Abstract
The bacterial plant pathogen Agrobacterium fabrum uses periplasmic-binding proteins (PBPs) along with ABC transporters to import a wide variety of plant molecules as nutrients. Nonetheless, how A. fabrum acquires plant metabolites is incompletely understood. Using genetic approaches and affinity measurements, we identified here the PBP MelB and its transporter as being responsible for the uptake of the raffinose family of oligosaccharides (RFO), which are the most widespread d-galactose–containing oligosaccharides in higher plants. We also found that the RFO precursor galactinol, recently described as a plant defense molecule, is imported into Agrobacterium via MelB with nanomolar range affinity. Structural analyses and binding mode comparisons of the X-ray structures of MelB in complex with raffinose, stachyose, galactinol, galactose, and melibiose (a raffinose degradation product) revealed how MelB recognizes the nonreducing end galactose common to all these ligands and that MelB has a strong preference for a two-unit sugar ligand. Of note, MelB conferred a competitive advantage to A. fabrum in colonizing the rhizosphere of tomato plants. Our integrative work highlights the structural and functional characteristics of melibiose and galactinol assimilation by A. fabrum, leading to a competitive advantage for these bacteria in the rhizosphere. We propose that the PBP MelB, which is highly conserved among both symbionts and pathogens from Rhizobiace family, is a major trait in these bacteria required for early steps of plant colonization.
Keywords: crystal structure, sugar transport, bacteria, ABC transporter, microbiology, agrobacterium, galactinol, periplasmic binding protein, RFOs, plant defense, Agrobacterium fabrum
Introduction
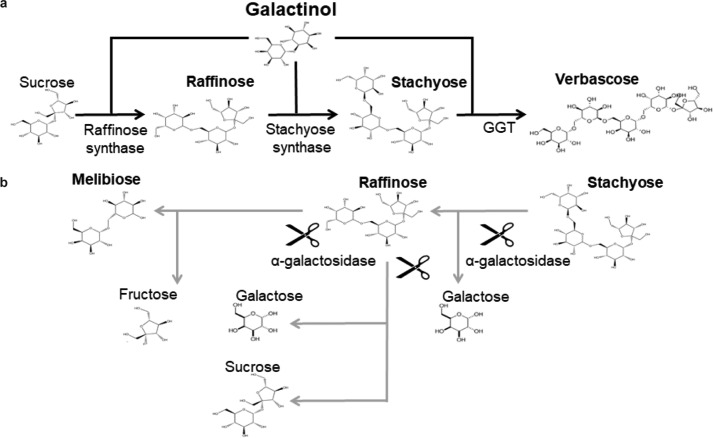
The plant–rhizospheric microbial population interaction is dynamic and largely influenced by root exudates, with either beneficial or harmful consequences for plant growth development and health (1). The germinating seeds in contact with the surrounding soil and microorganisms have strong influences on the rhizosphere composition and favor fast-growing microorganisms able to exploit carbon released, to resist to antimicrobial compounds, and to outcompete other surrounding bacteria (2–4). Raffinose and stachyose from the raffinose family of oligosaccharides (RFO)5 accumulate in plant seeds as energy-storage metabolites, and are released during plant germination (2, 5). The precursor of RFO synthesis, namely galactinol, a d-galactose bound to an inositol, is produced by the plant enzyme galactinol synthase (GolS) (5, 6) (Fig. 1). Galactinol plays an important role in plant health, being involved in plant resistance against abiotic (drought and temperature) (7, 8) and biotic stresses (9–11). Indeed, this molecule, which accumulates in plants in response to bacterial inoculation, is involved in the induced systemic resistance to phytopathogens (9). Raffinose and stachyose are synthesized from sucrose by the subsequent addition of activated galactose moieties donated by galactinol using plant raffinose and stachyose synthases, respectively (Fig. 1). Therefore, RFOs are α-(1,6)-galactosyl extensions of sucrose.
Figure 1.
RFO synthesis and degradation. Reactions for synthesis (a) are represented with black arrows whereas those for degradation (b) are with gray arrows. The first step of RFOs biosynthesis starts with the formation of galactinol from UDP-galactose and inositol catalyzed by the plant enzyme galactinol synthase. Raffinose, stachyose, and verbascose are synthesized from sucrose by the subsequent addition of activated galactose moieties donated by galactinol using plant raffinose and stachyose synthases and galactan:galactan galactosyltransferase (GGT), respectively. Cleavage, shown by scissors, of raffinose by an α-galactosidase leads to the formation of either melibiose and fructose or sucrose and galactose. For RFOs of higher degree of polymerization (DP), an α-galactosidase activity results in RFOs of lower DP and galactose.
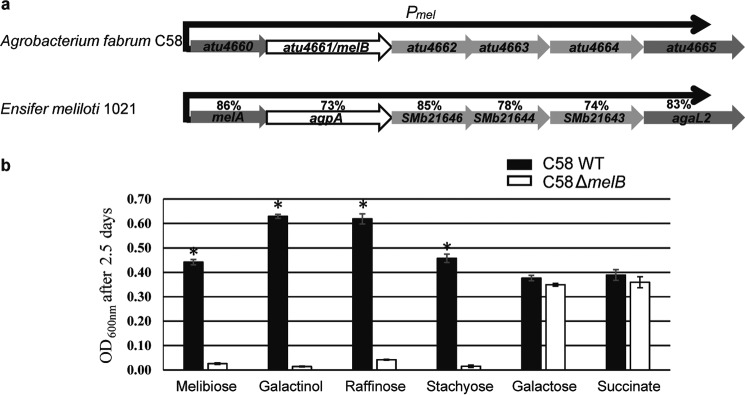
Periplasmic-binding proteins (PBPs) associated with their ATP-binding cassette (ABC) transporter are essential for transport (12). A PBP-mediated transport system is responsible for RFO uptake from seed exudates into bacterial cells as previously shown in Ensifer meliloti 1021 (13, 14). RFOs, which are degraded by α-galactosidases in this latter strain, are used as nutrients, and their assimilation may be involved in bacterial survival in plant rhizosphere (13, 14). In more detail, raffinose and stachyose can be degraded into melibiose and fructose, and raffinose and galactose, respectively (15, 16). Bacterial assimilation of RFOs and melibiose was associated with trophic advantage in plant–bacteria interaction (14, 17), whereas nothing was known for galactinol. Agrobacteria are telluric and rhizosphere bacteria, commonly isolated from roots of numerous plants as commensal bacteria. They can also be pathogenic with the presence of the tumor-inducing plasmid (18, 19). They are then able to create their own ecological niche after plant cell transformation that leads to tumor formation in a wide range of plants (18). An in silico analysis of α-galactosidases distribution in bacteria indicated that Agrobacterium fabrum C58 strain contains an operon putatively involved in RFO transport and degradation (20–22). This operon that we named mel is similar to the agp operon of E. meliloti (13, 14), and encodes the PBP MelB (Atu4661) which shares 73% sequence identity with the PBP AgpA, its associated ABC transporter (Atu4662–Atu4664), and two α-galactosidases (Atu4660 and Atu4665). All these latter proteins display between 74 and 86% sequence identity with their E. meliloti corresponding homologues (Fig. 2a).
Figure 2.
The mel operon structure and MelB involvement in galactinol, melibiose, and RFO consumption. a, atu4660-atu4665 genes belong to the same transcription unit (operon prediction by Westover et al. (22) that we called the mel operon). The mel promoter Pmel indicates the gene transcription direction. Both atu4660 and atu4665 genes are annotated as two α-galactosidases, atu4661 as the PBP MelB and atu4662-atu4664 genes as the associated ABC transporter. Comparison is between the mel operon genes and their homologues in E. meliloti 1021 (13, 14). the percentages of sequence identity between each homologous protein are indicated; for example, the PBPs AgpA and MelB share 73% sequence identity. b, 2.5 days growth (A at 600 nm) of A. fabrum C58 WT strain (in white) and the C58ΔmelB mutant (in black) in AT minimal medium supplemented with different carbon sources. Standard deviations were calculated from five technical and two biological replicates. Asterisks indicated significant differences (Mann-Whitney p value = 0.05).
We hypothesized that this mel operon was responsible for the transport and assimilation of α-galactosides in A. fabrum. Here, we focused on its transport function, and investigated the genetic and molecular role of the PBP MelB through an integrative approach using a defective mutant in cellulo and in planta, crystallography, and affinity measurements. We showed that MelB was the PBP responsible for RFO, melibiose, and galactinol import into A. fabrum C58, displaying the highest affinity for galactinol in nanomolar range and preferring to bind a 2-unit ligand. We structurally characterized the binding mode of MelB for its different ligands. Overall, our work highlights how the capacity of agrobacteria to assimilate plant α-galactosides confers on them an advantage in colonizing efficiently the plant tomato rhizosphere, explaining why the PBP MelB is highly conserved among symbionts and pathogen rhizobiales.
Results
The PBP MelB is responsible for galactinol, melibiose, and RFOs (raffinose and stachyose) uptake
The growth profiles of A. fabrum C58 wildtype (WT) and C58ΔmelB-defective mutant for MelB were compared in rich (YPG) and minimal medium containing RFOs, their derivatives or succinate (as control) as the sole source of carbon. The C58ΔmelB mutant has the same growth rate as the WT strain in minimum medium with succinate and in rich medium. However, the C58ΔmelB mutant did not grow on galactinol, melibiose, raffinose, and stachyose, in contrast to WT (Fig. 2b). Therefore, MelB associated to its ABC transporter is the transport system responsible for the uptake and is necessary for the assimilation of these four molecules in pure culture.
MelB exhibits a high affinity for galactinol
Binding of galactinol, melibiose, raffinose, and stachyose to the purified recombinant mature protein MelB was explored using tryptophan fluorescence spectroscopy (MelB possesses 16 tryptophans) and isothermal titration microcalorimetry (ITC). Intrinsic protein fluorescence titration experiments yielded apparent dissociation constant KD values of 10 ± 1 nm and 72 ± 4 nm with galactinol and melibiose, respectively, showing that MelB is very efficient for galactinol binding. Reducing the ligand size to a monosaccharide (galactose) or increasing it resulted in a substantial affinity reduction compared with galactinol: KD values of 35-fold higher for the raffinose, and over 1000-fold higher for both galactose and stachyose, respectively (Table 1 and Fig. S1). The KD values were slightly higher using ITC than those determined by autofluorescence, but this increased KD was consistent (Table 1 and Fig. S1). Because MelB was not stable at high concentration during the time course of ITC experiment, we were not able to measure an interpretable signal for the stachyose binding. The ITC data confirmed the 1:1 binding stoichiometry for all ligands and revealed a high enthalpy of binding for galactinol and melibiose meaning that both ligands use the same binding mechanism mainly involving polar interactions. In contrast, the binding mode of raffinose was characterized by an unfavorable enthalpy contribution and a strong entropy term indicating that hydrophobic interactions may play a predominant role and/or a displacement of water molecules occurs upon ligand binding. The galactose interaction adopts an intermediate behavior with a high entropy term, accompanied by a weakly favorable enthalpy of binding, suggesting that polar bonds are less important for galactose alone compared with a 2-unit ligand, resulting in a lower enthalpy, thus a weaker affinity. MelB is specific for α-(1,6)-galactosides (RFOs). Indeed, no interaction could be measured with glucose, sucrose, cellobiose, lactose, and α-(1,6)-glucosides.
Table 1.
Affinity measurement for MelB
KD values were measured by intrinsic protein fluorescence titration (Fluorescence) and by isothermal titration microcalorimetry (ITC).
| Fluorescence |
ITC |
|||||||
|---|---|---|---|---|---|---|---|---|
| KD | R2 | KD | n | Enthalpy (ΔH) | Entropy (ΔS) | Entropic contribution (−TΔS) | Free enthalpy (ΔG) | |
| μm | μm | cal/mol | cal/mol/deg | cal/mol | cal/mol | |||
| Galactinol | 0.010 ± 0.001 | 0.99 | 0.12 ± 0.03 | 0.83 | −6908 | 8 | −2356 | −9264 |
| Melibiose | 0.072 ± 0.004 | 0.99 | 0.76 ± 0.11 | 0.94 | −6412 | 6.1 | −1794 | −8206 |
| Raffinose | 0.347 ± 0.42 | 0.99 | 2.9 ± 0.5 | 0.97 | 2115 | 32.5 | −9527 | −7412 |
| Galactose | 13.8 ± 3 | 0.99 | 24 ± 2 | 1 | −797 | 18.4 | −5393 | −6190 |
| Stachyose | 24.6 ± 2.4 | 0.99 | Not determined | |||||
Thermal denaturation experiments revealed a contribution of more than 3 °C for two ligands for protein stability (Fig. S2). Indeed, adding galactinol or melibiose led to a melting temperature (Tm) of over 46 °C compared with the 43 °C for the unliganded protein in agreement with the measured KD values. Galactose or stachyose binding did not stabilize MelB whereas the raffinose binding produced a slight effect with a Tm of 45 °C.
MelB is a PBP from cluster C
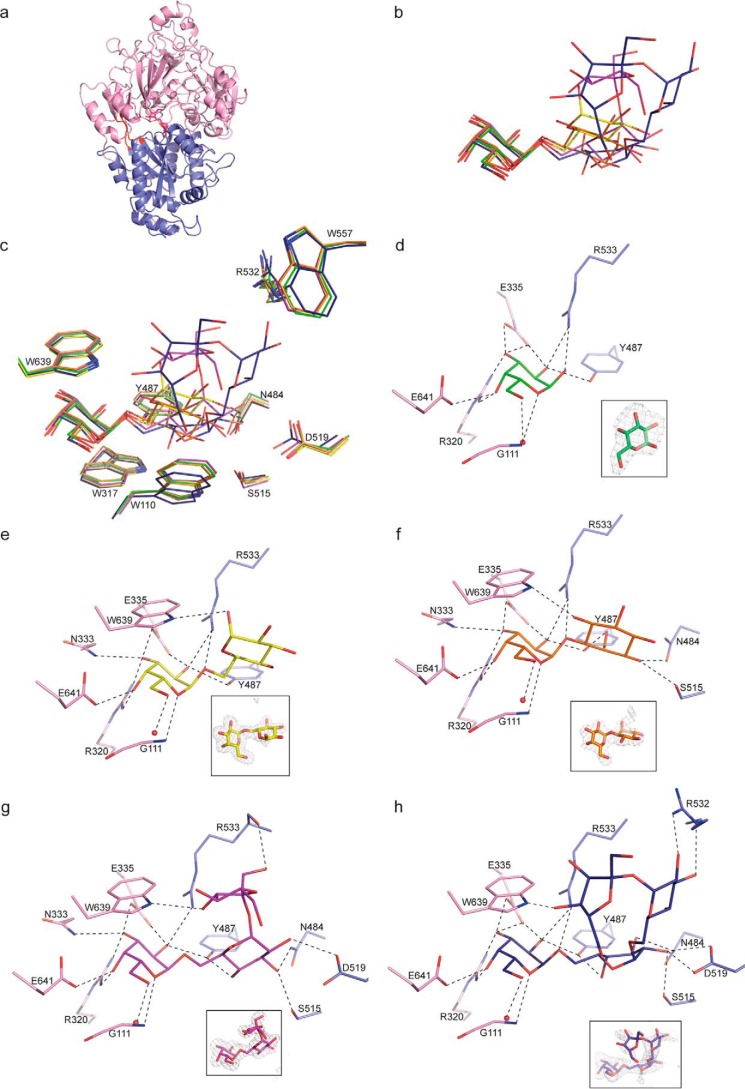
The mature MelB expression plasmid was a synthetic gene lacking the first 18 signal sequence residues that serve for localization to bacterial periplasm. The numbering used for the description of residues corresponds to the mature protein of 677 amino acids. Because MelB is the biggest PBP so far and shares low sequence identity (around 20%) compared with PBPs with known three-dimensional structures, we first solved the structure of seleniated MelB in complex with raffinose at 2 Å resolution by single wavelength anomalous dispersion method. The asymmetric unit is composed of four similar MelB-raffinose complexes (average root mean square deviation (RMSD) value of 0.4 Å). By the molecular replacement method, we then solved the structure of MelB in complex with galactinol, melibiose, galactose, and stachyose at 2.2, 1.8, 2.5, and 2.1 Å resolution, respectively (Table 2). The asymmetric unit of the galactinol and melibiose complexes also contains four very similar molecules with RMSD values between monomers ranging from 0.2 to 0.4 Å whereas that of the galactose and stachyose complexes possesses two identical molecules. Moreover, the five ligand-bound structures are very similar with an average RMSD value of 0.4 Å. They all adopt a closed conformation. MelB fold is monomeric, composed of two lobes, each formed by a central β-sheet flanked by α-helices (Fig. 3a). The biggest lobe (lobe 1) consists of residues 8–354 and 619–678 and the smallest (lobe 2) comprises the residues 364–610. Two short segments (Fig. 3a) define the hinge region connecting the two lobes. MelB possesses a typical fold of cluster C within the PBP structural classification (12) as SSM-EBI (http://www.ebi.ac.uk/msd-srv/ssm)6 reports: RMSD between MelB and similar PBP structures binding oligopeptide are over 2.6 Å for 450 Cα atoms. Nonetheless, a detailed structural comparison is irrelevant because MelB presents a distinct ligand-binding site.
Table 2.
Crystallographic data and refinement parameters
Values for the highest resolution shell are in parentheses.
| MelB |
|||||
|---|---|---|---|---|---|
| Raffinose (SeMet) | Galactinol | Melibiose | Galactose | Stachyose | |
| PDB code | 6EPY | 6EQ8 | 6EPZ | 6EQ0 | 6EQ1 |
| Space group | C2 | C2 | C2 | C2 | C2 |
| Cell parameters (Å, °) | a = 354.3 | a = 355.3 | a = 351.6 | a = 107.8 | a = 108.2 |
| b = 74.3 | b = 73.7 | b = 73.7 | b = 73.9 | b = 74 | |
| c = 108.2 | c = 108.1 | c = 107.6 | c = 171.1 | c = 171.4 | |
| β = 105.5 | β = 105.5 | β = 105.4 | β = 92.5 | β = 92.4 | |
| Resolution (Å) | 50–2 (2.17–2) | 50–2.2 (2.3–2.2) | 48–1.8 (1.9–1.8) | 50–2.5 (2.59–2.5) | 50–2.1 (2.2–2.1) |
| No. of observed reflections | 1,143,584 (175,894) | 725,141 (106,000) | 1,633,125 (250,861) | 314,407 (48,297) | 541,503 (82,382) |
| No. of unique reflections | 333,472 (51,505) | 137,636 (20,717) | 246,319 (38,629) | 49,700 (7682) | 79,391 (12,494) |
| Rsym (%) | 7 (52.8) | 17.1 (112.5) | 10 (77.2) | 18.2 (100) | 15.4 (200) |
| Completeness (%) | 99.8 (94.4) | 98.7 (92.6) | 99.4 (97.1) | 99.2 (95.7) | 99.6 (97.9) |
| I/σ | 10.8 (2) | 7.5 (1.5) | 11.4 (1.9) | 9 (1.6) | 8.12 (0.7) |
| CC1/2 | 99.8 (80.1) | 99.3 (57.9) | 99.8 (80) | 99.1 (51.2) | 99.7 (50.1) |
| Rcryst (%) | 17.5 | 18.5 | 17.7 | 19.4 | 19.1 |
| Rfree (%) | 19.9 | 21.7 | 19.5 | 24.2 | 22.5 |
| RMS bond deviation (Å) | 0.01 | 0.01 | 0.01 | 0.01 | 0.01 |
| RMS angle deviation (°) | 1.0 | 1.09 | 1 | 1.16 | 1.13 |
| Average B (Å2) | |||||
| protein | 44.3 | 45.6 | 33.7 | 55.9 | 52.6 |
| ligand | 41.2 | 36.6 | 30.5 | 40.5 | 44.7 |
| solvent | 51.7 | 49.6 | 39.2 | 55 | 52.2 |
CC1/2 = percentage of correlation between intensities from random half-dataset (40).
Figure 3.
Ribbon representation of MelB structures and ligand-binding site. a, raffinose in magenta is located in the cleft between lobes 1 and 2 shown in slate and in pink, respectively, and the hinge region is in red. b, superposition of the bound galactose, melibiose, galactinol, raffinose, and stachyose shown in green, yellow, orange, magenta, and blue sticks, respectively, in the binding site of MelB. c, same figure as in b showing the stacking between ligands and tryptophan (Trp639, Trp317, and Trp110). Except Trp639 and Trp317, all the other labeled amino acids mainly from lobe 2 can move up to 1 Å upon ligand binding. d–h, galactose (d), melibiose (e), galactinol (f), raffinose (g), and stachyose (h) bound to the binding site of MelB are shown in the same code color as in b. Hydrogen bonds between MelB and each ligand are shown as dashed lines in black (distances are up to 3.2 Å). A water molecule forming a hydrogen bond with each ligand is shown as a red circle. Each ligand is shown in its annealing Fo-Fc omit map contoured at 4σ.
Ligand-binding site of MelB
All ligands are bound between the two closed lobes of MelB. The size and the volume of the ligand-binding site constrain the conformation of bound RFOs. Indeed, raffinose and stachyose bind in a very compact form (Fig. 3b), and the addition of a galactosyl moiety at the nonreducing end of stachyose corresponding to verbascose will abolish its binding, because the pocket is not large enough to accommodate a pentasaccharide. All ligands are well defined in their electron density maps except the fructose moiety of the stachyose likely responsible for its low affinity (Fig. 3, c–h). They share a buried nonreducing end galactosyl unit wedged between two aromatic residues (Trp317 and Trp639) which superimposes very well with the bound galactose alone (Fig. 3, c–h). These galactoses at position 1 make 10 similar protein contacts involving the main chain amino group of Gly111 and the side chains of Arg320, Asn333, Glu335, and Glu641 from lobe 1 and both side chains of Tyr487 and Arg533 from lobe 2. The O6 atom of this pyranose interacts with a conserved water molecule observed in each complexed structure (except in molecule B of MelB-galactose complex), which in turn makes hydrogen bonds with the amino group of Gly112 and the side chain of Asp114. In both structures of MelB in complex with galactose and stachyose, the galactose at position 1 does not interact with Asn333 but because of two hydrogen bonds with the side chain of Glu335, it conserves 10 interactions with MelB. Modeling a glucosyl unit at the nonreducing end (position 1) creates steric hindrance between the equatorial C4-OH and the Trp639 indol explaining the specificity of MelB for a galactosyl unit at position 1 as shown by the affinity measurement.
In contrast to position 1, at position 2, the glucosyl moieties of melibiose and raffinose, the inositol moiety of galactinol and the galactose moiety of stachyose do not superimpose and can shift up to 3.5 Å to allow, for example, the fructose or the sucrose accommodation of raffinose and stachyose in the ligand-binding site, respectively (Fig. 3, b and c). A conformational change less than 1 Å for Trp110 and Tyr487 side chains is observed to accommodate a glucosyl unit at position 2 (Fig. 3c). More arrangements from amino acids of lobe 2 can occur to accommodate the glucosyl units at positions 3 and 4 (Fig. 3c). For example, Trp557 is pushed away by more than 1 Å compared with the other liganded structures to find a room for the fructose at position 4. All units at position 2 have in common the stacking onto the aromatic indol of Trp110, with the optimum one for the inositol because of the shorter link (one carbon shorter) between the two subunits of galactinol compared with melibiose and RFOs. The melibiose's glucose and the inositol share only one protein contact: an oxygen interacts with the NH of the Trp639 side chain (Fig. 3, e and f). The inositol makes three additional hydrogen bonds with Asn484, Tyr487, and Ser515 from lobe 2 whereas the rest of the oxygen of the melibiose's glucose interacts with the protein side chains via water molecules only. In contrast, the glucose moiety of raffinose and the galactose moiety of stachyose at position 2 make an additional H-bond compared with inositol involving the side chains of Asp519 (Fig. 3, g and h). The fructose at position 3 in raffinose interacts with the main chain of Arg533 from lobe 2 and the NH of the Trp639 indol from lobe 1 (Fig. 3g). In contrast, Arg532 side chain makes two H-bonds with the glucose moiety at position 3 in stachyose whereas only one is present between the fructose moiety at position 4 and the Trp639 side chain (Fig. 3h).
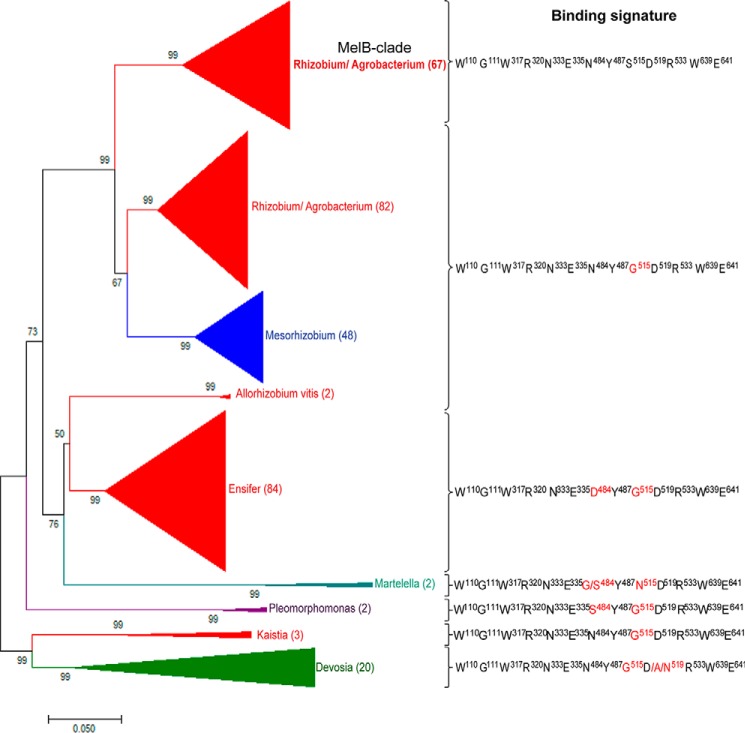
MelB is highly conserved among rhizobiales
Searching for MelB conservation in the bacterial kingdom (protein database at NCBI), and subsequent phylogenetic analysis revealed >310 PBPs above 65% sequence identity (Fig. 4). Galactinol-, melibiose-, and raffinose-binding signatures share 10 amino acids Trp110–Gly111–Trp317–Arg320–Asn333–Glu335–Tyr487–Arg533–Trp639–Glu641. Stachyose binding shares only nine of these latter because Asn333 does not interact with stachyose. Two additional residues (Asn484 and Ser515) are involved in galactinol, raffinose, and stachyose binding compared with melibiose binding. One additional residue Asp519 belongs to the raffinose and stachyose signatures. A last additional residue Arg532 defines the stachyose-binding signature composed of 13 residues in total. Members of the MelB subgroup (67 PBPs) display more than 90% identity sequence with the conserved binding signature. They all belong to Rhizobium and Agrobacterium genera. Outside the MelB cluster, the signature slightly degenerates for galactinol and raffinose binding. Nonetheless, modeling indicates that their binding would not be affected. Remarkably, all these PBPs belong to soil- and plant-interacting genera.
Figure 4.
MelB phylogeny and binding signature. For each protein clade, the residues, which are identical to (black) and different from (red) those involved in the galactinol/melibiose/raffinose binding of A. fabrum C58 MelB are indicated. Number in bracket represents the number of MelB-relative PBPs per clade.
Galactinol and melibiose are inducers of mel operon genes
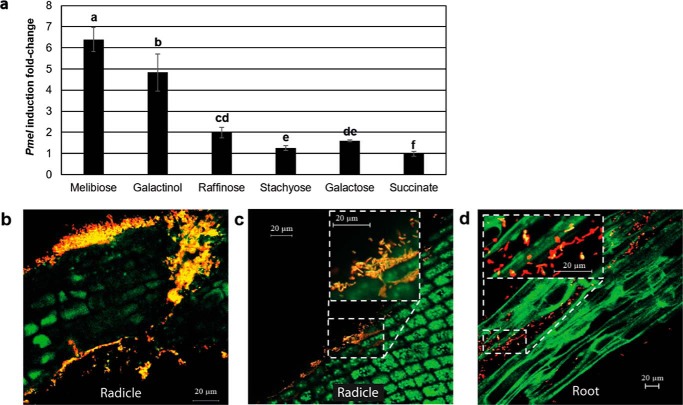
We constructed the C58 pOT1e-Pmel reporter fusion strain to study in cellulo gene expression of mel operon in the presence of commercial compounds. Compared with succinate, slight but significant inductions were observed with raffinose, galactose, and stachyose (2-, 1.6-, and 1.25-fold change values, respectively) (Fig. 5a). In contrast, galactinol and melibiose are efficient inducers with 4.8- and 6.5-fold change values, respectively.
Figure 5.
Expression of mel operon genes in cellulo and in tomato radicle and root. a, comparison of mel operon gene expression in AT minimal medium supplemented with different carbon sources. Standard deviations were obtained from four technical and two biological replicates. Letters above histograms indicate significant different -fold change values (Tukey's test, p value = 0.05). b–d, bacterial mel operon gene expression at two early stages of plant growth. Gene expression was monitored using the pOT1eM-Pmel transcriptional reporter fusion by confocal microscopy at 2 (b and c) and 14 (d) dpi, corresponding to radicle emergence and root elongation stages, respectively. Representative pictures from five plants per stages are shown. Red fluorescence from M-cherry indicates the presence of bacteria, whereas yellow fluorescence shows bacteria that were both active and able to express Pmel-egfp. Plant auto fluorescence, represented in green allows distinguishing different types of plant cells, small compact cells from the radicle and elongated cells from the growing root. The scale is represented in white. Most cells expressed Pmel-egfp at 2 dpi, whereas at 14 dpi, few cells only expressed the transcriptional fusion.
mel operon genes are expressed in early plant colonization
At two early stages of plant colonization, in planta expression of mel operon has been studied in the WT strain harboring the pOT1eM-Pmel plasmid reporter fusion (m-cherry constitutive expression and egfp inducible expression). 48 h after seed imbibition and inoculation, most bacteria in contact with the radicle cells expressed mel operon, as shown by the yellow cells in Fig. 5, b and c. 14 days after seed imbibition and inoculation, among bacterial cells present on plant roots (red and yellow cells), some of them were still expressing RFO uptake and degradation genes (yellow cells in Fig. 5d). Thus, mel operon was more expressed at the beginning of plant colonization.
The PBP MelB confers a competitive advantage in colonizing tomato rhizosphere
The colonization of plant rhizosphere by A. fabrum C58 WT and C58ΔmelB mutant was evaluated at 2 days post inoculation (dpi) and 14 dpi (Fig. 6). When tomato seeds were inoculated with each strain individually, the bacterial colonization level did not significantly differ at 2 dpi (Mann-Whitney p value of 0.09) whereas at 14 dpi, this was slightly higher for the WT strain (Mann-Whitney p value <2.2e–16e) (Fig. 6a). When A. fabrum C58 WT and C58ΔmelB mutant were co-inoculated, a slight and a drastic reduced fitness was observed for the C58ΔmelB mutant at 2 and 14 dpi, respectively (Fig. 6b), revealing a selective advantage conferred by galactinol/melibiose/RFO exploitation under a competitive challenge.
Figure 6.
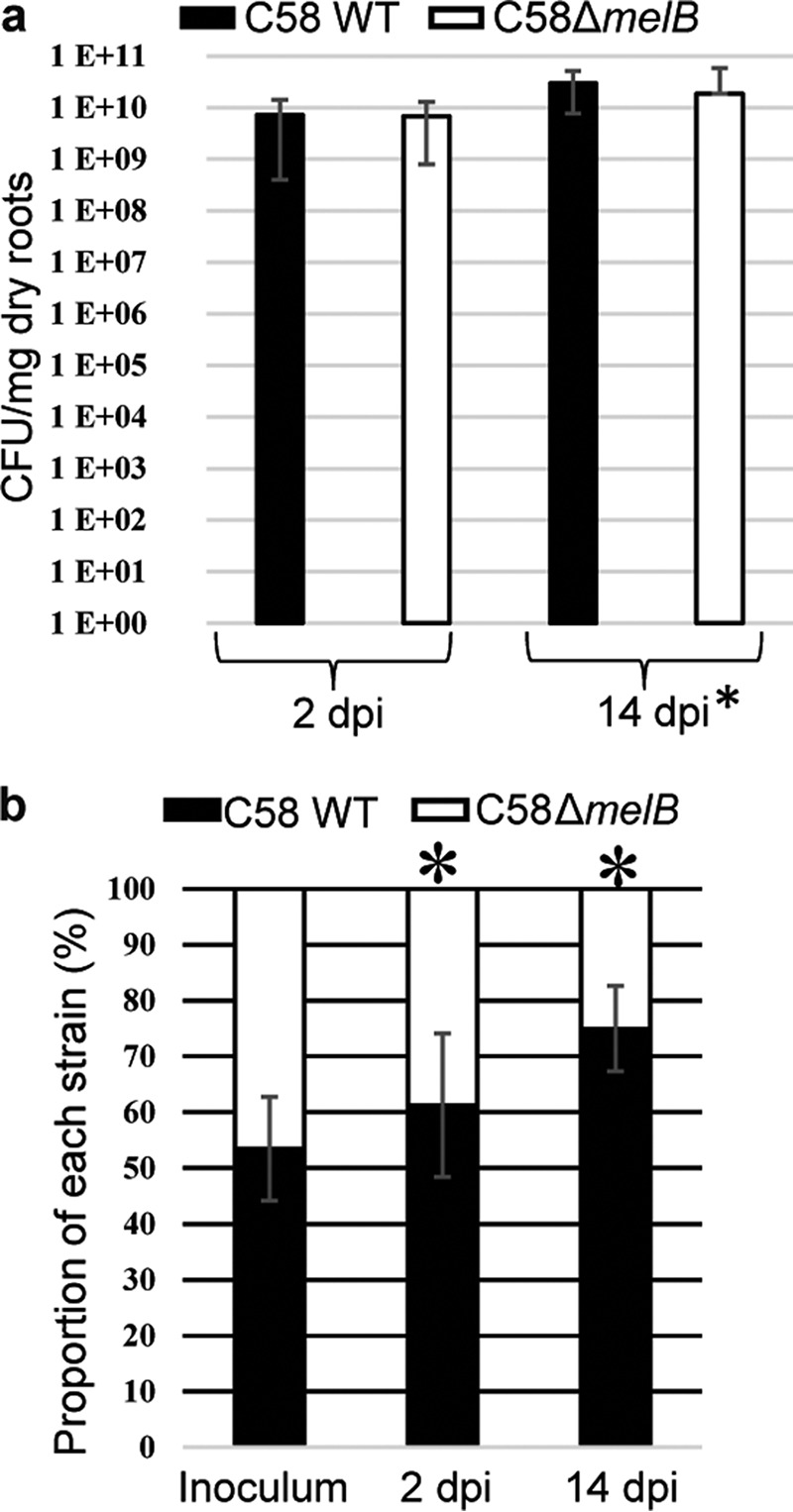
MelB confers a competitive advantage in tomato roots. a, A. fabrum bacterial concentration (cfu/mg dry roots) in tomato roots (at 2 and 14 dpi) infected with either A. fabrum C58 WT or C58ΔmelB mutant. Standard deviations were calculated from three technical and five biological replicates. b, proportion of A. fabrum genotypes (%) in inoculum and tomato root at 2 and 14 dpi infected with a mixture (1:1 ratio) of A. fabrum C58 WT and C58ΔmelB mutant. Standard deviations were calculated from 4 biological replicates and 10 independent assays. Asterisks indicate significant differences (Mann-Whitney test).
Discussion
This work reveals the molecular and ecological roles, and structural basis of the PBP MelB, encoded by the linear chromosome of A. fabrum C58.
At the beginning of the study, we made the straightforward assumption that MelB was behaving like its homologous PBP AgpA from E. meliloti described as an α-galactoside transporter (13). Our gene expression analyses and growth assays experiments showed that similarly to what has been reported for expression of agp operon genes in E. meliloti, α-galactosides induce expression of mel operon genes in A. fabrum, and are used as carbon sources after being imported by MelB. Indeed, in contrast to the WT strain, a MelB defective mutant is unable to grow on α-galactosides. Moreover, using two different biophysical methods, we demonstrated that MelB can bind melibiose, raffinose, and stachyose. Because of the small volume cavity of the ligand binding, stachyose displays a weak affinity for MelB (micromolar range) compared with melibiose and raffinose (nanomolar range). The accommodation of stachyose requires drastic conformational constraints on the ligand when bound to the protein. Indeed, only small local rearrangements of few protein side chains (Trp110 and Trp557) forming the binding site can occur. With an affinity in the micromolar range, MelB also binds galactose, which is the sugar common to all α-galactosides present at the nonreducing end. Nonetheless, the low affinity of MelB for galactose prevents galactose from competing with melibiose and raffinose. From our results, MelB can appear as an alternative galactose transporter suspected by Kemner et al. (23), which allowed a chvE-gguABC defective mutant to grow on galactose (23, 24). Conversely, the presence of the ChvE-GguABC sugar transporter can explain why melB defective mutant was still able to grow on galactose.
An unexpected outcome of our study was that galactinol, which is the precursor of α-galactosides production in plants, was uptaken into agrobacteria via the MelB-mediated transport system. Moreover, MelB displays a preference for galactinol with high affinity (nanomolar range) indicating that this molecule must be efficiently imported into A. fabrum, in line with the gene expression results. Remarkably, MelB recognizes similarly the nonreducing end galactose common to all tested α-galactosides and galactinol. Overall, using genetic, structural, and affinity data, this work demonstrates that the MelB-mediated transport system contributes to the import of α-galactosides with a strong preference for a 2-unit ligand (melibiose) and mainly contributes to that of galactinol. To our knowledge, this is the first description of a bacterial PBP allowing galactinol import.
The imported sugars are used to sustain bacterial growth. Galactinol- and α-galactoside–rich environments would facilitate the settlement of bacteria capable to assimilate these plant compounds efficiently. From in planta competition assays on tomato between the A. fabrum WT and melB defective mutant, we showed that MelB confers a marked selective advantage in colonizing tomato rhizosphere, and since early time. This observation correlates with the presence of melibiose and raffinose in the plant rhizosphere, as they are highly abundant in the seeds (25) and known to be released during seed germination (14). Hence, the competitive advantage of the WT in the tomato rhizosphere could be because of a trophic advantage of the strain during seed germination. A germinating seed can indeed be considered as a new environment to be colonized. The community composition of the mature plant is influenced by historical contingency (timing and order of arrival) of the seed community members and their ability to efficiently settle in that environment (26), as the first establishing species are known to affect the ability of potential immigrants to establish. This is called the priority effect (27). Thus, the ability of bacteria to compete and settle in germinating seed environment by growing on galactinol or α-galactosides released at this time could have long effect on their ability to persist and colonize plant rhizosphere. This is consistent with our findings that at 14 dpi, the competitive advantage of the WT strain is even higher than the one measured at 2 dpi.
Besides the trophic advantage linked to the mel operon, this operon could be associated to another aspect of bacterial plant interactions, linked to plant defense signaling and protection against pathogens. Indeed galactinol is a plant compound involved in plant defense (9–11). For example, Pseudomonas chlororaphis O6-mediated induced systemic resistance was shown associated with an elevation of galactinol content within plants, which conferred disease resistance against pathogen attack (9). The disease resistance was associated with induction of the expression of a set of pathogen-responsive genes (9, 10). Moreover, in Arabidopsis, deletion of enzymes that decrease the galactinol and/or raffinose content has been shown to increase plant resistance against the phytopathogenic nematode Heterodera schachtii (11). Similarly, Agrobacterium mel-mediated activity could modify the level of plant galactinol and/or raffinose, which could either drive bacterial recognition by the plant or reduce plant defense signaling through the fall of galactinol content. Agrobacterium is known to be able to bypass and overcome plant defenses (28). It would thus be of interest to study the involvement of the mel operon in that situation.
In this study, we defined the galactinol-/melibiose-/raffinose-binding signature and found out that this is strictly conserved in MelB homologues in Agrobacterium and Rhizobium, which share a high sequence identity over 90% with MelB. Moreover, phylogenetic and structural data showed that this wide occurrence is extended among Rhizobiaceae, all plant-interacting genera (Mesorhizobium, Allorhizobium, Ensifer, Martelella, Pleomorphomonas, Kaistia, and Devosia). Therefore, whatever advantage it gives, it is tempting to speculate that galactinol/melibiose (and to a lesser extent raffinose) may be associated with a selective pressure toward the acquisition of binding, transport, and degradation functions in microorganisms, making the PBP MelB a major trait in the first step of tomato colonization and likely of other plant species.
Experimental procedures
Bacterial culture conditions
Bacteria and plasmids used in this study are shown in Table S1. Escherichia coli strains were grown at 37 °C in LB medium supplemented when it was necessary with appropriate antibiotics (tetracycline 10 μg/ml, gentamicin 15 μg/ml, ampicillin 100 μg/ml). A. fabrum C58 strain and its derivatives were cultivated at 28 °C in YPG (yeast extract, 5 g per liter, peptone, 5 g per liter, glucose, 10 g per liter, and pH adjusted to 7.2) rich medium supplemented when required with neomycin (25 μg/ml), kanamycin (25 μg/ml), and/or gentamycin (20 μg/ml). In growth assays, AT minimal medium supplemented with 10 mm ammonium sulfate and 10 mm carbon sources was used. 200 μl were inoculated in Bioscreen honeycomb 100-well sterile plates and incubated in a Bioscreen C Reader (Labsystems, Helsinki, Finland) at 28 °C during 5 days. Cell growth was measured every 20 min. Analyses were performed in five technical replicates and in three biological replicates.
Construction of melB defective mutant in A. fabrum C58 and transcriptional fusion
The A. fabrum C58ΔmelB defective mutant was constructed as described previously (29) without marker exchange. Briefly, the recombinant region containing the upstream and downstream region flanking the melB gene (amplified by PCR using primers listed in Table S2) was inserted into pJQ200sk vector (30) leading to a nonpolar mutant. The resulting plasmid was introduced into A. fabrum C58 by electroporation. Bacteria were spread on YPG medium plates containing gentamicin (20 μg/ml) for the first selection and gentamicin-resistant colonies were spread on YPG plates supplemented with 5% sucrose for the second selection. The deletion of melB was verified by sequencing (GenoScreen, Lille, France).
The C58 pOT1e-Pmel transcriptional reporter fusion strain was obtained as follows: the promoter region of atu4660-atu4665 genes named Pmel was PCR amplified (using primer listed in Table S2) and the PCR fragment obtained was ligated into ClaI-SalI digested vector pOT1e. pOT1eM-Pmel plasmid was obtained by cloning Pmel into SpeI digested pOT1eM vector as described previously (31). Transcriptional reporter constructions were introduced into A. fabrum by electroporation.
Cloning, expression, and purification of mature MelB
The mature MelB expression plasmid was chemically synthesized using codon optimization for the expression in E. coli and inserted into pET-9a plasmid using NdeI and BamHI restriction enzyme (GenScript, Piscataway, NJ). E. coli BL21 competent cells transformed with pET9a-MelB were grown in LB media at 37 °C until A600 of 0.6. 0.5 mm of isopropyl β-d-thiogalactopyranoside (IPTG) was added to the culture for overnight expression at 20 °C. The cells were pelleted by centrifugation at 4000 × g for 15 min at 4 °C, resuspended in 50 mm Tris-HCl, pH 8, 300 mm NaCl, and 20 mm imidazole, and disrupted by sonication. After centrifugation at 25,000 × g for 30 min, the filtered supernatant was injected on a nickel affinity column (HiTrap 5 ml, GE Healthcare). After a washing step of 6% 50 mm Tris-HCl, pH 8, 300 mm NaCl, and 300 mm imidazole (Buffer B), the protein was eluted with Buffer B and injected on a gel filtration Superdex 200 26/60 (GE Healthcare) using 50 mm Tris-HCl, pH 8, and 150 mm NaCl. The protein fractions were pooled, concentrated at 10.7 mg/ml, and stored at −80 °C.
Expression and purification of mature seleniated MelB
The E. coli BL21 cells transformed with the plasmid pET9a-MelB were grown overnight at 28 °C in M9 media supplemented with 0.4% glucose; 2 mm MgSO4; 1 μm CaCl2; 100 mg/liter of lysine, threonine, and phenylalanine; and 50 mg/liter of leucine, valine, isoleucine, and methionine. The pelleted cells were resuspended in fresh M9 media (same as above) with 100 mg/liter of selenomethionine instead of methionine for 1 h at 37 °C before inducing the expression with 0.5 mm isopropyl β-d-thiogalactopyranoside overnight at 20 °C. The cells were centrifuged at 4000 × g for 15 min at 4 °C. The purification protocol was the same as described above.
Crystallization and data collection of MelB
Crystallization conditions for seleniated MelB in the presence of 2 mm raffinose were screened using Qiagen kits (Valencia, CA) with a Cartesian NanoDrop robot (Genomic Solutions). The crystals were manually reproduced in hanging drops experiments by mixing equal volumes of protein solution and the precipitant solution 25% PEG 4000, 0.2 m NaCl, 0.1 m Mes, pH 6.5, and 0.2 m CaCl2. For the four other complexes, a similar condition without CaCl2 and 0.6 m NaCl was used. Crystals were transferred to a cryoprotectant solution (mother liquor supplemented with 25% PEG 400) and flash-frozen in liquid nitrogen. X-ray diffraction data sets were collected at 100 K on the Proxima 1 or 2 beamlines (SOLEIL synchrotron, Saint-Aubin, France). Data processing was performed using the XDS package (32) (Table 2).
Structure determination and refinement of MelB
The crystal structure of the MelB-raffinose complex was determined by SAD method from selenomethionine-labeled protein and refined at 2 Å resolution. Solvent content analysis using CCP4 (Collaborative Computational Project, Number 4) indicated the presence of four monomers in the asymmetric unit. The positions of 12 over 15 selenium atoms per monomer were found using SHELX suite program (33) The phases were calculated using PHASER (34) and density modification was performed by PARROT (CCP4 suite). An iterative process of manual building in COOT combined with phase calculation where a partial model was used as input, allowed the modeling of the complete polypeptide chain. The structures of all other liganded MelB were solved using the SeMet-MelB monomer as a search model. Refinement of each structure was performed with BUSTER-2.10 (34), NCS restraints, and TLS group. Because of the strong anisotropy of the crystals of MelB-stachyose, the DEBYE and STARANISO programs developed by Global Phasing Ltd. were applied to the data scaled with AIMLESS using the STARANISO server (http://staraniso.globalphasing.org).6 These programs perform an anisotropic cut-off of merge intensity data on the basis of an analysis of local I/s(I); compute Bayesian estimates of structure amplitudes, taking into account their anisotropic fall-off; and apply an anisotropic correction to the data. The corrected anisotropic amplitudes were used for further refinement of the MelB-stachyose structure with BUSTER-2.10. Inspection of the density maps and manual rebuilding were performed using COOT (35). The three-dimensional models of stachyose and galactinol were generated with the ProDRG webserver (36), whereas those of melibiose and raffinose were found in the Protein Data Bank. Refinement details of each structure are shown in Table 2. Molecular graphics images were generated using PyMOL.
Fluorescence titration measurements of MelB
Each ligand bound to MelB was monitored by autofluorescence by exciting the protein at a wavelength of 295 nm and monitoring the quenching of fluorescence emission of tryptophans at 335 nm. All experiments were performed at 22 °C in 96-well plates (1/2 Area Plate-96F, PerkinElmer Life Sciences) using Tecan Infinite M1000 (Tecan, Männedorf, Switzerland) in 25 mm Tris-HCl, pH 8.0, and 150 mm NaCl with a fixed amount of proteins (1 μm) and increasing concentrations of ligand. Each ligand has no emission signal at 335 nm. The data were analyzed using Origin® 7 software and fitted to the following equation.
| (1) |
Isothermal titration microcalorimetry measurements of MelB
Isothermal titration microcalorimetry experiments were performed with an ITC200 isothermal titration calorimeter from MicroCal (GE Healthcare). The experiments were carried out at 20 °C. Protein concentration in the microcalorimeter cell (0.2 ml) varied from 10 to 300 μm. Nineteen injections of 2 μl of ligand solution (raffinose, stachyose, melibiose, galactose, and galactinol) concentration from 0.1 to 2.8 mm were performed at intervals of 180 s while stirring at 500 rpm. The experimental data were fitted to theoretical titration curves with software supplied by MicroCal (ORIGIN®). This software uses the relationship between the heat generated by each injection and ΔH (enthalpy change in kcal mol−1), Ka (the association binding constant in mol−1), n (the number of binding sites), total protein concentration, and free and total ligand concentrations (37).
Differential scanning calorimetry
Thermal stability of the WT and liganded MelB (13 μm and 50 μm for protein and ligand, respectively) was studied by differential scanning calorimetry (DSC) on a MicroCal model VP-DSC in a standard buffer. Each measurement was preceded by a baseline scan with the standard buffer. All solutions were degassed just before loading into the calorimeter. Scans were performed at 1 K·min−1 between 20 and 90 °C. The heat capacity of the buffer was subtracted from that of the protein sample before analysis. Thermodynamic parameters were determined by fitting the data to the following equation,
| (2) |
where Kd is the equilibrium constant for a two-state process, ΔHvH is the enthalpy calculated on the basis of a two-state process, and ΔHcal is the measured enthalpy.
Phylogenetic analysis
Sequences were analyzed using BlastP from NCBI (https://blast.ncbi.nlm.nih.gov/) and MicrosScope (https://www.genoscope.cns.fr/).6 Alignments of MelB and related sequences were conducted using ClustalW software. Relationship tree was build using Mega software, version 7. The bootstrap consensus tree inferred from 1000 replicates was taken to represent the evolutionary history of the taxa analyzed. The evolutionary distances were computed using the Poisson correction method and are in units of the number of amino acid substitutions per site.
Measurement of mel operon gene expression
Expression of mel operon genes was measured in the C58 pOT1e-Pmel strain. Quantification of fluorescence was carried out in a microplate filled with 200 μl of AT medium, supplemented with different carbon sources at a final concentration of 10 mm. Microplate wells were inoculated with overnight cultures to obtain an A600 of 0.2. A TECAN apparatus (Tecan SparkTM 15 m, Männedorf, Switzerland) was used to read microplates after 24 h of incubation at 28 °C. The following parameters were used: absorbance at 600 nm, fluorescence excitation at 488 nm, and emission at 510 nm. Results were normalized by the A600 and -fold change values were obtained by dividing the fluorescence by the corresponding value obtained from the empty pOT1e vector. The fluorescence level comparison was carried out using the Tukey's test (p value = 0.05) and computed with the “vegan” package in the R v3.1.3 statistical software environment (R Core Team, 2014).
Plant inoculation
For bacterial colonization, competition assays, and confocal observation studies, tomato seeds (Solanum lycopersicum “Marmande”) were sterilized as described (38). Seeds were plated on 0.8% agar plant cell culture supplemented with 1.5 g/liter of the Plant-Prod 15-15-30 High K nutrient solution (Master Plant-Prod Inc., Brampton, Ontario, Canada). They were inoculated with 10 μl of overnight culture (106 cfu/ml) of a single strain (A. fabrum C58 pTiatu6148:Km derivative of the WT strain (39) or C58ΔmelB mutant), or with a mixture of both at 1:1 ratio (competition). Petri dishes were placed 2 days in the dark and then in a climatic chamber at 24 °C with 18/8 h for light/dark and 65% of humidity. To determine bacterial colonization level, roots were ground at 2 and 14 dpi. Serial dilutions of crushed roots were plated on the YPG medium and colonies were counted after 2 days of incubation at 28 °C. Significant difference in the population level resulting from five plants per strain, with enumeration of three Petri dishes for each plant, was evaluated with Mann-Whitney test (p value = 0.05) performed with the R v3.1.3 statistical software environment.
In competition experiments, the colonized bacteria were nonselectively recovered at 2 and 14 dpi. To that end, crushed roots were first plated with a spiral plater (EasySpiral®, Interscience, Saint-Nom-la-Bretèche, France) on YPG medium without antibiotics to enable a biologically unbiased recovery of both C58 pTiatu6148:Km WT strain and C58ΔmelB mutant (kanamycin sensitive). Two hundred individual colonies were then plated in YPG medium with kanamycin/neomycin to determine the relative proportions of C58 pTiatu6148:Km and C58ΔmelB mutant strains (output ratio). The determination of the initial strains ratio of the inoculum was realized using the same protocol. The experimental assays were performed with 10 independent assays and repeated four times. The proportions of WT strains between the initial strain ratio and in planta output ratios were compared with the test of equal or given proportions (p value = 0.05).
Confocal microscopy analyses
Visualization of reporter bacterial cells harboring pOT1eM-Pmel on tomato radicles and roots was performed using a confocal laser scanning microscope (LSM 800 Meta Confocal Microscope, Zeiss, Oberkochen, Germany). In the reporter strain, M-Cherry (red color) is constitutively expressed and enhanced GFP (eGFP) (green color) is expressed under Pmel control. The red color indicates the bacteria presence (red cells) whereas eGFP reports the induction of Pmel shown as yellow-green cells. At 2 and 14 dpi, tomato radicles and roots were mounted between a slide and a coverslip in a commercial mounting fluid (Aqua Poly/Mount, Polysciences, Inc., Warrington, PA). The eGFP and the M-cherry were excited with argon laser at 488 nm and 584 nm, respectively, and fluorescence was captured at 528 nm and 607 nm. Analyses of images (five plants per condition) were performed thanks to LSM 800 software (Zeiss, Oberkochen, Germany).
Coordinates
The atomic coordinates and structure factors have been deposited at the Protein Data Bank (PDB) under accession codes 6EPY (seleniated MelB with raffinose), 6EQ1 (MelB with stachyose), 6EQ8 (MelB with galactinol), 6EPZ (MelB with melibiose) and 6EQ0 (MelB with galactose).
Author contributions
T. M., L. V., and C. L. performed all the microbiology work. A. V. and S. M. performed all the crystallography work. A. V. performed the fluorescence assays. A. V. and M. A. N. performed the microcalorimetry experiments. A. V., T. M., C. L., and S. M. performed the phylogenetic analysis. S. M. and C. L. wrote the manuscript. All the authors discussed the results and contributed to the writing of the manuscript.
Supplementary Material
Acknowledgments
This work has benefited from the I2BC crystallization and microcalorimetry platforms supported by FRISBI ANR-10-INSB-05-01 as well as from the “Centre Technologique des Microstructures” and the “Serre et chambres climatiques” platforms, supported by the FR BioEnviS Research Federation. We acknowledge SOLEIL for provision of synchrotron radiation facilities (proposals ID 20130869, 20140774, and 20160782) in using Proxima beamlines. We thank Andrew Saurin for critical reading of the manuscript.
This work was supported by CNRS (Mission pour l'interdisciplinarité, Agromics 2014–2016) (to T. M., A. V., S. M., and C. L.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 6EPY, 6EQ1, 6EQ8, 6EQ0, and 6EPZ) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- RFOs
- raffinose family of oligosaccharides
- PBP
- periplasmic-binding protein
- YPG medium
- yeast peptone glucose medium
- ITC
- isothermal titration microcalorimetry
- RMSD
- root mean square deviation
- dpi
- days post inoculation
- eGFP
- enhanced GFP.
References
- 1. Bais H. P., Weir T. L., Perry L. G., Gilroy S., and Vivanco J. M. (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57, 233–266 10.1146/annurev.arplant.57.032905.105159 [DOI] [PubMed] [Google Scholar]
- 2. Nelson E. B. (2004) Microbial dynamics and interactions in the spermosphere. Annu. Rev. Phytopathol. 42, 271–309 10.1146/annurev.phyto.42.121603.131041 [DOI] [PubMed] [Google Scholar]
- 3. Nelson E. B. (2018) The seed microbiome: Origins, interactions, and impacts. Plant Soil 422, 7–34 10.1007/s11104-017-3289-7 [DOI] [Google Scholar]
- 4. Barret M., Briand M., Bonneau S., Préveaux A., Valière S., Bouchez O., Hunault G., Simoneau P., and Jacquesa M.-A. (2015) Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 81, 1257–1266 10.1128/AEM.03722-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sengupta S., Mukherjee S., Basak P., and Majumder A. L. (2015) Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 6, 656 10.3389/fpls.2015.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishizawa A., Yabuta Y., and Shigeoka S. (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147, 1251–1263 10.1104/pp.108.122465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., and Shinozaki K. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 29, 417–426 10.1046/j.0960-7412.2001.01227.x [DOI] [PubMed] [Google Scholar]
- 8. Ibáñez C., Collada C., Casado R., González-Melendi P., Aragoncillo C., and Allona I. (2013) Winter induction of the galactinol synthase gene is associated with endodormancy in chestnut trees. Trees 27, 1309–1316 10.1007/s00468-013-0879-8 [DOI] [Google Scholar]
- 9. Kim M. S., Cho S. M., Kang E. Y., Im Y. J., Hwangbo H., Kim Y. C., Ryu C.-M., Yang K. Y., Chung G. C., and Cho B. H. (2008) Galactinol is a signaling component of the induced systemic resistance caused by Pseudomonas chlororaphis O6 root colonization. Mol. Plant Microbe Interact. 21, 1643–1653 10.1094/MPMI-21-12-1643 [DOI] [PubMed] [Google Scholar]
- 10. Cho S. M., Kang E. Y., Kim M. S., Yoo S. J., Im Y. J., Kim Y. C., Yang K. Y., Kim K. Y., Kim K. S., Choi Y. S., and Cho B. H. (2010) Jasmonate-dependent expression of a galactinol synthase gene is involved in priming of systemic fungal resistance in Arabidopsis thaliana. Botany 88, 452–461 10.1139/B10-009 [DOI] [Google Scholar]
- 11. Siddique S., Endres S., Sobczak M., Radakovic Z. S., Fragner L., Grundler F. M. W., Weckwerth W., Tenhaken R., and Bohlmann H. (2014) Myo-inositol oxygenase is important for the removal of excess myo-inositol from syncytia induced by Heterodera schachtii in Arabidopsis roots. New Phytol. 201, 476–485 10.1111/nph.12535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berntsson R. P.-A., Smits S. H. J., Schmitt L., Slotboom D.-J., and Poolman B. (2010) A structural classification of substrate-binding proteins. FEBS Lett. 584, 2606–2617 10.1016/j.febslet.2010.04.043 [DOI] [PubMed] [Google Scholar]
- 13. Gage D. J., and Long S. R. (1998) α-Galactoside uptake in Rhizobium meliloti: Isolation and characterization of agpA, a gene encoding a periplasmic binding protein required for melibiose and raffinose utilization. J. Bacteriol. 180, 5739–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bringhurst R. M., Cardon Z. G., and Gage D. J. (2001) Galactosides in the rhizosphere: Utilization by Sinorhizobium meliloti and development of a biosensor. Proc. Natl. Acad. Sci. U.S.A. 98, 4540–4545 10.1073/pnas.071375898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liljeström P. L., and Liljeström P. (1987) Nucleotide sequence of the melA gene, Coding for α-galactosidase in Escherichia coli K-12. Nucleic Acids Res. 15, 2213–2220 10.1093/nar/15.5.2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charaoui-Boukerzaza S., and Hugouvieux-Cotte-Pattat N. (2013) A family 3 glycosyl hydrolase of Dickeya dadantii 3937 is involved in the cleavage of aromatic glucosides. Microbiology. 159, 2395–2404 10.1099/mic.0.071407-0 [DOI] [PubMed] [Google Scholar]
- 17. Liu Y., Chen L., Wu G., Feng H., Zhang G., Shen Q., and Zhang R. (2017) Identification of root-secreted compounds involved in the communication between cucumber, the beneficial Bacillus amyloliquefaciens, and the soil-borne pathogen Fusarium oxysporum. Mol. Plant Microbe Interact. 30, 53–62 10.1094/MPMI-07-16-0131-R [DOI] [PubMed] [Google Scholar]
- 18. Nester E. W. (2014) Agrobacterium: nature's genetic engineer. Front. Plant Sci. 5, 730 10.3389/fpls.2014.00730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abarca-Grau A. M., Penyalver R., López M. M., and Marco-Noales E. (2011) Pathogenic and non-pathogenic Agrobacterium tumefaciens, A. rhizogenes and A. vitis strains form biofilms on abiotic as well as on root surfaces: Biofilms formed by Agrobacterium spp. Plant Pathol. 60, 416–425 10.1111/j.1365-3059.2010.02385.x [DOI] [Google Scholar]
- 20. Hall B. G., Pikis A., and Thompson J. (2009) Evolution and biochemistry of family 4 glycosidases: Implications for assigning enzyme function in sequence annotations. Mol. Biol. Evol. 26, 2487–2497 10.1093/molbev/msp162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wood D. W., Setubal J. C., Kaul R., Monks D. E., Kitajima J. P., Okura V. K., Zhou Y., Chen L., Wood G. E., Almeida N. F. Jr., Woo L., Chen Y., Paulsen I. T., Eisen J. A., Karp P. D., et al. (2001) The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 294, 2317–2323 10.1126/science.1066804 [DOI] [PubMed] [Google Scholar]
- 22. Westover B. P., Buhler J. D., Sonnenburg J. L., and Gordon J. I. (2005) Operon prediction without a training set. Bioinformatics 21, 880–888 10.1093/bioinformatics/bti123 [DOI] [PubMed] [Google Scholar]
- 23. Kemner J. M., Liang X., and Nester E. W. (1997) The Agrobacterium tumefaciens virulence gene chvE is part of a putative ABC-type sugar transport operon. J. Bacteriol. 179, 2452–2458 10.1128/jb.179.7.2452-2458.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cornish A., Greenwood J. A., and Jones C. W. (1989) Binding-protein-dependent sugar transport by Agrobacterium radiobacter and A. tumefaciens grown in continuous culture. J. Gen. Microbiol. 135, 3001–3013 10.1099/00221287-135-11-3001 [DOI] [PubMed] [Google Scholar]
- 25. Andersen K. E., Bjergegaard C., Møller P., Sørensen J. C., and Sørensen H. (2005) Compositional variations for α-galactosides in different species of leguminosae, brassicaceae, and barley: A chemotaxonomic study based on chemometrics and high-performance capillary electrophoresis. J. Agric. Food Chem. 53, 5809–5817 10.1021/jf040471v [DOI] [PubMed] [Google Scholar]
- 26. Kristin A., and Miranda H. (2013) The root microbiota—a fingerprint in the soil? Plant Soil 370, 671–686 10.1007/s11104-013-1647-7 [DOI] [Google Scholar]
- 27. Fukami T., Martijn Bezemer T., Mortimer S. R., and van der Putten W. H. (2005) Species divergence and trait convergence in experimental plant community assembly. Ecol. Lett. 8, 1283–1290 10.1111/j.1461-0248.2005.00829.x [DOI] [Google Scholar]
- 28. Veena Jiang H., Doerge R. W., and Gelvin S. B. (2003) Transfer of T-DNA and Vir proteins to plant cells by Agrobacterium tumefaciens induces expression of host genes involved in mediating transformation and suppresses host defense gene expression. Plant J. 35, 219–236 10.1046/j.1365-313X.2003.01796.x [DOI] [PubMed] [Google Scholar]
- 29. Lassalle F., Campillo T., Vial L., Baude J., Costechareyre D., Chapulliot D., Shams M., Abrouk D., Lavire C., Oger-Desfeux C., Hommais F., Gueguen L., Daubin V., Muller D., and Nesme X. (2011) Genomic species are ecological species as revealed by comparative genomics in Agrobacterium tumefaciens. Genome Biol. Evol. 3, 762–781 10.1093/gbe/evr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quandt J., and Hynes M. F. (1993) Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21 10.1016/0378-1119(93)90611-6 [DOI] [PubMed] [Google Scholar]
- 31. Meyer T., Renoud S., Vigouroux A., Miomandre A., Gaillard V., Kerzaon I., Prigent-Combaret C., Comte G., Moréra S., Vial L., and Lavire C. (2018) Regulation of hydroxycinnamic acid degradation drives Agrobacterium fabrum lifestyles. Mol. Plant Microbe Interact. 10.1094/MPMI-10-17-0236-R [DOI] [PubMed] [Google Scholar]
- 32. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A. 64, 112–122 10.1107/S0108767307043930 [DOI] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 36. Schüttelkopf A. W., and van Aalten D. M. F. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 10.1107/S0907444904011679 [DOI] [PubMed] [Google Scholar]
- 37. Wiseman T., Williston S., Brandts J. F., and Lin L. N. (1989) Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 179, 131–137 10.1016/0003-2697(89)90213-3 [DOI] [PubMed] [Google Scholar]
- 38. Vacheron J., Moënne-Loccoz Y., Dubost A., Gonçalves-Martins M., Muller D., and Prigent-Combaret C. (2016) Fluorescent Pseudomonas strains with only few plant-beneficial properties are favored in the maize rhizosphere. Front. Plant Sci. 7, 1212 10.3389/fpls.2016.01212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lang J., Planamente S., Mondy S., Dessaux Y., Moréra S., and Faure D. (2013) Concerted transfer of the virulence Ti plasmid and companion At plasmid in the Agrobacterium tumefaciens-induced plant tumour. Mol. Microbiol. 90, 1178–1189 10.1111/mmi.12423 [DOI] [PubMed] [Google Scholar]
- 40. Karplus P. A., and Diederichs K. (2012) Linking crystallographic model and data quality. Science. 336, 1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.