Abstract
p53 is the major cellular gatekeeper involved in proliferation, cell death, migration, and homeostasis. The role of p53 in pathogenesis of drug-induced liver injury is unknown. We investigated the role of p53 in liver injury and regeneration after acetaminophen (APAP) overdose, the most common cause of acute liver failure in the Western world. Eight-week-old male wild-type (WT) and p53 knockout (p53KO) mice were treated with 300 mg/kg APAP, and the dynamics of liver injury and regeneration were studied over a time course of 0 to 96 hours. Deletion of p53 resulted in a threefold higher liver injury than in WT mice. Interestingly, despite higher liver injury, p53KO mice recovered similarly as the WT mice because of faster liver regeneration. Deletion of p53 did not affect APAP bioactivation and initiation of injury. Microarray analysis revealed that p53KO mice had disrupted metabolic homeostasis and induced inflammatory and proliferative signaling. p53KO mice showed prolonged steatosis correlating with prolonged liver injury. Initiation of liver regeneration in p53KO mice was delayed, but once initiated, cell cycle was significantly faster than WT mice because of sustained AKT, extracellular signal–regulated kinase, and mammalian target of rapamycin signaling. These studies show that p53 plays a pleotropic role after APAP overdose, where it prevents progression of liver injury by maintaining metabolic homeostasis and also regulates initiation of liver regeneration through proliferative signaling.
p53 Functions as a cellular gatekeeper involved in regulation of various stress responses, including cell proliferation, migration, metabolism, autophagy, DNA repair, senescence, immune response, and stem cell reprogramming. Because of these multifaceted functions, deregulation of p53 is implicated in various diseases, such as cancer, obesity, diabetes, ischemia, and aging.1, 2, 3, 4, 5, 6 Recent studies in rodents using partial hepatectomy model have depicted role of p53 in regulation of ploidy and cell proliferation during liver regeneration.7, 8, 9, 10, 11 Liver regeneration is essential for patient survival after surgical resection or acute and chronic liver injury secondary to drugs, toxins, and viruses.12 However, role of p53 in liver homeostasis and regeneration after drug-induced liver injury is unknown.
Overdose of acetaminophen (APAP), the commonly used antipyretic and analgesic agent, is the leading cause of acute liver failure in the Western world.13, 14, 15 The mechanism of liver injury after APAP overdose involves formation of the reactive metabolite N-acetyl-p-quinoneimine, which depletes cellular glutathione and covalently binds to protein. These initial events lead to oxidative stress, activation of c-Jun N-terminal kinase (JNK), mitochondrial damage, and release of cytochrome c, apoptosis-inducing factor, and endonuclease that cause nuclear DNA fragmentation and eventually hepatocyte necrosis.16 The replacement of necrotic cells and restoration of liver function occur through liver regeneration. Several studies have shown that enhanced liver regeneration after APAP-induced liver injury improves final outcome.17, 18
Although recent studies have implicated p53 in pathogenesis of APAP-induced liver injury, the exact role of p53 in initiation and progression of injury and the subsequent liver regeneration after APAP overdose is unclear.19, 20 In this study, we investigated the role of p53 in liver injury and regeneration using APAP overdose in wild-type (WT) and p53 knockout (p53KO) mice. Our findings reveal that p53 regulates metabolic homeostasis, and initiation of cell proliferation and disruption of p53 signaling culminate in progression of liver injury, yet lead to faster recovery.
Materials and Methods
Animals, Treatments, and Tissue Harvesting
Two-month-old male WT (p53+/+) and p53KO (p53−/−) mice on C57BL/6J background, purchased from Jackson Laboratory (Bar Harbor, ME), were used in these studies. All mice were housed in Association for Assessment and Accreditation of Laboratory Animal Care–accredited facilities at the University of Kansas Medical Center (Kansas City) under a standard 12-hour light/dark cycle, with access to chow and water ad libitum. All animal experiments were performed with approved Institutional Animal Care and Use Committee protocols at University of Kansas Medical Center. Acute liver injury was induced by injecting 300 mg/kg APAP intraperitoneally (dissolved in saline) after 12 hours fasting, as described previously.17 After APAP treatment, mice (n = 3 to 5) were sacrificed at 0, 1, 12, 24, 48, and 96 hours, and blood and livers were collected. Serum samples obtained from blood were used for alanine aminotransferase (ALT) and glucose level measurement using the Infinity ALT (GPT) and the Infinity Glucose kit (Thermo Fisher Scientific, Middletown, VA), as per the manufacturer's protocol. Parts of liver tissue were processed separately to obtain paraffin sections, frozen sections, RNA samples, and nuclear, cytoplasmic, and total protein extracts, as described previously.21
Immunohistochemistry and Staining Procedures
Hepatocyte proliferation was determined using proliferating cell nuclear antigen (PCNA) immunohistochemical staining, and liver necrosis was determined by hematoxylin and eosin staining, as previously described.21 Fresh-frozen liver sections (5 μm thick) were used to stain lipid droplets using Oil Red O, as described before.21
Western Blotting and Real-Time PCR
Protein estimation and Western blot analysis were performed using pooled protein extracts, as described before.21 The antibodies used in this study are listed in Table 1. Total RNA isolation from WT and p53KO liver, reverse transcription to cDNA, and real-time PCR analysis by SYBR Green (Life Technologies, Carlsbad, CA) technology were performed as previously described.17 18s gene expression in the same samples was used for data normalization, and mRNA levels were determined by comparing post-APAP treatment time points with 0 hour control of respective group. Primers used for real-time PCR are listed in Table 2. Densitometric analysis of the Western blots was conducted using the UN-SCAN-IT software version 5.3 (Silk Scientific Corporation, Salt Lake City, UT).
Table 1.
Antibodies Used in this Study
| Name | Manufacturer | Catalog number |
|---|---|---|
| PCNA | Cell Signaling (Danvers, MA) | 2586 |
| Phospho-JNK | Cell Signaling | 4668 |
| JNK | Cell Signaling | 3708 |
| Cyclin D1 | Cell Signaling | 2978 |
| Phospho-pRb | Cell Signaling | 9308 |
| p21 | Santa Cruz Biotechnology (Dallas, TX) | 6246 |
| GAPDH | Cell Signaling | 2118 |
| CYP2E1 | Abcam (Cambridge, MA) | 19140 |
| SREBP2 | Thermo Fisher Scientific | PA1338 |
| TFAM | Cell Signaling | 8076 |
| NRF1 | Cell Signaling | 12381 |
| Mitochondrial complex | Mitosciences (Eugene, OR) | MS604 |
| Phospho-AKT | Cell Signaling | 9271 |
| AKT | Cell Signaling | 4691 |
| Phospho-ERK | Cell Signaling | 4376 |
| Phospho-EGFR | Cell Signaling | 1068 |
| EGFR | Cell Signaling | 4267 |
| Phospho-mTOR Ser 2448 | Cell Signaling | 2971 |
| Phospho-mTOR Ser 2481 | Cell Signaling | 2974 |
| mTOR | Cell Signaling | 2983 |
| Phospho-P70S6K | Cell Signaling | 9234 |
| P70S6K | Cell Signaling | 9202 |
CYP2E1, cytochrome P450 2E1; EGFR, epidermal growth factor receptor; ERK, extracellular signal–regulated kinase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; JNK, c-Jun N-terminal kinase; mTOR, mammalian target of rapamycin; NRF1, nuclear respiratory factor 1; P70S6K, ribosomal protein S6 kinase B1; PCNA, proliferating cell nuclear antigen; phospho-pRb, phospho-retinoblastoma protein; SREBP2, sterol regulatory element-binding protein 2; TFAM, transcription factor A, mitochondrial.
Table 2.
Primers Used in this Study
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Acot1 | 5′-ATACCCCCTGTGACTATCCTGA-3′ | 5′-CAAACACTCACTACCCAACTGT-3′ |
| Cd14 | 5′-CTCTGTCCTTAAAGCGGCTTAC-3′ | 5′-GTTGCGGAGGTTCAAGATGTT-3′ |
| Cyp7a1 | 5′-GGGATTGCTGTGGTAGTGAGC-3′ | 5′-GGTATGGAATCAACCCGTTGTC-3′ |
| Fasn | 5′-GGAGGTGGTGATAGCCGGTAT-3′ | 5′-TGGGTAATCCATAGAGCCCAG-3′ |
| Fdps | 5′-GGAGGTCCTAGAGTACAATGCC-3′ | 5′-AAGCCTGGAGCAGTTCTACAC-3′ |
| Fos | 5′-CGGGTTTCAACGCCGACTA-3′ | 5′-TTGGCACTAGAGACGGACAGA-3′ |
| Hmox1 | 5′-AAGCCGAGAATGCTGAGTTCA-3′ | 5′-GCCGTGTAGATATGGTACAAGGA-3′ |
| Elovl3 | 5′-TTCTCACGCGGGTTAAAAATGG-3′ | 5′-GAGCAACAGATAGACGACCAC-3′ |
| Ly6d | 5′-GCCTGGGCACTTCGATGTC-3′ | 5′-TGAGTTTGCACACTCTTTCC-3′ |
| Igfbp3 | 5′-CCAGGAAACATCAGTGAGTCC-3′ | 5′-GGATGGAACTTGGAATCGGTCA-3′ |
| Acot2 | 5′-GTTGTGCCAACAGGATTGGAA-3′ | 5′-GCTCAGCGTCGCATTTGTC-3′ |
| Afp | 5′-CTTCCCTCATCCTCCTGCTAC-3′ | 5′-ACAAACTGGGTAAAGGTGATGG-3′ |
| Cd36 | 5′-GCGCATGATTAATGGCACAG-3′ | 5′-GATCCGAACACAGCGTAGATAG-3′ |
| Scd1 | 5′-CTACAAGCCTGGCCTCCTGC-3′ | 5′-GGACCCCAGGGAAACCAGGA-3′ |
| Pdk4 | 5′-AGGGAGGTCGAGCTGTTCTC-3′ | 5′-GGAGTGTTCACTAAGCGGTCA-3′ |
| Stat1 | 5′-TCACAGTGGTTCGAGCTTCAG-3′ | 5′-GCAAACGAGACATCATAGGCA-3′ |
| Insig2 | 5′-GGAGTCACCTCGGCCTAAAAA-3′ | 5′-CAAGTTCAACACTAATGCCAGGA-3′ |
| G0s2 | 5′-TAGTGAAGCTATACGTTCTGGGC-3′ | 5′-GTCTCAACTAGGCCGAGCA-3′ |
| Fgf21 | 5′-CTGCTGGGGGTCTACCAAG-3′ | 5′-CTGCGCCTACCACTGTTCC-3′ |
| Lpin1 | 5′-CATGCTTCGGAAAGTCCTTCA-3′ | 5′-GGTTATTCTTTGGCGTCAACCT-3′ |
| Gadd45α | 5′-CCGAAAGGATGGACACGGTG-3′ | 5′-TTATCGGGGTCTACGTTGAGC-3′ |
| Gadd45β | 5′-CAACGCGGTTCAGAAGATGC-3′ | 5′-GGTCCACATTCATCAGTTTGGC-3′ |
| Gadd45γ | 5′-GGGAAAGCACTGCACGAACT-3′ | 5′-AGCACGCAAAAGGTCACATTG-3′ |
| Bax | 5′-TGAAGACAGGGGCCTTTTTG-3′ | 5′-AATTCGCCGGAGACACTCG-3′ |
| Birc5 | 5′-AAGGAATTGGAAGGCTGGG-3′ | 5′-TTCTTGACAGTGAGGAAGGC-3′ |
| Btg2 | 5′-ATGAGCCACGGGAAGAGAAC-3′ | 5′-GCCCTACTGAAAACCTTGAGTC-3′ |
| Cpt1a | 5′-CTCCGCCTGAGCCATGAAG-3′ | 5′-CACCAGTGATGATGCCATTCT-3′ |
| Gpx1 | 5′-AGTCCACCGTGTATGCCTTCT-3′ | 5′-GAGACGCGACATTCTCAATGA-3′ |
| Gpx3 | 5′-CCTTTTAAGCAGTATGCAGGCA-3′ | 5′-CAAGCCAAATGGCCCAAGTT-3′ |
| Nanog | 5′-TCTTCCTGGTCCCCACAGTTT-3′ | 5′-GCAAGAATAGTTCTCGGGATGAA-3′ |
| Sco2 | 5′-AGCTCTCTCAGTTCAAACCCC-3′ | 5′-GCAGTCTAGTTCTTAGCCCAGG-3′ |
| Sod2 | 5′-TGCTCTAATCAGGACCCATTG-3′ | 5′-CATTCTCCCAGTTGATTACATTCC-3′ |
| Tgfβ | 5′-AGCTGGTGAAACGGAAGCG-3′ | 5′-GCGAGCCTTAGTTTGGACAGG-3′ |
| Il1β | 5′-GAAATGCCACCTTTTGACAGTG-3′ | 5′-CTGGATGCTCTCATCAGGACA-3′ |
| Il10 | 5′-GCTCTTACTGACTGGCATGAG-3′ | 5′-CGCAGCTCTAGGAGCATGTG-3′ |
Measurement of Hepatic GSH and APAP Protein Adducts
Total hepatic glutathione (GSH) was measured in liver homogenates using the commercially available colorimetric assay kit (Millipore Sigma, St. Louis, Mo), as described before.22 Liver GSH and oxidized GSH levels were measured using a modified Tietze assay, as described.23 APAP protein adducts were measured in liver and serum samples using high-performance liquid chromatography method, as previously described.23
Transcriptome and Pathway Analysis
Total RNA samples isolated from WT and p53KO livers at 12, 24, and 48 hours after APAP treatment were used for global transcriptomic analysis. Equal amount of RNA was pooled from three representative samples at each time point from both groups. RNA integrity was determined on the Agilent Bioanalyzer 2100 (Agilent Technologies, Sanata Clara, CA) using the RNA6000 Nano assay kit VII (Agilent Technologies), and global transcriptomic analysis was performed using Affymetrix Clariom D (Thermo Fisher Scientific) array at the Genomics Core Facility of the University of Kansas Medical Center. Hierarchical clustering and heat map analyses of differential expression data of transcripts were performed as described previously.24 Ingenuity Pathway Analysis (Qiagen, Redwood City, CA) was used for functional pathway analysis of differentially expressed transcripts.
Mitochondrial DNA Measurement
Genomic and mitochondrial DNA was isolated using commercially available kit (Qiagen Inc.). Real-time PCR analysis was used to quantify relative copy numbers of genomic DNA and mitochondrial DNA (mito DNA) in WT and p53KO mice livers from a 0- to 96-hour time course. lipoprotein lipase as nuclear DNA target and NADH dehydrogenase complex 1 as mito DNA target were used for mito DNA content measurement. Exponential amplification efficiency of each primer pair was confirmed using series of dilution, and 5 ng of DNA was used in final PCR. Mito DNA content in each sample was calculated as described previously.25 Relative mito DNA fold change was calculated by dividing mito DNA value of a sample by average mito DNA value in WT at respective time point.
Statistical Analysis
All data are shown as means ± SEM. One-way analysis of variance and t-test were used for statistical analysis. Difference between groups was considered statistically significant at P < 0.05.
Results
p53 Deletion Results in Progression of APAP-Induced Injury and Faster Recovery
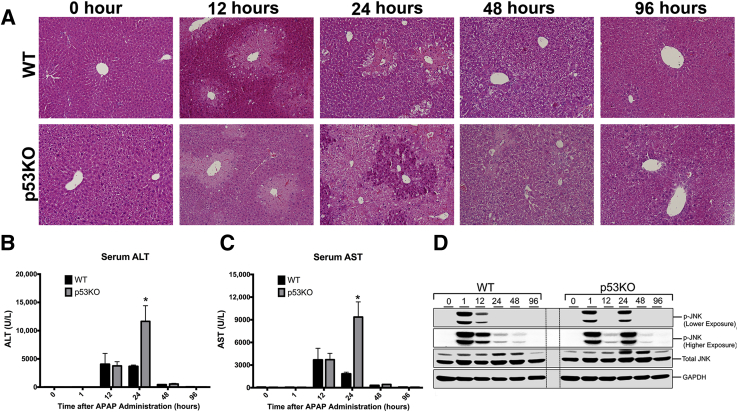
APAP treatment resulted in significant liver injury in both WT and p53KO mice, as demonstrated by histopathological (Figure 1A), as well as serum ALT and aspartate aminotransferase (Figure 1, B and C), analysis. Liver sections stained with hematoxylin and eosin showed necrotic cell injury at 12 and 24 hours in WT animals, which reduced by 48 hours. Serum ALT levels corroborated the histopathological findings. Liver injury was similar in WT and p53KO mice until 12 hours. However, at 24 hours, p53KO mice exhibited significantly higher liver injury, as demonstrated by increase in necrotic area and threefold higher serum ALT and aspartate aminotransferase. Interestingly, despite high liver injury, p53KO mice recovered similarly to WT mice at 48 hours after APAP treatment. c-Jun N-terminal kinase activation (phosphorylation) is a critical step in APAP pathophysiology. Western blot analysis of phospho-c-Jun N-terminal kinase revealed c-Jun N-terminal kinase activation was prolonged until 24 hours after APAP treatment in p53KO than WT group (Figure 1D).
Figure 1.
Deletion of p53 aggravates APAP-induced acute liver injury. A: Representative photomicrographs of hematoxylin and eosin–stained liver sections. B and C: Serum alanine aminotransferase (ALT; B) and serum aspartate aminotransferase (AST; C) levels. D: Western blot analysis of phospho-c-Jun N-terminal kinase (JNK) and JNK. ∗P < 0.05 versus WT. Original magnification, ×200 (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
p53 Deletion Results in Delayed but Faster Liver Regeneration
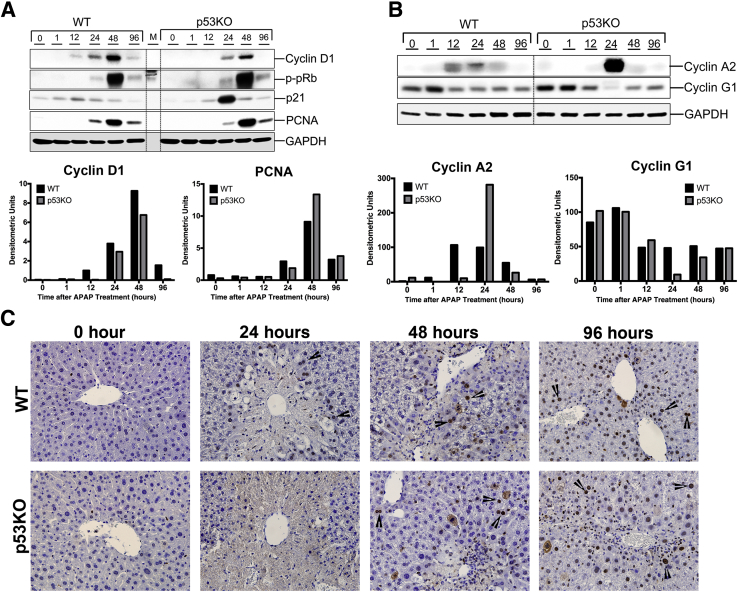
WT mice showed moderate increase in cell proliferation by 24 hours, accompanied by increased expression of cell cycle proteins cyclin D1, cyclin A2, phospho-pRb, and PCNA (Figure 2, A and B). PCNA staining of liver sections also revealed increased cell proliferation surrounding necrotic zone by 24 hours in WT group (Figure 2C). On the contrary, p53KO animals showed delayed onset of cell proliferation, as demonstrated by delay in appearance of PCNA-positive cells (Figure 2C) and delayed increase in cyclin D1 and cyclin A2 expression after APAP treatment (Figure 2, A and B). At 48 hours after APAP, cyclin D1, cyclin A2, phospho-pRb, and PCNA protein levels were comparable to WT (Figure 2, A and B). Interestingly, expression of cell cycle inhibitors was as follows: p21 was higher, whereas cyclin G1 was lower, in p53KO group at 24 hours than WT (Figure 2A).
Figure 2.
Deletion of p53 promotes faster recovery despite higher injury. A: Western blot analysis of cyclin D1, phospho-pRb, p21, and proliferating cell nuclear antigen (PCNA) and corresponding densitometric analysis. B: Western blots of cyclin A2 and cyclin G1 and corresponding densitometric analysis. All Western blots were performed using total liver extracts. C: Representative photomicrographs of PCNA-stained livers. Arrowheads point to PCNA-positive cells. Original magnification, ×400 (C). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
p53 Deletion Does Not Change APAP Bioactivation but Delays APAP-Adduct Secretion in Serum
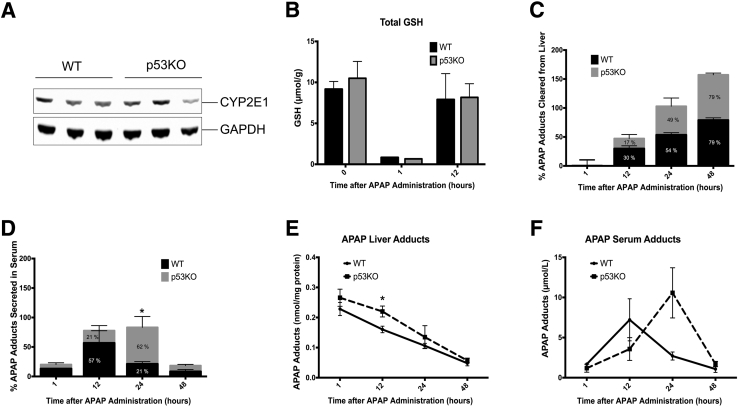
Because higher injury in p53KO mice was seen, total hepatic GSH, cytochrome P450 2E1, and APAP adducts,23, 26 which are involved in initiation of APAP toxicity, were measured to determine whether changes in bioactivation of APAP can explain the higher injury. CYP2E1 protein expression was not different between two groups (Figure 3A). Total GSH levels before APAP treatment were similar and depleted GSH to same extent 1 hour after APAP administration in both groups. Interestingly, GSH replenishment, known to be involved in injury progression, was not altered by p53 deletion as well (Figure 3B). Also, no difference was observed in APAP adduct formation in liver at 1 hour after 300 mg/kg APAP (APAP300) treatment in p53KO mice compared with WT mice (Figure 3E). All these data indicate that deletion of p53 in mice does not affect APAP bioactivation. Although adduct formation at 1 hour was same in p53KO and WT mice, liver adducts were significantly high in p53KO mice at 12 hours compared with WT mice (Figure 3E), suggesting prolonged accumulation of adducts in liver. However, adduct clearance from liver was not significantly different in both groups at 12 hours (Figure 3C). Interestingly, prolonged accumulation of liver adducts at 12 hours was accompanied by delay in peak serum adduct secretion in p53KO mice (Figure 3F). In WT mice, maximum (57% of total serum adducts secreted from 1 through 48 hours) adduct secretion in serum was observed at 12 hours; however, in p53KO mice, maximum adduct secretion (62%) was delayed up until 24 hours after APAP treatment (Figure 3D).
Figure 3.
p53 Deletion does not change APAP bioactivation but delays APAP-adduct secretion in the serum. A: Western blot of cytochrome P450 2E1 (CYP2E1) using total liver extract from untreated WT and p53KO mice. B–D: Bar graph showing hepatic total glutathione (GSH) levels (B), clearance of APAP-cysteine adducts from liver (C), and APAP-cysteine adduct secretion in serum (D). E and F: Line graph shows actual liver adduct levels (E) and adduct levels in serum (F). ∗P < 0.05 WT versus p53KO. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Transcriptomics Analysis Reveals Expression Patterns within Each Group
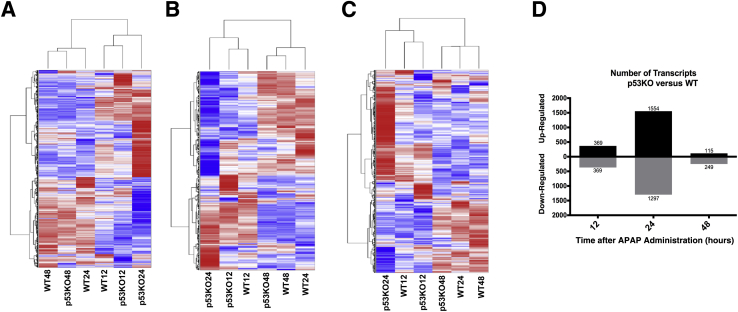
Global transcriptomic analysis was performed to delineate mechanisms through which p53 regulates progression of liver injury and initiation of regeneration. No difference was observed in liver injury between WT and p53KO mice until 12 hours. However, significant difference was observed at 24 hours in injury progression and initiation of liver regeneration after APAP treatment. Therefore, three time points, including 12, 24, and 48 hours, were chosen for transcriptome analysis. p53KO transcript expression was normalized to WT at respective time points. All (n = 3495) transcripts with twofold or greater up-regulated or down-regulated expression at least at one time point were selected. Next, coding (n = 676) and noncoding (n = 1,439) transcripts individually among 3495 transcripts were sorted, and hierarchical clustering analysis was performed on their differential expression data (Figure 4, A–C). Dendrograms generated by cluster analysis of all selected transcripts (Figure 4A), coding transcripts (Figure 4B), and noncoding transcripts (Figure 4C) revealed two clusters of expression pattern. Cluster 1 consisted of WT 48, p53KO 48, and WT 24, whereas cluster 2 consisted of WT 12, p53KO 12, and p53KO 24. Within cluster 1, transcript expression pattern of WT 48 and p53KO 48 was closely related, indicating similarity of both groups at pathophysiological level, which is consistent with injury and recovery data. Interestingly, WT 24 transcript expression pattern varied slightly but was similar to WT 48 and p53KO 48. Therefore, WT group begins to express cluster 1 pattern at 24 hours, and this continues until 48 hours after APAP treatment. However, deletion of p53 delayed transcript expression pattern occurs from 24 to 48 hours, which is also consistent with observations of liver regeneration. Within cluster 2, expression pattern of WT 12 and p53KO 12 was closely related, indicating similar pathophysiological condition in both groups. p53KO 24 expression pattern is related to WT 12 and p53KO 12 but varied highly in magnitude, which is also consistent with observations of liver injury. Finally, transcript expression pattern of WT 24 and p53KO 24 was significantly contrasting in all transcripts (Figure 4A), coding transcripts (Figure 4B), and noncoding transcripts (Figure 4C). This is consistent with the threefold difference in liver injury and delayed initiation of liver regeneration observed in p53KO mice compared with WT mice at 24-hour time point. Furthermore, selection of up-regulated and down-regulated transcripts at individual time point with expression twofold or greater in p53KO than WT revealed that most transcripts were significantly affected at 24 hours (Figure 4D). These data show that p53KO mice respond similarly to WT mice until 12- and 48-hour time points after APAP treatment but differ significantly at 24 hours after APAP administration.
Figure 4.
Transcriptomic expression patterns in WT and p53KO mice liver after 300 mg/kg APAP (APAP300) treatment. Global transcriptome analysis was performed using Affymetrix Clariom D array. A–C: Biweight (log2) values were used for cluster analysis and presented as heat maps of all selected transcripts (A), only coding transcripts (B), and only noncoding transcripts (C). Every group title represents the genotype and the time point after APAP300 treatment. D: Number of transcripts in p53KO mice at respective time point with twofold or greater change with respect to WT mice.
Functional Analysis of Transcriptomic Data Reveals Changes in Metabolic Homeostasis, Inflammation, and Proliferation
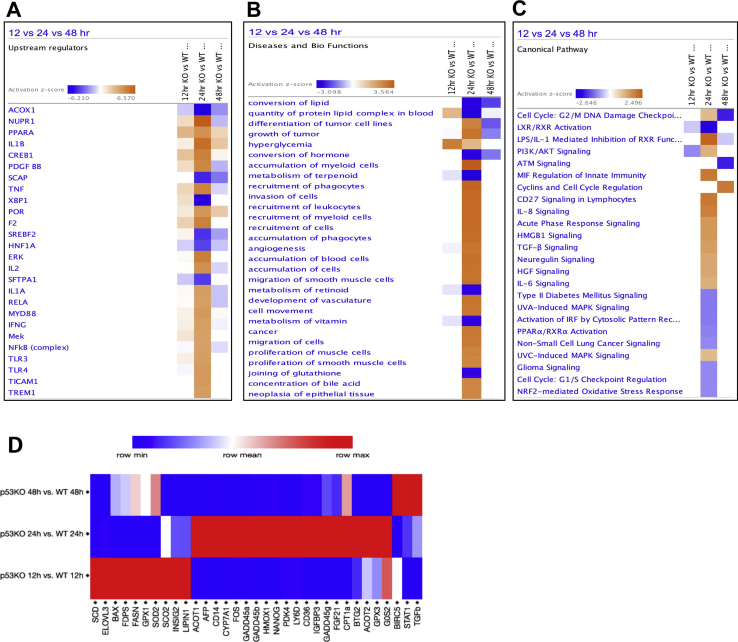
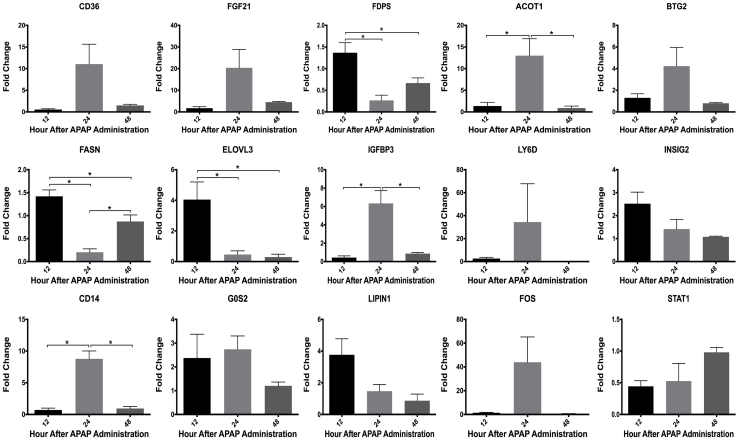
The transcript expression data were further analyzed using Core Analysis in Ingenuity Pathway Analysis, followed by comparison analysis between three time points. This analysis revealed differentially regulated upstream regulators, biofunctions, and canonical pathways (Supplemental Figure S1, A–C, and Supplemental Table S1). The analysis categorized the transcript expression changes into three groups: disrupted metabolic homeostasis [upstream regulators: Acyl CoA oxidase 1, peroxisome proliferator-activated receptor α, and sterol regulatory element-binding protein 2 (SREBP2)], increased inflammatory signaling (upstream regulators: IL-1β, tumor necrosis factor-α, IL-1α, NF-κB complex, and toll-like receptors), and proliferative signaling (upstream regulators: cAMP responsive element binding protein 1, extracellular signal–regulated kinase, tumor necrosis factor-α, and nuclear protein 1, transcription regulator). Next, transcriptomic gene expression data of selected genes chosen from these three categories were validated using real-time PCR analysis (Supplemental Figures S1D and S2).
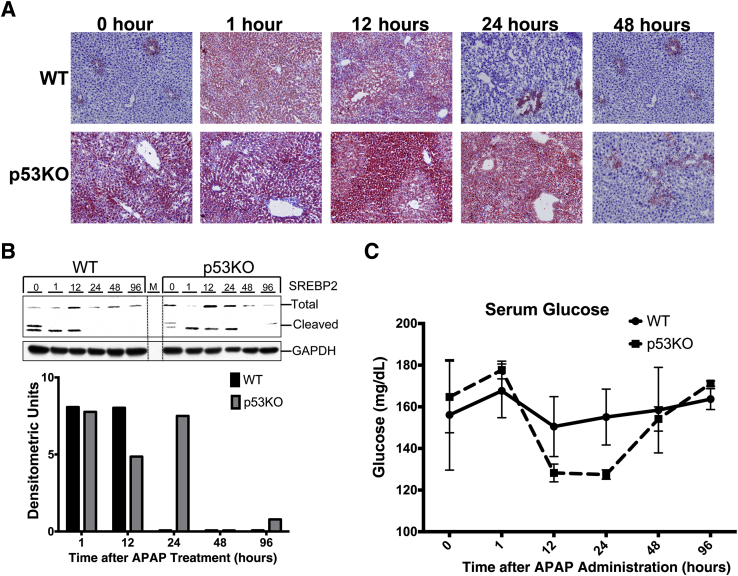
Deletion of p53 Induces Prolonged Steatosis after APAP300 Treatment in Mice
The transcriptomic data indicated that p53KO mice had changes in metabolic homeostasis after APAP administration. Therefore, hepatic lipid content and serum glucose levels were analyzed. Oil Red O staining of liver sections revealed that APAP treatment induced steatosis at 1 and 12 hours, and it was resolved by 24 hours, in WT mice (Figure 5A). Livers of p53KO mice showed higher steatosis at 0 hour than livers of WT mice, which further increased after APAP treatment, remained markedly higher than WT mice until 24 hours, and disappeared at 48 hours. Steatosis resolution was delayed by 24 hours in p53KO mice compared with WT mice (Figure 5A). SREBP2 is the major lipid sensor in the hepatocytes. The prolonged steatosis in p53KO mice was accompanied by prolonged SREBP2 cleavage (Figure 5B). However, serum glucose levels in p53KO mice were comparable to WT mice throughout the time course (Figure 5C).
Figure 5.
Prolonged steatosis in p53KO mice liver after 300 mg/kg APAP treatment. A: Representative photomicrographs of Oil Red O–stained frozen liver sections. B: Western blots of SREBP2 and cleaved SREBP2 using total liver extracts and bar graph of densitometry of the cleaved SREBP2 blot. C: Serum glucose levels from WT and p53KO mice over a time course of 0 to 96 hours after APAP administration. Original magnification, ×200 (A). GAPDH, glyceraldehyde-3-phosphate dehydrogenase; M, marker lane; SREBP2, sterol regulatory element-binding protein 2.
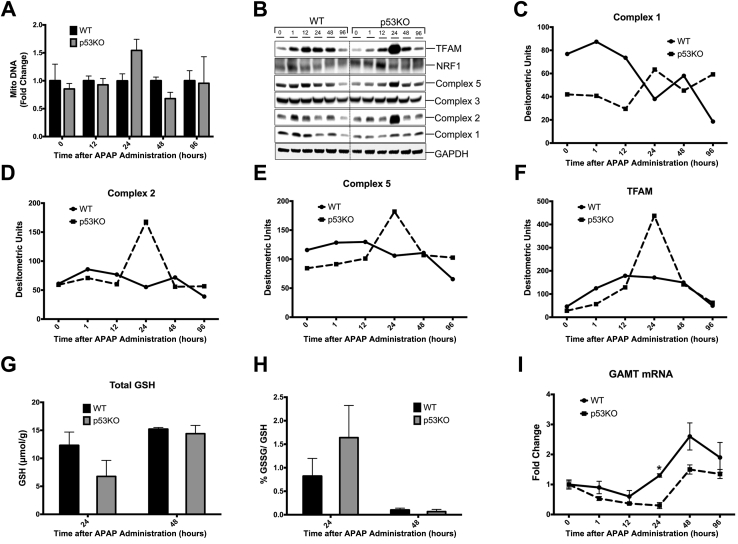
Deletion of p53 Reduces Expression of Mitochondrial Complexes
Mitochondria are central to APAP-induced injury.27 Studies have shown role of p53 in mitochondrial biogenesis and function.28 To investigate effect of p53 deletion after APAP treatment on number of mitochondria, mitochondrial DNA content was measured in mice liver. No significant difference was observed in mitochondrial DNA content between WT and p53KO mice liver (Figure 6A). Furthermore, Western blot analysis of mitochondrial complexes revealed that p53KO mice exhibited markedly low expression of complexes 1, 2, and 5 up to 24 hours than WT mice (Figure 6, B–E). Transcription factor A, mitochondrial and nuclear respiratory factor 1 are the two transcription factors that regulate expression of mitochondrial complexes.29 Protein expression of nuclear respiratory factor 1 was similar between two groups; however, transcription factor A, mitochondrial levels were low initially during the first 12 hours after APAP treatment but were induced at 24 hours after APAP treatment in p53KO than WT mice, correlating with induction of complexes 1, 2, and 5 at 24 hours (Figure 6, B and F).
Figure 6.
Reduced mitochondrial complexes during injury phase after 300 mg/kg APAP treatment in p53KO mice. A: Bar graph showing mitochondrial DNA (Mito DNA) content. B: Western blot of mitochondrial complexes 1, 2, 3, and 5 and their regulatory transcription factors TFAM and NRF1 using total liver extracts from the respective groups. C–F: Line graphs showing densitometric analysis of Western blots of complex 1 (C), complex 2 (D), complex 5 (E), and TFAM (F). G and H: Bar graph showing hepatic total glutathione (GSH) levels (G) and %oxidized GSH (GSSG)/GSH ratio (H). I: Real-time PCR analysis of Gamt mRNA. ∗P < 0.05 versus p53KO. GAPDH, glyceraldehyde-3-phosphate dehydrogenase NRF1, nuclear respiratory factor 1; TFAM, transcription factor A, mitochondrial.
It was further studied if p53KO mice have increased oxidative stress at 24 hours by measuring hepatic total GSH level and %oxidized GSH/GSH value. However, there was no significant difference in total GSH levels and %oxidized GSH/GSH value at 24 hours (Figure 6, G and H). One of the target genes of p53 is an enzyme called guanidinoacetate methyltransferase (Gamt), which is involved in GSH synthesis by maintaining cysteine pool.30 mRNA levels of Gamt expression were measured, which were significantly down-regulated at 24 hours after APAP treatment in p53KO mice, correlating with higher injury at 24 hours than WT mice (Figure 6I).
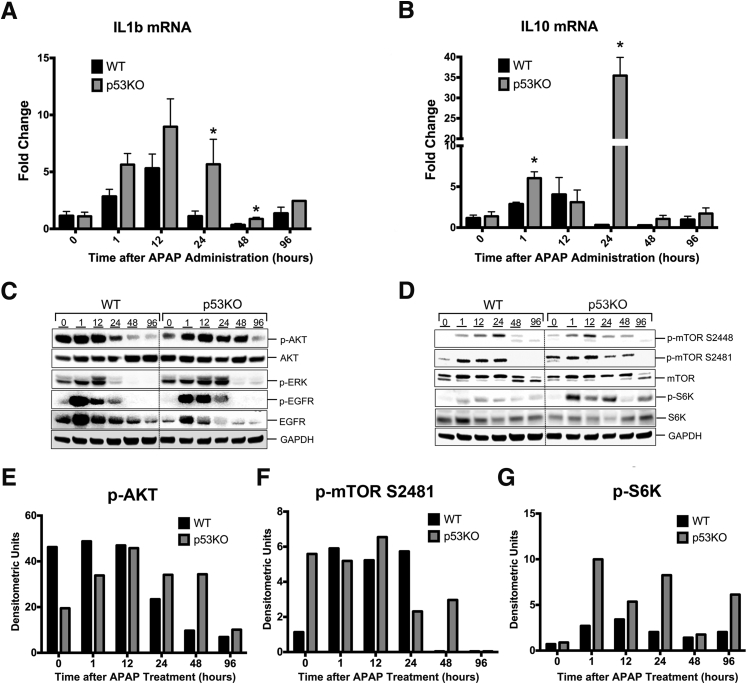
p53KO Mice Exhibit Increased Inflammation after APAP Treatment
The data on liver injury (serum ALT and hematoxylin and eosin) and repair (PCNA staining and cell cycle markers) revealed that despite threefold higher liver injury, p53KO mice still recover at the same time as WT mice. On the basis of the transcriptome analysis, we hypothesized that faster recovery, despite high injury, in p53KO mice is because of higher inflammatory and proliferative signaling. Consistently, p53KO mice exhibited significantly higher Il1b mRNA levels at 24 and 48 hours after APAP treatment compared with WT mice (Figure 7A). Likewise, Il10 mRNA levels were significantly induced at 1 and 24 hours after APAP treatment in p53KO mice than WT mice (Figure 7B). Previous studies have shown importance of both cytokines IL-1β and IL-10 in liver injury repair.31 These data indicate that p53 deletion augments cytokine signaling, which may result in faster recovery.
Figure 7.
Deletion of p53 augments inflammatory and proliferative signaling during recovery phase after APAP overdose. Real-time PCR analysis of Il1β (A) and Il10 (B). Western blot analysis of phospho-AKT, total AKT, phospho-extracellular signal–regulated kinase (ERK), phospho-epidermal growth factor receptor (EGFR), and total EGFR (C), and phospho-mammalian target of rapamycin (mTOR) Ser2448, phospho-mTOR Ser2481, total mTOR, phospho-p70S6K, and total p70S6K (D) using total liver extracts from the respective groups. Densitometric analysis of Western blots of phospho-AKT (E), phospho-mTOR2481 (F), and phospho-S6K (G). ∗P < 0.05 versus WT. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Enhanced Liver Regeneration in p53KO Mice after APAP Overdose
The mechanisms of the enhanced regenerative response in p53KO mice were further studied by measuring activation of key pathways involved in liver regeneration. Western blot analysis of total and activated (phosphorylated) AKT showed significantly higher and prolonged activation in p53KO mice after APAP treatment (Figure 7, C and E). Likewise, deletion of p53 resulted in prolonged phosphorylation of extracellular signal–regulated kinase until 24 hours after APAP300 treatment. p53 Is known to regulate epidermal growth factor receptor at transcriptional level.32 Consistent with this reduced expression of epidermal growth factor receptor, total protein was observed in p53KO mice versus WT mice at all time points. However, p53KO mice showed sustained epidermal growth factor receptor phosphorylation after APAP300 treatment (Figure 7C). These data indicate that p53 deletion resulted in sustained proliferative signaling via AKT and extracellular signal–regulated kinase, which may enhance faster regeneration.
Finally, mammalian target of rapamycin (mTOR) pathway was studied, which plays a critical role in lipid homeostasis, cell proliferation, mitochondrial biogenesis, and autophagy. p53 Is known to inhibit mTOR through AMP activated protein kinase, phosphatase and tensin homolog, and TSC complex subunit 2 expression33; hence, we hypothesized that deletion of p53 will increase activation of mTOR pathway. Western blot analysis of total mTOR protein showed no significant difference at all time points in both WT and p53KO groups. APAP300 treatment induced phosphorylation of mTOR (activation) in both groups; however, phosphorylation was prolonged in p53KO group (Figure 7, D and F). Phosphorylation of ribosomal protein S6 kinase, a downstream target of mTOR, was significantly higher and sustained in p53KO mice than WT mice throughout time course, indicating higher and prolonged activation of mTOR pathway (Figure 7, D and G).
Discussion
p53 Is primarily known as a tumor suppressor because of its ability to inhibit cell proliferation. However, there is increasing evidence that p53 can regulate various cell responses, such as migration, metabolism, oxidation-reduction balance, senescence, and stem cell reprogramming.1, 2, 3, 4, 5, 6 Recently, Huo et al20 have shown protective role of p53 in APAP-induced liver injury, but the exact mechanisms of this protection are not known. Our study strengthened this finding and further revealed various mechanisms by which p53 regulates injury progression and initiation of liver regeneration.
The pathogenesis of APAP-induced acute liver failure can be divided into three phases, including initiation of injury phase (formation of a reactive metabolite N-acetyl-p-quinoneimine and protein adducts), progression of injury phase (cellular signaling, oxidative stress, and mitochondrial damage, leading to cell death), and regeneration/recovery phase (cell proliferation, angiogenesis, and wound healing).12 Our data show that liver injury in WT and p53KO mice was similar until first 12 hours, indicating that p53 is not involved in initiation of liver injury by APAP. However, liver injury progressed rapidly in p53KO mice, resulting in threefold higher liver injury at 24 hours after APAP. These data indicate that p53 protects from progression of APAP-induced liver injury. Furthermore, we observed that, despite threefold higher injury, the p53KO mice recovered at the same time as the WT mice because of a robust regeneration. These data suggest that deletion of p53 stimulates liver regeneration after APAP overdose. Although the mechanisms of initiation of injury are well studied, the mechanisms of progression of APAP-initiated liver injury and the mechanisms of liver regeneration and recovery are not completely known. Our study indicates that p53 activation is a crucial component of these mechanisms.
No difference was observed in mechanisms of initiation of injury (CYP2E1, APAP adduct formation, GSH depletion, or recovery),16, 34 a result consistent with no difference in initial injury. Interestingly, p53KO mice exhibited prolonged adduct accumulation in the liver, accompanied by delayed adduct secretion in serum. APAP adduct formation has been shown to damage mitochondria, and it is known that autophagy helps in removal of those adducts from liver.23, 26 Accumulating evidence supports role of p53 in facilitating autophagy under stress condition.2, 3, 35 It is plausible that reduced autophagy may have resulted in prolonged adduct accumulation in liver, leading to delayed serum secretion and progression of injury in p53KO mice.
To determine the underlying mechanisms of increased injury and rapid repair in p53KO mice, global transcriptomic analysis was performed on three crucial time points (namely, 12, 24, and 48 hours after APAP treatment). The transcriptomic analysis revealed disruption in metabolic homeostasis and increased inflammatory and proliferative signaling in p53KO mice. Consistently, p53KO mice exhibited higher, sustained, steatosis and SREBP2 cleavage after APAP300 treatment, correlating with progression of injury until 24 hours after APAP treatment. Malnutrition and lipid imbalance are known predisposing factors for increased risk and severity of APAP-induced injury.36, 37 Several studies in cancer cells have shown that p53 can limit glycolytic flux and promote fatty acid oxidation.3, 38, 39 These data further indicate that p53-mediated metabolic homeostasis is disrupted in p53KO mice, making them more vulnerable to APAP-induced liver injury.
Another aspect that can contribute to progression of APAP-induced injury and lipid homeostasis is the number of functional mitochondria.27 Our data show that p53KO mice have reduced mitochondrial complexes until 24 hours. APAP overdose causes mitochondrial damage, reducing capacity of liver to maintain homeostasis, and reduced mitochondrial complexes could further affect the homeostasis. Interestingly, deficiency in hepatic transsulfuration pathway was also observed, as demonstrated by reduced GAMT levels, a key enzyme in replenishing cellular cysteine pool required for GSH synthesis.30, 40 The decrease in GAMT in p53KO mice is consistent with the observation that it is a p53 target gene. Taken together, increased steatosis, reduced mitochondrial complexes, and deficient GSH transsulfuration pathway during progression phase result in higher progression of injury in p53KO mice.
Previous studies have shown that damage associated molecular patterns released from necrotic cells after APAP overdose activate Kupffer cells through toll-like receptors, leading to increased cytokine and chemokine production.31 Proinflammatory cytokines and chemokines (eg, IL-1β, tumor necrosis factor-α, and monocyte chemoattractant protein 1) activate and recruit neutrophils and monocytes.41 Monocyte-derived M2 macrophages get recruited in liver and secrete anti-inflammatory cytokines, such as IL-10. These activated phagocytes remove dead cells from liver and initiate wound healing process.42, 43 Interestingly, Ingenuity Pathway Analysis revealed significant activation of inflammatory pathways in p53KO mice. Indeed, IL-1β and IL-10 cytokine expression was significantly higher in p53KO group than WT group. It is plausible that higher liver injury results in higher IL-1β secretion, which further stimulates increased inflammatory cell infiltration in the liver. Increased expression of proresolving cytokine IL-10 may induce higher M2 macrophage activation, which will further aid in rapid recovery.
Interestingly, despite high injury, recovery in p53KO animals was similar to WT animals. PCNA and cell cycle protein data demonstrated delayed, but rapid, liver regeneration in p53KO mice, indicating p53's role in initiation of liver regeneration. This is a significant observation because several acute toxicity studies with APAP and other toxic chemicals, including thioacetamide and carbon tetrachloride, have shown that high injury results in extensive cellular stress and delay in liver regeneration.12, 17, 18, 22 For example, previous study from our laboratory17 using incremental dose model (300 mg/kg or the regenerating dose and 600 mg/kg or the nonregenerating dose of APAP) showed that resolution of injury after 600 mg/kg dose in WT mice was significantly impaired. In fact, ALT levels in the mice treated with 600 mg/kg APAP, the nonregenerating dose in that study, were almost equal to levels observed in p53KO mice treated with 300 mg/kg dose in this study. However, the p53KO mice still recovered by 48 hours in contrast to the WT mice with similar injury. These data suggest that deletion of p53 improves recovery by boosting liver regeneration. Interestingly, p53KO mice showed an initial delay in cell cycle secondary to lower cyclin D1 levels and possibly because of increase in p53-independent p21 activation. However, once the cells entered cell cycle in p53KO livers, they completed the cell cycle much faster than in WT livers. However, hepatocyte proliferation/liver regeneration in p53KO mice is much faster than in WT mice, as demonstrated by higher PCNA levels at 48 hours. The moderate deficiency in cyclin D1 expression is outweighed by significant increase in cyclin A2, a G1/S transition cyclin, and a substantial decrease in cyclin G1, a G2/M phase arrest cyclin and a known p53 target. Thus, the mechanism behind the faster cell cycle (once initiated) proliferation/liver regeneration in p53KO mice seems to be cyclin A2 and G1 mediated.
Liver regeneration in response to injury has been shown to improve survival in rodent and human studies.17, 18 Several growth factor signaling pathways are known to stimulate liver regeneration. To study mechanism of rapid liver regeneration despite higher injury in p53KO animals, differential activation of several promitogenic pathways involved in liver regeneration was studied. These studies revealed significantly higher AKT, extracellular signal–regulated kinase, and epidermal growth factor receptor signaling in p53KO mice, all of which have been shown to stimulate hepatocyte proliferation and liver regeneration. In addition, mTOR pathway was highly activated in p53KO mice versus WT mice. mTOR is known to inhibit autophagy process,44 increase lipid synthesis through ribosomal protein S6 kinase–mediated SREBP cleavage, and increase cell proliferation.45 Previous studies have shown that p53 inhibits mTOR activity through direct transcriptional regulation of AMP activated protein kinase, TSC complex subunit 2, phosphatase and tensin homolog, and insulin like growth factor binding protein 3.33 It is plausible that during the injury progression phase after APAP overdose in WT mice, p53 activation leads to up-regulation of these negative regulators of mTOR. Deletion of p53 led to higher and sustained activation of mTOR activity, which may result in down-regulation of autophagy-mediated APAP adduct removal and disruption of lipid homeostasis, leading to higher progression of APAP-induced injury. It is also plausible that, during recovery phase, sustained mTOR and AKT activation may help faster liver regeneration in p53KO mice. Further studies are needed to delineate the exact role of mTOR signaling in p53KO mice after APAP.
Taken together, these data indicate that transient activation of p53 after APAP-induced liver injury is beneficial to prevent progression of injury; however, sustained activation of p53 may affect liver regeneration and recovery. Future studies using stage-specific activation (during injury phase) or inhibition of p53 (during regeneration phase) are required to further establish function of p53 in regulating injury progression and regeneration. Our studies have revealed novel pleiotropic role of p53 in linking the injury progression response to liver regeneration, and intervention of these mechanisms may have a therapeutic benefit.
Footnotes
Supported by NIH Centers of Biomedical Research Excellence grant P20 RR021940-03, National Institute of Environmental Health Sciences Toxicology Training grant T32ES007079-34, and National Institute of Diabetes and Digestive and Kidney Diseases grant R01DK98414 (U.A.).
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2018.03.006.
Supplemental Data
Supplemental Figure S1.
Functional analysis on transcriptomic data using Ingenuity Pathway Analysis (IPA). A–C: Comparison analysis in the form of heat map of top upstream regulators (A), diseases and biofunctions (B), and canonical pathways (C), after 300 mg/kg APAP treatment in p53KO mice versus WT mice. D: Validation of transcriptomic data by real-time PCR analysis of representative genes. The networks and functional analyses were generated through the use of IPA.
Supplemental Figure S2.
Individual graphs of gene expression fold-change values in p53KO mice with respect to WT mice, determined by real-time PCR. ∗P < 0.05 (analysis of variance).
References
- 1.Aloni-Grinstein R., Shetzer Y., Kaufman T., Rotter V. p53: The barrier to cancer stem cell formation. FEBS Lett. 2014;588:2580–2589. doi: 10.1016/j.febslet.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 2.Kenzelmann Broz D., Attardi L.D. TRP53 activates a global autophagy program to promote tumor suppression. Autophagy. 2013;9:1440–1442. doi: 10.4161/auto.25833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruiswijk F., Labuschagne C.F., Vousden K.H. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16:393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 4.Menendez D., Shatz M., Resnick M.A. Interactions between the tumor suppressor p53 and immune responses. Curr Opin Oncol. 2013;25:85–92. doi: 10.1097/CCO.0b013e32835b6386. [DOI] [PubMed] [Google Scholar]
- 5.Muller P.A., Vousden K.H., Norman J.C. p53 and its mutants in tumor cell migration and invasion. J Cell Biol. 2011;192:209–218. doi: 10.1083/jcb.201009059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyner S.D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., Lu X., Soron G., Cooper B., Brayton C., Park S.H., Thompson T., Karsenty G., Bradley A., Donehower L.A. p53 Mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 7.Jin J., Hong I.H., Lewis K., Iakova P., Breaux M., Jiang Y., Sullivan E., Jawanmardi N., Timchenko L., Timchenko N.A. Cooperation of C/EBP family proteins and chromatin remodeling proteins is essential for termination of liver regeneration. Hepatology. 2015;61:315–325. doi: 10.1002/hep.27295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurinna S., Stratton S.A., Coban Z., Schumacher J.M., Grompe M., Duncan A.W., Barton M.C. p53 Regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology. 2013;57:2004–2013. doi: 10.1002/hep.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurinna S., Stratton S.A., Tsai W.W., Akdemir K.C., Gu W., Singh P., Goode T., Darlington G.J., Barton M.C. Direct activation of forkhead box O3 by tumor suppressors p53 and p73 is disrupted during liver regeneration in mice. Hepatology. 2010;52:1023–1032. doi: 10.1002/hep.23746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stepniak E., Ricci R., Eferl R., Sumara G., Sumara I., Rath M., Hui L., Wagner E.F. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 2006;20:2306–2314. doi: 10.1101/gad.390506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Liu L., He Z., Li G., Liu J., Song Z., Jin H., Rudolph K.L., Yang H., Mao Y., Zhang L., Zhang H., Xiao Z., Ju Z. Inhibition of wild-type p53-induced phosphatase 1 promotes liver regeneration in mice by direct activation of mammalian target of rapamycin. Hepatology. 2015;61:2030–2041. doi: 10.1002/hep.27755. [DOI] [PubMed] [Google Scholar]
- 12.Mehendale H.M. Tissue repair: an important determinant of final outcome of toxicant-induced injury. Toxicol Pathol. 2005;33:41–51. doi: 10.1080/01926230590881808. [DOI] [PubMed] [Google Scholar]
- 13.Larson A.M., Polson J., Fontana R.J., Davern T.J., Lalani E., Hynan L.S., Reisch J.S., Schiodt F.V., Ostapowicz G., Shakil A.O., Lee W.M. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005;42:1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 14.Bernal W., Wendon J. Acute liver failure. N Engl J Med. 2013;369:2525–2534. doi: 10.1056/NEJMra1208937. [DOI] [PubMed] [Google Scholar]
- 15.Shehab N., Lovegrove M.C., Geller A.I., Rose K.O., Weidle N.J., Budnitz D.S. US emergency department visits for outpatient adverse drug events, 2013–2014. JAMA. 2016;316:2115–2125. doi: 10.1001/jama.2016.16201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaeschke H., Bajt M.L. Intracellular signaling mechanisms of acetaminophen-induced liver cell death. Toxicol Sci. 2006;89:31–41. doi: 10.1093/toxsci/kfi336. [DOI] [PubMed] [Google Scholar]
- 17.Bhushan B., Walesky C., Manley M., Gallagher T., Borude P., Edwards G., Monga S.P., Apte U. Pro-regenerative signaling after acetaminophen-induced acute liver injury in mice identified using a novel incremental dose model. Am J Pathol. 2014;184:3013–3025. doi: 10.1016/j.ajpath.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt L.E., Dalhoff K. Alpha-fetoprotein is a predictor of outcome in acetaminophen-induced liver injury. Hepatology. 2005;41:26–31. doi: 10.1002/hep.20511. [DOI] [PubMed] [Google Scholar]
- 19.Stamper B.D., Garcia M.L., Nguyen D.Q., Beyer R.P., Bammler T.K., Farin F.M., Kavanagh T.J., Nelson S.D. p53 Contributes to differentiating gene expression following exposure to acetaminophen and its less hepatotoxic regioisomer both in vitro and in vivo. Gene Regul Syst Bio. 2015;9:1–14. doi: 10.4137/GRSB.S25388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huo Y., Yin S., Yan M., Win S., Aung Than T., Aghajan M., Hu H., Kaplowitz N. Protective role of p53 in acetaminophen hepatotoxicity. Free Radic Biol Med. 2017;106:111–117. doi: 10.1016/j.freeradbiomed.2017.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borude P., Edwards G., Walesky C., Li F., Ma X., Kong B., Guo G.L., Apte U. Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology. 2012;56:2344–2352. doi: 10.1002/hep.25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhushan B., Borude P., Edwards G., Walesky C., Cleveland J., Li F., Ma X., Apte U. Role of bile acids in liver injury and regeneration following acetaminophen overdose. Am J Pathol. 2013;183:1518–1526. doi: 10.1016/j.ajpath.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGill M.R., Lebofsky M., Norris H.R., Slawson M.H., Bajt M.L., Xie Y., Williams C.D., Wilkins D.G., Rollins D.E., Jaeschke H. Plasma and liver acetaminophen-protein adduct levels in mice after acetaminophen treatment: dose-response, mechanisms, and clinical implications. Toxicol Appl Pharmacol. 2013;269:240–249. doi: 10.1016/j.taap.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walesky C., Edwards G., Borude P., Gunewardena S., O'Neil M., Yoo B., Apte U. Hepatocyte nuclear factor 4 alpha deletion promotes diethylnitrosamine-induced hepatocellular carcinoma in rodents. Hepatology. 2013;57:2480–2490. doi: 10.1002/hep.26251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rooney J.P., Ryde I.T., Sanders L.H., Howlett E.H., Colton M.D., Germ K.E., Mayer G.D., Greenamyre J.T., Meyer J.N. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol Biol. 2015;1241:23–38. doi: 10.1007/978-1-4939-1875-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni H.M., McGill M.R., Chao X., Du K., Williams J.A., Xie Y., Jaeschke H., Ding W.X. Removal of acetaminophen protein adducts by autophagy protects against acetaminophen-induced liver injury in mice. J Hepatol. 2016;65:354–362. doi: 10.1016/j.jhep.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGill M.R., Sharpe M.R., Williams C.D., Taha M., Curry S.C., Jaeschke H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest. 2012;122:1574–1583. doi: 10.1172/JCI59755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park J.Y., Wang P.Y., Matsumoto T., Sung H.J., Ma W., Choi J.W., Anderson S.A., Leary S.C., Balaban R.S., Kang J.G., Hwang P.M. p53 Improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105:705–712. doi: 10.1161/CIRCRESAHA.109.205310. 11 p following 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly D.P., Scarpulla R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 30.Lu S.C. Regulation of glutathione synthesis. Curr Top Cell Regul. 2000;36:95–116. doi: 10.1016/s0070-2137(01)80004-2. [DOI] [PubMed] [Google Scholar]
- 31.Jaeschke H., Williams C.D., Ramachandran A., Bajt M.L. Acetaminophen hepatotoxicity and repair: the role of sterile inflammation and innate immunity. Liver Int. 2012;32:8–20. doi: 10.1111/j.1478-3231.2011.02501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludes-Meyers J.H., Subler M.A., Shivakumar C.V., Munoz R.M., Jiang P., Bigger J.E., Brown D.R., Deb S.P., Deb S. Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol. 1996;16:6009–6019. doi: 10.1128/mcb.16.11.6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Z., Hu W., de Stanchina E., Teresky A.K., Jin S., Lowe S., Levine A.J. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 34.Reid A.B., Kurten R.C., McCullough S.S., Brock R.W., Hinson J.A. Mechanisms of acetaminophen-induced hepatotoxicity: role of oxidative stress and mitochondrial permeability transition in freshly isolated mouse hepatocytes. J Pharmacol Exp Ther. 2005;312:509–516. doi: 10.1124/jpet.104.075945. [DOI] [PubMed] [Google Scholar]
- 35.Crighton D., Wilkinson S., O'Prey J., Syed N., Smith P., Harrison P.R., Gasco M., Garrone O., Crook T., Ryan K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 36.Fromenty B. Drug-induced liver injury in obesity. J Hepatol. 2013;58:824–826. doi: 10.1016/j.jhep.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Michaut A., Moreau C., Robin M.A., Fromenty B. Acetaminophen-induced liver injury in obesity and nonalcoholic fatty liver disease. Liver Int. 2014;34:e171–e179. doi: 10.1111/liv.12514. [DOI] [PubMed] [Google Scholar]
- 38.Bensaad K., Tsuruta A., Selak M.A., Vidal M.N., Nakano K., Bartrons R., Gottlieb E., Vousden K.H. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 39.Parrales A., Iwakuma T. p53 As a regulator of lipid metabolism in cancer. Int J Mol Sci. 2016;17:2074. doi: 10.3390/ijms17122074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnackenberg L.K., Chen M., Sun J., Holland R.D., Dragan Y., Tong W., Welsh W., Beger R.D. Evaluations of the trans-sulfuration pathway in multiple liver toxicity studies. Toxicol Appl Pharmacol. 2009;235:25–32. doi: 10.1016/j.taap.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 41.Williams C.D., Farhood A., Jaeschke H. Role of caspase-1 and interleukin-1beta in acetaminophen-induced hepatic inflammation and liver injury. Toxicol Appl Pharmacol. 2010;247:169–178. doi: 10.1016/j.taap.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woolbright B.L., Jaeschke H. Role of the inflammasome in acetaminophen-induced liver injury and acute liver failure. J Hepatol. 2017;66:836–848. doi: 10.1016/j.jhep.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bourdi M., Masubuchi Y., Reilly T.P., Amouzadeh H.R., Martin J.L., George J.W., Shah A.G., Pohl L.R. Protection against acetaminophen-induced liver injury and lethality by interleukin 10: role of inducible nitric oxide synthase. Hepatology. 2002;35:289–298. doi: 10.1053/jhep.2002.30956. [DOI] [PubMed] [Google Scholar]
- 44.Martina J.A., Chen Y., Gucek M., Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.