Abstract
Bromodomain and extraterminal (BET) proteins recruit key components of basic transcriptional machinery to promote gene expression. Aberrant expression and mutations in BET genes have been identified in many malignancies. Small molecule inhibitors of BET proteins such as JQ1 have shown efficacy in preclinical cancer models, including affecting growth of hepatocellular carcinoma. BET proteins also regulate cell proliferation in nontumor settings. We recently showed that BET proteins regulate cholangiocyte-driven liver regeneration. Here, we studied the role of BET proteins in hepatocyte-driven liver regeneration in partial hepatectomy (PHx) and acetaminophen-induced liver injury models in mice and zebrafish. JQ1 was injected 2 or 16 hours after PHx in mice to determine effect on hepatic injury, regeneration, and signaling. Mice treated with JQ1 after PHx displayed increased liver injury and a near-complete inhibition of hepatocyte proliferation. Levels of Ccnd1 mRNA and Cyclin D1 protein were reduced in animals injected with JQ1 16 hours after PHx and were even further reduced in animals injected with JQ1 2 hours after PHx. JQ1-treated zebrafish larvae after acetaminophen-induced injury also displayed notably impaired hepatocyte proliferation. In both models, Wnt signaling was prominently suppressed by JQ1. Our results show that BET proteins regulate hepatocyte proliferation–driven liver regeneration, and Wnt signaling is particularly sensitive to BET protein inhibition.
The liver is a highly regenerative organ that is able to recover from repeated bouts of injury through proliferation of the main epithelial cell type of the liver, the hepatocytes.1 Alternatively, in conditions of extreme liver injury when proliferation of hepatocytes is impaired, biliary epithelial cells (BECs) can give rise to liver progenitor cells that can subsequently differentiate into hepatocytes to restore lost cell mass.2, 3 Despite this innate capacity for regeneration, chronic liver disease and cirrhosis is the 12th leading cause of death in the United States.4 Patients with chronic liver disease and cirrhosis are at great risk of developing hepatocellular carcinoma (HCC), the fifth most common cancer in men worldwide and the second leading cause of cancer-related death.5 The prognosis for HCC is exceedingly poor, with an overall 5-year survival rate of 5.1% in the United States, because there are currently no curative therapies for advanced HCC.6 Clearly, there is a great need to develop new therapies for HCC and underlying chronic liver disease. Because failure of endogenous liver regeneration is thought to drive progression in chronic liver disease,7, 8 understanding the mechanisms of liver regeneration may lead to the development of new therapies.
Recently, a class of drugs has been developed which inhibits bromodomain and extraterminal (BET) proteins and shows promise in the treatment of cancer and inflammation.9 The BET protein family consists of Brd2, Brd3, Brd4, and Brdt, which share conserved bromodomains, chromatin interaction modules that bind to acetylated lysine residues on histone tails.10 Brd4 in particular has been identified as a component of Mediator, a multiprotein complex that promotes transcription by interacting with transcription factors to recruit and activate RNA polymerase II.9, 11 Moreover, Brd4 has been found to occupy the promoters or enhancers of virtually all actively transcribed genes in a variety of cells types.12, 13, 14 The recently developed drug JQ1 displays potent and specific inhibition of Brd4 through competitively binding its bromodomains.15 Interestingly, JQ1 has shown potent anticancer effects by selectively inhibiting oncogenic gene expression in various cancer cell lines and murine models.12, 16, 17 The dosage of JQ1 required to inhibit tumors is well tolerated in mice, despite widespread expression of Brd4 in mouse tissues.12, 15, 16
In addition, recent reports have implicated a role of Brd4 in liver pathologic processes. Brd4 was found to be overexpressed in HCC cell lines compared with normal liver, and knockdown of Brd4 expression impaired proliferation, migration, and invasion of HCC cells.18, 19, 20 Furthermore, treatment with JQ1 reduced cell growth in both HCC cell lines and a xenograft tumor model.20 JQ1 treatment was also shown to be protective in a mouse model of carbon tetrachloride–induced liver fibrosis through abrogation of cytokine-induced activation of hepatic stellate cells.21 These reports would suggest Brd4 inhibitors as a viable treatment option for certain liver diseases. However, Brd4 has also been implicated in the promotion of liver regeneration. JQ1 inhibits BEC-driven liver regeneration in both zebrafish and mice.22 However, the role of Brd4 in the more common hepatocyte-driven liver regeneration has not been assessed. Here, we determine the effect of JQ1 treatment in two independent models of hepatocyte-driven liver regeneration: partial hepatectomy (PHx) in mice and acetaminophen-overdose in zebrafish. We show that JQ1 is a potent inhibitor of hepatocyte proliferation after liver injury. Therefore, the potential use of JQ1 in treatment of liver disease must be carefully considered to avoid negatively affecting normal liver regeneration.
Materials and Methods
Animals and Surgery
All animal experiments and procedures were performed under the NIH's Guide for the Care and Use of Laboratory Animals23 and after approval by the Institutional Animal Use and Care Committee at the University of Pittsburgh, School of Medicine. Male C57BL6/J mice (The Jackson Laboratory, Bar Harbor, ME) underwent PHx as previously described.24 Either 2 or 16 hours after surgery animals were injected intraperitoneally with 50 mg/kg JQ1 (ApexBio, Taiwan; A1910). A stock solution of 50 mg/mL JQ1 was prepared in dimethyl sulfoxide (DMSO) and subsequently diluted to a working concentration of 5 mg/mL in a solution of 10% hydroxypropyl β-cyclodextrin in sterile water (vehicle solution). As a control, mice were injected with the vehicle solution alone at the indicated time point after surgery. Animals were also injected with either JQ1 or vehicle solution after sham surgeries (when surgery was performed but no liver was removed). At the time of harvest, either 40 to 45 hours or 72 hours after surgery, animals were anesthetized with isoflurane, and blood was drawn from the inferior vena cava. Subsequently, anesthetized mice were euthanized by cervical dislocation. Livers were washed in phosphate buffered saline (PBS), flash-frozen in liquid nitrogen, and stored at −80°C until subsequent protein and RNA analyses, or alternatively were drop-fixed in 10% buffered formalin for 48 hours before paraffin embedding. Serum aspartate aminotransferase (AST) levels were determined by automated methods at the University of Pittsburgh Medical Center clinical chemistry laboratory.
Immunohistochemistry
Tissue samples embedded in paraffin were cut into 4-μm sections. Tissue sections were deparaffinized in xylene and hydrated through changes of 100% ethanol, followed by 95% ethanol, washed in distilled water, and finally washed in PBS. Antigen retrieval was performed by microwaving slides in pH 6 sodium citrate buffer (Cyclin D1) or zinc sulfate buffer [proliferating cell nuclear antigen (PCNA)]. After cooling, endogenous peroxidase activity was blocked through incubation in 3% H2O2 in distilled water for 10 minutes. Slides were then washed in PBS and blocked with Super Block (ScyTek Laboratories, Logan, UT; AAA500) for 10 minutes (Cyclin D1) or 20 minutes (PCNA). Primary antibodies were diluted in 1% bovine serum albumin (Fisher BioReagents, Pittsburgh, PA; BP1605100) with 0.1% Tween 20 (Fisher BioReagents; BP337-500) in PBS as follows: Cyclin D1 (Abcam, Cambridge, UK; ab134175; dilution 1:200) and PCNA (Santa Cruz Biotechnology, Dallas, TX; sc-56; dilution 1:5000) and incubated on the slides for 1 hour at room temperature. Sections were washed three times in PBS, and the correct biotinylated anti-rabbit (Vector Laboratories, Burlingame, CA; BA-1000; dilution 1:500) or biotinylated anti-mouse (Millipore, Burlington, MA; AP181B; dilution 1:700) secondary antibodies were incubated on the tissue sections for 30 minutes. After washing with PBS three times, slides were sensitized with the Vectastain ABC kit (Vector Laboratories; PK-6101) for 30 minutes. Sections were washed three times with PBS, and color was developed with the DAB (diaminobenzidine) Peroxidase Substrate Kit (Vector Laboratories; SK-4100), followed by quenching in distilled water for 5 minutes. Sections were counterstained in hematoxylin (Thermo Scientific, Pittsburgh, PA; 7211) for approximately 8 to 20 seconds and washed under running tap water for 5 minutes. Slides were then dehydrated in changes of alcohol (70%, 95%, 100%), followed by xylene, and coverslips were applied with Cytoseal XYL (Thermo Scientific; 8312-4). Images were taken on Axioskop 40 (Zeiss, Oberkochen, Germany) inverted brightfield microscope.
Protein Extraction and Western Blot Analysis
Whole-cell lysates of liver tissue were prepared with RIPA buffer that contained fresh protease and phosphatase inhibitors (Thermo Scientific; 1861282) as described previously.25 Gel electrophoresis was performed with 30 μg of protein run on precast 7.5% polyacrylamide gels (Bio-Rad, Hercules, CA; 456-1025) and transferred onto nitrocellulose membranes with the use of the Trans-Blot Turbo Transfer kit (Bio-Rad; 170-4270). Membranes were blocked in 5% milk (LabScientific, Highlands, NJ; M0841) in Blotto blocking buffer (0.15 mol/L NaCl, 0.02 mol/L Tris pH 7.5, 0.1% Tween in dH2O) for 30 minutes at room temperature. Primary antibodies were diluted in 5% milk/Blotto as follows: Cyclin D1 (Thermo Scientific; RB-9041-P; dilution 1:200), β-catenin (BD Biosciences, San Jose, CA; 610154; dilution 1:1000), and glyceraldehyde-3-phosphate dehydrogenase (Santa Cruz Biotechnology; sc-25778; dilution 1:1000) and incubated on membranes at 4°C overnight. Membranes were washed in Blotto for 15 minutes before incubation with the correct horseradish peroxidase–conjugated mouse (Millipore, Billerica, MA; AP308P; dilution 1:10,000) or rabbit (Thermo Scientific; 31460; dilution 1:20,000) secondary antibodies diluted in 5% milk/Blotto for 1 hour at room temperature. Membranes were then washed for 15 minutes in Blotto before exposure with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific; 34080) for 3 minutes at room temperature. The resulting bands were viewed by autoradiography.
RNA Isolation and Real-Time PCR
Cellular RNA was isolated by homogenizing liver tissue in TRIzol Reagent (Thermo Scientific; 15596026), followed by a chloroform extraction, precipitating nucleic acid with isopropanol at −20°C, and dissolving the resulting dried nucleic acid pellet in nuclease-free water (Qiagen, Hilden, Germany; 129117). DNase treatment was performed with the DNA-free Kit (Ambion, Pittsburgh, PA; AM1906) according to the manufacturer's instructions. Approximately 1 μg of RNA was reverse-transcribed with SuperScript III (Invitrogen, Carlsbad, CA; 18080-044). Real-time PCR was performed on a StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA; 4376600) by using the Power SYBR Green PCR Master Mix (Applied Biosystems; 4367660) with the mouse-specific primers listed in Table 1. Fold-changes in gene expression were calculated after normalizing target gene expression to the average of two housekeeping genes (Gapdh and Rn18s) by using the ΔΔ-Ct method.
Table 1.
Sequence of Real-Time PCR Primers Used in the Study in Mice
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Ccnd1 | 5′-TTTCTTTCCAGAGTCATCAAGTGT-3′ | 5′-TGACTCCAGAAGGGCTTCAA-3′ |
| Ctnnb1 | 5′-GGGTCCTCTGTGAACTTGCTC-3′ | 5′-TTCTTGTAATCCTGTGGCTTGTCC-3′ |
| Axin2 | 5′-GAGAGTGAGCGGCAGAGC-3′ | 5′-CGGCTGACTCGTTCTCCT-3′ |
| Regucalcin | 5′-CGATTCAATGATGGGAAGGT-3′ | 5′-CGTTTCCTCAGCCATGGTA-3′ |
| Lect2 | 5′-CCCACAACAATCCTCATTTCA-3′ | 5′-GTTAGCCCATGGTCCTGCTA-3′ |
| Brd4 | 5′-TTCAGCACCTCACTTCGACC-3′ | 5′-CTGGTGTTTTTGGCTCCTGC-3′ |
| Gapdh | 5′-AACTTTGGCATTGTGGAAGG-3′ | 5′-ACACATTGGGGGTAGGAACA-3′ |
| Rn18s | 5′-GTAACCCGTTGAACCCCATT-3′ | 5′-CCATCCAATCGGTAGTAGCG-3′ |
Cell Culture
HepG2 liver tumor cells were grown to approximately 30% to 40% confluency in Eagle's Minimum Essential Medium (ATCC, Manassas, VA; 30-2003) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA; 100-106) and penicillin-streptomycin (ATCC; 30-2300) and then serum-starved overnight. Cells were transfected with 800 ng of TopFlash (Millipore; 21-170) or the p65 reporter (gift from Dr. David Geller, Department of Surgery, University of Pittsburgh Medical Center, Pittsburgh, PA26) and 200 ng of Renilla in Opti-MEM media (Gibco, Gaithersburg, MD; 31985-070) by using the Lipofectamine 3000 Transfection Kit (Invitrogen; L3000-008) according to the manufacturer's instructions. After 5 hours of incubation with transfection reagents, 0.25 μm, 0.5 μm, or 1 μm JQ1, or DMSO alone as a control, in Eagle's Minimum Essential Medium supplemented with 4% FBS and penicillin-streptomycin, was added to the cells. Treated cells were harvested 24 hours later, and luciferase activity was measured by using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI; E1960) and normalized to Renilla levels.
For siRNA experiments, HepG2 or Hep3B HCC cells were seeded at a density of 200,000 cells/well in 6-well plates. After 48 hours of culture, cells were transfected with Silencer Select BRD4 siRNA S23901 (Ambion; 4390824) or Silencer Select Negative Control #2 siRNA (Ambion; 4390847) in Opti-MEM media (Gibco; 31985-070) by using the Lipofectamine RNAiMAX kit (Invitrogen; 13778-150) and incubated for 24 hours at 37°C. Next, TopFlash transfections were performed as described earlier, and luciferase activity was measured 24 hours after TopFlash transfection.
Zebrafish Studies
Experiments were performed with approval of the Institutional Animal Care and Use Committee at the University of Pittsburgh. Embryos and adult fish were raised and maintained under standard laboratory conditions.27 The following transgenic lines: Tg(fabp10a:CFP-nfsB)s931,28 Tg(EPV.Tp1-Mmu.Hbb:hist2h2l-mCherry)s939,29 and Tg(fabp10a:mAGFP-gmnn,cryaa:ECFP)pt608,22 [referred to here as Tg(fabp10a:CFP), Tg(Tp1:H2B-mCherry), and Tg(fabp10a:mAGFP-gmnn), respectively] and Tg(OTM:d2EGFP)kyu230 were used. Liver injury was induced by treating Tg(fabp10a:CFP) larvae with 10 mmol/L acetaminophen (N-acetyl-p-aminophenol; APAP) from 3.5 to 5 days after fertilization for 36 hours. The 10 mmol/L APAP working solution was freshly made by dissolving APAP powder in egg water supplemented with 0.2% DMSO and 0.2 mmol/L 1-phenyl 2-thiourea. Hepatocyte ablation was performed by treating Tg(fabp10a:CFP) larvae with 10 mmol/L metronidazole in egg water supplemented with 0.2% DMSO and 0.2 mmol/L 1-phenyl 2-thiourea. For BET inhibition, larvae were treated with 3 μmol/L JQ1 or 50 μmol/L iBET151 from 0 hour of recovery (R0h) to 48 hours of recovery (R48h), and fabp10a:CFP expression was imaged by using the M205 FA epifluorescence microscope (Leica, Wetzlar, Germany).22 To quantify liver size and Wnt/β-catenin activity, corrected total cell fluorescence of fabp10a:CFP and OTM:d2EGFP, respectively, was measured with the ImageJ software version 1.51 (NIH, Bethesda, MD) as previously described.31
Zebrafish Whole-Mount Confocal Microscopy
Whole-mount immunostaining was performed as previously described,32 using the following antibodies: mouse anti-Alcam (dilution 1:20; ZIRC, Eugene, OR) and Alexa Fluor 647-conjugated secondary antibodies (dilution 1:500; Life Technologies, Grand Island, NY). Zeiss LSM700 confocal microscope (Zeiss, Thornwood, NY) was used to obtain image data, and confocal stacks were analyzed with the Zen 2009 software (Zeiss, Berlin, Germany). All figures, labels, arrows, scale bars, and outlines were assembled or drawn with Illustrator software version CS5 (Adobe, San Jose, CA).
Zebrafish qPCR
Total RNA was extracted from 50 dissected livers by using the RNeasy Mini Kit (Qiagen, Valencia, CA); cDNA was synthesized from the RNA by using the SuperScript III First-Strand Synthesis SuperMix (Life Technologies) according to the kit protocols. Real-time quantitative PCR (qPCR) was performed as previously described,33 using the Bio-Rad iQ5 qPCR machine with the iQ SYBR Green Supermix (Bio-Rad). Fold changes were calculated after normalizing target gene expression to eef1a1l1 by using the ΔΔ-Ct method. At least three independent experiments were performed. The zebrafish-specific primers used for qPCR are listed in Table 2.
Table 2.
Sequence of Real-Time PCR Primers Used in the Study in Zebrafish
| Gene | Forward primer | Reverse primer |
|---|---|---|
| brd4 | 5′-GGCCCCTTCTTCCTTCAACC-3′ | 5′-TGTAGAATGCGCTCTCTAGGC-3′ |
| ccnd1 | 5′-GGAACTGCTGGCGCTTAAATA-3′ | 5′-GACTTGCGAGAGGAAGTTGG-3′ |
| pcna | 5′-ATCTGGATGTGGAGCAGCTT-3′ | 5′-TGTGACCGTCTTGGACAGAG-3′ |
| myca | 5′-TATTGTTAACCCGCCCCCAC-3′ | 5′-ATGCATTCACCAGCAGTCCA-3′ |
| axin2 | 5′-GGCTACAGGTCCTACAGAC-3′ | 5′-GCTTGTAAGGAGGAATGGC-3′ |
| eef1α1l1 | 5′-CTGGAGGCCAGCTCAAACAT-3′ | 5′-ATCAAGAAGAGTAGTACCGCTAGCATTAC-3′ |
Statistical Analysis
Statistical analysis was performed with Prism software version 7 (GraphPad Inc., San Diego, CA). For analyses concerning only two groups, a two-tailed t-test was performed, with P < 0.05 considered significant. For analyses concerning more than two groups, a one-way analysis of variance test was performed, with P < 0.05 considered significant. For survival curves, a Gehan-Breslow-Wilcoxon test was performed, with P < 0.05 considered significant.
Results
JQ1 Administration 16 Hours after PHx Impairs Cyclin D1 Expression and Hepatocyte Proliferation at 40 Hours
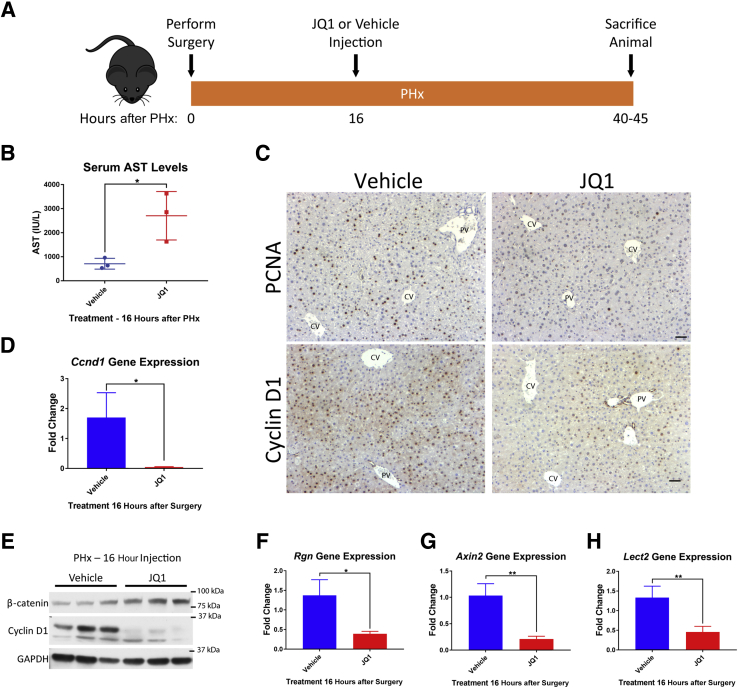
To determine whether JQ1 would have an impact on hepatocyte proliferation–driven liver regeneration, the mouse PHx model, a common model of liver regeneration whereby two-thirds of the liver is surgically removed and the remaining hepatocytes proliferate to restore the lost liver mass, was used.34 To allow time for mice to recover from the surgery before injection, a single injection of JQ1 was administered at 16 hours after PHx. Mice were harvested within the 40-to 45-hour window after PHx, which is the peak of DNA synthesis in mouse hepatocytes after PHx35, 36 (Figure 1A). JQ1-injected mice at 40 hours after PHx displayed significantly elevated levels of serum liver injury marker AST compared with control mice injected with the vehicle solution (10% hydroxypropyl β-cyclodextrin in distilled water) (Figure 1B). Assessment of hepatocyte proliferation by immunohistochemistry for PCNA37 revealed several PCNA-positive hepatocytes in vehicle-injected animals and very few PCNA-positive hepatocytes in JQ1-injected animals (Figure 1C). Cyclin D1, which is involved in promoting the G1/S phase transition in the cell cycle, is known to be highly up-regulated to drive hepatocyte proliferation after PHx in mice.38, 39 Robust hepatocyte expression of Cyclin D1 was detected in vehicle-injected mice after PHx, whereas JQ1-injected mice displayed a marked reduction in Cyclin D1 staining (Figure 1C). Cyclin D1 was also dramatically reduced at both the gene expression (Figure 1D) and protein (Figure 1E) levels in JQ1-injected animals.
Figure 1.
Injection of JQ1 16 hours after partial hepatectomy (PHx) impaired liver regeneration. A: Schematic for experimental plan for injection of JQ1 (50 mg/kg) or vehicle solution 16 hours after PHx, followed by harvest at 40 to 45 hours after PHx. B: Serum aspartate aminotransferase (AST) levels were significantly elevated in JQ1-injected mice compared with vehicle-injected controls 40 hours after PHx. C: Immunohistochemistry for proliferating cell nuclear antigen (PCNA) and Cyclin D1 revealed significantly reduced hepatocyte proliferation and Cyclin D1 expression in JQ1-injected mice. D: Significantly reduced Ccnd1 gene expression was found in JQ1-injected mice. E: No reduction in total β-catenin levels but a significant reduction in Cyclin D1 protein levels was evident in JQ1-injected mice. F: Significant reduction in Rgn gene expression was found in JQ1-injected mice. G: Significant reduction in Axin2 gene expression was found in JQ1-injected mice. H: Significant reduction in Lect2 gene expression was found in JQ1-injected mice. Data are expressed as means ± SD. ∗P < 0.05, ∗∗P < 0.01 (t-test). Scale bars = 50 μm. CV, central vein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PV, portal vein.
JQ1 Administration 16 Hours after PHx Impairs Expression of Additional Wnt Target Genes in Addition to Cyclin D1
Cyclin D1 is a known target of the Wnt/β-catenin signaling pathway,40, 41 and mice with liver-specific deletion of β-catenin display dramatically reduced Cyclin D1 levels and a delay in liver regeneration after PHx.25 Therefore, we hypothesized that JQ1 may be inhibiting Wnt/β-catenin signaling in mice after PHx. Indeed, in JQ1-injected mice there were significant decreases in gene expression of β-catenin targets regucalcin (Rgn)42 (Figure 1F), Axin243 (Figure 1G), and leukocyte cell–derived chemotaxin 2 (Lect2)44 (Figure 1H) compared with vehicle-injected controls at 40 hours after PHx. However, there was no reduction in total protein levels of β-catenin in JQ1-injected mice (Figure 1E), suggesting JQ1 was inhibiting transcription of β-catenin target genes rather than directly inhibiting expression of β-catenin.
An Earlier Administration of JQ1 at 2 Hours after PHx Completely Prevents Cyclin D1 Expression and Hepatocyte Proliferation Leading to Enhanced Injury and Mortality at 40 Hours
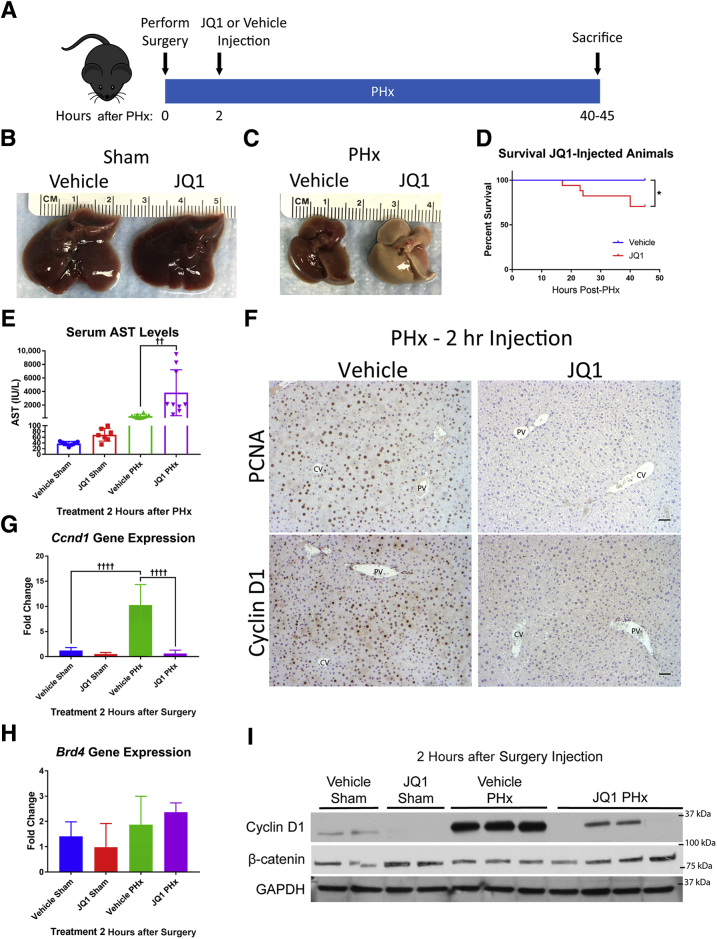
Cyclin D1 can be induced as early as 12 hours after PHx in mice,45, 46 so JQ1 was administered at 2 hours after PHx (Figure 2A) rather than 16 hours after PHx to investigate if this would completely abrogate any increases in Cyclin D1. Tissue was harvested at the peak of hepatocyte proliferation (40 to 45 hours after PHx). As a control, mice were injected with either JQ1 or the vehicle solution 2 hours after a sham surgery and harvested at 40 hours after surgery to assess the effects of JQ1 on a noninjured liver. Injection of JQ1 had no effect on the gross appearance of sham animal livers (Figure 2B). However, JQ1 injection 2 hours after PHx resulted in severe liver injury, with the livers from JQ1-injected animals 40 hours after PHx exhibiting striking discoloration (Figure 2C). Animals injected with JQ1 2 hours after PHx had significantly reduced survival compared with vehicle-injected controls, with 30% of mice exhibiting notable morbidity that required euthanasia at or before 45 hours after PHx (Figure 2D). Although there was no increase in serum AST in JQ1-injected animals that underwent sham surgery, JQ1-injected PHx mice displayed significantly elevated serum AST levels compared with vehicle-injected PHx animals (Figure 2E). Although there was a robust hepatocyte expression of PCNA and Cyclin D1 in vehicle-injected animals, in mice injected with JQ1 2 hours after PHx there were almost no PCNA-positive hepatocytes in addition to extremely little Cyclin D1 staining (Figure 2F). In accordance with these results, Ccnd1 gene expression was significantly up-regulated in vehicle-injected PHx mice compared with sham controls, but this up-regulation was completely absent in mice injected with JQ1 2 hours after PHx (Figure 2G). In addition, levels of Cyclin D1 protein were dramatically lower in JQ1-injected mice than in vehicle-injected controls after PHx, whereas levels of β-catenin were not reduced (Figure 2I). Expression of Brd4 was not significantly altered in JQ1-injected mice (Figure 2H), demonstrating that JQ1 did not induce compensatory up-regulation of Brd4.
Figure 2.
Injection of JQ1 2 hours after partial hepatectomy (PHx) impaired liver regeneration. A: Schematic for experimental design of injecting JQ1 (50 mg/kg) or vehicle solution 2 hours after PHx and harvesting at 40 to 45 hours after PHx. B: No effect was found on the gross morphology of livers from JQ1-injected sham surgery animals. C: Striking discoloration of the livers of JQ1-injected animals was found 40 hours after PHx. D: Reduced survival of animals injected with JQ1 2 hours after PHx was found. E: No increase in serum aspartate aminotransferase (AST) was found in JQ1-injected sham animals, but a significant increase was found in serum AST in JQ1-injected PHx animals compared with vehicle-injected PHx mice. F: Immunohistochemical staining for proliferating cell nuclear antigen (PCNA) and Cyclin D1 revealed robust staining in vehicle-injected controls but virtually no staining in JQ1-injected mice 40 hours after PHx. G: Dramatic up-regulation of Ccnd1 gene expression was found in vehicle-injected animals after PHx, but no up-regulation in JQ1-injected animals. H: No change in Brd4 expression was found after either PHx or JQ1 injection. I: Reduced Cyclin D1 protein was found in JQ1-injected animals compared with vehicle-injected controls, but no reduction in total β-catenin protein levels. Data are expressed as means ± SD. n = 12 vehicle-injected mice (D); n = 17 JQ1-injected mice (D). ∗P < 0.05 (Gehan-Breslow-Wilcoxon test); ††P < 0.01, ††††P < 0.0001 (one-way analysis of variance). Scale bars = 50 μm. CV, central vein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PV, portal vein.
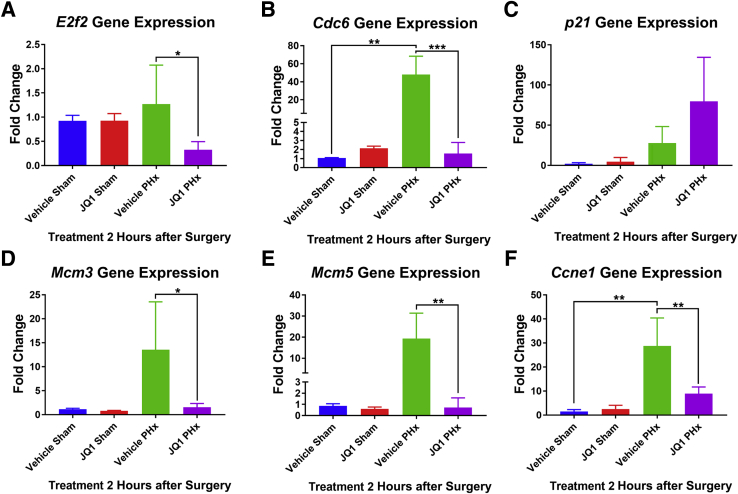
Another factor known to play a critical role in promoting cell cycle progression was the E2f2 transcription factor, which has been shown to be required for hepatocyte proliferation after PHx.47 In addition, E2F2 has been shown to be a direct target of JQ1-mediated BRD4 inhibition in human liver cancer cells.48 Therefore, it was investigated if E2f2 signaling was inhibited in JQ1-injected mice after PHx. Although there was no increase in E2f2 expression in vehicle-injected mice after PHx compared with sham controls, a decrease in E2f2 expression in JQ1-injected animals was observed after PHx (Figure 3A).
Figure 3.
E2f2-driven transcription was inhibited in JQ1-injected animals after partial hepatectomy (PHx). A: Expression of E2f2 was significantly reduced in JQ1-injected mice after PHx. B: Expression of Cdc6 was induced in vehicle-injected but not JQ1-injected mice after PHx. C: Expression of p21 was further increased in JQ1-injected mice after PHx. D: Expression of Mcm3 was induced in vehicle-injected but not JQ1-injected mice after PHx. E: Expression of Mcm5 was induced in vehicle-injected but not JQ1-injected mice after PHx. F: Expression of Ccne1 was induced in vehicle-injected but not JQ1-injected mice after PHx. Data are expressed as means + SD. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (one-way analysis of variance).
The expression of E2f2 target genes known to be involved in cell cycle progression was assessed next. Expression of E2f2 targets Cdc6 and Mcm3, which are induced in late G1 phase,49 were dramatically induced in vehicle-injected mice after PHx, and this increase was blocked in JQ1-injected animals (Figure 3, B and D). Disruption of E2f2 signaling can lead to the induction of p21,49 and indeed there was an increase in p21 expression in JQ1-injected mice after PHx compared with vehicle-injected controls (Figure 3C). Additional E2f2 targets mcm5 and ccne147, 49 were induced in vehicle-injected animals after PHx, but this induction was blocked in JQ1-injected animals (Figure 3, E and F). Interestingly, expression of E2f2 target genes was largely unaffected in JQ1-injected mice which underwent sham surgery (Figure 3). Thus, the data indicate that JQ1, in addition to inhibiting β-catenin target gene expression, was inhibiting expression of E2f2-driven target genes after PHx, resulting in cell cycle arrest. Collectively, these results showed that injection of JQ1 2 hours after PHx nearly completely abrogated hepatocyte proliferation and liver regeneration, but JQ1 injection was well tolerated in animals in the absence of liver injury.
Earlier JQ1 Administration after Hepatectomy Impairs Expression of Additional Wnt Target Genes, which Was Validated in Vitro
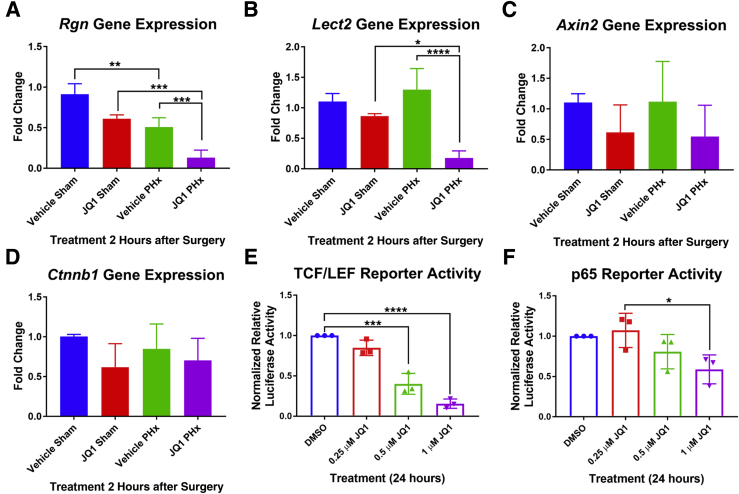
The hepatic expression of β-catenin target genes that were down-regulated in animals injected with JQ1 16 hours after PHx was determined in mice injected with JQ1 2 hours after PHx. Expression of Rgn was significantly reduced in JQ1-injected PHx animals compared with both JQ1-injected sham and vehicle-injected PHx mice (Figure 4A). Similarly, gene expression of Lect2 was significantly reduced in JQ1-injected PHx mice compared with both JQ1-injected sham and vehicle-injected PHx animals (Figure 4B). Expression of Axin2 tended to be lower in JQ1-injected PHx mice than in vehicle-injected PHx animals (Figure 4C). There was no significant change in Ctnnb1 gene expression in mice injected with JQ1 2 hours after PHx compared with mice injected with the vehicle solution (Figure 4D).
Figure 4.
β-Catenin–driven transcription was sensitive to JQ1 inhibition. A:Rgn gene expression was significantly reduced in JQ1-injected mice compared with vehicle-injected controls after partial hepatectomy (PHx). B:Lect2 gene expression was significantly reduced in JQ1-injected mice after PHx. C: Expression of Axin2 tended to be lower in JQ1-injected mice after PHx. D: No change in the gene expression of Ctnnb1 was observed in JQ1-injected mice. E: HepG2 cells transfected with a T-cell factor (TCF)/LEF luciferase reporter and treated with JQ1 for 24 hours showed a dose-dependent decrease in reporter activity after JQ1 treatment. F: HepG2 cells transfected with a p65 luciferase reporter and treated with JQ1 for 24 hours showed a reduction of reporter activity after treatment with 1 μm of JQ1. Data are pooled results. Data are expressed as means + SD. n = 3 independent experiments (A–F). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 (one-way analysis of variance).
To determine whether JQ1 could directly inhibit β-catenin-driven transcriptional activity, HepG2 cells, which have constitutively active β-catenin owing to a deletion that affects exon-3 of the CTNNB1 gene, were transfected with a TopFlash luciferase reporter. JQ1 dose dependently significantly inhibited β-catenin–T-cell factor (TCF)/LEF reporter activity (Figure 4E). To determine whether β-catenin–driven transcriptional activity was especially sensitive to inhibition by JQ1 treatment, the effect of JQ1 was tested on the NF-κB signaling pathway, another pathway implicated in liver regeneration after PHx because of the observed activation of the NF-κB subunit p65/RelA within 30 minutes of PHx in mice and rats.50, 51 When HepG2 cells transfected with a p65 luciferase reporter were treated with JQ1, at the highest concentration of JQ1 there was only a 50% reduction in reporter activity (Figure 4F), compared with up to a 90% reduction in β-catenin-TCF/LEF reporter activity. These results suggest that β-catenin–driven transcriptional activity was particularly sensitive to inhibition by JQ1.
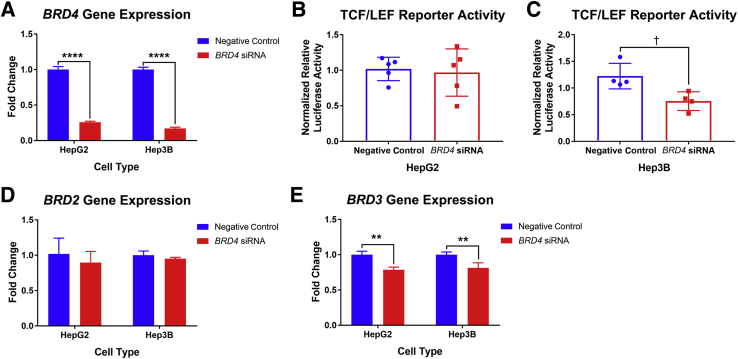
To determine whether the JQ1-mediated reduction in β-catenin-TCF/LEF reporter activity was specifically because of inhibition of BRD4 activity, two liver tumor cell lines, HepG2 and Hep3B, were treated with BRD4 siRNA and TCF/LEF reporter activity was measured subsequently. It was first validated that robust knockdown of BRD4 expression was achieved in the siRNA-treated cells (Figure 5A). Interestingly, HepG2 cells treated with BRD4 siRNA showed no reduction in TCF/LEF reporter activity (Figure 5B). Because HepG2 cells have constitutively active mutant β-catenin, reduction in BRD4 expression levels may not be sufficient to reduce signaling driven by mutant β-catenin protein. Therefore, Hep3B cells, a HCC cell line with wild-type β-catenin, were treated with BRD4 siRNA. A modest but significant reduction in TCF/LEF reporter activity was seen in Hep3B cells (Figure 5C), which was roughly on-par with the reduction of TCF/LEF reporter activity seen in HepG2 cells with low-dose JQ1 treatment (Figure 4E). Because JQ1 was known to inhibit the bromodomains of BET proteins15 and the BET family proteins share conserved bromodomains,10 the expression levels of BRD2 and BRD3 were assessed in liver cancer cells to determine whether there was compensatory up-regulation of other members of the BET protein family on BRD4 knockdown. The expression of BRD2 was unchanged by BRD4 siRNA treatment (Figure 5D), whereas the expression level of BRD3 was slightly reduced in both HeG2 and Hep3B cells (Figure 5E). BRD2 and BRD3 may partially compensate for BRD4 transcriptional activity, whereas all BET proteins would be inhibited by high-dose JQ1 treatment, offering a potential explanation for the relatively minor effects of BRD4 siRNA on TCF/LEF reporter activity. Overall, data suggest that JQ1-inhibition of BRD4 was at least partially responsible for the reduction in β-catenin–driven transcriptional activity in a wild-type β-catenin setting.
Figure 5.
BRD4 siRNA reduced T-cell factor (TCF)/LEF transcriptional activity in Hep3B cells. A:BRD4 siRNA treatment dramatically reduced BRD4 expression in both HepG2 and Hep3B cells. B: There was no reduction in TCF/LEF reporter activity in HepG2 cells treated with BRD4 siRNA. C: Significant reduction was found in TCF/LEF reporter activity in Hep3B cells treated with BRD4 siRNA. D: No change was found in expression of BRD2 in cells treated with BRD4 siRNA. E: Slight reduction was found in the expression of BRD3 in cells treated with BRD4 siRNA. Data are expressed as means ± SD (A–E). Data are pooled results. n = 5 independent experiments (B); n = 4 independent experiments (C). ∗∗P < 0.01, ∗∗∗∗P < 0.0001 (two-way analysis of variance); †P < 0.05 (t-test).
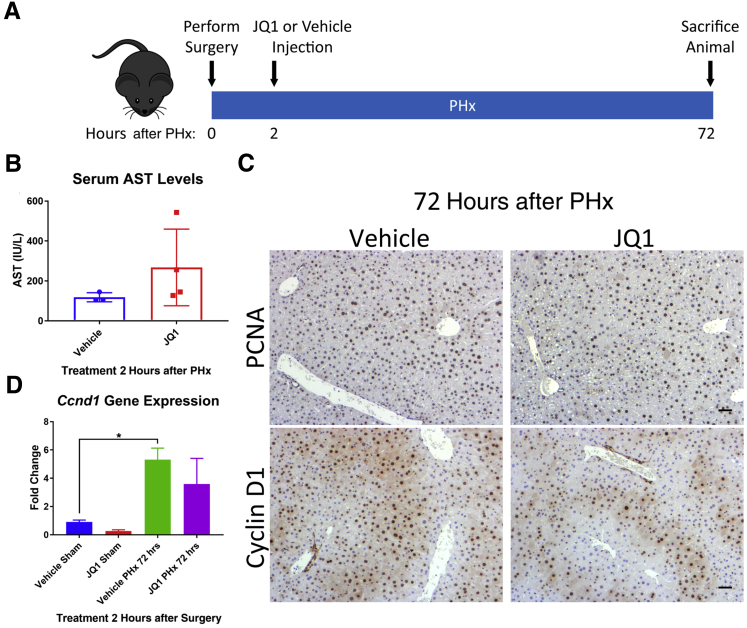
A Single Early Dose of JQ1 2 Hours after Hepatectomy Is Insufficient to Have an Impact on Cyclin D1 Expression and Hepatocyte Proliferation at 72 Hours after Hepatectomy
To determine the extent to which a single dose of JQ1 can block liver regeneration, mice were injected with JQ1 or the vehicle solution 2 hours after PHx, and the level of liver regeneration was assessed at 72 hours after PHx (Figure 6A). The 70% of JQ1-injected animals that survived to 72 hours after PHx showed serum liver injury marker AST levels that were comparable with vehicle-injected animals (Figure 6B). Robust proliferation of hepatocytes was evident via PCNA immunohistochemistry in both vehicle and JQ1-injected mice (Figure 6C). There was also the appearance of Cyclin D1 staining in the hepatocytes of JQ1-injected mice at this time point (Figure 6C), which was accompanied by an up-regulation of Ccnd1 gene expression in the JQ1-injected mice (Figure 6D). Most likely this induction of liver regeneration coincided with the clearance of JQ1 from the mice, because liver regeneration in zebrafish resumed after washout of JQ1.22
Figure 6.
Induction of liver regeneration in JQ1-injected animals 72 hours after partial hepatectomy (PHx). A: Schematic of experimental design, with a 50 mg/kg JQ1 injection (or vehicle injection) 2 hours after PHx and harvest at 72 hours after PHx. B: Serum aspartate aminotransferase (AST) levels normalized in both JQ1- and vehicle-injected mice 72 hours after PHx. C: Proliferating cell nuclear antigen (PCNA) staining revealed robust hepatocyte proliferation in both vehicle- and JQ1-injected mice 72 hours after PHx, and Cyclin D1 staining was present in hepatocytes in both vehicle- and JQ1-injected animals 72 hours after PHx. D: Up-regulation of Ccnd1 gene expression was present in both vehicle- and JQ1-injected animals 72 hours after PHx. Data are expressed as means ± SD. ∗P < 0.05 (one-way analysis of variance). Scale bars = 50 μm.
BET Protein Inhibition Also Impairs Hepatocyte Proliferation and Liver Size in a Zebrafish Model of Hepatic Injury and Hepatocyte-Mediated Regeneration
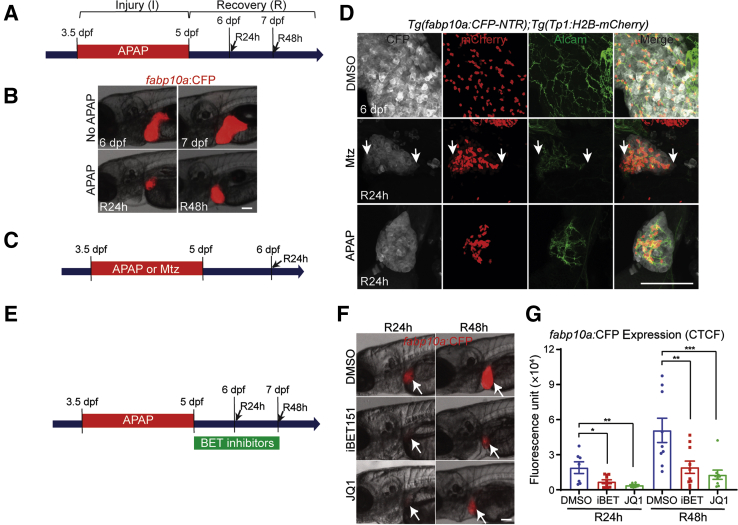
To further validate the effect of BET protein inhibition on hepatocyte-driven liver regeneration, a zebrafish model in which APAP overdose induced liver damage and subsequent regeneration, was used.52, 53 The APAP overdose model has been widely used in rodents54 and zebrafish,52, 53, 55 because it mimics human APAP-induced acute liver failure (ALF), the most common cause of ALF in the United States.54, 56 As previously reported,52, 53 treating zebrafish larvae with APAP greatly reduced liver size, and after APAP washout liver size gradually recovered (Figure 7, A and B). It was first determined whether hepatocytes solely or together with BECs contribute to regenerating hepatocytes in the APAP-induced injury model. As a positive control for BEC contribution to regenerating hepatocytes, previous model of BEC-driven liver regeneration,28 in which Tg(fabp10a:CFP-NTR) zebrafish, which express nitroreductase (NTR) fused with cyan fluorescent protein (CFP) specifically in hepatocytes via the hepatocyte-specific fabp10a promotor, was used (Figure 7C). NTR metabolized the prodrug metronidazole into a cytotoxic compound, resulting in the ablation of NTR-expressing cells. Thus, treating Tg(fabp10a:CFP-NTR) larvae with metronidazole induced hepatocyte-specific cell death. In this model, near-complete ablation of hepatocytes induced the dedifferentiation of BECs into liver progenitor cells, which subsequently differentiated into either hepatocytes or BECs.28 Given the BEC-restricted Notch activity in the zebrafish liver,57 to mark BECs, a Notch reporter line, Tg(Tp1:H2B-mCherry), which expressed H2B-mCherry fusion proteins under the promoter containing the Notch-responsive element, was used.29 Because of the prolonged stability of H2B-mCherry proteins, this line also revealed BEC-derived cells even after Notch signaling was turned off. In metronidazole-treated larvae after R24h, all hepatocytes weakly expressed Tp1:H2B-mCherry (Figure 7D) but not mature BEC marker Alcam, as previously reported,28 indicating hepatocytes with BEC origin. However, in APAP-treated larvae at R24h no hepatocytes expressed Tp1:H2B-mCherry (Figure 7D), revealing that liver regeneration in the zebrafish APAP-induced injury model was hepatocyte- but not BEC-driven. To inhibit BET proteins, two different BET inhibitors, JQ115 and iBET151,58 which both block BET proteins by specifically binding to acetyl-recognizing BET pockets, were used. Treatment with JQ1 or iBET151 immediately after 36-hour APAP treatment (R0h) significantly reduced liver size after R24h and R48h, respectively, as assessed by hepatocyte-specific fabp10a:CFP expression (Figure 7, E–G), supporting the positive role of BET proteins in hepatocyte-driven liver regeneration.
Figure 7.
Bromodomain and extraterminal (BET) inhibition impaired hepatocyte-driven liver regeneration in zebrafish. A: Scheme illustrating the periods of acetaminophen (APAP) treatment (I, injury) and liver regeneration (R). Arrows indicate analysis stage. B: Epifluorescence images showing fabp10a:CFP expression in the untreated larvae or larvae treated with APAP. C: Scheme illustrating the periods of APAP or metronidazole (Mtz) treatment and liver regeneration. Arrow indicates analysis stage. D: Confocal images showing the expression of Alcam (green), Tp1:H2B-mCherry (red), and fabp10a:CFP-NTR (gray) in regenerating livers at 24 hours of recovery (R24h). Arrows point to biliary epithelial cell–derived hepatocytes, which weakly expressed Tp1:H2B-mCherry. E: Scheme illustrating the periods of APAP and BET inhibitor treatments. Arrows indicate analysis stages. F: Epifluorescence images showing fabp10a:CFP expression in the regenerating larvae treated with two different BET inhibitors, iBET151 and JQ1. Arrows point to the liver. G: Quantification of fabp10a:CFP expression in the regenerating larvae treated with dimethyl sulfoxide (DMSO) or BET inhibitors, as shown in D. Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001 (one-way analysis of variance). Scale bars = 100 μm (B, D, and F). CFP, cyan fluorescent protein; CTCF, corrected total cell fluorescence; dpf, days after fertilization.
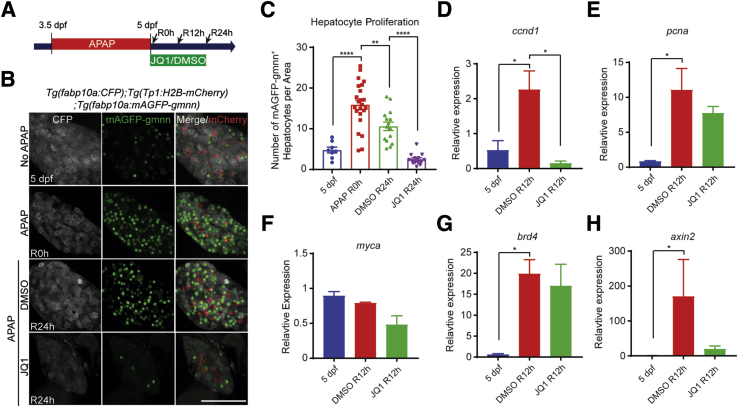
Given the reduced liver size in BET-inhibited larvae (Figure 7G) and the role of BET proteins in proliferation,22 hepatocyte proliferation was examined by using the Tg(fabp10a:mAGFP-gmnn) line that expressed geminin fused with monomeric Azami green fluorescent proteins in hepatocytes (Figure 8A).22 Because geminin was degraded in G0 and G1 phases,59 this line revealed proliferating hepatocyte in S/G2/M phases.22 Hepatocyte proliferation was greatly increased in APAP-treated larvae compared with control larvae at 5 days after fertilization (Figure 8, B and C). JQ1 treatment significantly reduced the hepatocyte proliferation induced by APAP overdose (Figure 8, B and C), suggesting the crucial role of BET proteins in hepatocyte proliferation during regeneration. This proliferation phenotype was further supported by the reduced expression of ccnd1 (Figure 8D), consistent with the mouse data (Figure 1D). However, other proliferation-related genes, such as pcna and myca, were not significantly reduced in JQ1-treated livers (Figure 8, E and F). brd4 expression was significantly up-regulated in APAP-treated livers compared with untreated control livers (Figure 8G), as observed in a zebrafish BEC-driven liver regeneration model.22 In addition, similar to the mouse PHx model (Figure 1G), expression of Wnt/β-catenin signaling target axin2 tended to be lower in JQ1-treated larvae after APAP overdose (Figure 8H). Altogether, these zebrafish data together with mouse data strongly support the observation that BET proteins regulated hepatocyte-driven liver regeneration.
Figure 8.
Bromodomain and extraterminal (BET) inhibition reduced hepatocyte proliferation in the zebrafish acetaminophen (APAP)-induced injury model. A: Scheme illustrating the periods of APAP and JQ1 treatments and analysis stages (arrows). B: Confocal images showing the hepatic expression of fabp10a:CFP (gray, hepatocytes), Tp1:H2B-mCherry (red, biliary epithelial cells), and fabp10a:mAGFP-gmnn (green, proliferating hepatocytes). C: Quantification of the number of mAGFP-gmnn+ cells, as shown in B. D–H: Real-time quantitative PCR data showing the relative expression levels of ccnd1, pcna, myca, brd4, and axin2 among uninjured control livers at 5 days after fertilization (dpf) and dimethyl sulfoxide (DMSO)- and JQ1-treated regenerating livers at 12 hours of recovery (R12h). Data are expressed as means ± SEM. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001 (one-way analysis of variance). Scale bar = 100 μm. CFP, cyan fluorescent protein.
BET Inhibition Represses Wnt/β-Catenin Signaling in APAP-Treated Zebrafish Livers
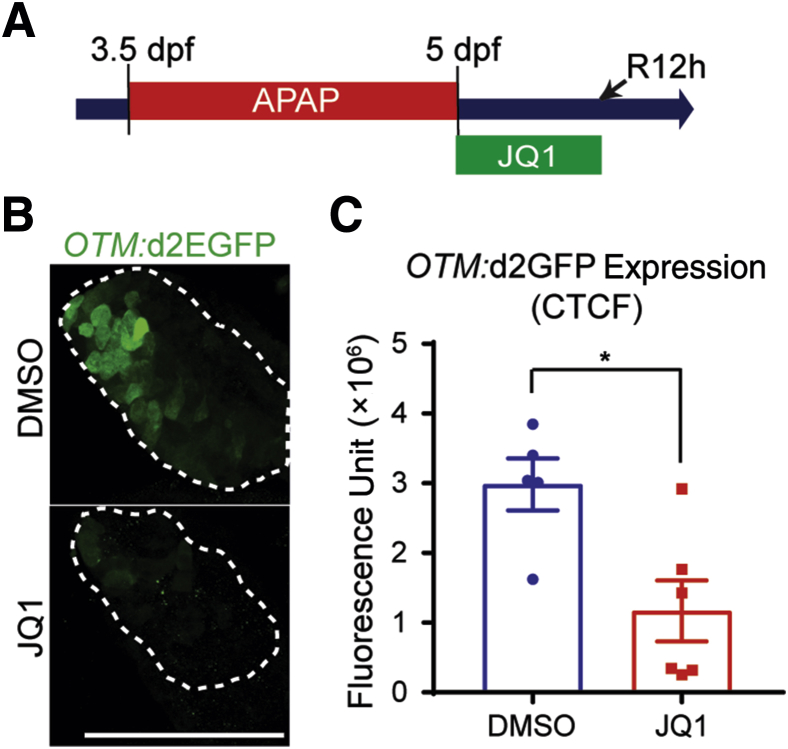
Given the crucial role of Wnt/β-catenin signaling in liver regeneration induced by APAP overdose52, 60 and the reduced expression of Wnt target genes ccnd1 and axin2 in JQ1-treated regenerating zebrafish livers (Figure 8, D and H), we hypothesized that BET inhibition may suppress Wnt/β-catenin signaling in livers injured by APAP treatment. To test this hypothesis, a Wnt reporter zebrafish line, Tg(OTM:d2EGFP), which expressed destabilized green fluorescent protein (GFP) under a promoter that contained six copies of Tcf/Lef binding site,30 meaning that the level of GFP expression was driven by β-catenin signaling, was used. GFP expression was significantly reduced in JQ1-treated regenerating livers compared with DMSO-treated control livers (Figure 9). These zebrafish data together with the in vitro data (Figures 4 and 5) suggested that BET proteins regulate hepatocyte-driven liver regeneration by regulating Wnt/β-catenin signaling in addition to other signaling pathways.
Figure 9.
Bromodomain and extraterminal inhibition reduced Wnt/β-catenin signaling in the zebrafish acetaminophen (APAP)-induced injury model. A: Scheme illustrating the periods of APAP and JQ1 treatments and analysis stage (arrow). B: Confocal images showing the hepatic expression of OTM:d2EGFP (green, T-cell factor/Lef reporter activity) in regenerating larvae at 12 hours of recovery (R12h). Dashed lines outline the liver. C: Quantification of OTM:d2EGFP expression in the regenerating larvae treated with dimethyl sulfoxide (DMSO) or JQ1, as shown in B. Data are expressed as means ± SEM. ∗P < 0.05 (t-test). Scale bar = 100 μm. CTCF, corrected total cell fluorescence; dpf, days after fertilization.
Discussion
BET protein inhibitors, such as JQ1, have been proposed for use in the treatment of a variety of conditions because of their ability to inhibit oncogenic gene expression,12, 16, 17 reduce inflammation,61, 62 and protect against liver fibrosis.21 Adding to their therapeutic potential, effective antineoplastic doses of JQ1 are well tolerated in animals, despite the widespread activity of Brd4 in nearly every tissue type.12, 15, 16 JQ1 has also been shown to be effective in reducing cell growth in a tumor xenograft HCC model,20 which may suggest BET protein inhibitors to be a potential treatment of liver cancer. However, BET proteins also play a role in biliary-driven liver regeneration.22 In the present study, we show that JQ1-mediated inhibition of BET proteins significantly blocks hepatocyte proliferation in two independent models of hepatocyte-driven liver regeneration. Because most HCC cases occur in the patients with chronic liver diseases, which in turn are characterized by ongoing regeneration, inadvertent suppression of regeneration during HCC treatment may adversely affect hepatic function. Hence, our findings suggest the use of BET proteins in the treatment of liver pathologic processes must be carefully considered, because BET protein inhibitors are liable to inhibit the regeneration of normal liver tissue.
Two-thirds PHx in rodents is widely used to study the mechanisms of hepatocyte proliferation during liver regeneration34 and is a useful model for the study of temporal changes in gene expression during liver regeneration. In this study, either early (2 hours after PHx) or late (16 hours after PHx) injections of JQ1 were used to study the effects of BET protein inhibition on hepatocyte proliferation. In both instances, injection of JQ1 resulted in a near total block of hepatocyte proliferation with significant reduction in expression of cell cycle–regulator Cyclin D1, suggesting that transcriptional activation of pro-regeneration genes occurs even many hours after the initial injury. Impaired induction of E2f2 downstream targets involved in cell cycle progression was also observed, further demonstrating the complete block of hepatocyte proliferation in JQ1-injected mice after PHx.
The second model to study the role of BET proteins in hepatocyte proliferation was APAP overdose, because APAP-induced ALF is the most common cause of ALF in the United States.54, 56 APAP overdose experiments were performed in zebrafish, because their large clutch sizes, growth in water, optical transparency, and fully developed livers by 5 days after fertilization63 make them ideal for drug treatment studies. Consistent with the results from the mouse PHx study, JQ1-treated zebrafish larvae displayed significant reductions in hepatocyte proliferation and Cyclin D1 expression during recovery from APAP overdose. Interestingly, in comparison with the mouse PHx model, a large increase in brd4 expression was detected in zebrafish larvae during recovery from APAP overdose. This finding suggests that BET proteins are important regulators of liver regeneration in this model, although there are presently few studies on the role of BET proteins in liver regeneration after acetaminophen injury. Thus, this work presents an opportunity to study signaling pathways under the control of BET proteins during liver regeneration.
A signaling pathway well known to be active in promoting liver regeneration25, 60, 64 in addition to being an oncogenic driver in subsets of liver cancer65, 66, 67, 68 is the Wnt/β-catenin signaling pathway. These results demonstrate that Wnt/β-catenin signaling targets, such as Cyclin D1, are significantly reduced in JQ1-treated animals after liver injury. In this respect, mice injected with JQ1 after PHx are reminiscent of liver-specific β-catenin knockout animals subjected to PHx, which show an early lag in hepatocyte proliferation in conjunction with reduced Cyclin D1 levels.25 Furthermore, activation of Wnt/β-catenin signaling is thought to promote liver regeneration after APAP-induced liver injury,52, 60 and the zebrafish results demonstrate that JQ1 is a potent inhibitor of hepatocyte proliferation and Wnt/β-catenin signaling after APAP overdose. In addition, in vitro experiments revealed that Wnt/β-catenin signaling was sensitive to inhibition by JQ1. In vitro data showed that knockdown of BRD4 in Hep3B cells with wild-type β-catenin leads to a reduction in TCF/LEF reporter activity, suggesting that the ability of JQ1 to impair β-catenin signaling is mediated at least partially through inhibition of BRD4. However, treatment of HepG2 cells with high doses of JQ1 led to a robust decrease in TCF/LEF reporter activity, suggesting that other BET proteins such as BRD2 and BRD3 may also play a role in regulating β-catenin signaling, although this hypothesis remains to be tested. Together, these data suggest JQ1 as a potent inhibitor of Wnt/β-catenin signaling. However, these results must be considered in a cell type-specific context, because JQ1 had minimal effect on β-catenin/TCF-mediated transcription in colorectal cancer cells.69 This raises the interesting question of whether JQ1 would be especially effective in treating liver cancer with activating β-catenin mutations, because these cancers have been shown to be sensitive to inhibition of β-catenin.68 Although we have demonstrated that JQ1 is capable of inhibiting normal liver regeneration, with careful dose titration BET inhibitors may be capable of inhibiting oncogenic Wnt signaling without impinging signaling pathways required for normal hepatic function. Because tumors tend to be addicted to several oncogenic signaling pathways, such as Met and Wnt/β-catenin signaling, whereas normal liver regeneration occurs through many redundant signaling mechanisms, low doses of BET inhibitors may be able to selectively inhibit oncogenic Wnt/β-catenin signaling while sparing compensatory regeneration mechanisms in normal liver.
The sensitivity of Wnt/β-catenin signaling to JQ1 may be due in part to proficiency of BET inhibitors to reduce BRD4 enhancer occupancy at super enhancers,12 or large enhancers with multiple transcription factor binding sites, high levels of Mediator occupancy, and high levels of associated gene expression.70, 71, 72 Of interest, super enhancers in colorectal cancer were found to be occupied by TCF4 and thus were sensitive to perturbation of oncogenic Wnt signaling,73 and transcription factor targets of Wnt signaling were found to be enriched for super enhancers in embryonic stem cells.72 In addition, Cyclin D1 was found to be regulated by a super enhancer in a model of Ewing sarcoma.74 These data suggest that super enhancers under the control of the Wnt/β-catenin signaling pathway may regulate the expression of genes activated during hepatocyte-driven liver regeneration, such as Cyclin D1, although this hypothesis remains to be tested. Little is known about the role of super enhancer-driven gene expression during liver regeneration, and our model of JQ1 treatment after PHx may provide a useful platform for the identification of these important signaling mechanisms.
Conclusions
This work has identified the key role of BET proteins in multiple models of hepatocyte-driven liver regeneration. In addition, we have identified the Wnt/β-catenin signaling pathway, a known important driver of liver regeneration,25, 64 to be sensitive to JQ1-mediated BET protein inhibition, resulting in an abrogation of hepatocyte proliferation. Although more work remains to be done to identify the mechanism behind JQ1-mediated inhibition of Wnt/β-catenin signaling, these results have important implications for the clinical use of BET inhibitors. Because these drugs are herein demonstrated to be potent inhibitors of normal liver regeneration, their potential use to treat liver pathologic processes must be carefully considered, because patients may become more susceptible to liver failure under a regimen of BET protein inhibitors.
Acknowledgment
We thank Dr. David Geller for p65 reporter.
Footnotes
Supported by NIH grants 1R01DK62277 (S.P.M.), 1R01DK100287 (S.P.M.), R01CA204586 (S.P.M.), DK101426 (D.S.), T32EB0010216 (J.O.R.) and 1F31DK115017-01 (J.O.R.), and Endowed Chair for Experimental Pathology (S.P.M.).
J.O.R. and S.K. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.02.006.
Contributor Information
Donghun Shin, Email: donghuns@pitt.edu.
Satdarshan P. Monga, Email: smonga@pitt.edu.
Supplemental Data
References
- 1.Thorgeirsson S.S. Hepatic stem cells in liver regeneration. FASEB J. 1996;10:1249–1256. [PubMed] [Google Scholar]
- 2.Michalopoulos G.K., Khan Z. Liver stem cells: experimental findings and implications for human liver disease. Gastroenterology. 2015;149:876–882. doi: 10.1053/j.gastro.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan A.W., Dorrell C., Grompe M. Stem cells and liver regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kochanek K.D., Murphy S.L., Xu J., Tejada-Vera B. Deaths: final data for 2014. Natl Vital Stat Rep. 2016;65:1–122. [PubMed] [Google Scholar]
- 5.Mittal S., El-Serag H.B. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2–S6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Njei B., Rotman Y., Ditah I., Lim J.K. Emerging trends in hepatocellular carcinoma incidence and mortality. Hepatology. 2015;61:191–199. doi: 10.1002/hep.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pellicoro A., Ramachandran P., Iredale J.P., Fallowfield J.A. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14:181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 8.Luedde T., Kaplowitz N., Schwabe R.F. Cell death and cell death responses in liver disease: mechanisms and clinical relevance. Gastroenterology. 2014;147:765–783. doi: 10.1053/j.gastro.2014.07.018. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J., Vakoc C.R. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y.W., Veschambre P., Erdjument-Bromage H., Tempst P., Conaway J.W., Conaway R.C., Kornberg R.D. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proc Natl Acad Sci U S A. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovén J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anand P., Brown J.D., Lin C.Y., Qi J., Zhang R., Artero P.C., Alaiti M.A., Bullard J., Alazem K., Margulies K.B., Cappola T.P., Lemieux M., Plutzky J., Bradner J.E., Haldar S.M. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., Prakash C., Sum C., Gong Y., Li Y., Kwok J.J., Thiessen N., Pettersson S., Jones S.J., Knapp S., Yang H., Chin K.C. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., Philpott M., Munro S., McKeown M.R., Wang Y., Christie A.L., West N., Cameron M.J., Schwartz B., Heightman T.D., La Thangue N., French C.A., Wiest O., Kung A.L., Knapp S., Bradner J.E. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delmore J.E., Issa G.C., Lemieux M.E., Rahl P.B., Shi J., Jacobs H.M., Kastritis E., Gilpatrick T., Paranal R.M., Qi J., Chesi M., Schinzel A.C., McKeown M.R., Heffernan T.P., Vakoc C.R., Bergsagel P.L., Ghobrial I.M., Richardson P.G., Young R.A., Hahn W.C., Anderson K.C., Kung A.L., Bradner J.E., Mitsiades C.S. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockwood W.W., Zejnullahu K., Bradner J.E., Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci U S A. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y.H., Sui X.M., Sui Y.N., Zhu Q.W., Yan K., Wang L.S., Wang F., Zhou J.H. BRD4 induces cell migration and invasion in HCC cells through MMP-2 and MMP-9 activation mediated by the Sonic hedgehog signaling pathway. Oncol Lett. 2015;10:2227–2232. doi: 10.3892/ol.2015.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang P., Dong Z., Cai J., Zhang C., Shen Z., Ke A., Gao D., Fan J., Shi G. BRD4 promotes tumor growth and epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Immunopathol Pharmacol. 2015;28:36–44. doi: 10.1177/0394632015572070. [DOI] [PubMed] [Google Scholar]
- 20.Li G.Q., Guo W.Z., Zhang Y., Seng J.J., Zhang H.P., Ma X.X., Zhang G., Li J., Yan B., Tang H.W., Li S.S., Wang L.D., Zhang S.J. Suppression of BRD4 inhibits human hepatocellular carcinoma by repressing MYC and enhancing BIM expression. Oncotarget. 2016;7:2462–2474. doi: 10.18632/oncotarget.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding N., Hah N., Yu R.T., Sherman M.H., Benner C., Leblanc M., He M., Liddle C., Downes M., Evans R.M. BRD4 is a novel therapeutic target for liver fibrosis. Proc Natl Acad Sci U S A. 2015;112:15713–15718. doi: 10.1073/pnas.1522163112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko S., Choi T.Y., Russell J.O., So J., Monga S.P., Shin D. Bromodomain and extraterminal (BET) proteins regulate biliary-driven liver regeneration. J Hepatol. 2016;64:316–325. doi: 10.1016/j.jhep.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Committee for the Update of the Guide for the Care and Use of Laboratory Animals; National Research Council . National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals: Eighth Edition. [Google Scholar]
- 24.Ye H., Holterman A.X., Yoo K.W., Franks R.R., Costa R.H. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan X.P., Behari J., Cieply B., Michalopoulos G.K., Monga S.P.S. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 26.Du Q., Zhang X., Cardinal J., Cao Z., Guo Z., Shao L., Geller D.A. Wnt/beta-catenin signaling regulates cytokine-induced human inducible nitric oxide synthase expression by inhibiting nuclear factor-kappaB activation in cancer cells. Cancer Res. 2009;69:3764–3771. doi: 10.1158/0008-5472.CAN-09-0014. [DOI] [PubMed] [Google Scholar]
- 27.Westerfield M. ed 5. University of Oregon Press; Eugene, OR: 2007. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) [Google Scholar]
- 28.Choi T.Y., Ninov N., Stainier D.Y., Shin D. Extensive conversion of hepatic biliary epithelial cells to hepatocytes after near total loss of hepatocytes in zebrafish. Gastroenterology. 2014;146:776–788. doi: 10.1053/j.gastro.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninov N., Borius M., Stainier D.Y. Different levels of Notch signaling regulate quiescence, renewal and differentiation in pancreatic endocrine progenitors. Development. 2012;139:1557–1567. doi: 10.1242/dev.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimizu N., Kawakami K., Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/beta-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370:71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 31.McCloy R.A., Rogers S., Caldon C.E., Lorca T., Castro A., Burgess A. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle. 2014;13:1400–1412. doi: 10.4161/cc.28401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi T.Y., Khaliq M., Ko S., So J., Shin D. Hepatocyte-specific ablation in zebrafish to study biliary-driven liver regeneration. J Vis Exp. 2015:e52785. doi: 10.3791/52785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Groh E.D., Swanhart L.M., Cosentino C.C., Jackson R.L., Dai W., Kitchens C.A., Day B.W., Smithgall T.E., Hukriede N.A. Inhibition of histone deacetylase expands the renal progenitor cell population. J Am Soc Nephrol. 2010;21:794–802. doi: 10.1681/ASN.2009080851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalopoulos G.K., DeFrances M.C. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 35.Weglarz T.C., Sandgren E.P. Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proc Natl Acad Sci U S A. 2000;97:12595–12600. doi: 10.1073/pnas.220430497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 37.Hall P.A., Levison D.A., Woods A.L., Yu C.C., Kellock D.B., Watkins J.A., Barnes D.M., Gillett C.E., Camplejohn R., Dover R. Proliferating cell nuclear antigen (PCNA) immunolocalization in paraffin sections: an index of cell proliferation with evidence of deregulated expression in some neoplasms. J Pathol. 1990;162:285–294. doi: 10.1002/path.1711620403. [DOI] [PubMed] [Google Scholar]
- 38.Albrecht J.H., Hu M.Y., Cerra F.B. Distinct patterns of cyclin D1 regulation in models of liver regeneration and human liver. Biochem Biophys Res Commun. 1995;209:648–655. doi: 10.1006/bbrc.1995.1548. [DOI] [PubMed] [Google Scholar]
- 39.Núñez K.G., Gonzalez-Rosario J., Thevenot P.T., Cohen A.J. Cyclin D1 in the liver: role of noncanonical signaling in liver steatosis and hormone regulation. Ochsner J. 2017;17:56–65. [PMC free article] [PubMed] [Google Scholar]
- 40.Shtutman M., Zhurinsky J., Simcha I., Albanese C., D'Amico M., Pestell R., Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tetsu O., McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 42.Nejak-Bowen K.N., Zeng G., Tan X., Cieply B., Monga S.P. Beta-catenin regulates vitamin C biosynthesis and cell survival in murine liver. J Biol Chem. 2009;284:28115–28127. doi: 10.1074/jbc.M109.047258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jho E.H., Zhang T., Domon C., Joo C.K., Freund J.N., Costantini F. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ovejero C., Cavard C., Périanin A., Hakvoort T., Vermeulen J., Godard C., Fabre M., Chafey P., Suzuki K., Romagnolo B., Yamagoe S., Perret C. Identification of the leukocyte cell-derived chemotaxin 2 as a direct target gene of beta-catenin in the liver. Hepatology. 2004;40:167–176. doi: 10.1002/hep.20286. [DOI] [PubMed] [Google Scholar]
- 45.Albrecht J.H., Poon R.Y., Ahonen C.L., Rieland B.M., Deng C., Crary G.S. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene. 1998;16:2141–2150. doi: 10.1038/sj.onc.1201728. [DOI] [PubMed] [Google Scholar]
- 46.Rickheim D.G., Nelsen C.J., Fassett J.T., Timchenko N.A., Hansen L.K., Albrecht J.H. Differential regulation of cyclins D1 and D3 in hepatocyte proliferation. Hepatology. 2002;36:30–38. doi: 10.1053/jhep.2002.33996. [DOI] [PubMed] [Google Scholar]
- 47.Delgado I., Fresnedo O., Iglesias A., Rueda Y., Syn W.K., Zubiaga A.M., Ochoa B. A role for transcription factor E2F2 in hepatocyte proliferation and timely liver regeneration. Am J Physiol Gastrointest Liver Physiol. 2011;301:G20–G31. doi: 10.1152/ajpgi.00481.2010. [DOI] [PubMed] [Google Scholar]
- 48.Hong S.H., Eun J.W., Choi S.K., Shen Q., Choi W.S., Han J.W., Nam S.W., You J.S. Epigenetic reader BRD4 inhibition as a therapeutic strategy to suppress E2F2-cell cycle regulation circuit in liver cancer. Oncotarget. 2016;7:32628–32640. doi: 10.18632/oncotarget.8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timmers C., Sharma N., Opavsky R., Maiti B., Wu L., Wu J., Orringer D., Trikha P., Saavedra H.I., Leone G. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27:65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cressman D.E., Greenbaum L.E., Haber B.A., Taub R. Rapid activation of post-hepatectomy factor/nuclear factor kappa B in hepatocytes, a primary response in the regenerating liver. J Biol Chem. 1994;269:30429–30435. [PubMed] [Google Scholar]
- 51.FitzGerald M.J., Webber E.M., Donovan J.R., Fausto N. Rapid DNA binding by nuclear factor kappa B in hepatocytes at the start of liver regeneration. Cell Growth Differ. 1995;6:417–427. [PubMed] [Google Scholar]
- 52.North T.E., Babu I.R., Vedder L.M., Lord A.M., Wishnok J.S., Tannenbaum S.R., Zon L.I., Goessling W. PGE2-regulated wnt signaling and N-acetylcysteine are synergistically hepatoprotective in zebrafish acetaminophen injury. Proc Natl Acad Sci U S A. 2010;107:17315–17320. doi: 10.1073/pnas.1008209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox A.G., Saunders D.C., Kelsey P.B., Jr., Conway A.A., Tesmenitsky Y., Marchini J.F., Brown K.K., Stamler J.S., Colagiovanni D.B., Rosenthal G.J., Croce K.J., North T.E., Goessling W. S-nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep. 2014;6:56–69. doi: 10.1016/j.celrep.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaeschke H., Xie Y., McGill M.R. Acetaminophen-induced liver injury: from znimal models to humans. J Clin Transl Hepatol. 2014;2:153–161. doi: 10.14218/JCTH.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He J.H., Guo S.Y., Zhu F., Zhu J.J., Chen Y.X., Huang C.J., Gao J.M., Dong Q.X., Xuan Y.X., Li C.Q. A zebrafish phenotypic assay for assessing drug-induced hepatotoxicity. J Pharmacol Toxicol Methods. 2013;67:25–32. doi: 10.1016/j.vascn.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Nourjah P., Ahmad S.R., Karwoski C., Willy M. Estimates of acetaminophen (Paracetomal)-associated overdoses in the United States. Pharmacoepidemiol Drug Saf. 2006;15:398–405. doi: 10.1002/pds.1191. [DOI] [PubMed] [Google Scholar]
- 57.Lorent K., Moore J.C., Siekmann A.F., Lawson N., Pack M. Reiterative use of the notch signal during zebrafish intrahepatic biliary development. Dev Dyn. 2010;239:855–864. doi: 10.1002/dvdy.22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dawson M.A., Prinjha R.K., Dittmann A., Giotopoulos G., Bantscheff M., Chan W.I., Robson S.C., Chung C.W., Hopf C., Savitski M.M., Huthmacher C., Gudgin E., Lugo D., Beinke S., Chapman T.D., Roberts E.J., Soden P.E., Auger K.R., Mirguet O., Doehner K., Delwel R., Burnett A.K., Jeffrey P., Drewes G., Lee K., Huntly B.J., Kouzarides T. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sugiyama M., Sakaue-Sawano A., Iimura T., Fukami K., Kitaguchi T., Kawakami K., Okamoto H., Higashijima S.I., Miyawaki A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A. 2009;106:20812–20817. doi: 10.1073/pnas.0906464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Apte U., Singh S., Zeng G., Cieply B., Virji M.A., Wu T., Monga S.P. Beta-catenin activation promotes liver regeneration after acetaminophen-induced injury. Am J Pathol. 2009;175:1056–1065. doi: 10.2353/ajpath.2009.080976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nicodeme E., Jeffrey K.L., Schaefer U., Beinke S., Dewell S., Chung C.W., Chandwani R., Marazzi I., Wilson P., Coste H., White J., Kirilovsky J., Rice C.M., Lora J.M., Prinjha R.K., Lee K., Tarakhovsky A. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu Y., Vakoc C.R. Brd4 is on the move during inflammation. Trends Cell Biol. 2014;24:615–616. doi: 10.1016/j.tcb.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chu J., Sadler K.C. New school in liver development: lessons from zebrafish. Hepatology. 2009;50:1656–1663. doi: 10.1002/hep.23157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J., Mowry L.E., Nejak-Bowen K.N., Okabe H., Diegel C.R., Lang R.A., Williams B.O., Monga S.P. beta-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation! Hepatology. 2014;60:964–976. doi: 10.1002/hep.27082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tao J., Calvisi D.F., Ranganathan S., Cigliano A., Zhou L., Singh S., Jiang L., Fan B., Terracciano L., Armeanu-Ebinger S., Ribback S., Dombrowski F., Evert M., Chen X., Monga S.P. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tao J., Xu E., Zhao Y., Singh S., Li X., Couchy G., Chen X., Zucman-Rossi J., Chikina M., Monga S.P. Modeling a human hepatocellular carcinoma subset in mice through coexpression of met and point-mutant beta-catenin. Hepatology. 2016;64:1587–1605. doi: 10.1002/hep.28601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tao J., Zhang R., Singh S., Poddar M., Xu E., Oertel M., Chen X., Ganesh S., Abrams M., Monga S.P. Targeting beta-catenin in hepatocellular cancers induced by coexpression of mutant beta-catenin and K-Ras in mice. Hepatology. 2017;65:1581–1599. doi: 10.1002/hep.28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delgado E., Okabe H., Preziosi M., Russell J.O., Alvarado T.F., Oertel M., Nejak-Bowen K.N., Zhang Y., Monga S.P. Complete response of Ctnnb1-mutated tumours to beta-catenin suppression by locked nucleic acid antisense in a mouse hepatocarcinogenesis model. J Hepatol. 2015;62:380–387. doi: 10.1016/j.jhep.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tögel L., Nightingale R., Chueh A.C., Jayachandran A., Tran H., Phesse T., Wu R., Sieber O.M., Arango D., Dhillon A.S., Dawson M.A., Diez-Dacal B., Gahman T.C., Filippakopoulos P., Shiau A.K., Mariadason J.M. Dual targeting of bromodomain and extraterminal domain proteins, and WNT or MAPK signaling, inhibits c-MYC expression and proliferation of colorectal cancer cells. Mol Cancer Ther. 2016;15:1217–1226. doi: 10.1158/1535-7163.MCT-15-0724. [DOI] [PubMed] [Google Scholar]
- 70.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Parker S.C., Stitzel M.L., Taylor D.L., Orozco J.M., Erdos M.R., Akiyama J.A., van Bueren K.L., Chines P.S., Narisu N., NISC Comparative Sequencing Program. Black B.L., Visel A., Pennacchio L.A., Collins F.S., National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program Authors. NISC Comparative Sequencing Program Authors Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hnisz D., Schuijers J., Lin C.Y., Weintraub A.S., Abraham B.J., Lee T.I., Bradner J.E., Young R.A. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kennedy A.L., Vallurupalli M., Chen L., Crompton B., Cowley G., Vazquez F., Weir B.A., Tsherniak A., Parasuraman S., Kim S., Alexe G., Stegmaier K. Functional, chemical genomic, and super-enhancer screening identify sensitivity to cyclin D1/CDK4 pathway inhibition in Ewing sarcoma. Oncotarget. 2015;6:30178–30193. doi: 10.18632/oncotarget.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.