx1.2) (Fig. 3B).
These observations indicate that LiCl and UV treatment had the expected effect on transcription, regulating the choice between dorso-anterior and ventro-posterior specification, respectively.
2.3. UV- and LiCl-induced perturbations show DV-related gene signatures
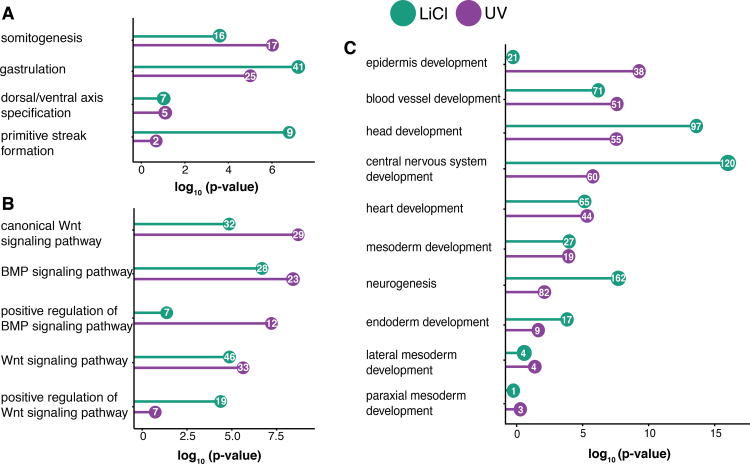
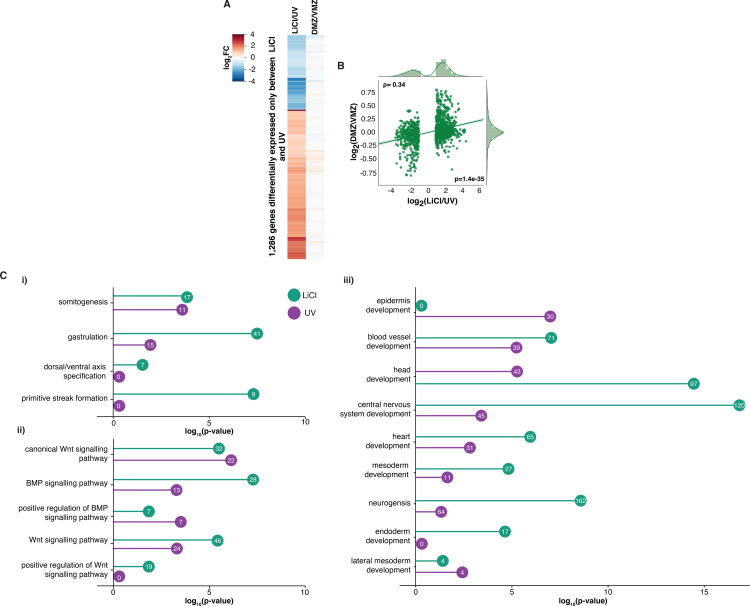
To characterise the transcriptomes of UV- and LiCl-treated embryos in detail, differentially expressed genes were analysed for enriched biological processes (BPs) of the Gene Ontology (GO) project (Fig. 4). LiCl-triggered gene upregulations were preferentially enriched for the BPs of gastrulation and primitive streak formation (known as blastopore lip formation in Xenopus) (Fig. 4A). For example, these BPs were associated with the upregulation of gsc, cer1, nodal, otx2, otx1, lhx1 and hhex upon LiCl treatment. By contrast, the BP of somitogenesis was overrepresented among upregulated genes in UV-treated embryos, many of which were markers of paraxial mesoderm such as msgn1, mesp2, mespA, cdx4 and t. The enrichment level of genes associated with DV axis specification was similar among upregulated genes in UV- and LiCl-treated embryos.
Fig. 4.
LiCl and UV treatments affect DV axis determination and neural induction. Plots show the number of upregulated genes and associated hypergeometric p-values of enrichment for specific biological processes (BPs) of the Gene Ontology (GO) project. Selected BPs refer to early developmental processes (A), signalling (B) and differentiation of germ layers and their derivatives (C).
Many of the genes whose expression were changed in UV- and LiCl-treated embryos were involved in the BMP and Wnt signalling pathways (Fig. 4B). This was consistent with the known effects of these pathways on DV specification. The BMP-related genes overrepresented in UV-treated embryos encode several BMP ligands (bmp4, bmp7.1) as well as several transcription factors (TFs) of the helix-loop-helix (hes3.3, hes5.1, hes6.1 and hes8), ventx (ventx1.1, ventx1.2 and ventx3.2) family and some other families (msx1 and foxj1). Interpretation of changes in expression of components of the Wnt signalling pathway was more complicated because Wnt signalling normally occurs in two waves in the early embryo: an early maternal wave at the dorsal side and a later zygotic wave, predominantly mediated by Wnt8a, on the ventro-lateral side of the embryo. Upregulated genes in LiCl-treated embryos associated with positive regulation of the pathway included admp, hhex and the TF family of zic genes (zic1–4).
At later stages, genes overrepresented in LiCl-treated embryos included those involved in neurogenesis, head development (e.g. chrd) and central nervous system development (e.g. zic TFs and the blimp-associated TF prdm12). Genes overexpressed in UV-treated embryos included those involved in epidermis development (e.g. keratin and grainyhead like TF 3). Endoderm development was slightly enriched in LiCl-treated embryos (e.g. Nodal ligand nodal and TFs hhex, otx2, foxa2 and nkx2-1). This was consistent with reports that lithium ions potentiate mesendoderm formation in response to Nodal signalling (Cooke and Smith, 1987, Cooke et al., 1989). Expression of genes related to mesoderm formation did not differ between UV- and LiCl-treated embryos.
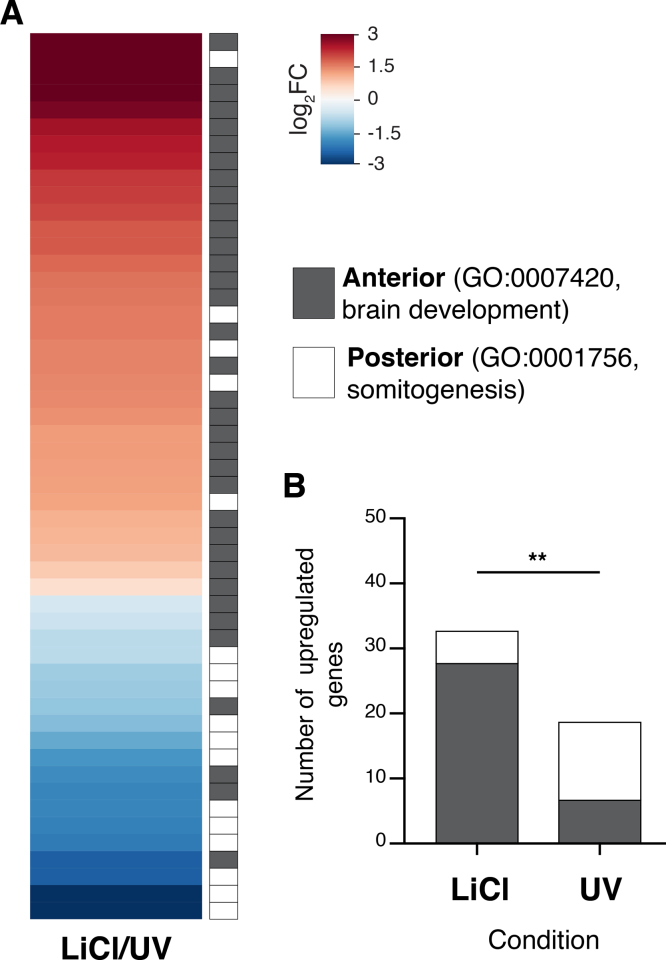
Although there is no GO term describing anterior or posterior identity we observed that genes involved in anterior (brain) or posterior (somitogenesis) development are upregulated in LiCl- or UV-treated embryos respectively (Fisher's exact test, p-value 0.007) (Fig. S2). This association is consistent with the fate map of the Xenopus early gastrula, where prospective anterior and dorsal structures overlap, as do ventral and posterior structures (Lane and Sheets, 2000, Martinez Arias and Steventon, 2018).
2.4. DV axis perturbation versus manual dissection of DV tissue
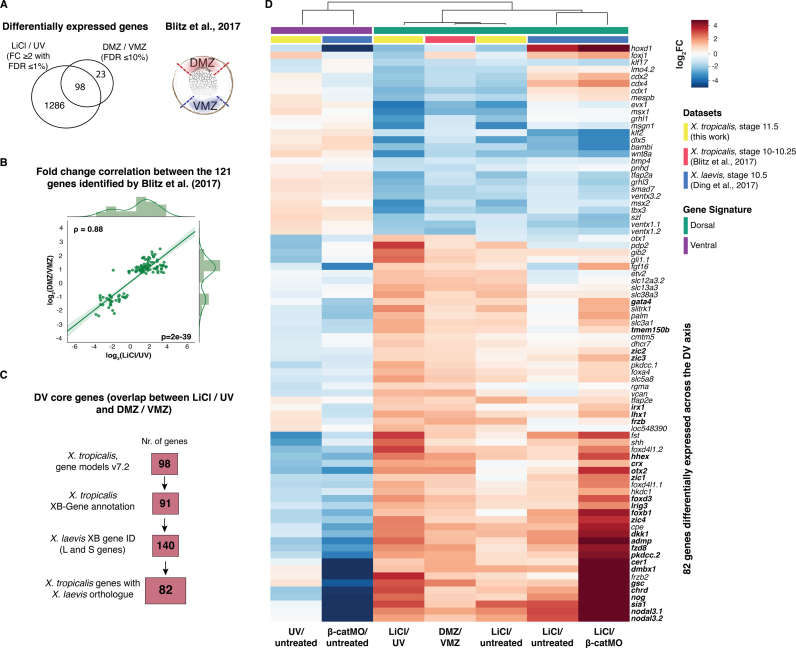
In an effort to verify our results we compared data from UV- and LiCl-treated embryos with the transcriptional profiles of dorsal and ventral marginal zone (DMZ and VMZ) tissue from untreated embryos (Blitz et al., 2017) (Fig. 5A).
Fig. 5.
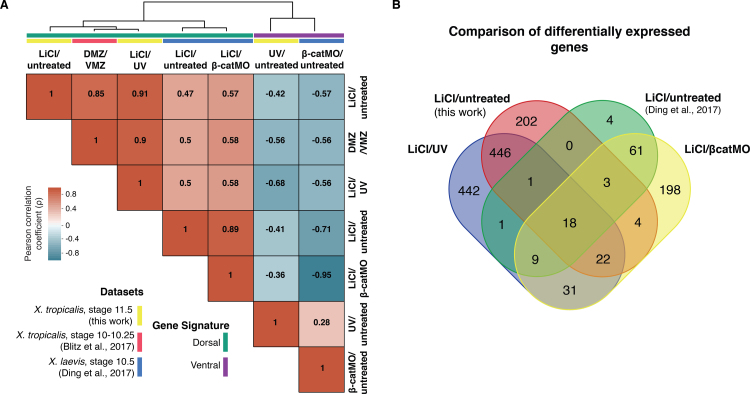
Comparison with differential gene expression across DV axis confirms that LiCl and UV treatments generate dorso-neuralised and ventro-epidermalised embryos. (A) Venn diagram of differential gene expression between LiCl and UV (LiCl vs UV, stage 11.5) and across DV axis (DMZ vs VMZ, stage 10–10.25; Blitz et al., 2017). (B) Regression plot of indicated transcriptional fold changes for differential gene expression across the DV axis (Blitz et al., 2017). Pearson coefficient, ρ = 0.88. Student's two-tailed t-test, p = 2e-39. (C) Procedure to find X. laevis orthologues of the X. tropicalis DV core genes via Xenbase (XB) gene identities. L and S refer to gene's location on either the long or short chromosome. If both S and L genes were present, the latter were selected since the L subgenome has been shown to better resemble the ancestral condition (Session et al., 2016). (D) Heatmap of transcriptional fold changes (FC) of DV core genes between indicated conditions. Gene names in bold are part of the early dorsal β-catenin gene signature (Ding et al., 2017).
Our pairwise comparison of DMZ and VMZ transcriptomes identified 121 differentially expressed genes (FDR ≤10%), of which 86 were upregulated in DMZ and 35 in VMZ (Fig. 5A,B). Manual dissections thus identified significantly fewer differentially expressed genes than did our UV and LiCl perturbations (1384 genes), but we note that the ratio of upregulated genes in DMZ (~ 71%) versus VMZ (~ 29%) resembles the ratio of upregulated genes in LiCl- versus UV-treated embryos (~ 70% versus ~ 30%). The difference in numbers may derive from the different experimental approaches; the different stages of analysis (stage 10–10.25 for dissection versus st. 11.5 in our experiments); and the number of biological replicates (two for dissection versus five in our experiments).
About 80% (98 out of 121) of the genes whose expression differed between DMZ and VMZ regions had differences in transcript level of at least two-fold between LiCl- and UV-treated embryos. Pairwise correlation revealed that in most cases (117 out of 121) where a gene was upregulated in DMZ or VMZ regions, upregulation was also observed in LiCl- or UV-treated embryos, respectively (Fig. 5B,D). The discrepancies (e.g., myf5) might derive from differences in experimental approaches, in that dissected DMZs might have contained BMP-exposed lateral marginal zone (LMZ) by the stage of analysis (de Souza et al., 1999, Hopwood et al., 1991; Kim et al., 1998; Wilm and Solnica-Krezel, 2005). We studied transcriptional fold changes between DMZ and VMZ of the 1286 genes identified as differentially expressed only between LiCl- and UV-treated embryos (Fig. S3A). There was a weakly positive correlation between the transcriptional fold changes of the two experiments (Fig. S3B). Some of the genes were involved in DV axis determination while others reflected UV- and LiCl-induced perturbations also affecting antero-posterior patterning and neural induction (Fig. S3C).
Ding et al., 2017a, Ding et al., 2017b recently defined a dorsal and β-catenin-dependent gene signature for X. laevis embryos at early gastrula stage. This was based on the integrative analysis of various transcriptomes from embryos treated with LiCl, embryos injected with β-catenin morpholino (β-catMO), as well as embryos bisected into dorsal and ventral halves. To compare this data with our results, we first defined a core set of 98 DV genes based on the overlapping differential expression between LiCl/UV and DMZ/VMZ in X. tropicalis (Fig. 5A,C). This core set was reduced to 82 genes for which orthologues between the two Xenopus species and experiments could be found (Fig. 5C). Despite the lower Pearson correlation between X. laevis and X. tropicalis experiments (Fig. S4A), the transcriptional behaviour of most DV core genes was in line with the treatment and consistent between Xenopus species (Fig. 5D). Some inconsistencies of expression level between experiments might be explained by the differences in stage, species, embryo manipulation and library preparation. The genes with highest correlation among all the experiments were immediate-early targets of maternal β-catenin such as nodal 3.1, nodal 3.2 and sia1 (Fig. 5D).
2.5. Validation and characterisation of novel DV markers identified by axis perturbation
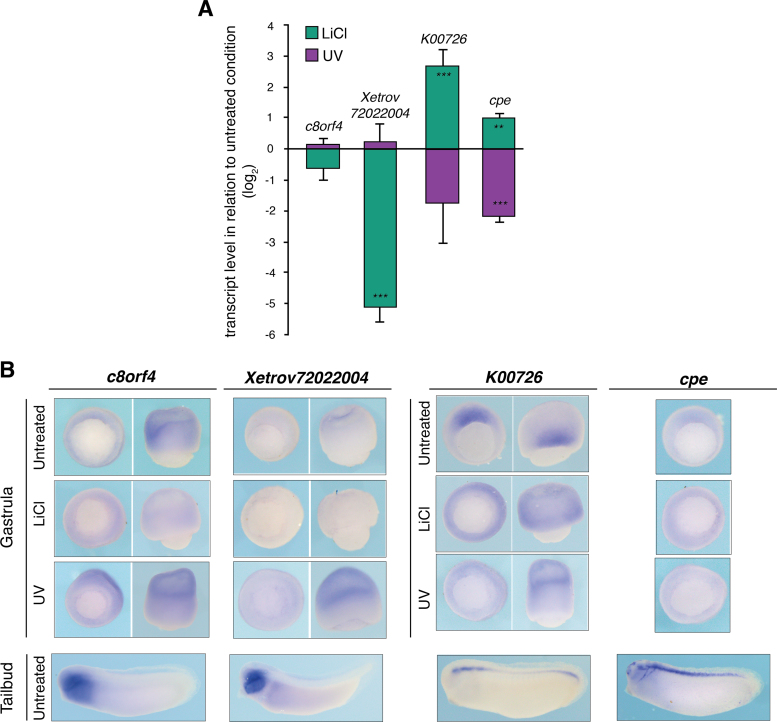
To validate our transcriptome dataset, we selected two genes that were upregulated in LiCl-treated embryos (cpe and K00726) and two that were upregulated in response to UV radiation (Xetrov72022004 and c8orf4). More uncharacterised genes identified by DV axis perturbation are listed in Table S4. The cpe gene has recently been identified as part of the dorsal gene signature in X. laevis (Ding et al., 2016).
Quantitative transcript profiling at the gastrula stage confirmed that cpe and K00726 were upregulated in LiCl-treated embryos and downregulated in UV-treated embryos, while Xetrov72022004 and c8orf4 were downregulated in LiCl-treated embryos and unaffected in UV-treated embryos (Fig. 6A). Whole mount in situ hybridisation revealed that cpe and K00726 were expressed in the DMZ at gastrula stages and in the neural tube by tailbud stages. As expected, their expression expanded around the blastopore in LiCl-treated embryos and was reduced in UV-treated embryos. Expression of both Xetrov72022004 and c8orf4 was weak and ectodermal in unperturbed embryos (Fig. 6B). By the tailbud stage, expression of both was restricted to the head ectoderm. LiCl treatment significantly reduced their ectodermal expression, consistent with its ability to promote dorsal and neural structures.
Fig. 6.
Expression analysis of uncharacterised genes identified by the means of nongenetic axis perturbation and comparative transcriptomics. RT-qPCR (A) and WMISH (B) of c8orf4, Xetrov72022004, K00726 and cpe. (A) Error bars, SEM of biological triplicates. Student's two-tailed t-test: *, p < 0.1; **, p < 0.05; and ***, p < 0.01. (B) Mid-gastrula stages, vegetal and lateral view except for cpe with vegetal view only; and mid-tailbud stage, lateral view.
2.6. β-catenin activates dorsal marker K00726, a Rnf220-like ubiquitin ligase
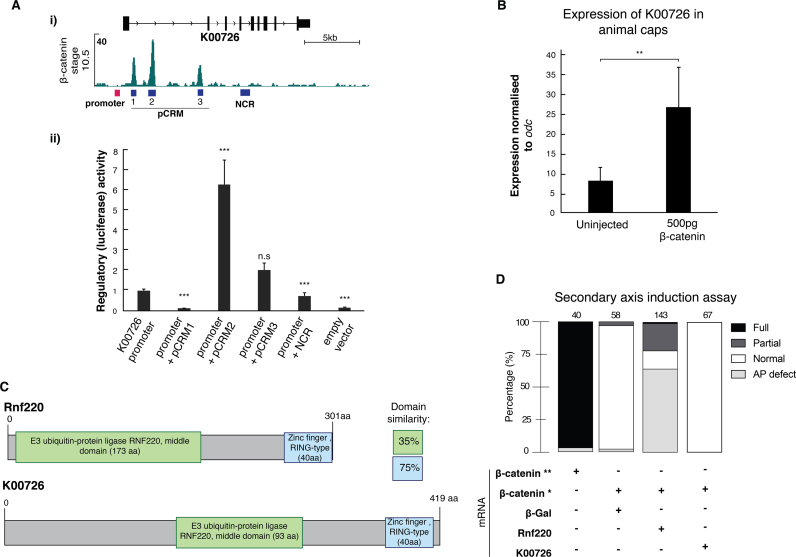
The spatial expression pattern of K00726, and its response to UV and LiCl treatment, suggested that it might be regulated by Wnt signalling. With this in mind, we noted that chromatin profiling of early gastrula embryos (Nakamura et al., 2016) revealed that β-catenin binds three putative cis-regulatory modules within the first intron of K00726 (pCRM1, pCRM2 and pCRM3) (Fig. 7A.i). The transcriptional activities of the three pCRMs, and a negative control region (NCR), were assayed individually at the early gastrula stage in a dual luciferase reporter assay using the endogenous promoter of K00726. The luciferase assay showed that the promoter by itself was capable of increasing reporter expression six-fold compared to a promoterless luciferase vector (Fig. 7A.ii). Addition of pCRM2 or pCRM3 both enhanced promoter activity, with pCRM2, which showed the highest level of β-catenin DNA occupancy, increasing luciferase activity six-fold (Fig. 7A.ii). Interestingly, pCRM1 completely repressed the promoter, while the NCR had no effect. Consistent with these observations overexpression of β-catenin enhanced the transcription of K00726 in animal caps cultured until the start of gastrulation (Fig. 7B).
Fig. 7.
RNF220-like ubiquitin ligaseK00726is a β-catenin-responsive dorsal marker. (A) (i) Genomic snapshot of DNA occupancy of β-catenin (Nakamura et al., 2016) at the K00726 locus in early gastrula embryos. (ii) Luciferase assay for indicated constructs containing K00726 promoter, putative cis-regulatory modules (pCRM) 1–3 or negative control region (NCR). Genomic position of these genomic elements are shown in (i). Reporter activity was normalised to basal activity of the K00726 promoter. Error bars, SEM of biological duplicates. Student's two-tailed t-test: ***, p < 0.01; n.s, not significant. (B) RT-qPCR of K00726 in animal caps derived from embryos injected with and without 500 pg of β-catenin mRNA. Error bars, SEM of biological triplicates. Student's two-tailed t-test: **, p < 0.05. (C) Representation of Rnf220 and K00726 protein including known domains. Comparison of the domain sequences revealed that the E3 ubiquitin ligase and zinc-finger RING-type domain have 35% and 75% conservation, respectively. (D) Morphological outcomes of the secondary axis induction assay: AP defect, smaller or kinked antero-posterior axis; partial, secondary axis formed without cement gland; full, secondary axis formed with cement gland. Amount of injected mRNA: β-catenin**, 250–500 pg; β-catenin*, 5–25 pg (subthreshold); β-Gal, K00726 and Rnf220, 500–1000 pg. At the top of each bar is the total number of analysed embryos.
K00726 protein shared sequence similarity with Rnf220, a protein that may stabilise β-catenin via its C-terminal RING-type domain, in an interaction mediated by the ubiquitin-specific protease 7 (USP7) (Ma et al., 2014). The conservation between Rnf220 and K00726 is relatively high within the zinc finger RING-type domain (75%), but low within the E3 ubiquitin protein ligase domain (35%) (Fig. 7C). With this in mind, we asked whether K00726 could enhance Wnt signalling. We used an assay based on that of Ma et al. (2014), in which subthreshold amounts of β-catenin mRNA, incapable of inducing a secondary axis, were co-injected with mouse Rnf220 or K00726 mRNA into ventral blastomeres of X. laevis. In contrast to Rnf220, K00726 was unable to induce a secondary axis or to cause any other axial defects (Fig. 7D).
3. Discussion
Amphibian embryos provide a powerful model system for the study of early vertebrate development because they are available in large numbers and because they are so easy to manipulate. Here, we use the classic techniques of UV and LiCl treatment to enrich the X. tropicalis embryo for ventral or dorsal cell types, and we then explore the transcriptomes of these embryos in an effort to discover new genes involved in the specification of DV asymmetry.
While both UV- and LiCl-treated embryos display gastrulation defects, only the transcriptome of LiCl-treated embryos deviates substantially from that of unperturbed embryos. This is at least in part because UV-treated embryos lack only the rather small dorsal organiser region, whereas the marginal zone of LiCl-treated embryos consists almost entirely of dorsal organiser cells at the expense of any lateral and ventral cell types. However, the transcriptome-wide comparison of LiCl- and UV-treated embryos also indicates that the treatments significantly affect the binary choice of ectodermal cells towards epidermis or neural tissue. UV-treated embryos fail to produce BMP antagonists in the DMZ and consequently, dorsally expanded BMP signals both ventralise the marginal zone and generate an excess of epidermis at the expense of neural tissue, creating an ‘aneural’ phenotype (Scharf and Gerhart, 1980). For the most part, our results are in line with previous transcriptome-wide studies exploring DV asymmetry by manual dissections or axis perturbation (Blitz et al., 2017, Ding et al., 2017a, Ding et al., 2017b)
Our transcriptomics of DV perturbation has identified low-expression genes whose transcript levels are increased upon LiCl (cpe and K00726) or UV (Xetrov72022004 and c8orf4) treatment. Further experiments suggest that K00726, a Rnf220-like ubiquitin ligase, forms part of a β-catenin-dependent early dorsal gene signature in Xenopus. We have identified two β-catenin+ cis-regulatory modules in the first intron of K00726 that are capable of significantly enhancing transcription.
Overall, our analysis suggests that perturbation-mediated cell type enrichment provides a powerful technique to reveal rare transcripts differentially expressed across specific body axes. We believe the datasets generated in this work will be useful for researchers studying DV axis determination. The differential expression of such genes between LiCl- and UV-treated embryos may predict their spatial expression and their contributions to cell fate decisions.
4. Material and methods
4.1. Embryo culture and manipulation
All animal procedures were performed under licence as required by the Animals (Scientific Procedures) Act 1986 (UK). Standard procedures were used for Xenopus ovulation (Khokha et al., 2002, Sive et al., 2007). X. tropicalis and X. laevis eggs were fertilised in vitro. The embryos were cultured in 0.05x Marc's Modified Ringer's solution (MMR) at 25–28 °C or 0.1x Normal Amphibian Medium (NAM) at 14–20 °C, respectively. Approximately 10 min after fertilisation, embryos were dejellied with 2.2% [w/v] L-cysteine diluted in the appropriate culture medium (pH 8.0). Injections were carried out with X. laevis embryos equilibrated in 4% Ficoll PM-400 (Sigma)/0.1x NAM. Injected embryos were transferred to 0.1x NAM without Ficoll once they reached about stage 8. Embryos were staged according to Nieuwkoop and Faber (1994).
4.1.1. UV irradiation
Immediately after removing their jelly coats, 50–100×X. tropicalis fertilised eggs were transferred into a 50-ml conical centrifuge tube filled to the top with 0.05x MMR. The tube was closed by wrapping Saran cling film around the top using rubber bands and turned upside down allowing the embryos to settle on the film. The tube was swirled gently to position the zygotes to the centre of the film wrapping. The tube was placed onto a styrofoam hole whose depth of 2 cm kept vegetal poles at the appropriate distance from a compact UV lamp (UVP, UVGL-25). The embryos were irradiated with 254 nm shortwave UV for 2 min. After that, the tube was kept upside down and transferred to the incubator without disturbing the embryos until the completion of the first cell division. The embryos were then transferred to an agarose-coated petri dish containing 0.05x MMR and cultured to the developmental stage of interest.
4.1.2. LiCl treatment
At the 32-cell stage, dejellied X. tropicalis embryos were transferred to an agarose-coated petri dish containing 0.3 M LiCl/0.05x MMR. They were gently swirled in the dish and incubated for 5 min. LiCl treatment was terminated by rinsing the embryos in 0.05x MMR five times. The embryos were then cultured in 0.05x MMR to the desired developmental stage.
4.1.3. Microinjection
A total volume of 10 nl of mRNA and/or plasmid DNA was injected into X. laevis embryos at the 1-cell, 2-cell or 4-cell stage using a microinjector (Narishige IM-300). Microinjection needles were produced from borosilicate glass capillaries (Harvard Apparatus, GC120-15) using a micropipette puller (Sutter p97).
4.1.4. Animal caps
Approximately 500 pg of β-catenin mRNA was injected into the animal side of each blastomere of X. laevis 2-cell embryos. At stage 8, embryos were transferred into 0.7x MMR and animal caps were cut using 13 µm wire tip electrodes (Protech International, 13-Y1) at medium voltage (setting 2 on the MC-2010 micro cautery instrument, Protech International). After dissection, caps were transferred to a fresh agarose-coated plate containing 0.7x MMR and incubated at 18 °C alongside whole sibling embryos until stage 15. Animal caps were processed for total RNA extraction.
4.2. Cloning
Forward and reverse primers were designed based on the genomic sequence of X. tropicalis (assembly v7.1) to clone cpe, Xetrov72022004, K00726 and c8orf4 from gastrula-staged cDNA (see Table S5). GoTaq-amplified gene fragments were cloned into pGEM-T Easy vector (Promega).
For luciferase assays, the K00726 promoter, three putative cis-regulatory modules (pCRM1–3), and a negative control region (NCR) were amplified from X. tropicalis genomic DNA using KAPA HiFi HotStart ReadyMix (KAPA Biosystems) and reverse and forward primers containing 5′ HindIII (promoter) or BamHI (enhancer) restriction sites (Table S5). The K00726 promoter was inserted into the HindIII site of promoterless pGL3-Basic vector. Single pCRMs were inserted into the BamHI site of the pGL3 vector equipped with the endogenous promoter.
4.3. Whole mount in situ hybridisation
Whole mount in situ hybridisation (WMISH) was carried out as described (Monsoro-Burq, 2007) with digoxigenin-labelled RNA probes. Antisense (AS) probes were generated from linearised cDNA clones in vitro with the appropriate RNA polymerase (Roche) and digoxigenin-11-UTP. The following AS probes were transcribed with T7 RNA polymerase from plasmids linearised with EcoRI unless otherwise specified: pGEM-5Zf(-)[noggin] (W. C. Smith and Harland, 1992); pBS-SK(-)[goosecoid] (Cho et al., 1991); pBS-SK(-)[chordin] (Sasai et al., 1994); pGEM1[wnt8a] linearised with BamHI (Christian et al., 1991); pBS-SK(+)[bmp4]; pCS2(+)[not1] linearised with XhoI (Dassow et al., 1993); pCMV-Sport6.ccdB[msgn1] (Gentsch et al., 2013); pGEM-T[c8orf4] linearised with NdeI; pGEM-T[Xetrov72022004] linearised with SacI; pGEM-T[K00726] linearised with SacI; and pGEM-T[cpe] linearised with ApaI and transcribed with SP6 RNA polymerase. After DNase treatment, probes were precipitated with LiCl overnight at − 20 °C and rinsed with 80% EtOH. The precipitate was dissolved with RNase-free water. The WMISH probes were diluted with hybridisation buffer (50% [v/v] formamide, 5x SSC, 1x Denhardt's, 10 mM EDTA, 1 mg/ml torula RNA, 100 μg/ml heparin, 0.1% [v/v] Tween-20% and 0.1% [w/v] CHAPS) to a final concentration of 10 ng/µl (10x stock).
4.4. Synthesis of capped mRNA for microinjection
Approximately 1 μg of linearised (with NotI unless otherwise specified) expression vector was transcribed using the mMESSAGE mMACHINE SP6 Transcription Kit (Thermo Fisher Scientific): pCS2+[3xHA-K00726] linearised with ApaI; pCS2+[β-catenin-GFP] (Addgene, #16839); pSP64T[nlacZ] (nuclear β-galactosidase); pCS2+[rluc] (Renilla luciferase) (Demagny et al., 2014); and pCS2+[mRNF220-FLAG].
4.5. Total RNA extraction and cDNA synthesis
10–20 embryos or 15–20 animal caps were homogenised in 200 µl of TRIzol (Thermo Fisher Scientific) by vortexing for 10 min at room temperature. The homogenate was mixed with 40 µl of chloroform and centrifuged for 5 min at 4 °C at full speed. The upper phase was transferred to a clean 1.5-ml Eppendorf tube. RNA was precipitated using the RNA Zymo Clean and Concentrator-5 or -25 kit (Zymo Research). Approximately 1 μg of total RNA was converted into first strand cDNA using the SuperScript III reverse transcriptase (Thermo Fisher Scientific).
4.6. Quantification of transcript levels
Gene expression levels were determined by absolute quantification using SYBR Green I reagents (Roche) and the LightCycler LC480 II (Roche). Each cDNA sample was run in technical triplicates. The standard curve for each primer pair was calculated from eight crossing points (Ct-values) reflecting amplifications from a 1:3 serial dilution of wild-type cDNA. The primer sequences are listed in Table S5. Microsoft Excel was used to normalise quantifications to the housekeeping gene odc1, design plots and perform paired Student's two-tailed t-tests.
4.7. Library preparation, sequencing and biocomputational analysis
Total RNA representing 10 X. tropicalis embryos (15–20 μg) was processed according to the low sample protocol of the TruSeq RNA sample preparation guide version 2 (Illumina). The concentration of the library was determined by fluorometry using Qubit dsDNA HS reagents (Thermo Fisher Scientific). The quality of the library was verified by micro-capillary electrophoresis using the Agilent 2100 Bioanalyzer. Libraries were read paired-end along 50 bases on the HiSeq. 2500 machine (Illumina). The reads were mapped to the X. tropicalis gene models v7.2 using the splice-aware aligner Tophat2 (D. Kim et al., 2013) disabling any search for novel splice junctions. Reads were counted per gene using the python script HTSeq-count in ‘intersection-nonempty’ mode (Anders et al., 2015). The raw read counts from each experiment were merged before running the R package DESeq. 2 (Love et al., 2014) to determine differential gene expression between selected conditions (Table S6). The gene set enrichment analysis was carried out using previously assembled GO annotations (Gentsch et al., 2015) and the R package GOstats (Falcon and Gentleman, 2007). All sequencing reads and raw read counts are available at the Gene Expression Omnibus (GEO) database under the accession number GSE107424.
4.8. Dual luciferase reporter assay
Embryos were injected with 50 pg of various firefly luciferase reporter constructs (pGL3-Basic, pGL3[K00726 promoter], pGL3[K00726 promoter + pCRM1], pGL3[K00726 promoter + pCRM2], pGL3[K00726 promoter + pCRM3], pGL3[K00726 promoter + NCR]) and 5 pg of Renilla luciferase mRNA. Embryos were harvested at early gastrula stage 10. We used the reagents of the Dual-Luciferase Reporter Assay System kit (Promega) to perform the luciferase assay. Embryos were homogenised in 100 µl of 1xPassive Lysis Buffer. The homogenate was spun for 1 min at 16,000 g and 4 °C. 20 µl of the supernatant was transferred to 96-well plates (solid bottom, white). 50 µl of LARII was added to the supernatant. Firefly luminescence was measured twice for each sample with a Perkin Elmer ENVision 2102 Multilabel plate reader. After these readings, 50 µl of 1x Stop&Glo reagent was added and Renilla luminescence was recorded twice for each sample. The Renilla signal was used to normalise firefly measurements.
Acknowledgements
We thank the Francis Crick Aquatics Facility for maintenance of frogs and Abdul Sesay and Leena Bhaw from the Advanced Sequencing Facility for helping with sequencing the poly(A) RNA libraries. We also thank Pengcheng Ma for the Rnf220 clone, Edward de Robertis for the pCS2+[rluc] plasmid, and Praneet Chaturvedi for the read count matrix of GEO dataset GSE93195. We are grateful for advice from James Briscoe, Kathy Niakan and members of the Smith lab. This research was supported by DevCom, a European Initial Training Network for Developmental and Computational Biology (Project Reference 607142). G.E.G. and J.C.S. are supported by the Francis Crick Institute (Cancer Research UK FC001157, the UK Medical Research Council FC001157, Wellcome Trust FC001157).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2018.04.022.
Contributor Information
Rita S. Monteiro, Email: rita.monteiro@crick.ac.uk.
James C. Smith, Email: jim.smith@crick.ac.uk.
Appendix A. Supplementary material
Fig. S1.
Verification of UV- and LiCl-induced DV axis perturbation. (A,B) WMISH of dorsal (chordin and goosecoid), ventral (wnt8a) and mesoderm markers (not and mesogenin1 [msgn1]) under indicated conditions at mid-gastrula stage. (C) RT-qPCR of several mesodermal markers under indicated conditions in mid-gastrula embryos. Error bars, SEM of biological triplicates. Student's two-tailed t-test: *, p < 0.1; **, p < 0.05; and ***, p < 0.01.
Fig. S2.
Linking LiCl/UV-induced transcriptional changes to antero-posterior development. (A) Heatmap of transcriptional fold changes between LiCl- and UV-treated embryos of genes associated with anterior development (brain) and posterior (somitogenesis) development. (B) Stacked bar graph showing the number of genes that are upregulated in either LiCl- or UV-treated embryos and the GO term associated with the gene. Fisher’s exact test (p = 0.007) suggests that genes upregulated in LiCl- or UV-treated embryos influence anterior and posterior development, respectively.
Fig. S3.
Gene set enrichment analysis of differentially expressed genes between LiCl- and UV-treated embryos not captured by the comparative transcriptomics of DMZ and VMZ. (A-C) Analysis of genes differentially expressed between LiCl- and UV-treated embryos, but not captured by the transcriptional comparison of DMZ and VMZ. Heatmap (A) and regression plot (B) of indicated transcriptional comparisons. (B) Pearson coefficient, ρ = 0.34. Student’s two-tailed t-test, p = 1.4e-35. (C) Plots show the number of upregulated genes and associated hypergeometric p-values of enrichment for specific biological processes (BPs) of the Gene Ontology (GO) project. Selected BPs refer to early developmental processes (i), signalling (ii), and differentiation of germ layers and their derivatives (iii).
Fig. S4.
Comparative transcriptomics of DV axis determination. (A) Pairwise Pearson correlations of indicated transcriptional comparisons of the DV core genes. This includes work from Blitz et al. (2017) and Ding et al., 2017a, Ding et al., 2017b. Conditions were clustered according to Euclidean distances. (B) Venn diagram comparing genes identified as differentially expressed between indicated conditions (transcriptional fold change ≥ 2 with FDR ≤ 1%). LiCl/untreated (stage 11.5), LiCl/UV (stage 11.5), LiCl/untreated (stage 10.5; Ding et al., 2017) and LiCl/βcatMO (stage 10.5; Ding et al., 2017).
Supplementary materialTable S1. Summary of RNA-Seq read alignments. Paired-end reads were aligned to the X. tropicalis gene models v7.2.
Supplementary materialTable S2. Upregulated genes in LiCl-treated embryos at gastrula stage. The list includes genes with transcriptional fold changes ≥ 2 (FDR ≤ 1%).
Supplementary materialTable S3. Upregulated genes in UV-treated embryos at gastrula stage. The list includes genes with transcriptional fold changes ≥ 2 (FDR ≤ 1%).
Supplementary materialTable S4. Shortlist of mostly uncharacterised genes upregulated upon UV or LiCl treatment. The list includes expression data from the X. tropicalis gene expression database (http://genomics.crick.ac.uk/apps/profiles/) (Collart et al., 2014, Owens et al., 2016) and further details from Xenbase (http://www.xenbase.org) (Karpinka et al., 2015).
Supplementary materialTable S5. List of primer sequences for cloning and RT-qPCR.
Supplementary materialTable S6. Unfiltered output of differential gene expression analysis between LiCl- and UV-treated embryos using DESeq2.
References
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz I.L., Paraiso K.D., Patrushev I., Chiu W.T.Y., Cho K.W.Y., Gilchrist M.J. A catalog of Xenopus tropicalis transcription factors and their regional expression in the early gastrula stage embryo. Dev. Biol. 2017;426:409–417. doi: 10.1016/j.ydbio.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.W., Cho K.W., Blumberg B., Blumberg B., Steinbeisser H., Steinbeisser H., de Robertis E.M., De Robertis E.M. Molecular nature of Spemann's organizer: the role of the Xenopus homeobox gene goosecoid. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian J.L., McMahon J.A., McMahon A.P., Moon R.T. Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development. 1991;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- Collart C., Owens N.D.L., Bhaw-Rosun L., Cooper B., De Domenico E., Patrushev I., Sesay A.K., Smith J.N., Smith J.C., Gilchrist M.J. High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development. 2014;141:1927–1939. doi: 10.1242/dev.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J., Smith J.C. The midblastula cell cycle transition and the character of mesoderm in u.v.-induced nonaxial Xenopus development. Development. 1987;99:197–210. doi: 10.1242/dev.99.2.197. [DOI] [PubMed] [Google Scholar]
- Cooke J., Symes K., Smith E.J. Potentiation by the lithium ion of morphogenetic responses to a Xenopus inducing factor. Development. 1989;105:549–558. doi: 10.1242/dev.105.3.549. [DOI] [PubMed] [Google Scholar]
- von Dassow G., Schmidt J.E., Kimelman D. Induction of the Xenopus organizer: expression and regulation of Xnot, a novel FGF and activin-regulated homeo box gene. Genes Dev. 1993;7:355–366. doi: 10.1101/gad.7.3.355. [DOI] [PubMed] [Google Scholar]
- De Domenico E., Owens N.D.L., Grant I.M., Gomes-Faria R., Gilchrist M.J. Molecular asymmetry in the 8-cell stage Xenopus tropicalis embryo described by single blastomere transcript sequencing. Dev. Biol. 2015;408:252–268. doi: 10.1016/j.ydbio.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demagny H., Araki T., De Robertis E.M. The tumor suppressor Smad4/DPC4 is regulated by phosphorylations that integrate FGF, Wnt, and TGF-β signaling. CellReports. 2014;9:688–700. doi: 10.1016/j.celrep.2014.09.020. [DOI] [PubMed] [Google Scholar]
- Ding Y., Colozza G., Zhang K., Moriyama Y., Ploper D., Sosa E.A., Benitez M.D.J., De Robertis E.M. Genome-wide analysis of dorsal and ventral transcriptomes of the Xenopus laevis gastrula. Dev. Biol. 2017;426:176–187. doi: 10.1016/j.ydbio.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Ploper D., Sosa E.A., Colozza G., Moriyama Y., Benitez M.D.J., Zhang K., Merkurjev D., De Robertis E.M. Spemann organizer transcriptome induction by early beta-catenin, Wnt, Nodal, and Siamois signals in Xenopus laevis. Proc. Natl. Acad. Sci. USA. 2017;114:E3081–E3090. doi: 10.1073/pnas.1700766114. 〈 https://doi.org/10.1073/pnas.1700766114〉. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinson R.P. Site of sperm entry and a cortical contraction associated with egg activation in the frog Rana pipiens. Dev. Biol. 1975;44:257–268. doi: 10.1016/0012-1606(75)90281-x. [DOI] [PubMed] [Google Scholar]
- Elinson R.P., Manes M.E. Morphology of the site of sperm entry on the frog egg. Dev. Biol. 1978;63:67–75. doi: 10.1016/0012-1606(78)90114-8. [DOI] [PubMed] [Google Scholar]
- Falcon S., Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- Gentsch G.E., Owens N.D.L., Martin S.R., Piccinelli P., Faial T., Trotter M.W.B., Gilchrist M.J., Smith J.C. In vivo T-box transcription factor profiling reveals joint regulation of embryonic neuromesodermal bipotency. Cell Rep. 2013;4:1185–1196. doi: 10.1016/j.celrep.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch G.E., Patrushev I., Smith J.C. Genome-wide snapshot of chromatin regulators and states in Xenopus embryos by ChIP-Seq. J. Vis. Exp. 2015;96 doi: 10.3791/52535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T.S. The mode of action of lithium salts in amphibian development. J. Exp. Zool. Part A: Ecol. Genet. Physiol. 1942;89:1–35. [Google Scholar]
- Hellsten U., Harland R.M., Gilchrist M.J., Hendrix D., Jurka J., Kapitonov V., Ovcharenko I., Putnam N.H., Shu S., Taher L., Blitz I.L., Blumberg B., Dichmann D.S., Dubchak I., Amaya E., Detter J.C., Fletcher R., Gerhard D.S., Goodstein D., Graves T., Grigoriev I.V., Grimwood J., Kawashima T., Lindquist E., Lucas S.M., Mead P.E., Mitros T., Ogino H., Ohta Y., Poliakov A.V., Pollet N., Robert J., Salamov A., Sater A.K., Schmutz J., Terry A., Vize P.D., Warren W.C., Wells D., Wills A., Wilson R.K., Zimmerman L.B., Zorn A.M., Grainger R., Grammer T., Khokha M.K., Richardson P.M., Rokhsar D.S. The genome of the Western clawed frog Xenopus tropicalis. Science. 2010;328:633–636. doi: 10.1126/science.1183670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood N.D., Pluck A., Gurdon J.B. Xenopus Myf-5 marks early muscle cells and can activate muscle genes ectopically in early embryos. Development. 1991;111:551–560. doi: 10.1242/dev.111.2.551. [DOI] [PubMed] [Google Scholar]
- Itoh K., Jacob J., Sokol S.Y. A role for Xenopus Frizzled 8 in dorsal development. Mech. Dev. 1998;74:145–157. doi: 10.1016/s0925-4773(98)00076-8. [DOI] [PubMed] [Google Scholar]
- Kao K.R., Elinson R.P. The entire mesodermal mantle behaves as Spemann's organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev. Biol. 1988;127:64–77. doi: 10.1016/0012-1606(88)90189-3. [DOI] [PubMed] [Google Scholar]
- Kao K.R., Masui Y., Elinson R.P. Lithium-induced respecification of pattern in Xenopus laevis embryos. Nature. 1986;322:371–373. doi: 10.1038/322371a0. [DOI] [PubMed] [Google Scholar]
- Karpinka J.B., Fortriede J.D., Burns K.A., James-Zorn C., Ponferrada V.G., Lee J., Karimi K., Zorn A.M., Vize P.D. Xenbase, the Xenopus model organism database; new virtualized system, data types and genomes. Nucleic Acids Res. 2015;43:D756–D763. doi: 10.1093/nar/gku956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha M.K., Chung C., Bustamante E.L., Gaw L.W.K., Trott K.A., Yeh J., Lim N., Lin J.C.Y., Taverner N., Amaya E., Papalopulu N., Smith J.C., Zorn A.M., Harland R.M., Grammer T.C. Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes inthe presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Yamamoto A., Bouwmeester T., Agius E., Robertis E.M. The role of paraxial protocadherin in selective adhesion and cell movements of the mesoderm during Xenopus gastrulation. Development. 1998;125:4681–4690. doi: 10.1242/dev.125.23.4681. [DOI] [PubMed] [Google Scholar]
- Klein P.S., Melton D.A. A molecular mechanism for the effect of lithium on development. Proc. Natl. Acad. Sci. USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane M.C., Sheets M.D. Designation of the anterior/posterior axis in pregastrula Xenopus laevis. Dev. Biol. 2000;225:37–58. doi: 10.1006/dbio.2000.9803. [DOI] [PubMed] [Google Scholar]
- Larabell C.A., Torres M., Rowning B.A., Yost C., Miller J.R., Wu M., Kimelman D., Moon R.T. Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. J. Cell Biol. 1997;136:1123–1136. doi: 10.1083/jcb.136.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent M.N., Blitz I.L., Hashimoto C., Rothbacher U., Cho K.W. The Xenopus homeobox gene twin mediates Wnt induction of goosecoid in establishment of Spemann's organizer. Development. 1997;124:4905–4916. doi: 10.1242/dev.124.23.4905. [DOI] [PubMed] [Google Scholar]
- Lemaire P., Garrett N., Gurdon J.B. Expression cloning of Siamois, a xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell. 1995;81:85–94. doi: 10.1016/0092-8674(95)90373-9. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma P., Yang X., Kong Q., Li C., Yang S., Li Y., Mao B. The ubiquitin ligase RNF220 enhances canonical Wnt signaling through USP7-mediated deubiquitination of β-catenin. Mol. Cell. Biol. 2014;34:4355–4366. doi: 10.1128/MCB.00731-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manes M.E., Elinson R.P. Ultraviolet light inhibits grey crescent formation on the frog egg. Wilehm. Roux. Arch. Dev. Biol. 1980;189:73–76. doi: 10.1007/BF00848569. [DOI] [PubMed] [Google Scholar]
- Martinez Arias A., Steventon B. On the nature and function of organizers. Development. 2018;145 doi: 10.1242/dev.159525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.R., Rowning B.A., Larabell C.A., Yang-Snyder J.A., Bates R.L., Moon R.T. Establishment of the dorsal-ventral axis in Xenopus Embryos coincides with the dorsal enrichment of dishevelled that is dependent on cortical rotation. J. Cell Biol. 1999;146:427–438. doi: 10.1083/jcb.146.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsoro-Burq A.H. A rapid protocol for whole-mount in situ hybridization on Xenopus embryos. Cold Spring Harb. Protoc. 2007;2007 (pdb.prot4809) [Google Scholar]
- Nakamura Y., de Paiva Alves E., Veenstra G.J.C., Hoppler S. Tissue- and stage-specific Wnt target gene expression is controlled subsequent to β-catenin recruitment to cis-regulatory modules. Development. 2016;143:1914–1925. doi: 10.1242/dev.131664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop P.D., Faber J. Garland Publishing; New York: 1994. Normal table of Xenopus laevis. [Google Scholar]
- Owens N.D.L., Blitz I.L., Lane M.A., Patrushev I., Overton J.D., Gilchrist M.J., Cho K.W.Y., Khokha M.K. Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome kinetics in development. Cell Rep. 2016;14:632–647. doi: 10.1016/j.celrep.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteels J. Recherches sur l′action du LiCl sur les oeufs des Amphibiens. Arch. Biol. 1945 [Google Scholar]
- Piccolo S., Agius E., Leyns L., Bhattacharyya S., Grunz H., Bouwmeester T., de Robertis E.M. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov I.K., Kwon T., Crossman D.K., Crowley M.R., Wallingford J.B., Chang C. Identification of new regulators of embryonic patterning and morphogenesis in Xenopus gastrulae by RNA sequencing. Dev. Biol. 2017;426:429–441. doi: 10.1016/j.ydbio.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y., Lu B., Steinbeisser H., Geissert D., Gont L.K., de Robertis E.M. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf S.R., Gerhart J.C. Determination of the dorsal-ventral axis in eggs of Xenopus laevis: complete rescue of uv-impaired eggs by oblique orientation before first cleavage. Dev. Biol. 1980;79:181–198. doi: 10.1016/0012-1606(80)90082-2. [DOI] [PubMed] [Google Scholar]
- Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., van Heeringen S.J., Quigley I., Heinz S., Ogino H., Ochi H., Hellsten U., Lyons J.B., Simakov O., Putnam N., Stites J., Kuroki Y., Tanaka T., Michiue T., Watanabe M., Bogdanović O., Lister R., Georgiou G., Paranjpe S.S., van Kruijsbergen I., Shu S., Carlson J., Kinoshita T., Ohta Y., Mawaribuchi S., Jenkins J., Grimwood J., Schmutz J., Mitros T., Mozaffari S.V., Suzuki Y., Haramoto Y., Yamamoto T.S., Takagi C., Heald R., Miller K., Haudenschild C., Kitzman J., Nakayama T., Izutsu Y., Robert J., Fortriede J., Burns K., Lotay V., Karimi K., Yasuoka Y., Dichmann D.S., Flajnik M.F., Houston D.W., Shendure J., DuPasquier L., Vize P.D., Zorn A.M., Ito M., Marcotte E.M., Wallingford J.B., Ito Y., Asashima M., Ueno N., Matsuda Y., Veenstra G.J.C., Fujiyama A., Harland R.M., Taira M., Rokhsar D.S. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H., Grainger R.M., Harland R.M. Early Development of Xenopus laevis: A Laboratorial Manual. 2nd ed. Cold Spring Harbour Press; 1999. [Google Scholar]
- Sive H.L., Grainger R.M., Harland R.M. Dejellying Xenopus laevis Embryos. CSH Protoc. 2007;2007 doi: 10.1101/pdb.prot4731. (pdb.prot4731) [DOI] [PubMed] [Google Scholar]
- Smith W.C., Harland R.M. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- de Souza F.S., Gawantka V., Gómez A.P., Delius H., Ang S.L., Niehrs C. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann's organizer. EMBO J. 1999;18:6062–6072. doi: 10.1093/emboj/18.21.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambolic V., Ruel L., Woodgett J.R. Lithium inhibits glycogen synthase kinase-3 activity and mimics wingless signalling in intact cells. Curr. Biol. 1996;6:1664–1668. doi: 10.1016/s0960-9822(02)70790-2. [DOI] [PubMed] [Google Scholar]
- Tao Q., Yokota C., Puck H., Kofron M., Birsoy B., Yan D., Asashima M., Wylie C.C., Lin X., Heasman J. Maternal wnt11 activates the canonical wnt signaling pathway required for axis formation in Xenopus embryos. Cell. 2005;120:857–871. doi: 10.1016/j.cell.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Vincent J.P., Oster G.F., Gerhart J.C. Kinematics of gray crescent formation in Xenopus eggs: the displacement of subcortical cytoplasm relative to the egg surface. Dev. Biol. 1986;113:484–500. doi: 10.1016/0012-1606(86)90184-3. [DOI] [PubMed] [Google Scholar]
- Vitorino M., Silva A.C., Inácio J.M., Ramalho J.S., Gur M., Fainsod A., Steinbeisser H., Belo J.A. Xenopus Pkdcc1 and Pkdcc2 are two new tyrosine kinases involved in the regulation of JNK dependent Wnt/PCP signaling pathway. PLoS One. 2015;10:e0135504. doi: 10.1371/journal.pone.0135504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C., Farr G.H., Pan W., Rowning B.A., Wang J., Mao J., Wu D., Li L., Larabell C.A., Kimelman D. GBP binds kinesin light chain and translocates during cortical rotation in Xenopus eggs. Development. 2003;130:5425–5436. doi: 10.1242/dev.00737. [DOI] [PubMed] [Google Scholar]
- Wilm T.P., Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393–404. doi: 10.1242/dev.01572. [DOI] [PubMed] [Google Scholar]
- Zhang X., Abreu J.G., Yokota C., MacDonald B.T., Singh S., Coburn K.L.A., Cheong S.-M., Zhang M.M., Ye Q.-Z., Hang H.C., Steen H., He X. Tiki1 is required for head formation via Wnt cleavage-oxidation and inactivation. Cell. 2012;149:1565–1577. doi: 10.1016/j.cell.2012.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materialTable S1. Summary of RNA-Seq read alignments. Paired-end reads were aligned to the X. tropicalis gene models v7.2.
Supplementary materialTable S2. Upregulated genes in LiCl-treated embryos at gastrula stage. The list includes genes with transcriptional fold changes ≥ 2 (FDR ≤ 1%).
Supplementary materialTable S3. Upregulated genes in UV-treated embryos at gastrula stage. The list includes genes with transcriptional fold changes ≥ 2 (FDR ≤ 1%).
Supplementary materialTable S4. Shortlist of mostly uncharacterised genes upregulated upon UV or LiCl treatment. The list includes expression data from the X. tropicalis gene expression database (http://genomics.crick.ac.uk/apps/profiles/) (Collart et al., 2014, Owens et al., 2016) and further details from Xenbase (http://www.xenbase.org) (Karpinka et al., 2015).
Supplementary materialTable S5. List of primer sequences for cloning and RT-qPCR.
Supplementary materialTable S6. Unfiltered output of differential gene expression analysis between LiCl- and UV-treated embryos using DESeq2.