Abstract
Connexin-43 (Cx43, also known as GJA1) is the most ubiquitously expressed connexin isoform in mammalian tissues. It forms intercellular gap junction (GJ) channels, enabling adjacent cells to communicate both electrically and metabolically. Cx43 is a short-lived protein which can be quickly degraded by the ubiquitin-dependent proteasomal, endolysosomal, and autophagosomal pathways. Here, we report that the ubiquitin-specific peptidase 8 (USP8) interacts with and deubiquitinates Cx43. USP8 reduces both multiple monoubiquitination and polyubiquitination of Cx43 to prevent autophagy-mediated degradation. Consistently, knockdown of USP8 results in decreased Cx43 protein levels in cultured cells and suppresses intercellular communication, revealed by the dye transfer assay. In human breast cancer specimens, the expression levels of USP8 and Cx43 proteins are positively correlated. Taken together, these results identified USP8 as a crucial and bona fide deubiquitinating enzyme involved in autophagy-mediated degradation of Cx43.
Keywords: ubiquitin ligase, ubiquitylation (ubiquitination), autophagy, connexin, deubiquitylation (deubiquitination), USP8, Cx43, ubiquitin, degradation, autophagy
Introduction
The connexins represent a family of transmembrane proteins that form the intercellular gap junction (GJ)3 channels at the plasma membrane of cell–cell contacts between adjacent cells, enabling direct intercellular exchange of low-molecular-weight molecules which include nucleotides, amino acids, sugars, and signaling mediators such as inositol trisphosphate (IP3) and cAMP (1, 2). Intercellular communication via GJ channels plays an essential role in coordinating activities of individual cells in tissues. Its dysfunction has been implicated as a causative factor in many human diseases including heart failure, neuropathology, deafness, skin disorders, and cataracts (3). In humans, there are 21 known members of the connexin family protein, of which the best-characterized member is connexin-43 (Cx43, also known as GJA1) (4). Cx43 is ubiquitously expressed in many tissues, and Cx43-containing GJ channel is critical for various physiological functions such as cardiac excitability. Aberrant Cx43 expression is found in several types of tumors including liver, breast, and prostate, and has been suggested to play important role in tumor genesis, tumor cell proliferation, differentiation, and metastasis (5, 6). However, the molecular mechanism underlying the deregulated Cx43 expression in the malignant cells remains elusive.
GJ channels are highly dynamic, and the connexins are short-lived proteins with a half-life of only 1–5 h in cultured cells and in most types of tissues (7). The connexins including Cx43 have long been found to be posttranslationally regulated by the ubiquitin (Ub) system (8, 9). The conjugation of ubiquitin chains to Cx43 targets the protein for degradation through the proteasomal, endolysosomal, and autophagosomal pathways (10–12). Many earlier studies using proteasome inhibitors showed that Cx43 is degraded by proteasome (13, 14). However, recent studies confirmed that only the endoplasmic reticulum–dislocated unfolded Cx43 is subjected to degradation by proteasome (15). The proteasome inhibitor modulates Cx43 protein level and location at the plasma membrane mostly via indirect mechanisms. There is increasing evidence showing that cells appear to continuously internalize and turn over their GJs via a combined endocytic/exocytic process which utilizes clathrin-mediated endocytosis components (16). The endocytosed GJs form a double-membrane vacuole called annular gap junction (AGJ) or connexosome (17, 18). The AGJ may be recycled to the cell surface, or destined for degradation through the endolysosomal and/or autophagosomal pathways (19, 20). Ubiquitination of Cx43 may occur at several subcellular locations and is involved in multiple stages of degradation. At first, ubiquitination serves as a main signal for endocytosis of Cx43-based GJs from the plasma membrane and for the formation of cytoplasmic AGJs (18, 21). In addition, ubiquitination promotes Cx43 to be sorted into lysosomes or to be fused to autophagosomes (11, 17, 19, 22). The latter is eventually fused with lysosomes where the contents are degraded. Cx43 ubiquitination was reported to be regulated by several E3 ubiquitin ligases including NEDD4 (20), WWP1 (23), and TRIM21 (24). They may act at different stages/levels in the regulation of the intracellular trafficking and degradation of Cx43-containing GJs. For example, NEDD4 promotes endocytosis of Cx43, and controls its degradation via either the endolysosomal or autophagosomal pathway (20, 21).
Ubiquitination is a reversible posttranslational modification, and deubiquitination of proteins is catalyzed by a family of protein consisting of ∼100 enzymes known as deubiquitinases (DUBs) in human (25). Recently, the associated molecule with the Src homology 3 domain (AMSH) of the signal transducing adapter molecule (STAM), an endosome-associated DUB, was found to be capable of removing lysine 63–linked polyubiquitin chains from Cx43 to regulate the internalization and degradation of Cx43-containing GJ channels (26). However, other DUBs potentially involved in modulating the ubiquitination level and degradation of Cx43 need to be further elucidated.
USP8, also known as UBPY, is another endosome-associated DUB that belongs to the ubiquitin-specific protease (USP) family and plays an important role in the endosomal sorting of many proteins (27–31). USP8 can remove Lys48-, Lys63-, and Lys6-linked ubiquitin chains (32, 33). More recently, we and others reported that USP8 gene is recurrently mutated in human pituitary corticotrophin adenomas, and USP8 mutation causes disease via up-regulation of EGFR (34, 35).
Here, we show evidences that Cx43 protein levels are directly regulated by USP8. USP8 modulates multi-monoubiquitination as well as Lys48- and Lys63-linked polyubiquitination of Cx43 to prevent it from autophagy-mediated degradation. USP8 expression positively correlates with Cx43 expression in human breast cancers.
Results
USP8 interacts with Cx43
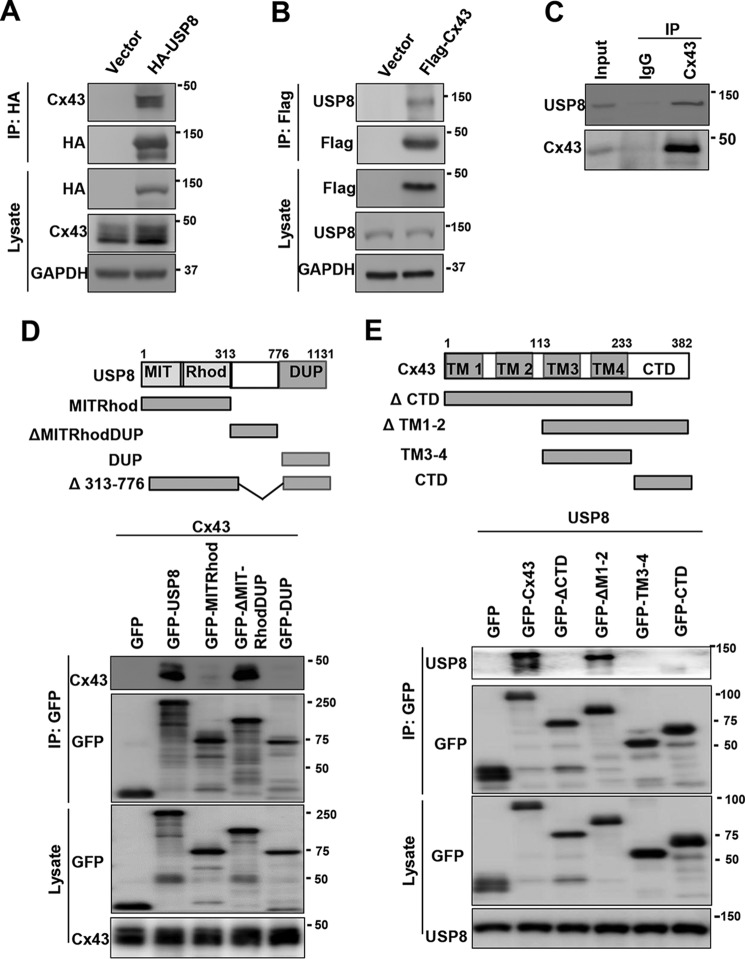
The ubiquitin-dependent degradation of Cx43 involves the endolysosomal and autophagosomal pathways, suggesting that endosomal DUBs may play a crucial role in controlling this process. We reasoned that USP8, an endosomal DUB, may regulate the ubiquitination and degradation of Cx43. We first examined whether USP8 could interact with Cx43. To this end, we transfected FLAG-USP8 or control vector into HEK293T cells which expresses endogenous Cx43 and USP8. We found that endogenous Cx43 was co-immunoprecipitated with anti-HA antibody only in HA-USP8 transfected cells (Fig. 1A). Similarly, endogenous USP8 was co-immunoprecipitated with anti-FLAG antibody only in FLAG-Cx43 transfected cells (Fig. 1B). Furthermore, endogenous USP8 was specifically immunoprecipitated by anti-Cx43 antibody but not control IgG (Fig. 1C). These results suggest that USP8 interacts with Cx43 in the cells. To determine which domain of USP8 interacts with Cx43, we constructed a panel of GFP-tagged USP8 deletion mutants, including the N-terminal MIT and Rhod domain (amino acids (aa) 1–313), central domain (aa 313–776), and C-terminal USP domain (aa 776–1131) mutants. We co-introduced GFP-USP8 or its deletion mutants with Cx43 into HEK293T cells, and performed immunoprecipitation assay (Fig. 1D, top panel). We found that the central domain of USP8 was necessary and sufficient for binding to Cx43 (Fig. 1D, lane 4). To test which domain of Cx43 interacts with USP8, we then constructed a panel of GFP-tagged Cx43 deletion mutants (Fig. 1E, top panel). We co-expressed GFP-Cx43 or its deletion mutants together with USP8 into HEK293T cells and performed immunoprecipitation assay. We found that the first and second transmembrane (TM) domains (TM1–2) of Cx43 were not required for interacting with USP8 (Fig. 1E). However, either the third and fourth transmembrane domains (TM3–4) or the C-terminal domain of Cx43 was indispensable, but not sufficient for the interaction with USP8 (Fig. 1E).
Figure 1.
USP8 interacts with Cx43. A, HEK293T cells were transfected with HA-USP8 expression or control vectors, immunoprecipitated with HA beads and immunoblotted with antibodies against Cx43 and HA. B, HEK293T cells were transfected with FLAG-USP8 expression or control vector, immunoprecipitated with FLAG beads and immunoblotted with antibodies against USP8 and FLAG. C, HEK293T cells were immunoprecipitated with anti-Cx43 antibody, and the precipitated complex was immunoblotted with antibodies against USP8 and Cx43. D, schematic representation of GFP-USP8 or its deletion mutants (top panel). HEK293T cells were co-transfected with Cx43 and GFP-USP8 or its deletion mutants, immunoprecipitated with GFP beads, and immunoblotted with antibodies against GFP and Cx43. E, schematic representation of GFP-Cx43 or its deletion mutants (top panel). HEK293T cells were co-transfected with USP8 and GFP-Cx43 or its deletion mutants, immunoprecipitated with GFP beads, and immunoblotted with antibodies against GFP and USP8.
USP8 deubiquitinates and stabilizes Cx43
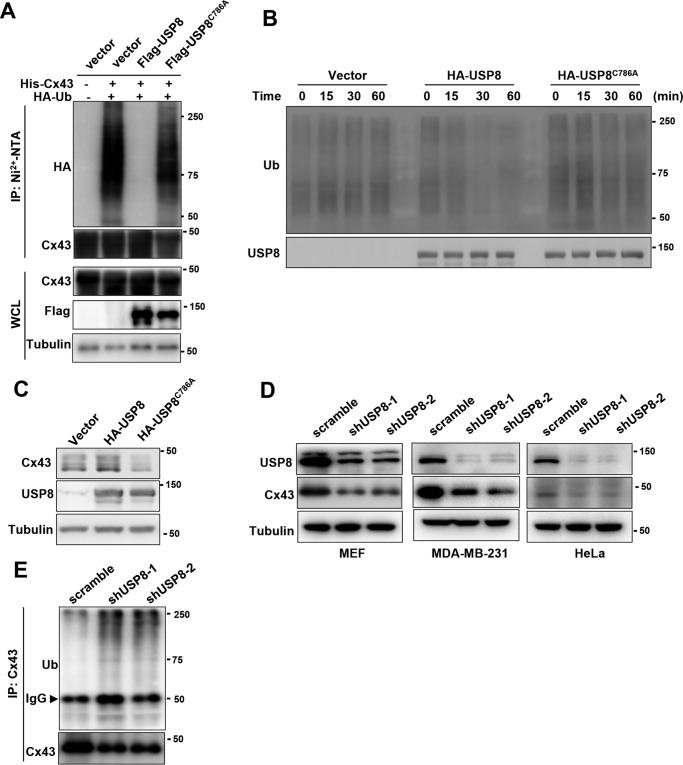
USP8 is a cysteine protease, which directly deubiquitinates and stabilizes many substrates. We next examined whether USP8 regulates the ubiquitination of Cx43. To this end, we performed an in vivo ubiquitination assay and found that USP8 significantly reduced the levels of ubiquitinated species of Cx43 (Fig. 2A). This effect required the DUB activity of USP8, as the catalytic inactive mutant (USP8C786A) failed to reduce the ubiquitination of Cx43 (Fig. 2A). To determine whether USP8 directly deubiquitinates Cx43 in vitro, we purified ubiquitinated Cx43 from HEK293T cells co-transfected with HA-Cx43 and His-ubiquitin using affinity purification with anti-HA agarose beads and HA-peptide elution. FLAG-USP8 or USP8C786A was purified from HEK293T cells transfected with FLAG-USP8 or USP8C786A expression vectors using affinity purification with anti-FLAG agarose beads and FLAG-peptide elution. The ubiquitinated Cx43 was incubated with purified WT USP8 or USP8C786A for indicated times, followed by immunoblotting with anti-ubiquitin antibody. As shown in Fig. 2B, WT USP8, but not the C786A mutant, markedly reduced the ubiquitinated species of Cx43, indicating that USP8 directly deubiquitinates Cx43. We next asked whether USP8 is able to regulate the protein level of Cx43. We found that overexpression of USP8 increased the levels of endogenous Cx43 in HEK293T cells. Of note, the C786A mutant suppressed the protein levels of endogenous Cx43, suggesting a domain-negative effect of this mutant (Fig. 2C). The central domain of USP8 is necessary for its interaction with Cx43 (Fig. 1D). USP8 mutant which lacks the central domain failed to deubiquitinate and stabilize Cx43 (Fig. S1). Furthermore, knockdown of endogenous USP8 in MEF cells significantly reduced Cx43 protein expression and enhanced its ubiquitination level (Fig. 2, D and E). USP8 knockdown also decreased Cx43 protein levels in two human cells lines (Fig. 2D). These results reveal that USP8 is a bona fide Cx43 deubiquitinase.
Figure 2.
USP8 deubiquitinates Cx43 and increases its protein level. A, HEK293T cells were co-transfected with His-Cx43, HA-ubiquitin (Ub), and FLAG-USP8 or FLAG-USP8C786A. After 48h, the cells were subjected to pulldown using the Ni2+-NTA beads under denaturation conditions, followed by immunoblotting. B, ubiquitinated FLAG-Cx43 was incubated with HA-USP8 or USP8C786A purified from HEK293T cells for indicated times. The reaction mixture was analyzed by immunoblotting using antibodies against Ub and USP8. C, immunoblotting of Cx43 and USP8 in HEK293T cells transfected with HA-USP8 or HA-USP8C786A expression vector. D, immunoblotting of USP8 and Cx43 in indicated cell lines stably transfected with scramble and USP8 shRNA. E, MEFs stably transfected with scramble or USP8 shRNA were immunoprecipitated with anti-Cx43 antibody and immunoblotted with antibodies against Cx43 and Ub.
USP8 protects Cx43 from autophagy-mediated degradation
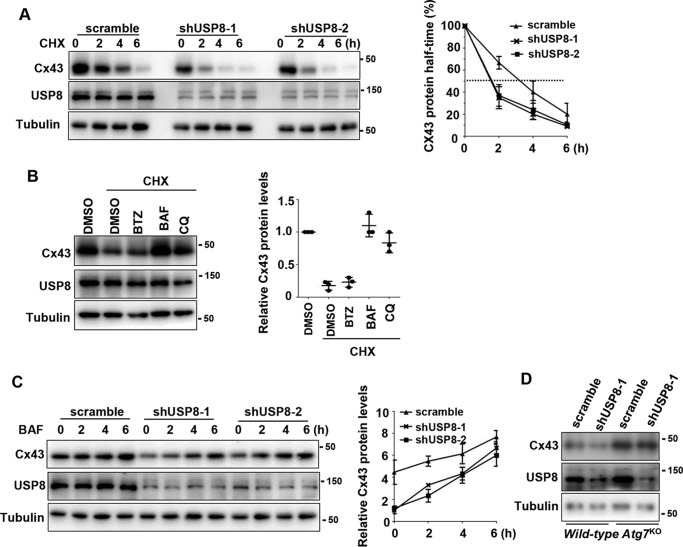
Based on the fact that Cx43 protein has a short half-time, and USP8 knockdown led to reduction of endogenous Cx43 protein levels, we proposed that USP8 could control the stability of Cx43 protein. To test this hypothesis, we treated control or USP8 knockdown MEF cells with a protein synthesis inhibitor cycloheximide (CHX). As shown in Fig. 3A, USP8 knockdown remarkably shortened the half-time of Cx43 protein. The similar results were observed in MDA-MB-231 cells (Fig. S2A). These results demonstrate a critical role of USP8 in maintaining the stability of Cx43 protein. Degradation of Cx43 in MEFs has previously been ascribed mainly to the autophagosomal pathway (19). Indeed, we found that treatment with bafilomycin A1 (BAF), an inhibitor which blocks the lysosome/autophagosome fusion and thereby prevents the autophagic degradation pathway, completely blocked the degradation of Cx43 protein in MEFs in the present of CHX. The lysosome inhibitor chloroquine was also effective, although to a lesser extent, in inhibiting Cx43 degradation (Fig. 3B). On the contrary, the proteasome inhibitor bortezomib failed to prevent Cx43 degradation (Fig. 3B). Furthermore, we found that BAF treatment significantly slowed down the degradation of Cx43 in USP8 knockdown MEFs (Fig. 3C). In MDA-MB-231 cells, we also found that Cx43 degradation was blocked by BAF, and BAF prevented Cx43 degradation caused by USP8 knockdown (Fig. S2, B and C). These results indicate that USP8 knockdown promotes the autophagosomal degradation of Cx43. To further confirm this idea, we examined the effect of USP8 knockdown on Cx43 protein levels in Atg7-null MEFs, which are deficient in autophagy. We found that USP8 knockdown did not lead to reduction of Cx43 protein in autophagy-deficient MEFs (Fig. 3D). Taken together, these results demonstrate that Cx43 is degraded mainly via the autophagic degradation pathway in MEFs and USP8 regulates this process.
Figure 3.
USP8 protects Cx43 from autophagy-mediated degradation. A, immunoblotting of Cx43 and USP8 protein in scramble or shUSP8-infected MEFs in the presence of CHX (50 mg/ml). B, immunoblotting of Cx43 protein in MEFs treated with bortezomib (BTZ) (50 nm), BAF (200 nm), or chloroquine (CQ) (50 μ m) for 6 h in the presence of CHX (50 mg/ml). C, immunoblotting of Cx43 protein in scramble or shUSP8-infected MEFs treated with BAF (200 nm) for indicated times. A–C, quantification of the immunoblots comes from three independent experiments (mean ± S.D.) and shown at the right. D, immunoblotting of Cx43 and USP8 protein in WT and Atg7-null MEFs stably infected with indicated lentivirus expressing scramble or USP8 shRNA.
USP8 reduces multi-monoubiquitination and polyubiquitination of Cx43
Autophagy modulates the dynamics of Cx43 at the plasma membrane in an ubiquitin-dependent manner. We next attempted to determine which types of ubiquitin moieties are attached to Cx43 and modified by USP8. It is generally thought that monoubiquitination, multi-monoubiquitination (monoubiquitination at multiple sites), and Lys63-linked polyubiquitin chains are responsible for the internalization and endocytic sorting of plasma membrane proteins (36), whereas Lys48-linked polyubiquitin chains target proteins for proteasome-mediated degradation (37). However, both Lys63- and Lys48-linked polyubiquitin chains can be recognized by the autophagy adaptor p62/SQSTM1 via its UBA-domain, leading to delivery of endocytosed polyubiquitinated protein complexes to the autophagic degradation pathway (38).
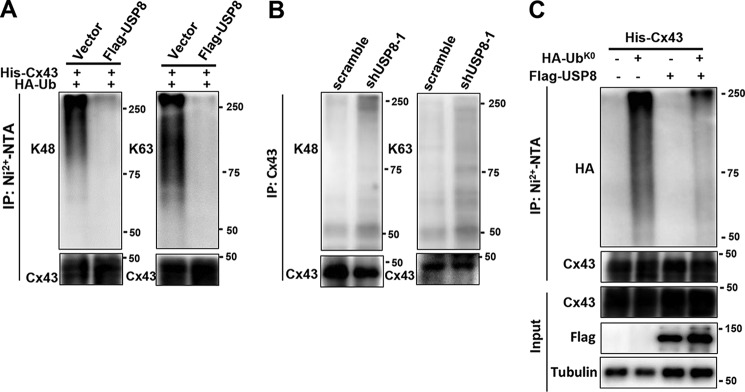
It was reported that Cx43 can be modified with Lys63-linked polyubiquitin chains, and to a lesser extent, Lys48-linked polyubiquitin chains (26). We first tested the effect of USP8 on Lys48- and Lys63-linked polyubiquitination of Cx43. To this end, we transfected His-Cx43, HA-ubiquitin, and FLAG-USP8 into HEK293T cells. His-Cx43 was purified using Ni2+-nitrilotriacetic acid (Ni2+-NTA) at the stringent condition to ensure no other protein was retained, and the attached ubiquitin chains were probed using antibodies specific for Lys63- or Lys48-linked polyubiquitin chain. We found that overexpression of FLAG-USP8 led to a significant reduction of Cx43 modified with Lys63- or Lys48-linked polyubiquitin chains (Fig. 4A). To further determine whether USP8 was able to remove Lys48- or Lys63-linked polyubiquitin chains from Cx43, endogenous Cx43 immunopurified from MEFs stably transfected with scramble or shUSP8 was probed with antibodies specific for Lys63- or Lys48-linked polyubiquitin chains. As shown in Fig. 4B, USP8 knockdown resulted in a robust accumulation of Cx43 modified with Lys63- and Lys48-linked polyubiquitin chains.
Figure 4.
USP8 regulates multi-monoubiquitination and polyubiquitination of Cx43. A, HEK293T cells were co-transfected with His-Cx43, HA-Ub, and FLAG-USP8, immunoprecipitated with Ni2+-NTA beads and immunoblotted antibodies against Cx43, Lys48- or Lys63-linked polyubiquitin chains. B, MEFs transfected with scramble or shUSP8 were immunoprecipitated with anti-Cx43 antibody and immunoblotted with antibodies against Cx43, Lys48- or Lys63-linked polyubiquitin chains. C, HEK293T cells were co-transfected with His-Cx43, HA-UbK0, and FLAG-USP8, immunoprecipitated with Ni2+-NTA beads and immunoblotted with antibodies against Cx43 and HA.
To determine whether Cx43 could be monoubiquitinated, FLAG-Cx43 was coexpressed with a HA-tagged mutant form of ubiquitin with all lysine residues mutated to arginine (HA-Ubk0) in HEK293T cells. In this case, addition of HA-Ubk0 prevents the formation of polyubiquitin chains, generating modified proteins with one or more sites monoubiquitinated. As shown in Fig. 4C, FLAG-Cx43 can be monoubiquitinated at multiple sites by HA-Ubk0. Importantly, the incorporation of HA-Ubk0 into FLAG-Cx43 was significantly reduced in cells co-transfected with USP8, suggesting that USP8 regulates multiple monoubiquitination of Cx43 (Fig. 4C). Collectively, these results demonstrate that Cx43 can be modified with multi-monoubiquitination and polyubiquitination, and both modifications can be regulated by USP8.
USP8 knockdown impairs intercellular communication via GJ
Cx43 is the most abundant connexin isoform expressed in multiple cell types including MEFs, and the levels of intercellular communication via GJs relies largely on the amount of Cx43 at the plasma membrane. Consistent with previous results that USP8 stabilizes Cx43 protein in MEFs, Triton X-100 fractionation assay showed that the levels of Cx43 incorporated into GJs decreased (Fig. S3). It is expected that USP8 knockdown could impair intercellular communication via GJs. We next compared GJ permeability between control and USP8 knockdown MEFs. GJ permeability was determined by the diffusion of a low-molecular-weight dye permeable through GJ channels, which was measured by the flow cytometry. USP8 knockdown in MEFs significantly reduced dye migration (Fig. 5), indicating USP8 is important for the intercellular communication via GJs.
Figure 5.
USP8 knockdown significantly reduced intercellular communication via GJs. A, flow cytometric analysis of unlabeled (top) and calcein AM–labeled (bottom) scramble or shUSP8-infected MEFs. B, unlabeled MEFs were co-cultured with calcein AM–labeled MEFs (1:1) for 3 h to allow the dye transfer between two populations, and then subjected for the flow cytometric analysis. In mixed culture, 50% cells with low fluorescence intensity present unlabeled population (top panel). The relative mean fluorescence intensity (MFI) of unlabeled cells in shUSP8-infected MEFs was calculated and normalized to that in scramble-infected MEFs (bottom panel). Each circle, square, or triangle represents one experiment. The data are shown as mean ± S.D. from four independent experiments. *, p < 0.05; Student's t test.
USP8 positively correlates with Cx43 protein levels in human breast tumors
Cx43 is a prevalent connexin in breast tissues, and its aberrant expression has been implicated in multiple stages of human breast cancers including initiation, development, and metastasis (6). However, the molecular basis underlying the aberrant regulation of Cx43 remains elusive. The identification of key deubiquitinases in regulating Cx43 protein levels in human breast tumors will be valuable for understanding the pathogenesis of this disease.
To determine the relevance of Cx43 regulation by USP8 in these patients, we performed immunohistochemical staining of USP8 and Cx43 proteins on human breast tumor samples. As expected, Cx43 protein levels were heterogeneous in these samples. Very interestingly, a significant positive correlation (r = 0.485; p < 0.01; Pearson score relation analysis) between USP8 and Cx43 protein levels was observed in these breast carcinomas (Fig. 6), suggesting that deregulation of USP8 may contribute to the aberrant expression of Cx43.
Figure 6.
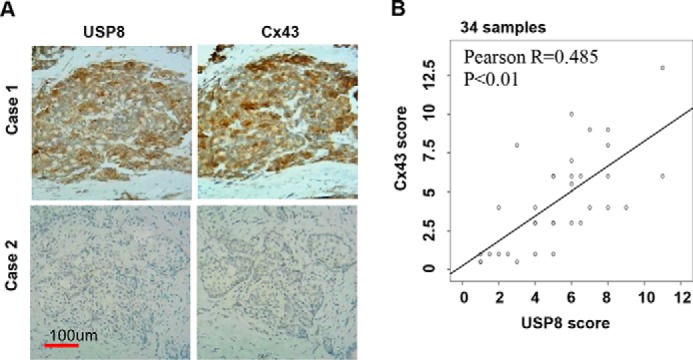
USP8 positively correlates with Cx43 protein levels in human breast cancer. A, representative primary human breast tumor specimen stained for USP8 and Cx43. Case 1 is strongly positive for both USP8 and Cx43; Case 2 is negative for both proteins. B, USP8 and Cx43 expression score are calculated as described in method. Pearson correlation between USP8 and Cx43 expression score in 34 human breast cancer samples is shown.
Discussion
Ubiquitination of Cx43 has long been known to modulate the internalization and degradation of Cx43-containing GJs (8). However, the DUBs which specifically deubiquitinate Cx43 and reverse its degradation pathway remain elusive. AMSH is the only DUB reported to deubiquitinate Cx43 and regulate its degradation (26). In the current work, we demonstrate that USP8 interacts with and deubiquitinates Cx43. USP8 cleaves not only monoubiquitin at multiple sites, but also Lys63- and Lys48-polyubiquitin chains from Cx43. USP8 knockdown results in accumulation of ubiquitinated Cx43, promoting Cx43 degradation and thereby inhibiting intercellular communication via GJs. Consistent with other previous reports that Cx43 degradation is mainly mediated by autophagy in resting cells (19), USP8 knockdown accelerated the autophagy-mediated degradation of Cx43. Finally, USP8 positively correlates with Cx43 protein levels in human breast tumors. Taken together, this study identifies USP8 as a novel DUB involved in regulating the ubiquitination and degradation of Cx43. This finding may have important implications for our understanding of the molecular mechanism underlying the aberrant expression of Cx43 during the pathogenesis of some Cx43-associated human diseases.
The importance of ubiquitination in controlling the endocytosis and degradation of Cx43 has been well-established. However, the types of ubiquitin moieties conjugated to Cx43 remains poorly understood. Here, we provide the evidence that Cx43 can be both multi-monoubiquitinated and polyubiquitinated. Multi-monoubiquitination is generally believed to be responsible for the internalization and clathrin-mediated endocytosis of membrane receptors (36). Using overexpression system in cells, we observed that exogenous Cx43 can be multi-monoubiquitinated, and this kind of modification was counteracted by USP8. Although we did not exclude the possibility that this effect is artificial, it is likely that USP8 prevents internalization of Cx43-containing GJs through inhibiting multi-monoubiquitination of Cx43. Further study is demanded to determine whether the endogenous Cx43 could be multi-monoubiquitinated and the exact positions of target lysines. Of note, it is possible that USP8 regulates the endocytosis of other receptors such as EGFR via a similar mechanism.
Cx43 undergoes modifications with Lys63- and Lys48-linked polyubiquitin chains in vivo, and AMSH is reported to specifically remove Lys63-linked polyubiquitin chains. Here, we find that USP8 is able to remove both Lys63- and Lys48-linked polyubiquitin chains from Cx43. Lys48-linked polyubiquitination primarily targets proteins for proteasome-mediated degradation. However, similar to Lys63-ubiquitin linkage, Lys48-ubiquitin linkage is also able to provide signals for the targets to be sorted to the lysosome. Moreover, both Lys63- and Lys48-polyubiquitin linkages can be recognized by the autophagy adaptor p62/SQSTM1, via its UBA-domain, to trigger autophagy-mediated degradation. It is well-established that endocytosed Cx43-containing GJs are degraded through the endolysosomal and autophagosomal pathways (19, 20). Therefore, it is expected that USP8 can prevent endocytosed Cx43-containing GJs from entering these two degradation pathways via removing both Lys63- and Lys48-polyubiquitin chains. In this study, we find that Cx43 protein is constitutively degraded mainly through the autophagosomal pathway in resting MEFs and USP8 inhibits this process. However, it is absolutely possible that USP8 can also inhibit the degradation of Cx43 mediated by the endolysosomal pathway in other cell types or under other conditions. Therefore, USP8 may act at different stages/levels in the regulation of the intracellular trafficking and degradation of Cx43-containing GJs. Further studies are needed to reveal these processes.
USP8 knockdown significantly reduced the intercellular communication via GJ. GJs are composed of different types of connexins and the ubiquitin system plays a crucial role in regulating the turnover of these connexin members (22). In addition to Cx43, other connexins might be deubiquitinated and stabilized by USP8. It is likely that USP8 promotes intercellular communication via GJ through modulating multiple connexin proteins.
Finally, USP8 positively correlates with Cx43 protein levels in human breast cancers, suggesting that aberrant expression of Cx43 in these tumors is in part caused by the deregulation of USP8. USP8 may play a very important role in the pathogenesis of human breast.
Experimental procedures
Cell culture and transfection
HEK293T, MDA-MB-231, HeLa, WT, and Atg7KO MEF cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (100 units/ml penicillin, 100 g/ml streptomycin) and maintained at 37 °C under 5% CO2. Transient transfections of cells were performed with PEI (Polysciences, Inc.) according to manufacturer's recommendations. All constructs were described in supporting information and the sequence of primers used to generate these constructs was listed in Table S1.
Immunoprecipitation, Triton X-100 fractionation assay, and immunoblotting
Immunoprecipitation experiments were carried out in HEK293T cells. In brief, cells were transfected with the indicated expression vector for 48 h and then lysed in a cell lysate buffer (20 mm Tris-HCL, 150 mm NaCL, 1% Nonidet P-40, 0.5 mm EDTA, protease inhibitor mixture) (Roche). After centrifugation, the supernatant was pulled down by the indicated antibodies and then analyzed by immunoblotting. The Triton X-100 fractionation assay was performed as described previously by others (26). Whole cell lysates were prepared in RIPA and proteins were resolved by SDS-PAGE and immunoblotted using indicated antibodies. The antibodies used were as follows: anti-Cx43 (Santa Cruz Biotechnology, sc-271837), anti-USP8 (Sigma, SAB4200527), anti-Ub (Abcam, ab7254), anti–Lys63-Ub (Abcam, ab179434), anti–Lys48-Ub (Abcam, ab140601), anti-GFP (Abmart, M20004S), and anti-tubulin (Proteintech Group, 10094–1-AP), anti-FLAG (Sigma, F102204), anti-HA (Sigma,), anti-FLAG resin (Sigma, M8823), and anti-HA resin (Sigma, IP0010).
In vivo ubiquitination assay
Cells were transfected with indicated plasmids and harvested at 48 h after transfection, 20% of the cells were used for direct immunoblotting and the rest of cells were harvested in a denaturation buffer (6 m guanidine-HCl, 0.1 m Na2HPO4/NaH2PO4, and 10 mm imidazole). The lysates were incubated with Ni2+-NTA–agarose beads (Qiagen) for 3 h, followed by four times of washing with denaturation buffer and two times of washing with a low-salt buffer (25 mm Tris-HCl and 20 mm imidazole). Bead-bound proteins were eluted by boiling SDS sample buffer in the presence of 200 mm imidazole. After centrifugation, the supernatants were analyzed by immunoblotting.
In vitro ubiquitination assay
Ubiquitinated Cx43 was generated from HEK293T cells transfected with FLAG-Cx43 and His-Ub and purified using anti-FLAG affinity purification. HA-USP8 and its C786A mutant proteins were expressed in HEK293T cells and purified using the anti-HA affinity purification. The ubiquitinated Cx43 was then incubated with purified HA-USP8 (WT or the C786A mutant) in deubiquitination buffer consisting of 50 mm Tris-HCl, pH 8.0, and 10 mm DTT at 37 °C for indicated times. The reactions were analyzed by immunoblotting.
Lentivirus-mediated gene knockdown
Lentivirus vectors encoding shRNA against USP8 were generated using the pLKO.1 vector with puromycin selection marker (Addgene). The shRNA sequences for mouse USP8 are 5′-TCAAGCAACAGCAGGATTATT-3′ (shUSP8–1) and 5′-CTCACATCTAATGCTTACAA-3′ (shUSP8–2). The shRNA sequences for human USP8 are 5′-TCAAGCAACAGCAGG ATTATT-3′ (shUSP8–1) and 5′-GCTGTGTTACTAGCACTATAT-3′ (shUSP8–2). To make a virus, targeting vectors were packaged with the helper plasmids dR8.9 and VSVG. After centrifugation, the virus supernatant was collected and used to infect MEFs. Three days post infection, MEFs were selected in the present of puromycin (1 μg/ml) for 5 days. USP8 knockdown in MEFs was confirmed by immunoblotting.
Flow cytometric measurement of fluorescent dye transfer
Intercellular communication via GJs was measured by dye transfer assay described previously with modifications (40). In brief, MEF cells were labeled with 1 μm calcein AM (Invitrogen) for 20 min. After washing, labeled cells were mixed with unlabeled cells at the ratio of 1:1 and co-cultured on the dishes for 3 h, and then the dye transfer was analyzed by flow cytometry.
Immunochemistry and quantification
Immunohistochemical staining was performed as described previously (39). 6-μm paraffin-embedded sections were routinely deparaffinized with xylene, rehydrated through a graded alcohol series, and incubated in 3% (v/v) hydrogen peroxide to block endogenous peroxidase activity. After antigen retrieving, sections were incubated with 10% (v/v) normal goat serum to block nonspecific staining. Sections were incubated with a primary antibody (USP8, 1:200; Cx43, 1:500) at 4 °C in a humidified chamber overnight, followed by incubation for 1 h with biotin-conjugate secondary antibody (Jackson ImmunoResearch Laboratories). After that, streptavidin–horseradish peroxidase was added, followed by incubation for 30 min. Horseradish peroxidase activity was detected with a DAB kit (Vector Laboratories). Images were captured with a FSX100 microscope equipped with a digital camera system (Olympus). Samples were examined using the German semi-quantitative scoring method. Each specimen was scored for staining intensity (no staining = 0; weak staining = 1; moderate staining = 2; strong staining = 3) and for extent of stained cells (0% = 0; 1–24% = 1; 25–49% = 2; 50–74% = 3; 75–100% = 4). Tissue sections were viewed with a light microscope (Axioskop 2; Carl Zeiss Microscopy, Thornwood, NY) using Plan-Neofluar lens at a 4×/0.50 air objective or 20×/0.50 air objective. Images were taken using a color camera (Axiocam; Carl Zeiss Microscopy) and were analyzed using Axiovision software (Carl Zeiss Microscopy).
Statistical analysis
Student's t test was used for statistical analysis. p < 0.05 was considered significant.
Author contributions
J. S., Q. H., H. P., C. P., and L. Z. data curation; J. S., Q. H., H. P., C. P., and L. Z. software; J. S. and C. H. formal analysis; J. S. investigation; J. S., Q. H., H. P., C. P., and L. Z. visualization; J. S., Q. H., H. P., C. P., and L. Z. methodology; J. L. and C. H. conceptualization; J. L. and C. H. supervision; J. L. and C. H. funding acquisition; J. L. and C. H. project administration; C. H. writing-original draft; C. H. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Guoqiang Hua for assistance with the analysis of the immunochemistry. We also acknowledge Dr. Ziping Wu and Dr. Jinglu Lu for collecting the breast cancer samples.
This work was supported in part by the 1000 Youth Elite Program, the startup fund by Shanghai Jiao Tong University School of Medicine and Shanghai Pujiang Scholarship 15PJ1407400 (to C. H.); Science and Technology Commission of Shanghai Municipality Grant 1402700 (to J. L.); National Natural Science Foundation of China Grant 81172505 (to J. L.); Municipality Cross Research Foundation of Shanghai Jiao Tong University Grant YG2017QN49 (to L. Z.); and Shanghai Municipality Commission of Health and Family Planning Grant 201640006 (to J. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Table S1.
- GJ
- gap junction
- Cx43
- connexin-43
- AGJ
- annular gap junction
- USP8
- ubiquitin-specific protease 8
- DUB
- deubiquitinase
- AMSH
- associated molecule with the Src homology 3 domain
- aa
- amino acids
- CHX
- cycloheximide
- MEF
- murine embryonic fibroblast
- BAF
- bafilomycin A1
- Ub
- ubiquitin
- Ni2+-NTA
- Ni2+-nitrilotriacetic acid.
References
- 1. Sáez J. C., Berthoud V. M., Brañes M. C., Martínez A. D., and Beyer E. C. (2003) Plasma membrane channels formed by connexins: Their regulation and functions. Physiol. Rev. 83, 1359–1400 10.1152/physrev.00007.2003 [DOI] [PubMed] [Google Scholar]
- 2. Goodenough D. A., Goliger J. A., and Paul D. L. (1996) Connexins, connexons, and intercellular communication. Annu. Rev. Biochem. 65, 475–502 10.1146/annurev.bi.65.070196.002355 [DOI] [PubMed] [Google Scholar]
- 3. Laird D. W. (2010) The gap junction proteome and its relationship to disease. Trends Cell Biol. 20, 92–101 10.1016/j.tcb.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 4. Söhl G., and Willecke K. (2003) An update on connexin genes and their nomenclature in mouse and man. Cell Commun. Adhes. 10, 173–180 [DOI] [PubMed] [Google Scholar]
- 5. Aasen T., Mesnil M., Naus C. C., Lampe P. D., and Laird D. W. (2016) Gap junctions and cancer: Communicating for 50 years. Nat. Rev. Cancer 16, 775–788 10.1038/nrc.2016.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grek C. L., Rhett J. M., Bruce J. S., Ghatnekar G. S., and Yeh E. S. (2016) Connexin 43, breast cancer tumor suppressor: Missed connections? Cancer Lett. 374, 117–126 10.1016/j.canlet.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 7. Laird D. W. (2006) Life cycle of connexins in health and disease. Biochem. J. 394, 527–543 10.1042/BJ20051922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leithe E., and Rivedal E. (2007) Ubiquitination of gap junction proteins. J. Membr. Biol. 217, 43–51 10.1007/s00232-007-9050-z [DOI] [PubMed] [Google Scholar]
- 9. Laing J. G., and Beyer E. C. (1995) The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 270, 26399–26403 10.1074/jbc.270.44.26399 [DOI] [PubMed] [Google Scholar]
- 10. Leithe E. (2016) Regulation of connexins by the ubiquitin system: Implications for intercellular communication and cancer. Biochim. Biophys. Acta 1865, 133–146 10.1016/j.bbcan.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 11. Falk M. M., Kells R. M., and Berthoud V. M. (2014) Degradation of connexins and gap junctions. FEBS Letters 588, 1221–1229 10.1016/j.febslet.2014.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carette D., Gilleron J., Denizot J. P., Grant K., Pointis G., and Segretain D. (2015) New cellular mechanisms of gap junction degradation and recycling. Biol. Cell 107, 218–231 10.1111/boc.201400048 [DOI] [PubMed] [Google Scholar]
- 13. Leithet E., and Rivedal E. (2004) Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J. Biol. Chem. 279, 50089–50096 10.1074/jbc.M402006200 [DOI] [PubMed] [Google Scholar]
- 14. Leithe E., and Rivedal E. (2004) Epidermal growth factor regulates ubiquitination, internalization and proteasome-dependent degradation of connexin43. J. Cell Sci. 117, 1211–1220 10.1242/jcs.00951 [DOI] [PubMed] [Google Scholar]
- 15. Su V., Nakagawa R., Koval M., and Lau A. F. (2010) Ubiquitin-independent proteasomal degradation of endoplasmic reticulum-localized connexin43 mediated by CIP75. J. Biol. Chem. 285, 40979–40990 10.1074/jbc.M110.170753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Piehl M., Lehmann C., Gumpert A., Denizot J. P., Segretain D., and Falk M. M. (2007) Internalization of large double-membrane intercellular vesicles by a clathrin-dependent endocytic process. Mol. Biol. Cell 18, 337–347 10.1091/mbc.E06-06-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leithe E., Sirnes S., Fykerud T., Kjenseth A., and Rivedal E. (2012) Endocytosis and post-endocytic sorting of connexins. Biochim. Biophys. Acta 1818, 1870–1879 10.1016/j.bbamem.2011.09.029 [DOI] [PubMed] [Google Scholar]
- 18. Falk M. M., Baker S. M., Gumpert A. M., Segretain D., and Buckheit Iii R. W. (2009) Gap junction turnover is achieved by the internalization of small endocytic double-membrane vesicles. Mol. Biol. Cell 20, 3342–3352 10.1091/mbc.E09-04-0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fong J. T., Kells R. M., Gumpert A. M., Marzillier J. Y., Davidson M. W., and Falk M. M. (2012) Internalized gap junctions are degraded by autophagy. Autophagy 8, 794–811 10.4161/auto.19390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Totland M. Z., Bergsland C. H., Fykerud T. A., Knudsen L. M., Rasmussen N. L., Eide P. W., Yohannes Z., Sørensen V., Brech A., Lothe R. A., and Leithe E. (2017) The E3 ubiquitin ligase NEDD4 induces endocytosis and lysosomal sorting of connexin 43 to promote loss of gap junctions. J. Cell Sci. 130, 2867–2882 10.1242/jcs.202408 [DOI] [PubMed] [Google Scholar]
- 21. Girão H., Catarino S., and Pereira P. (2009) Eps15 interacts with ubiquitinated Cx43 and mediates its internalization. Exp. Cell Res. 315, 3587–3597 10.1016/j.yexcr.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 22. Bejarano E., Girao H., Yuste A., Patel B., Marques C., Spray D. C., Pereira P., and Cuervo A. M. (2012) Autophagy modulates dynamics of connexins at the plasma membrane in a ubiquitin-dependent manner. Mol. Biol. Cell 23, 2156–2169 10.1091/mbc.E11-10-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Basheer W. A., Harris B. S., Mentrup H. L., Abreha M., Thames E. L., Lea J. B., Swing D. A., Copeland N. G., Jenkins N. A., Price R. L., and Matesic L. E. (2015) Cardiomyocyte-specific overexpression of the ubiquitin ligase Wwp1 contributes to reduction in Connexin 43 and arrhythmogenesis. J. Mol. Cell. Cardiol. 88, 1–13 10.1016/j.yjmcc.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen V. C., Kristensen A. R., Foster L. J., and Naus C. C. (2012) Association of connexin43 with E3 ubiquitin ligase TRIM21 reveals a mechanism for gap junction phosphodegron control. J. Proteome Res. 11, 6134–6146 10.1021/pr300790h [DOI] [PubMed] [Google Scholar]
- 25. Komander D., Clague M. J., and Urbé S. (2009) Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- 26. Ribeiro-Rodrigues T. M., Catarino S., Marques C., Ferreira J. V., Martins-Marques T., Pereira P., and Girão H. (2014) AMSH-mediated deubiquitination of Cx43 regulates internalization and degradation of gap junctions. FASEB J. 28, 4629–4641 10.1096/fj.13-248963 [DOI] [PubMed] [Google Scholar]
- 27. Naviglio S., Mattecucci C., Matoskova B., Nagase T., Nomura N., Di Fiore P. P., and Draetta G. F. (1998) UBPY: A growth-regulated human ubiquitin isopeptidase. EMBO J. 17, 3241–3250 10.1093/emboj/17.12.3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Niendorf S., Oksche A., Kisser A., Löhler J., Prinz M., Schorle H., Feller S., Lewitzky M., Horak I., and Knobeloch K. P. (2007) Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol. Cell. Biol. 27, 5029–5039 10.1128/MCB.01566-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Urbé S., McCullough J., Row P., Prior I. A., Welchman R., and Clague M. J. (2006) Control of growth factor receptor dynamics by reversible ubiquitination. Biochem. Soc. Trans. 34, 754–756 10.1042/BST0340754 [DOI] [PubMed] [Google Scholar]
- 30. Li S., Chen Y., Shi Q., Yue T., Wang B., and Jiang J. (2012) Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol. 10, e1001239 10.1371/journal.pbio.1001239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeates E. F., and Tesco G. (2016) The endosome-associated deubiquitinating enzyme USP8 regulates BACE1 enzyme ubiquitination and degradation. J. Biol. Chem. 291, 15753–15766 10.1074/jbc.M116.718023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Durcan T. M., Tang M. Y., Pérusse J. R., Dashti E. A., Aguileta M. A., McLelland G. L., Gros P., Shaler T. A., Faubert D., Coulombe B., and Fon E. A. (2014) USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 33, 2473–2491 10.15252/embj.201489729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alexopoulou Z., Lang J., Perrett R. M., Elschami M., Hurry M. E., Kim H. T., Mazaraki D., Szabo A., Kessler B. M., Goldberg A. L., Ansorge O., Fulga T. A., and Tofaris G. K. (2016) Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc. Natl. Acad. Sci. U.S.A. 113, E4688–E4697 10.1073/pnas.1523597113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ma Z. Y., Song Z. J., Chen J. H., Wang Y. F., Li S. Q., Zhou L. F., Mao Y., Li Y. M., Hu R. G., Zhang Z. Y., Ye H. Y., Shen M., Shou X. F., Li Z. Q., Peng H., et al. (2015) Recurrent gain-of-function USP8 mutations in Cushing's disease. Cell Res. 25, 306–317 10.1038/cr.2015.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reincke M., Sbiera S., Hayakawa A., Theodoropoulou M., Osswald A., Beuschlein F., Meitinger T., Mizuno-Yamasaki E., Kawaguchi K., Saeki Y., Tanaka K., Wieland T., Graf E., Saeger W., Ronchi C. L., Allolio B., Buchfelder M., Strom T. M., Fassnacht M., and Komada M. (2015) Mutations in the deubiquitinase gene USP8 cause Cushing's disease. Nat. Genet. 47, 31–38 10.1038/ng.3166 [DOI] [PubMed] [Google Scholar]
- 36. Mukhopadhyay D., and Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 10.1126/science.1127085 [DOI] [PubMed] [Google Scholar]
- 37. Glickman M. H., and Ciechanover A. (2002) The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82, 373–428 10.1152/physrev.00027.2001 [DOI] [PubMed] [Google Scholar]
- 38. Kirkin V., McEwan D. G., Novak I., and Dikic I. (2009) A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- 39. Pan X., Zhou T., Tai Y. H., Wang C., Zhao J., Cao Y., Chen Y., Zhang P. J., Yu M., Zhen C., Mu R., Bai Z. F., Li H. Y., Li A. L., Liang B., Jian Z., Zhang W. N., Man J. H., Gao Y. F., Gong W. L., Wei L. X., and Zhang X. M. (2011) Elevated expression of CUEDC2 protein confers endocrine resistance in breast cancer. Nat. Med. 17, 708–714 10.1038/nm.2369 [DOI] [PubMed] [Google Scholar]
- 40. Juul M. H., Rivedal E., Stokke T., and Sanner T. (2000) Quantitative determination of gap junction intercellular communication using flow cytometric measurement of fluorescent dye transfer. Cell Adhes. Commun. 7, 501–512 10.3109/15419060009040307 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.