Abstract
A central step in the pathogenesis of prion diseases is the conformational transition of the cellular prion protein (PrPC) into the scrapie isoform, denoted PrPSc. Studies in transgenic mice have indicated that this conversion requires a direct interaction between PrPC and PrPSc; however, insights into the underlying mechanisms are still missing. Interestingly, only a subfraction of PrPC is converted in scrapie-infected cells, suggesting that not all PrPC species are suitable substrates for the conversion. On the basis of the observation that PrPC can form homodimers under physiological conditions with the internal hydrophobic domain (HD) serving as a putative dimerization domain, we wondered whether PrP dimerization is involved in the formation of neurotoxic and/or infectious PrP conformers. Here, we analyzed the possible impact on dimerization of pathogenic mutations in the HD that induce a spontaneous neurodegenerative disease in transgenic mice. Similarly to wildtype (WT) PrPC, the neurotoxic variant PrP(AV3) formed homodimers as well as heterodimers with WTPrPC. Notably, forced PrP dimerization via an intermolecular disulfide bond did not interfere with its maturation and intracellular trafficking. Covalently linked PrP dimers were complex glycosylated, GPI-anchored, and sorted to the outer leaflet of the plasma membrane. However, forced PrPC dimerization completely blocked its conversion into PrPSc in chronically scrapie-infected mouse neuroblastoma cells. Moreover, PrPC dimers had a dominant-negative inhibition effect on the conversion of monomeric PrPC. Our findings suggest that PrPC monomers are the major substrates for PrPSc propagation and that it may be possible to halt prion formation by stabilizing PrPC dimers.
Keywords: neurodegenerative disease; prion disease; glycosylphosphatidylinositol; GPI anchor; dimerization; trafficking; neurological disease; amyloid; Creutzfeldt-Jakob disease; scrapie; PrPC, amyloid plaque
Introduction
Prion diseases in humans and other mammals are characterized by a conformational transition of the cellular prion protein (PrPC)5 into an aberrantly folded isoform, designated scrapie prion protein (PrPSc). PrPSc can form amyloid plaques in the diseased brain and is the major constituent of infectious prions (for reviews, see Refs. 1–4). Propagation of PrPSc is strictly dependent on the synthesis of PrPC by the host (5) and involves a direct interaction of the two conformers (6, 7). Both mature PrPC and PrPSc contain a glycosylphosphatidylinositol (GPI) anchor and two N-linked carbohydrate moieties of complex structure, indicating that the conversion takes place after trafficking of PrPC through the secretory pathway at the plasma membrane or within endocytic compartments (8–12). However, both post-translational modifications are not required for PrPSc formation. Studies in prion-infected cultured cells and transgenic mice indicated that PrPC devoid of N-linked glycans still supports PrPSc propagation and formation of infectious prions (13). Similarly, transgenic mice expressing a secreted version of PrPC by deleting the C-terminal GPI anchor signal peptide (PrPΔGPI) propagate infectious prions after infection (14, 15).
Based on transgenic studies published by Prusiner and co-workers (6, 7), in 1991 John Hardy (16) proposed a model for the propagation of prions involving the formation of a PrPC/PrPSc heterodimer as an initial and essential step. Furthermore, he speculated that PrPC exists under physiological conditions as a dimer that has to dissociate before monomeric PrPC can interact with and is converted by PrPSc (16). Indeed, we and others have provided experimental evidence for the existence of PrPC dimers in vitro and in vivo (17–20) with the internal hydrophobic domain (HD) as a putative dimerization domain (21). In this context, it might be interesting to note that in scrapie-infected cells only a small subfraction of PrPC is converted into PrPSc, indicating that not all PrPC molecules are suitable substrates for the conversion into PrPSc (12).
To address the possibility that alterations in dimerization of PrP might be implicated in the formation of pathogenic PrP conformers, we investigated the activity of pathogenic PrP mutants to dimerize and analyzed the conversion of PrPC dimers into PrPSc. Our study revealed that a pathogenic PrP mutant dimerizes similarly to WTPrPC; moreover, mutant PrP forms heterodimers with wildtype (WT) PrPC. Strikingly, stabilizing PrPC dimers prevented their conversion into PrPSc in scrapie-infected neuroblastoma cells and inhibited endogenous prion propagation in trans.
Results
A neurotoxic mutation does not interfere with homodimerization of PrP
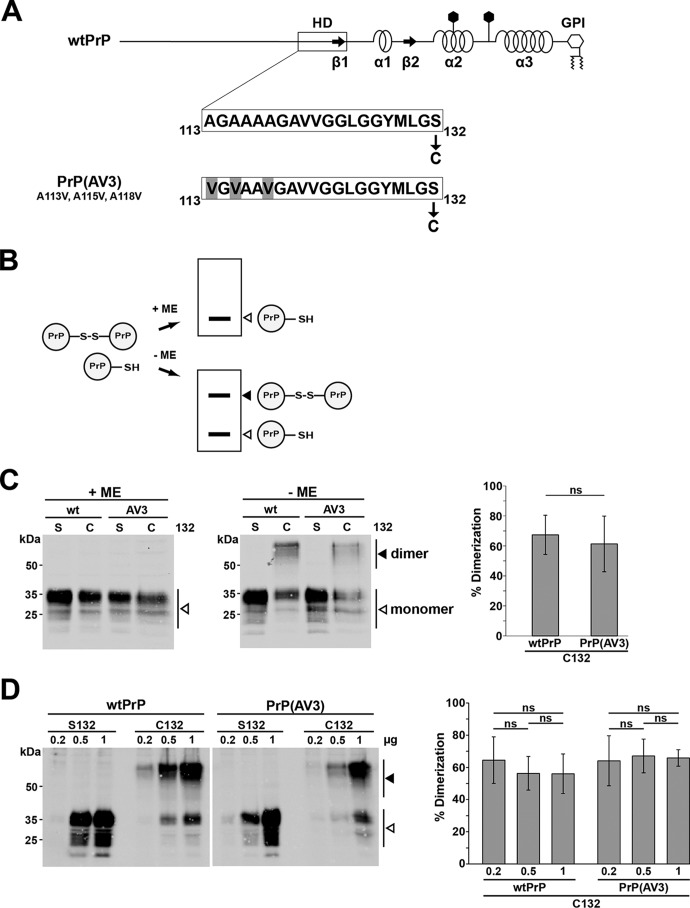
The formation of PrPC dimers was reported previously (17–19), and the internal HD has been identified as a putative dimerization domain (21). To address a possible role of PrP dimerization in the formation of a neurotoxic PrP conformer, we studied dimerization of PrP(AV3) in mouse neuroblastoma (N2a) cells. PrP(AV3) contains three alanine-to-valine changes within the HD (Fig. 1A) and causes early onset neurodegeneration upon expression in transgenic mice (22).
Figure 1.
A neurotoxic mutant of PrP forms homodimers similarly to WTPrPC. A, schematic presentation of the constructs analyzed. Straight line, intrinsically disordered regions; box, highly conserved HD; arrows, β-strands (β). α-Helical structure is indicated by helices, polygons represent N-linked glycosylation acceptor sites, and GPI indicates the GPI anchor. The amino acid sequences of the HD of WTPrPC and PrP(AV3) are shown in the detail magnification. In some constructs, serine 132 is replaced by cysteine (denoted C132) to allow formation of an intermolecular disulfide bond. Numbering of the amino acids refers to human prion protein. B, scheme of the experimental strategy. In case PrP-C132 forms an intermolecular disulfide bond, PrP dimers can be separated from PrP monomers on SDS-PAGE under oxidizing conditions (−ME). Under reducing conditions (Laemmli sample buffer containing ME), only PrP monomers are detected by Western blotting (+ME). C, PrP forms disulfide bond–stabilized homodimers in neuronal cells. N2a cells were transiently transfected with WTPrP or PrP(AV3) containing a serine (S132) or cysteine (C132) at amino acid 132. Cell lysates were prepared and denatured by boiling in Laemmli sample buffer with (+ME) or without reducing agent (−ME). White arrowheads represent monomeric PrP; the black arrowhead indicates PrP homodimers. Right panel, quantifications of the dimerization efficiency measured densitometrically. ns, not significant. Data represent mean ± S.D. of ≥3 independent experiments. D, homodimerization is not due to overexpression. N2a cells were transiently transfected with different DNA amounts of the PrP variants as indicated. Cell lysates were analyzed by Western blotting in the absence of reducing agents. A white arrowhead represents the monomeric prion protein; the black arrowhead represents the homodimer. Please note that a longer exposure compared with the blots in C is shown to visualize the bands in the 0.2-μg samples. Right panel, quantifications of the dimerization efficiency. ns, not significant. Data represent mean ± S.D. of five independent experiments. Error bars represent S.D.
To force formation of PrP dimers, we replaced serine 132 by cysteine in PrP(AV3) (Fig. 1A). In case PrP dimerizes, an intermolecular disulfide bond can be formed that is stable in SDS buffer under nonreducing conditions (Fig. 1B). This approach was successfully used to study dimerization of the transmembrane receptor ErbB-2/Her2 (23), the amyloid precursor protein (24), and WTPrPC (19). Indeed, Western blot analysis of transiently transfected N2a cells revealed that similarly to WTPrPC the neurotoxic PrP(AV3) mutant forms homodimers that disassemble in the presence of reducing agents, such as β-mercaptoethanol (ME) (Fig. 1C). The ratio of monomeric/dimeric PrP species was stable at various expression levels, indicating that dimer formation was not an artifact of PrP overexpression (Fig. 1D).
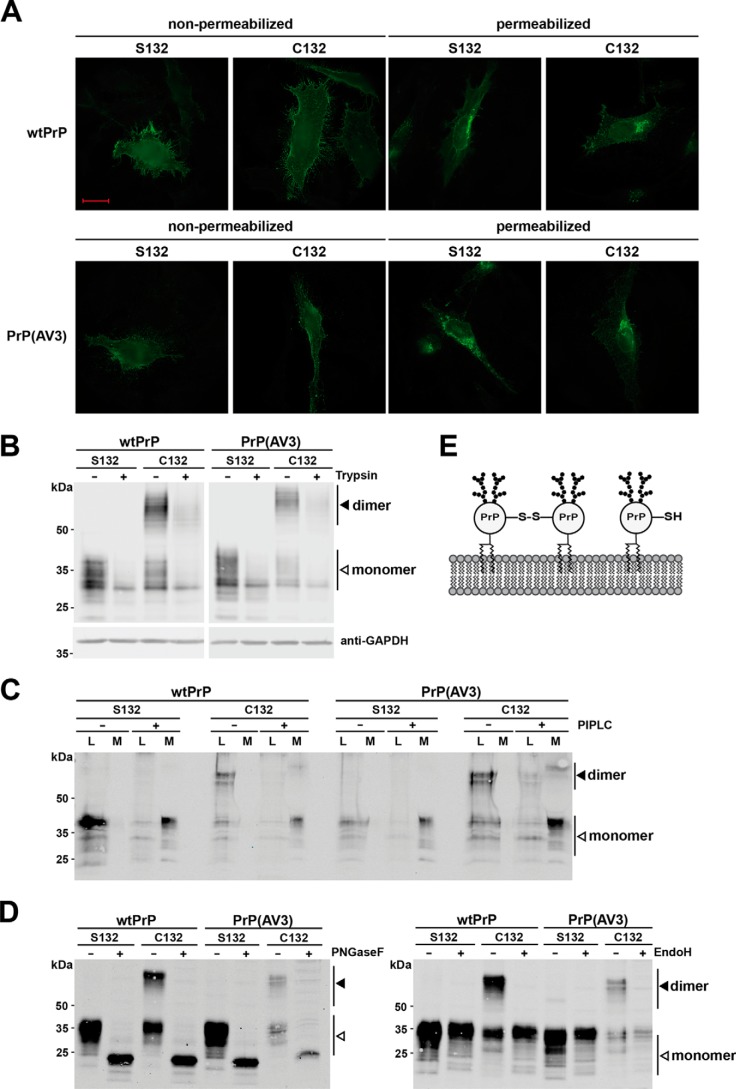
PrP is characterized by a series of post-translational modifications. It is modified by two N-linked glycans of complex structure, a C-terminal GPI anchor, and an internal disulfide bond (for a review, see Ref. 25). To study whether forced dimerization interferes with maturation and/or cellular trafficking, we performed an indirect immunofluorescence analysis of cells transiently transfected with our PrP constructs. The staining pattern did not reveal obvious differences between the PrP variants containing a cysteine or serine at position 132, indicating that disulfide bond–linked dimers of WTPrPC and PrP(AV3) are transported through the secretory pathway (Fig. 2A, permeabilized) and are localized at the outer leaflet of the plasma membrane (Fig. 2A, non-permeabilized). To analyze the cell surface localization of PrP dimers in more detail, transfected cells were treated either with trypsin to remove extracellular domains of membrane-anchored proteins in general or with phosphatidylinositol-specific phospholipase C (PIPLC) to specifically liberate GPI-anchored proteins. As shown by Western blot analysis, trypsin treatment of live cells significantly reduced the signals of both PrP monomers and dimers in cell lysates, confirming that PrP dimers had been located at the plasma membrane (Fig. 2B). The cytosolic protein GAPDH was not affected by trypsin, verifying that the protease only digested extracellular proteins (Fig. 2B). Similarly, after incubation with PIPLC, monomeric as well as dimeric PrP was present in the cell culture media (Fig. 2C), demonstrating that the PrP dimers were inserted in the plasma membrane via a GPI anchor. Another post-translational modification of PrP is the conversion of the two N-linked glycans into complex structures (11). To evaluate the glycosylation status of PrP in detail, cell lysates were treated with endoglycosidase H (EndoH), which only cleaves high-mannose N-linked glycans, or peptide:N-glycosidase F (PNGaseF), which cleaves high-mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins. Forced dimerization via the introduced cysteine residue did not interfere with complex glycosylation because the electrophoretic mobility of PrP was only increased after PNGaseF (Fig. 2D, left panel) but not after EndoH digestion (Fig. 2D, right panel). Notably, the enzyme reaction buffer contains reducing agent. Thus, only monomeric PrP is seen in Western blot analysis after EndoH or PNGaseF digestion. If PrP dimers were modified with high-mannose glycans only, an additional band would appear in the EndoH-treated samples.
Figure 2.
Covalently linked PrPC dimers are complex glycosylated and GPI-anchored at the outer leaflet of the plasma membrane. A–C, PrP dimers are GPI-anchored at the outer leaflet of the plasma membrane. A, HeLa cells were transiently transfected with the constructs indicated and analyzed by indirect immunofluorescence. Cells were either permeabilized or left nonpermeabilized. PrP constructs were detected with the 3F4 antibody. Scale bar, 20 μm. B, transiently transfected HeLa cells were treated with trypsin for 25 min at room temperature to digest proteins at the plasma membrane. Lysates of treated and untreated cells were analyzed by Western blotting. Detection of cytosolic GAPDH was used to verify that trypsin only digested extracellular proteins. C, transiently transfected N2a cells were incubated for 4 h with and without PIPLC in PBS at 37 °C. PrP present in the cell lysates (L) and the cell culture media (M) were analyzed by Western blotting under nonreducing conditions. PrP was detected by Western blotting using the 3F4 antibody. D, dimerization of PrP does not interfere with complex glycosylation. N2a cells were transiently transfected with the indicated constructs and analyzed by Western blotting. To determine the glycosylation status, lysates were treated either with PNGaseF (+; left panel) that cleaves high-mannose, hybrid, and complex oligosaccharides from N-linked glycoproteins or EndoH, which cleaves only mannose-rich oligosaccharides (+; right panel). Please note that the reaction buffer for PNGaseF and EndoH contains a reducing agent; therefore, only PrP monomers are seen in the PNGaseF- and EndoH-treated samples. White arrowhead, monomer; black arrowhead, dimer. E, schematic representation of monomeric and covalently linked dimeric PrPC located at the plasma membrane. Both fractions are complex glycosylated and inserted into the outer leaflet of plasma membrane via a GPI anchor.
This analysis revealed that neurotoxic mutations in the hydrophobic domain do not interfere with the dimerization of PrP. In addition, our data indicated that an engineered intermolecular disulfide bond between the hydrophobic domains of two PrP molecules does not impair maturation and cellular trafficking. Like WTPrPC, covalently linked PrP dimers are complex glycosylated and anchored to the outer leaflet of the plasma membrane via a GPI anchor.
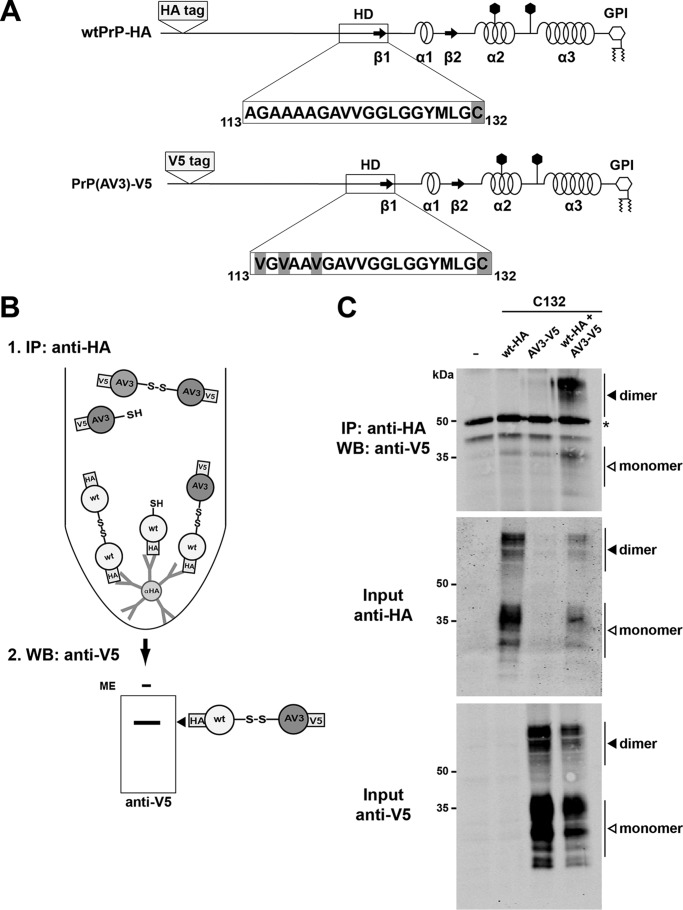
A neurotoxic PrP mutant forms heterodimers with WT PrPC
To analyze whether the mutations in the HD might interfere with the formation of PrP(AV3)/WTPrP heterodimers, we inserted an HA epitope tag into WTPrPC and a V5 epitope tag into PrP(AV3). Both proteins contain a cysteine at position 132 to allow intermolecular disulfide bond formation (Fig. 3A). N2a cells transiently coexpressing WTPrP-HA and PrP(AV3)-V5 were lysed and subjected to immunoprecipitation with anti-HA antibodies under nonreducing conditions. The immunopellet was then analyzed by Western blotting using anti-V5 antibodies. PrP(AV3)-V5 copurified with WTPrP-HA, indicative of the formation of PrP heterodimers (Fig. 3, B and C). As a control, anti-HA immunoprecipitations were performed in lysates from cells that express either HA-tagged WTPrPC or V5-tagged PrP(AV3). In neither case was V5-positive PrP detected in the immunopellets.
Figure 3.
The neurotoxic mutant PrP(AV3) forms disulfide bond–linked heterodimers with WTPrP. A, schematic representation of the mutants used. The cysteine variants of WTPrP and PrP(AV3) were modified with an HA (WTPrP-HA) or a V5 tag (PrP(AV3)-V5). The tags were inserted after amino acid 35. B, scheme of the experimental strategy. To specifically detect WT/AV3 heterodimers, HA-tagged WTPrP was first immunoprecipitated under nonreducing conditions using anti-HA-agarose beads. To detect copurified V5-tagged PrP(AV3), the immunopellet was then analyzed by Western blotting under nonreducing conditions using an anti-V5 antibody. C, N2a cells were transiently transfected with either HA-tagged WTPrP, V5-tagged PrP(AV3), or both. Cells were lysed, and WTPrP was immunoprecipitated under nonreducing conditions with HA-agarose beads. The immunopellet was analyzed by Western blotting (WB) under nonreducing conditions using an anti-V5 antibody. Western blot analysis of the inputs is shown below. White arrowhead, monomer; black arrowhead; dimer. Asterisk, signal corresponds to primary antibody used in the immunoprecipitation (IP).
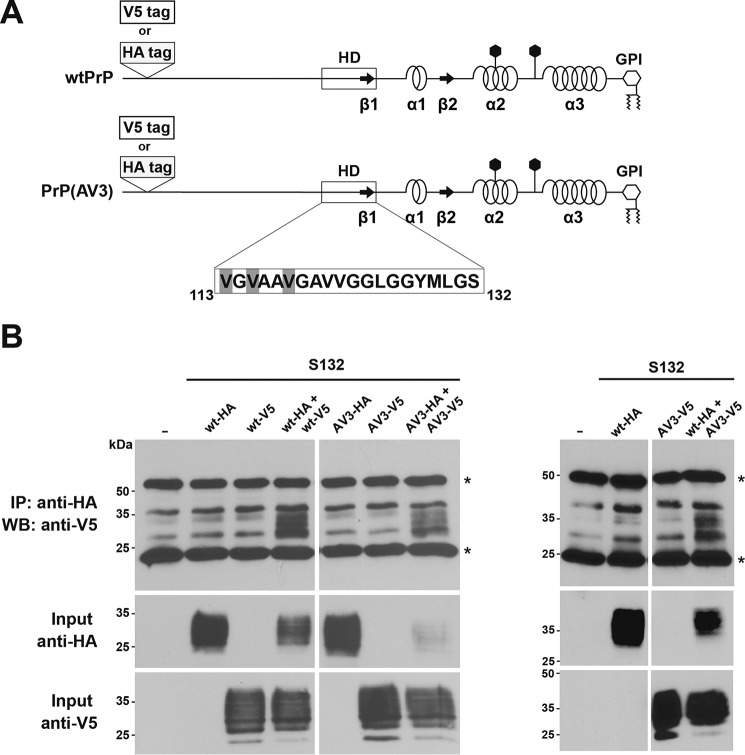
The formation of disulfide bond–stabilized PrP homo- and heterodimers indicated that PrPC has an intrinsic propensity to at least transiently undergo homotypic interactions in the secretory pathway of neuronal cells. To assess the dimerization under physiological conditions, we performed native immunoprecipitation assays with HA- and V5-tagged PrP constructs that did not include the Cys-132 mutation (Fig. 4A). Cells were cotransfected with an HA- together with a V5-tagged PrP construct, and cell lysates were subjected to immunoprecipitation under native conditions with anti-HA antibodies. The resulting immunopellet was then analyzed by Western blotting using anti-V5 antibodies. Using this approach, we could verify the formation of homodimers of WTPrPC and PrP(AV3) under physiological conditions (Fig. 4B, wt-HA + wt-V5 or AV3-HA + AV3-V5). Moreover, we could also detect heterodimers formed between WTPrPC and PrPC(AV3) (Fig. 4B, wt-HA + AV3-V5). In lysates prepared from cells that express only V5- or HA-tagged PrP, no specific signals were detectable in Western blot analysis, demonstrating the specificity of the assay.
Figure 4.
The neurotoxic mutant PrP(AV3) forms heterodimers with WTPrP under physiological conditions. A, schematic representation of the constructs used. The serine variants of WTPrP and PrP(AV3) were modified with an HA (WTPrP-HA and PrP(AV3)-HA) and a V5 tag (WTPrP-V5 and PrP(AV3)-V5). The tags were inserted after amino acid 35. B, HeLa cells were transiently transfected with the indicated constructs. Cells were lysed, and HA-tagged PrP was immunoprecipitated under nonreducing conditions with HA-agarose beads. The immunopellet was dissolved in Laemmli sample buffer containing β-mercaptoethanol and analyzed by Western blotting (WB) using an anti-V5 antibody. Western blot analysis of the inputs is shown below. Asterisk, signal corresponds to primary antibody used in the immunoprecipitation (IP).
Dimerization of PrPC blocks formation of PrPSc
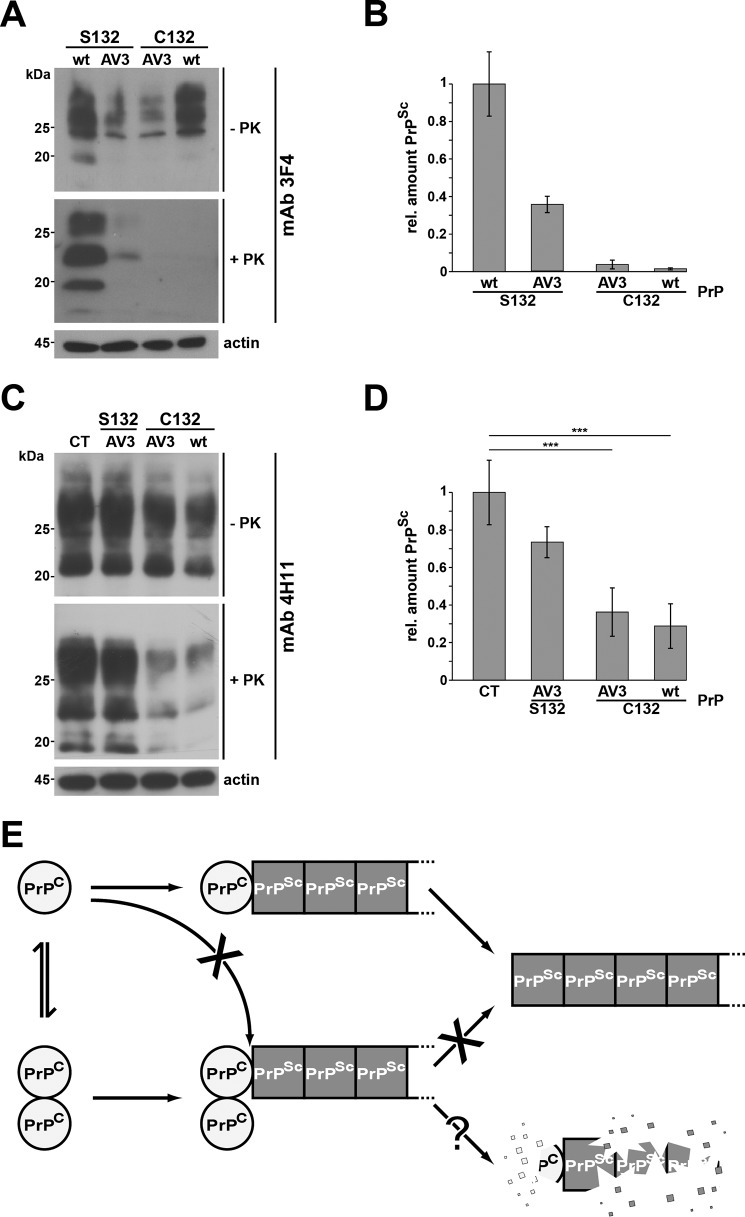
Encouraged by the finding that maturation and intracellular trafficking of covalently linked PrP dimers were comparable with those of native PrPC, we tested whether PrP dimers can be converted into PrPSc. To this end, we used scrapie-infected mouse neuroblastoma (ScN2a) cells that propagate proteinase K (PK)-resistant PrPSc and infectious mouse prions (26–28). After transient transfection with 3F4-tagged PrP constructs, the conversion of exogenous PrP is monitored by the appearance of 3F4-positive PK-resistant PrP species because endogenous mouse PrPSc is not detected by the 3F4 antibody (29). This conversion assay is illustrated in Fig. 5. Cell lysates were prepared from ScN2a cells transiently transfected with WTPrPC and treated with PK or left untreated prior to Western blot analysis. The 3F4-positive signals in PK-treated extracts demonstrated the conversion of transfected WTPrPC (Fig. 5A). Similarly, PrP(AV3) was converted into PrPSc upon expression in ScN2a cells (Fig. 5A). The cysteine variants of WTPrPC and PrP(AV3) were also expressed in ScN2a cells after transient transfection, illustrated by the 3F4-positive signals in lysates that have not been treated with PK prior to Western blot analysis (Fig. 5A, −PK). However, after PK treatment of the lysates, almost no transfected PrP constructs converted into PrPSc were detectable (Fig. 5A, +PK). Quantification of PrP conversion rates confirmed that less than 1.5% of the transfected cysteine variants had been converted into PrPSc (Fig. 5B). Interestingly, the monomeric fraction of the cysteine variants, which is around 40% (Fig. 1, C and D), was obviously also protected against conversion into prions. To investigate this phenomenon in more detail, we analyzed a possible effect of the forced PrPC dimers on the conversion of endogenous mouse PrPC, which lacks the Ser-to-Cys mutation. This approach allows to test whether the Cys point mutation itself and not the dimerization inhibits conversion by PrPSc. Using the anti-PrP antibody 4H11, which detects both endogenous mouse and the transfected 3F4-positive PrPC/PrPSc, we could show a significant reduction in the total amount of PK-resistant PrPSc in cells expressing the cysteine variant of PrP (Fig. 5, C and D). A similar dominant-negative effect on the conversion of WTPrPC was described earlier for certain deletion mutants of PrP (30) Importantly, expressing the serine variant of PrP(AV3) had no significant inhibitory effect on endogenous PrPSc propagation, indicating that the decrease in endogenous PrPSc levels are not due to the transfection and/or expression of a mutated PrP. Please note that one cannot expect a similar decrease in the amount of endogenous mouse PrPSc as seen for the transfected 3F4-positive PrP (Fig. 5, A and B) because ScN2a cells had already accumulated PrPSc prior to the expression of the PrPC dimers, and PrPSc has a half-life time >24 h. As another control, we generate an alanine variant of PrPC (PrP-A132). After transient expression in ScN2a cells, it was converted into PK-resistant PrPSc, indicating that the mutation of the serine residue does not prevent conversion into PrPSc (Fig. S1). In conclusion, the experiments in ScN2a cells revealed that dimeric PrPC is not converted into PrPSc and has a dominant-negative effect on PrPSc propagation in trans.
Figure 5.
Forced dimerization of PrPC interferes with propagation of PrPSc. A and C, persistently infected 22L-ScN2a cells were transiently transfected with the constructs indicated. Cell lysates were prepared and subjected to PK digestion (+PK) or left untreated (−PK) prior to immunoblot analysis using the monoclonal anti-PrP antibody 3F4 to exclusively detect the transfected PrP but not the endogenous PrPC (A) or using the monoclonal anti-PrP antibody 4H11 to detect endogenous mouse PrPC and PrPSc in addition (C). B and D, quantitative analysis of the amount of 3F4-positive (B) and total (D) PrPSc in transfected 22L-ScN2a cells. The relative (rel.) amount of PrP and PK-resistant PrPSc was measured densitometrically using ImageJ software. The relative amount of PK-resistant PrPSc present in cells expressing transfected WTPrPC (C) or control-transfected cells (D) was set as 1. Data represent mean ± S.D. of ≥3 independent experiments. E, putative model of the protective activity of PrPC dimers. Under physiological conditions, PrPC forms a dimer. Upon dissociation, PrPC monomers interact with and are converted by PrPSc. PrP dimers may interact with PrPSc, but conversion does not occur. In case PrPC dimers bind to PrPSc with a higher affinity than monomeric PrPC, interaction of monomeric PrPC with PrPSc is decreased, and its conversion is reduced. Alternatively or in addition, PrPSc in complex with PrPC dimers is subjected to increased intracellular degradation. Error bars represent S.D.
Discussion
Dimerization of cell surface receptors is often associated with their physiological function. Our study emphasizes a propensity of the cellular prion protein to form dimers at the plasma membrane. Furthermore, we show that neurotoxic mutations within the hydrophobic domain do not interfere with the formation of homodimers or heterodimers between mutant PrP and WTPrPC. However, in contrast to monomeric PrP, covalently linked PrP dimers are not converted into PrPSc in scrapie-infected neuroblastoma (ScN2a) cells and inhibit prion propagation in trans.
The biological function of PrPC still remains enigmatic, but various studies suggest a role of PrPC as a cell surface receptor in stress-protective and neurotoxic signaling pathways (19, 31–36). Because receptors often form dimers, it is interesting to note that dimerization of PrPC has been described in vitro and in vivo (17–20). Using disulfide bridge–mediated dimerization, we first corroborated these studies and showed that covalently linked dimers of PrP are complex glycosylated and GPI-anchored to the outer leaflet of the plasma membrane. The introduction of an artificial disulfide bond allowed us to study a possible impact of dimerization on PrP maturation in the secretory pathway and to compare dimer formation between WT and mutant PrP. Specifically, these approaches revealed that at least 60% of total PrP dimerizes. Moreover, maturation and cellular trafficking of covalently linked PrP dimers were not altered compared with WTPrPC. Thus, neither the quality control machinery nor the glycan-modifying enzymes in the secretory pathway regarded PrP dimers as nonphysiological conformers. Finally, native immunoprecipitation assays provided evidence for the formation of WTPrPC dimers under physiological conditions.
We observed that three mutations within the HD that induce the formation of neurotoxic PrP conformers (AV3) did not interfere with PrP dimerization, suggesting that the toxic potential of PrP(AV3) is not linked to alterations in dimer formation. However, we only analyzed dimer formation of the secreted form of PrP(AV3) and not of its transmembrane isoform, denoted PrPCtm (22). Our finding that PrP(AV3) interacts and forms stable heterodimers with WTPrPC might be relevant for the observation that WTPrPC can prevent the toxic activity of PrP mutants lacking the HD (PrPΔHD) (37). It is conceivable that WTPrPC via a direct interaction either blocks aberrant binding of PrPΔHD to cellular proteins or prevents formation of a channel-forming PrPΔHD conformer (38).
One of the most important findings of our study was the resistance of forced PrPC dimers to be converted into PrPSc and their activity to interfere with endogenous PrPSc propagation in trans. After transient expression in ScN2a cells, WTPrPC as well as PrP(AV3) was converted into PK-resistant PrPSc. The dimeric variants of both proteins were expressed; however, they were not converted into PrPSc. Moreover, the PrPC dimers inhibited the conversion of the monomeric fraction of the transfected PrP constructs and of endogenous mouse PrPC in trans. It was reported previously that a secreted artificial PrP-immunoglobulin Fc (PrP-Fc2) fusion protein that forms disulfide bond–stabilized dimers was not converted into PrPSc and delayed onset of prion disease in transgenic mice (39). However, this study did not include the analysis of a variant of PrP-Fc2 deficient in disulfide bond formation that would have been secreted as a monomeric PrP-Fc fusion protein. Thus, it remains unclear whether the observed effects were due to the dimerization of PrP-Fc2 or the expression of an artificial PrP-immunoglobulin fusion protein.
How can dimerization of GPI-anchored PrPC interfere with PrPSc propagation (Fig. 5E)? Seminal studies in transgenic mice strongly suggested that PrPC directly interacts with PrPSc to initiate propagation (6, 7). Based on these findings, John Hardy (16) proposed in 1991 that PrPC exists as a dimer in equilibrium with monomeric PrPC. He hypothesized that only PrPC monomers interact with PrPSc and are converted by PrPSc, whereas PrPC dimers are protected (16). Our study addressed this hypothesis experimentally. By stabilizing PrPC dimers via the intermolecular disulfide bond, dissociation is prevented, thereby decreasing the fraction of PrPC monomers that can serve as PrPSc substrates for conversion. However, the inhibitory effect of dimeric PrPC on the conversion of monomeric PrPC points to a more complex scenario. In particular, one can envision the following, mutually not exclusive pathways. (i) PrPC dimers do not bind to PrPSc and therefore are not converted. This mode of action does not provide an obvious explanation for the inhibitory effect of dimeric PrPC on the conversion of monomeric PrPC. (ii) Dimeric PrPC still interacts with PrPSc; however, such interactions do not lead to the de novo formation of PrPSc either because the energy barrier for the conversion of PrPC dimers is too high or because the interaction surface between the PrPC dimer and PrPSc is different and not suited to initiate or complete conversion. Because we artificially introduced a covalent linkage to stabilize PrPC dimers, one cannot rule out that this modification and not dimerization of PrPC blocked the conversion. In particular, the artificial cross-link could limit the structural mobility of the protein in ways that a native dimer may not, resulting in the inhibition of prion conversion. (iii) PrPC dimers bind to PrPSc with a higher affinity than monomeric PrPC. As a consequence, interaction of monomeric PrPC with PrPSc is decreased, and its conversion is reduced. (iv) PrPSc in complex with PrPC dimers is subjected to increased intracellular degradation. It will now be interesting to explore the therapeutic potential of substances that can stabilize native PrPC dimers and hence block the conversion process.
Experimental procedures
Plasmids
Plasmid amplification and maintenance were carried out in Escherichia coli TOP10® (Thermo Fisher Scientific). The murine prion protein (GenBankTM accession number M18070) was modified to express PrP-L108M/V111M (40), allowing detection by the mAb 3F4 (29). The amino acid numbers refer to the human prion protein. All constructs generated for this study were developed by standard PCR cloning techniques. In some constructs, serine 132 was mutated to alanine or cysteine to enable the formation of a disulfide-bonded dimer (denoted A132 or C132). PrP(AV3) (A113V/A115V/A118V) was cloned from a plasmid encoding PrP(AV3,L9R) kindly provided by David Harris (41). All constructs described above were inserted into pcDNA3.1/Neo (+) vector (Invitrogen). If indicated, PrP mutants were equipped with a V5 tag (GGTAAACCGATACCGAACCCGCTCCTCGGTCTCGATTCGACG) or HA tag (TACCCATACGATGTTCCAGATTACGCT) inserted in the unstructured N-terminal region between amino acids 35 and 36.
Antibodies and reagents
The following antibodies were used: anti-PrP monoclonal antibodies 3F4 (29) and 4H11 (42), mouse monoclonal anti-V5 antibody (mAb R960CUS, Thermo Fisher Scientific), anti-HA (mAb MMS-101R, Covance), mouse monoclonal anti-GAPDH (mAb AM4300, Thermo Fisher Scientific), horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Thermo Fisher Scientific), and IRDye-conjugated secondary antibody (IR-Dye 800CW donkey anti-mouse, LI-COR Biosciences). All standard chemicals and reagents were purchased from Sigma-Aldrich if not otherwise noted. The following reagents were used: EndoH (New England Biolabs), PNGaseF (New England Biolabs), PIPLC (Thermo Fisher Scientific), trypsin (Thermo Fisher Scientific), monoclonal anti-HA-agarose beads (Sigma-Aldrich), cOmplete® Mini EDTA-free Protease Inhibitor Mixture (Roche Applied Science).
Cell lines, transfection, and lysis
Human HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) GlutaMAX (Thermo Fisher Scientific), and murine N2a cells were cultured in minimum essential medium (MEM; Thermo Fisher Scientific), both with the addition of 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were grown in a humidified 5% CO2 atmosphere at 37 °C. Cells cultivated on a 3.5-cm cell culture dish (Nunc, Roskilde, Denmark) were transfected with plasmid DNA by a liposome-mediated method using Lipofectamine® LTX and PLUSTM reagent (Life Technologies) according to the manufacturer's instructions. After 24 h, cells were washed twice with cold phosphate-buffered saline (PBS), scraped off the plate, pelleted by centrifugation (5,000 × g, 5 min), and lysed in detergent buffer (0.5% Triton X-100, 0.5% sodium deoxycholate in PBS). The cell lysates were either analyzed directly or centrifuged (20,000 × g, 10 min) to analyze the postnuclear supernatant.
N2a cells persistently infected with the mouse prions strain 22L (22L-ScN2a) (28) were cultivated in Opti-MEM GlutaMAX (Gibco). 22L-ScN2a cells were transfected using Lipofectamine® LTX and PLUSTM reagent according to the manufacturer's protocol. Briefly, 1 × 106 cells per plate were cultured in 10-cm plates. Plasmid (3 μg) and PLUSTM reagent were incubated along with Opti-MEM medium for 5 min and then added to the mixture of Lipofectamine® LTX reagent and Opti-MEM. The solution was kept for 15 min at room temperature. This solution was added drop by drop to cells and gently mixed. Cells were incubated at 37 °C. After 48 h, a second transfection was done as described above. 96 h after the first transfection, cells were lysed in cold lysis buffer (10 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm EDTA, 0.5% Triton X-100, 0.5% sodium deoxycholate) for 10 min. One half of the lysate from each plate was incubated with PK at a final concentration of 20 μg/ml for 30 min at 37 °C. Proteinase inhibitor (0.5 mm Pefabloc) was added to inhibit PK digestion, and samples were precipitated in methanol. To the untreated sample, Pefabloc was added before methanol precipitation. Precipitated proteins were resuspended in TNE buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA).
Deglycosylation (PNGaseF and EndoH), phospholipase C treatment, and trypsin digestion
To deglycosylate proteins, cell lysates were treated with PNGaseF or endoglycosidase H for 1 h at 37 °C according to the manufacturer's instructions. For PIPLC treatment, cells were washed twice with PBS. PIPLC diluted in PBS was added to the cells for 4 h at 37 °C. Secreted PrP was precipitated by trichloroacetic acid (TCA) and analyzed by Western blotting. To analyze the localization of the prion protein and the generated mutants, cells were digested with trypsin, a member of the serine protease S1 family that digests cell surface proteins. Cells were washed twice with PBS and treated with trypsin for 25 min at room temperature. The reaction was terminated by the addition of cOmplete Mini EDTA-free Protease Inhibitor Mixture. Cell lysates were further analyzed by Western blotting.
Coimmunoprecipitation
To analyze formation of dimers, N2a or HeLa cells were cotransfected with the indicated constructs (V5- or HA-tagged) and lysed in detergent buffer (0.5% (v/v) Triton X-100, 0.5% (w/v) deoxycholate in PBS). Postnuclear supernatants were incubated with anti-HA-agarose beads under nonreducing conditions in the case of disulfide-stabilized coimmunoprecipitation and under reducing conditions for the native coimmunoprecipitation (overnight, 4 °C, rotating). The immunocomplex was washed with lysis buffer and PBS and further analyzed by Western blotting.
Western blotting
For Western blot analysis, lysates were boiled in Laemmli sample buffer with or without β-mercaptoethanol (4%, v/v). Following SDS-PAGE, proteins were transferred to nitrocellulose by electroblotting. Membranes were blocked by incubation in TBS-T (TBS with 0.1% (w/v) Tween 20) containing 5% skimmed milk for 1 h at room temperature and incubated with primary antibody in TBS-T + 5% skimmed milk for 18 h at 4 °C. After washing with TBS-T, blots were incubated with respective secondary antibody (IRDye-IR Technology, LI-COR Biosciences; or HRP) in TBS-T for 1 h at room temperature. Protein signals were visualized using an Odyssey® 9120 scanner. Peroxidase activity was detected by enhanced chemiluminescence (ECL) (Promega).
To analyze PrP in persistently scrapie-infected 22L-ScN2a cells, Western blot analysis was performed as described previously (43). Briefly, samples (±PK) were subjected to 12.5% SDS-PAGE and electroblotted on Hybond P 0.45-μm PVDF membranes (Amersham Biosciences). Anti-PrP mAb 3F4 or 4H11 was used as primary antibody, and goat anti-mouse HRP antibody was used as secondary antibody. The detection of signal in the immunoblot was done using Luminata Western chemiluminescent HRP substrate (Millipore).
Quantification
Dimerization efficiency of the constructs was measured densitometrically (Image Studio Lite) as the percentage of dimer fraction relative to total protein amount. In ScN2a cells, the amount of total PrP and PrPSc was also measured densitometrically (ImageJ software), and the relative amount of PrPSc present in cells expressing WTPrPC was set as 1. Data represent mean ± S.D. of ≥3 independent experiments. The standard deviation was determined using a Student's t test (*, p < 0.05; ns, not significant).
Immunofluorescence analysis
Transiently transfected HeLa cells were grown on glass coverslips and fixed 24 h after transfection with 4% paraformaldehyde (10 min). One set was permeabilized (0.2% Triton in PBS), and one set was left unpermeabilized. Both sets were blocked in PBS with 5% normal goat serum for 1 h and incubated with anti-3F4 antibody overnight at 4 °C (in PBS containing 5% normal goat serum). After washing with PBS, incubation with the Cy3-conjugated anti-mouse secondary antibody (Alexa Fluor 488) followed for 1 h. Cells were mounted onto glass slides (with Fluoromount-G, Thermo Fisher Scientific) and examined by fluorescence microscopy (Zeiss ELYRA PS.1 and LSM 880).
Author contributions
A. D. E., G. M., M. B., M. E., H. M. S., K. F. W., and J. T. conceptualization; A. D. E., G. M., M. B., H. M. S., K. F. W., and J. T. supervision; J. T. funding acquisition; A. D. E., A. G., S. T., S. J., S. U., R. P. S., S. B., and G. M. investigation; A. D. E., A. G., S. T., S. J., and S. U. visualization; A. D. E., K. F. W., and J. T. writing-original draft; A. D. E., A. G., S. T., S. J., S. U., R. P. S., S. B., G. M., M. B., M. E., H. M. S., and K. F. W. writing-review and editing; R. P. S. and S. B. methodology; M. B. and M. E. resources.
Supplementary Material
Acknowledgments
We are grateful to Petra Goldmann, Barbara Kachholz, and Andrea Roth-Sturm for technical support.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant TA167/6 (to J. T.), Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen Grant Az 233-1.08.03.03-031.68079, National Institutes of Health Grant R01 NS076853-01A1 (to H. M. S.), and Calgary Prion Research Unit, Alberta Prion Research Institute Grant 201600010 (to H. M. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Fig. S1.
- PrPC
- cellular prion protein
- ME
- β-mercaptoethanol
- HD
- hydrophobic domain
- GPI
- glycosylphosphatidylinositol
- PrPSc
- PrP scrapie
- PIPLC
- phosphatidylinositol-specific phospholipase C
- EndoH
- endoglycosidase H
- PNGaseF
- peptide:N-glycosidase F
- PK
- proteinase K.
References
- 1. Collinge J. (2001) Prion diseases of humans and animals: their causes and molecular basis. Annu. Rev. Neurosci. 24, 519–550 10.1146/annurev.neuro.24.1.519 [DOI] [PubMed] [Google Scholar]
- 2. Prusiner S. B., Scott M. R., DeArmond S. J., and Cohen F. E. (1998) Prion protein biology. Cell 93, 337–348 10.1016/S0092-8674(00)81163-0 [DOI] [PubMed] [Google Scholar]
- 3. Weissmann C., Fischer M., Raeber A., Büeler H., Sailer A., Shmerling D., Rülicke T., Brandner S., and Aguzzi A. (1996) The role of PrP in pathogenesis of experimental scrapie. Cold Spring Harb. Symp. Quant. Biol. 61, 511–522 10.1101/SQB.1996.061.01.051 [DOI] [PubMed] [Google Scholar]
- 4. Chesebro B. (2003) Introduction to the transmissible spongiform encephalopathies or prion diseases. Br. Med. Bull. 66, 1–20 10.1093/bmb/66.1.1 [DOI] [PubMed] [Google Scholar]
- 5. Büeler H., Aguzzi A., Sailer A., Greiner R.-A., Autenried P., Aguet M., and Weissmann C. (1993) Mice devoid of PrP are resistant to scrapie. Cell 73, 1339–1347 10.1016/0092-8674(93)90360-3 [DOI] [PubMed] [Google Scholar]
- 6. Prusiner S. B., Scott M., Foster D., Pan K.-M., Groth D., Mirenda C., Torchia M., Yang S.-L., Serban D., Carlson G. A., Hoppe P. C., Westaway D., and DeArmond S. J. (1990) Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63, 673–686 10.1016/0092-8674(90)90134-Z [DOI] [PubMed] [Google Scholar]
- 7. Scott M., Foster D., Mirenda C., Serban D., Coufal F., Wälchli M., Torchia M., Groth D., Carlson G., DeArmond S. J., Westaway D., and Prusiner S. B. (1989) Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59, 847–857 10.1016/0092-8674(89)90608-9 [DOI] [PubMed] [Google Scholar]
- 8. Stahl N., Borchelt D. R., Hsiao K., and Prusiner S. B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240 10.1016/0092-8674(87)90150-4 [DOI] [PubMed] [Google Scholar]
- 9. Stahl N., Borchelt D. R., and Prusiner S. B. (1990) Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry 29, 5405–5412 10.1021/bi00474a028 [DOI] [PubMed] [Google Scholar]
- 10. Endo T., Groth D., Prusiner S. B., and Kobata A. (1989) Diversity of oligosaccharide structures linked to asparagines of the scrapie prion protein. Biochemistry 28, 8380–8388 10.1021/bi00447a017 [DOI] [PubMed] [Google Scholar]
- 11. Haraguchi T., Fisher S., Olofsson S., Endo T., Groth D., Tarentino A., Borchelt D. R., Teplow D., Hood L., Burlingame A., Lycke E., Kobata A., and Prusiner S. B. (1989) Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch. Biochem. Biophys. 274, 1–13 10.1016/0003-9861(89)90409-8 [DOI] [PubMed] [Google Scholar]
- 12. Caughey B., and Raymond G. J. (1991) The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 266, 18217–18223 [PubMed] [Google Scholar]
- 13. Neuendorf E., Weber A., Saalmueller A., Schatzl H., Reifenberg K., Pfaff E., and Groschup M. H. (2004) Glycosylation deficiency at either one of the two glycan attachment sites of cellular prion protein preserves susceptibility to bovine spongiform encephalopathy and scrapie infections. J. Biol. Chem. 279, 53306–53316 10.1074/jbc.M410796200 [DOI] [PubMed] [Google Scholar]
- 14. Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., and Oldstone M. (2005) Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435–1439 10.1126/science.1110837 [DOI] [PubMed] [Google Scholar]
- 15. Stöhr J., Watts J. C., Legname G., Oehler A., Lemus A., Nguyen H. O., Sussman J., Wille H., DeArmond S. J., Prusiner S. B., and Giles K. (2011) Spontaneous generation of anchorless prions in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 21223–21228 10.1073/pnas.1117827108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hardy J. (1991) Prion dimers: a deadly duo. Trends Neurosci. 14, 423–424 10.1016/0166-2236(91)90038-V [DOI] [PubMed] [Google Scholar]
- 17. Priola S. A., Caughey B., Wehrly K., and Chesebro B. (1995) A 60-kDa prion protein (PrP) with properties of both the normal and scrapie-associated forms of PrP. J. Biol. Chem. 270, 3299–3305 10.1074/jbc.270.7.3299 [DOI] [PubMed] [Google Scholar]
- 18. Meyer R. K., Lustig A., Oesch B., Fatzer R., Zurbriggen A., and Vandevelde M. (2000) A monomer-dimer equilibrium of a cellular prion protein (PrPC) not observed with recombinant PrP. J. Biol. Chem. 275, 38081–38087 10.1074/jbc.M007114200 [DOI] [PubMed] [Google Scholar]
- 19. Rambold A. S., Müller V., Ron U., Ben-Tal N., Winklhofer K. F., and Tatzelt J. (2008) Stress-protective activity of prion protein is corrupted by scrapie-prions. EMBO J. 27, 1974–1984 10.1038/emboj.2008.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sakthivelu V., Seidel R. P., Winklhofer K. F., and Tatzelt J. (2011) Conserved stress-protective activity between prion protein and shadoo. J. Biol. Chem. 286, 8901–8908 10.1074/jbc.M110.185470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Warwicker J. (2000) Modeling a prion protein dimer: predictions for fibril formation. Biochem. Biophys. Res. Commun. 278, 646–652 10.1006/bbrc.2000.3829 [DOI] [PubMed] [Google Scholar]
- 22. Hegde R. S., Mastrianni J. A., Scott M. R., DeFea K. A., Tremblay P., Torchia M., DeArmond S. J., Prusiner S. B., and Lingappa V. R. (1998) A transmembrane form of the prion protein in neurodegenerative disease. Science 279, 827–834 10.1126/science.279.5352.827 [DOI] [PubMed] [Google Scholar]
- 23. Cao H., Bangalore L., Dompé C., Bormann B. J., and Stern D. F. (1992) An extra cysteine proximal to the transmembrane domain induces differential cross-linking of p185neu and p185neu. J. Biol. Chem. 267, 20489–20492 [PubMed] [Google Scholar]
- 24. Munter L. M., Voigt P., Harmeier A., Kaden D., Gottschalk K. E., Weise C., Pipkorn R., Schaefer M., Langosch D., and Multhaup G. (2007) GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 26, 1702–1712 10.1038/sj.emboj.7601616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tatzelt J., and Winklhofer K. F. (2004) Folding and misfolding of the prion protein in the secretory pathway. Amyloid 11, 162–172 10.1080/1350-6120400000723 [DOI] [PubMed] [Google Scholar]
- 26. Butler D. A., Scott M. R., Bockman J. M., Borchelt D. R., Taraboulos A., Hsiao K. K., Kingsbury D. T., and Prusiner S. B. (1988) Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol. 62, 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Race R. E., Fadness L. H., and Chesebro B. (1987) Characterization of scrapie infection in mouse neuroblastoma cells. J. Gen. Virol. 68, 1391–1399 10.1099/0022-1317-68-5-1391 [DOI] [PubMed] [Google Scholar]
- 28. Bach C., Gilch S., Rost R., Greenwood A. D., Horsch M., Hajj G. N., Brodesser S., Facius A., Schädler S., Sandhoff K., Beckers J., Leib-Mösch C., Schätzl H. M., and Vorberg I. (2009) Prion-induced activation of cholesterogenic gene expression by Srebp2 in neuronal cells. J. Biol. Chem. 284, 31260–31269 10.1074/jbc.M109.004382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., and Diringer H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Taguchi Y., Mistica A. M., Kitamoto T., and Schätzl H. M. (2013) Critical significance of the region between Helix 1 and 2 for efficient dominant-negative inhibition by conversion-incompetent prion protein. PLoS Pathog. 9, e1003466 10.1371/journal.ppat.1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Laurén J., Gimbel D. A., Nygaard H. B., Gilbert J. W., and Strittmatter S. M. (2009) Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 10.1038/nature07761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Resenberger U. K., Harmeier A., Woerner A. C., Goodman J. L., Müller V., Krishnan R., Vabulas R. M., Kretzschmar H. A., Lindquist S., Hartl F. U., Multhaup G., Winklhofer K. F., and Tatzelt J. (2011) The cellular prion protein mediates neurotoxic signalling of β-sheet-rich conformers independent of prion replication. EMBO J. 30, 2057–2070 10.1038/emboj.2011.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ferreira D. G., Temido-Ferreira M., Miranda H. V., Batalha V. L., Coelho J. E., Szegö É. M., Marques-Morgado I., Vaz S. H., Rhee J. S., Schmitz M., Zerr I., Lopes L. V., and Outeiro T. F. (2017) α-Synuclein interacts with PrPC to induce cognitive impairment through mGluR5 and NMDAR2B. Nat. Neurosci. 20, 1569–1579 10.1038/nn.4648 [DOI] [PubMed] [Google Scholar]
- 34. Mouillet-Richard S., Ermonval M., Chebassier C., Laplanche J. L., Lehmann S., Launay J. M., and Kellermann O. (2000) Signal transduction through prion protein. Science 289, 1925–1928 10.1126/science.289.5486.1925 [DOI] [PubMed] [Google Scholar]
- 35. Khosravani H., Zhang Y., Tsutsui S., Hameed S., Altier C., Hamid J., Chen L., Villemaire M., Ali Z., Jirik F. R., and Zamponi G. W. (2008) Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J. Cell Biol. 181, 551–565 10.1083/jcb.200711002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chiarini L. B., Freitas A. R., Zanata S. M., Brentani R. R., Martins V. R., and Linden R. (2002) Cellular prion protein transduces neuroprotective signals. EMBO J. 21, 3317–3326 10.1093/emboj/cdf324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shmerling D., Hegyi I., Fischer M., Blättler T., Brandner S., Götz J., Rülicke T., Flechsig E., Cozzio A., von Mering C., Hangartner C., Aguzzi A., and Weissmann C. (1998) Expression of animo-terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell 93, 203–214 10.1016/S0092-8674(00)81572-X [DOI] [PubMed] [Google Scholar]
- 38. Wu B., McDonald A. J., Markham K., Rich C. B., McHugh K. P., Tatzelt J., Colby D. W., Millhauser G. L., and Harris D. A. (2017) The N-terminus of the prion protein is a toxic effector regulated by the C-terminus. Elife 6, e23473 10.7554/eLife.23473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meier P., Genoud N., Prinz M., Maissen M., Rülicke T., Zurbriggen A., Raeber A. J., and Aguzzi A. (2003) Soluble dimeric prion protein binds PrP(Sc) in vivo and antagonizes prion disease. Cell 113, 49–60 10.1016/S0092-8674(03)00201-0 [DOI] [PubMed] [Google Scholar]
- 40. Winklhofer K. F., Heller U., Reintjes A., and Tatzelt J. (2003) Inhibition of complex glycosylation increases formation of PrPSc. Traffic 4, 313–322 10.1034/j.1600-0854.2003.00088.x [DOI] [PubMed] [Google Scholar]
- 41. Stewart R. S., Drisaldi B., and Harris D. A. (2001) A transmembrane form of the prion protein contains an uncleaved signal peptide and is retained in the endoplasmic reticulum. Mol. Biol. Cell 12, 881–889 10.1091/mbc.12.4.881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ertmer A., Gilch S., Yun S. W., Flechsig E., Klebl B., Stein-Gerlach M., Klein M. A., and Schätzl H. M. (2004) The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. J. Biol. Chem. 279, 41918–41927 10.1074/jbc.M405652200 [DOI] [PubMed] [Google Scholar]
- 43. Gilch S., Winklhofer K. F., Groschup M. H., Nunziante M., Lucassen R., Spielhaupter C., Muranyi W., Riesner D., Tatzelt J., and Schätzl H. M. (2001) Intracellular re-routing of prion protein prevents propagation of PrPSc and delays onset of prion diseases. EMBO J. 20, 3957–3966 10.1093/emboj/20.15.3957 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.