ABSTRACT
The YbeY endoribonuclease is one of the best-conserved proteins across the kingdoms of life. In the present study, we demonstrated that YbeY in Brucella abortus is linked to a variety of important activities, including proper cellular morphology, mRNA transcript levels, and virulence. Deletion of ybeY in B. abortus led to a small-colony phenotype when the bacteria were grown on agar medium, as well as to significant aberrations in the morphology of the bacterial cell as evidenced by electron microscopy. Additionally, compared to the parental strain, the ΔybeY strain was significantly attenuated in both macrophage and mouse models of infection. The ΔybeY strain also showed increased sensitivities to several in vitro-applied stressors, including bile acid, hydrogen peroxide, SDS, and paraquat. Transcriptomic analysis revealed that a multitude of mRNA transcripts are dysregulated in the ΔybeY strain, and many of the identified mRNAs encode proteins involved in metabolism, nutrient transport, transcriptional regulation, and flagellum synthesis. We subsequently constructed gene deletion strains of the most highly dysregulated systems, and several of the YbeY-linked gene deletion strains exhibited defects in the ability of the bacteria to survive and replicate in primary murine macrophages. Taken together, these data establish a clear role for YbeY in the biology and virulence of Brucella; moreover, this work further illuminates the highly varied roles of this widely conserved endoribonuclease in bacteria.
IMPORTANCE Brucella spp. are highly efficient bacterial pathogens of animals and humans, causing significant morbidity and economic loss worldwide, and relapse of disease often occurs following antibiotic treatment of human brucellosis. As such, novel therapeutic strategies to combat Brucella infections are needed. Ribonucleases in the brucellae are understudied, and these enzymes represent elements that may be potential targets for future treatment approaches. The present work demonstrates the importance of the YbeY endoribonuclease for cellular morphology, efficient control of mRNA levels, and virulence in B. abortus. Overall, the results of this study advance our understanding of the critical roles of YbeY in the pathogenesis of the intracellular brucellae and expand our understanding of this highly conserved RNase.
KEYWORDS: Brucella, ybeY, RNase, YbeY, ribonuclease
INTRODUCTION
Ribonucleases (RNases) are enzymes that catalyze the cleavage of myriad RNAs, be they mRNA, tRNA, rRNA, or small RNA (sRNA), and these enzymes are divided into two major classes called exoribonucleases and endoribonucleases depending on their ability to cleave RNA strands at terminal or nonterminal nucleotides, respectively (1, 2). The “day-to-day operations” of RNases include degradation of RNAs during housekeeping turnover processes, but RNases also process longer RNA transcripts into shorter, functional RNAs. A classic example of RNA processing is the generation of the three major rRNAs (i.e., 23S, 16S, and 5S) and of tRNAs from precursor RNAs, a process catalyzed by several different RNases in bacteria (3, 4). As such, bacteria encode an extensive array of RNases to perform a wide variety of degradation and processing functions.
One of the more recently described bacterial RNases is the YbeY endoribonuclease (5). Interestingly, the structure of YbeY was studied prior to the availability of any insights into its biological functions. The crystal structure of the Aquifex aeolicus YbeY ortholog revealed resemblances to metal-dependent proteinases such as collagenases (6), while crystallization of the Escherichia coli YbeY protein as part of an NIH-funded Protein Structure Initiative program led to the suggestion that it is a metal-dependent hydrolase (7). Subsequently, the Sinorhizobium meliloti YbeY ortholog was found to be required for symbiosis, while E. coli YbeY drew attention because of its regulation as a heat shock protein (8). YbeY was then shown to participate in the maturation of ribosomal RNAs and the biosynthesis of ribosomes, and, more recently, evidence has been reported that YbeY functions as an endoribonuclease in rRNA maturation activities and 70S ribosome quality control (5, 9–11). Additionally, YbeY plays a significant role in the regulation and stability of bacterial sRNAs (12, 13). Not only has YbeY been linked to the capacity of S. meliloti to form an effective symbiotic relationship with its plant host alfalfa, but ybeY has been shown to be required for the full virulence of Vibrio cholerae and Yersinia enterocolitica (14–16). While RNases, including YbeY, are known to be important virulence determinants for several bacterial pathogens, very little is known about the role of RNases in the Brucella spp. (17).
The brucellae are small Gram-negative bacteria that cause significant disease in both humans and animals globally (18), and these bacteria are intracellular pathogens of macrophages and dendritic cells, where they reside in a vacuole-bound niche in close proximity to the endoplasmic reticulum (19, 20). Interestingly, the brucellae do not produce classical virulence factors, such as toxins or endotoxic LPS; rather, these bacteria are stealthy pathogens whose ability to cause disease is directly related to their capacity to survive and replicate inside the cells of the host (21, 22). As noted above, little is known about RNases in Brucella spp., and in fact, only two published reports describe RNases in Brucella, and neither of the described RNases is required for the infectivity of the brucellae (23, 24). We have recently investigated the contribution of the RNase YbeY to Brucella biology, and among several interesting observations, we have determined that YbeY is required for normal cellular morphology and wild-type virulence in B. abortus 2308. Overall, the current report defines and characterizes the importance of YbeY in Brucella; moreover, these data shed light on the significance of this endoribonuclease for intracellular bacterial pathogens.
RESULTS
YbeY is required for normal growth and cellular morphology of Brucella abortus.
Brucella abortus 1_2156 (bab1_2156; also known as bab_rs26200) is located on chromosome I of Brucella melitensis biovar Abortus 2308 between bab1_2155 (phoH) and bab1_2157 (tlyC) (Fig. 1A). The YbeY protein exhibits 56% identity and 68% similarity to the YbeY endoribonuclease from Sinorhizobium meliloti 1021. For this reason, and due to the results outlined in this report, here we refer to bab1_2156 as ybeY.
FIG 1.
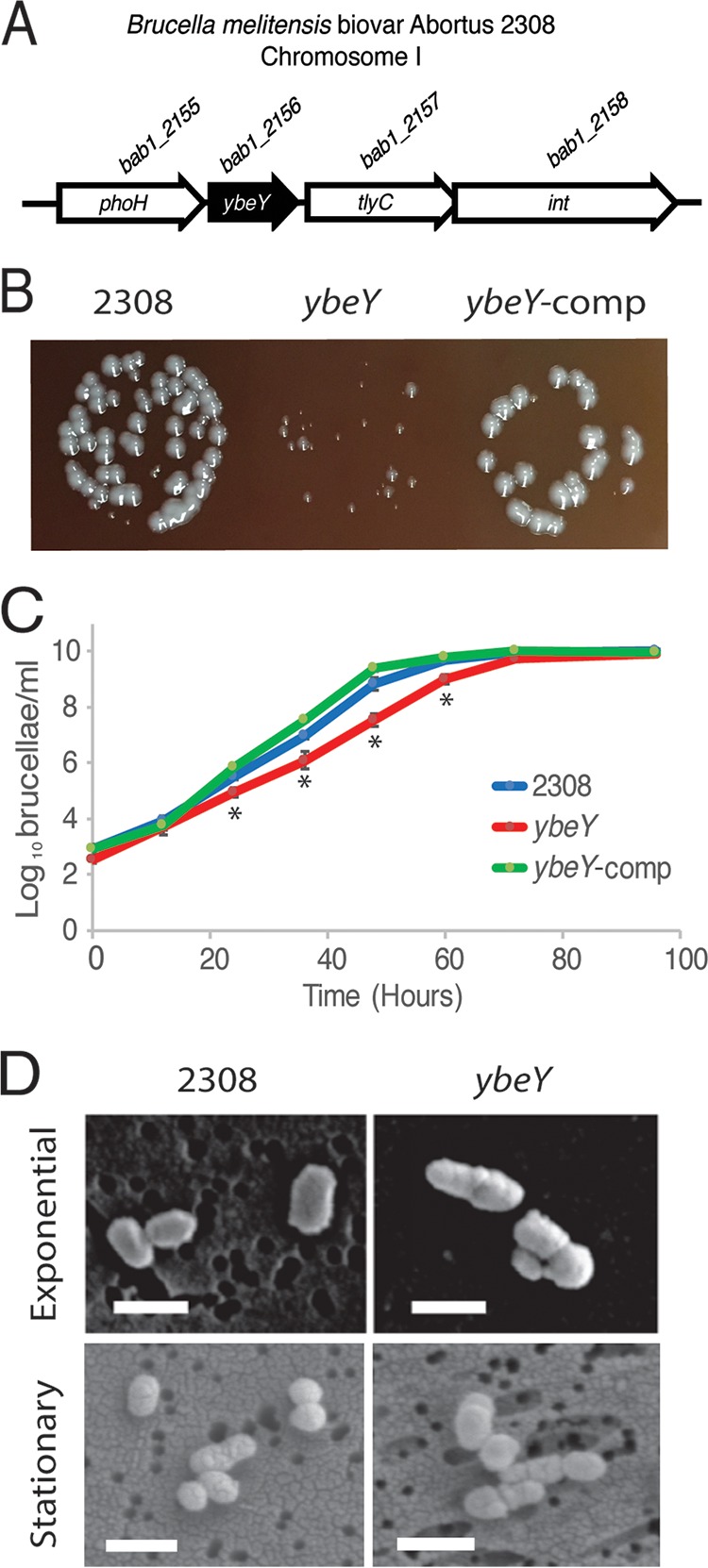
In vitro growth characteristics and cellular morphology of B. abortus 2308::ΔybeY. (A) Genomic context of ybeY. The ybeY gene (bab1_2156; also known as bab_rs26200) is located on chromosome I in B. abortus 2308. ybeY is flanked by phoH (bab1_2155; also known as bab_rs26195) and a gene encoding a putative hypothetical protein (bab1_2157; also known as bab_rs26205). (B) Photograph of B. abortus colonies on SBA after 72 h of growth. (C) Growth curve of Brucella abortus strains in rich medium. B. abortus 2308, B. abortus 2308::ΔybeY, and B. abortus 2308::ΔybeY-comp were grown in brucella broth, and CFU counts per milliliter were monitored by serial dilution. The asterisk denotes a statistically significant difference (P < 0.05; Student's t test) between the ybeY deletion strain and parental strain 2308. (D) Electron microscopy of Brucella abortus cells. Exponential- and stationary-phase cells of Brucella strains were fixed and viewed using scanning electron microscopy (magnification, ×30,000). Bars = 1 μm.
An isogenic deletion of ybeY in B. abortus 2308 resulted in impaired growth in vitro and abnormal cellular morphology compared to the parental strain (Fig. 1). The B. abortus ΔybeY deletion strain exhibited a small-colony phenotype when grown on agar medium, and this defect was genetically complemented when ybeY was provided in trans on plasmid pBBR-1MCS4 (Fig. 1B). When cultured in brucella broth (i.e., rich medium), the ybeY deletion strain was able to grow to similar maximum numbers of bacteria as the parental strain, but the ybeY deletion strain had a decreased rate of growth during the exponential-growth phase (Fig. 1C). During the exponential-growth phase, B. abortus 2308 had a generation time of 2.2 h whereas the ybeY deletion strain had a generation time of 2.8 h. Importantly, the growth rate of the B. abortus ΔybeY deletion strain was restored to a 1.9-h generation time by in trans complementation of ybeY.
Using scanning electron microscopy, the ybeY deletion strain was observed to have cellular morphology deformities when the bacteria were collected from the exponential and stationary phases of growth in brucella broth (Fig. 1D). As expected, B. abortus 2308 cells were coccobacilli in shape during the exponential phase of growth and cocci during the stationary phase of growth, with clear septa between dividing cells. The ybeY deletion strain, however, exhibited noticeable morphological irregularities, including occurrences of clusters of cells appearing to be unable to properly divide during both the exponential and stationary phases of growth. Taken together, these data demonstrate that YbeY is required for the efficient growth and cellular morphology of B. abortus.
YbeY contributes to B. abortus virulence in macrophages and experimentally infected mice.
To characterize the importance of YbeY for B. abortus virulence, the ybeY deletion strain was assessed for the ability to infect peritoneal macrophages in vitro and BALB/c mice in vivo (Fig. 2). Peritoneally derived macrophages isolated from BALB/c mice were infected with B. abortus 2308 or B. abortus 2308::ΔybeY or B. abortus 2308::ΔybeY-comp at a multiplicity of infection (MOI) of 100. The ybeY deletion strain was strikingly less able to survive and replicate within the macrophage than the parental strain at 24 and 48 h postinfection, and this decrease in survival and replication was restored to wild-type levels in the ybeY complemented strain (Fig. 2A). Similarly, the ybeY deletion strain exhibited a substantially reduced ability to infect BALB/c mice compared to parental strain 2308, as significantly fewer bacteria were recovered from the spleens of mice infected with the ybeY deletion strain after both 4 and 8 weeks of infection (Fig. 2B). These experiments indicate that YbeY is necessary for the ability of B. abortus to sustain infection in macrophages and mice.
FIG 2.
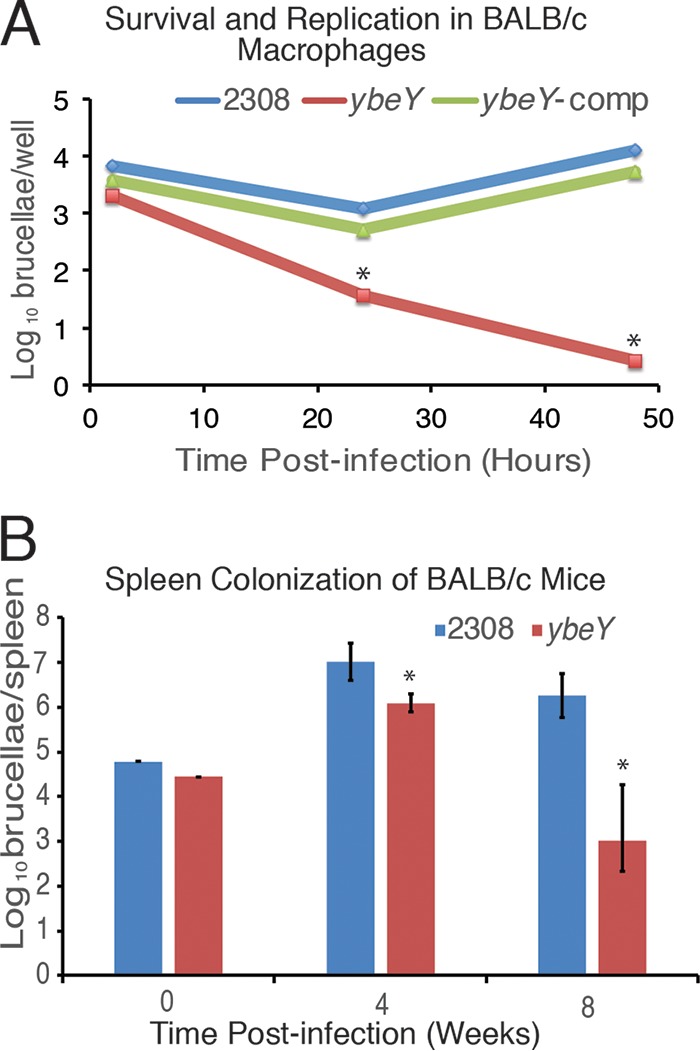
Virulence of B. abortus 2308 and ΔybeY in peritoneally derived macrophages and BALB/c mice. (A) Macrophage survival and replication experiments. Cultured peritoneal macrophages from BALB/c mice were infected with B. abortus 2308, the isogenic ybeY deletion strain (ybeY), and the ybeY complemented strain (ybeY-comp). At the indicated times postinfection, macrophages were lysed, and the number of intracellular brucellae present in these phagocytes was determined by serial dilution and plating on agar medium. The asterisk denotes a statistically significant difference (P < 0.05; Student's t test) between the ybeY deletion strain and parental strain 2308 and between the ybeY deletion strain and the complemented strain at 24 and 48 h postinfection. (B) Mouse infection experiments. BALB/c mice (5 per strain) were infected intraperitoneally with B. abortus 2308 and with the isogenic ybeY deletion strain (ybeY). Mice were sacrificed at weeks 4 and 8 postinfection, and levels of brucellae/spleen were determined. The data are presented as average numbers of brucellae ± standard deviations of results from the 5 mice colonized with a specific Brucella strain at each time point. The asterisk denotes a statistically significant difference (P < 0.05; Student's t test) between the ybeY deletion strain and parental strain 2308 at 4 and 8 weeks postinfection.
Deletion of ybeY in B. abortus leads to increased sensitivities to general stress and wide-ranging metabolic aberrations.
Due to the decreased growth rate, defect in cell morphology, and reduced ability to infect in the in vitro and in vivo models of the ybeY deletion strain, we sought to gain insight into the link between YbeY and general stress in B. abortus. To achieve this, we employed disk diffusion assays in which B. abortus strains were exposed to a variety of stressors, including deoxycholate (10%), H2O2 (30%), sodium dodecyl sulfate (SDS) (20%), polymyxin B (10 mg/ml), and paraquat (0.25 M) (Fig. 3). In these experiments, the ybeY deletion strain was more sensitive than parental strain 2308 to deoxycholate, H2O2, SDS, and paraquat, and genetic complementation of ybeY in the deletion strain restored the zones of inhibition to the levels observed for strain 2308. Interestingly, deletion of ybeY had no effect on the ability of B. abortus to withstand killing by polymyxin B. These data demonstrate that YbeY is important for the ability of B. abortus to cope with general stress conditions.
FIG 3.
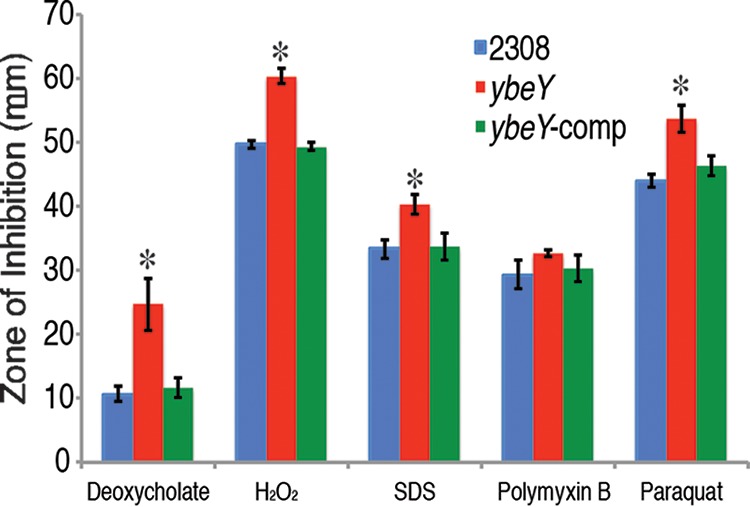
YbeY is required for optimal resistance to biologically relevant stresses. Sensitivity assay results are shown. Brucella abortus 2308, the ybeY deletion strain, and ybeY-comp complemented strain were assessed in a disk diffusion assay for their comparative susceptibilities to various sources of stress, including deoxycholate (10%), H2O2 (30%), SDS (20%), polymyxin B (10 mg/ml), and paraquat (0.25 M). The results are plotted as the average diameter (±standard deviation) of the zone of inhibition around a disk, and the results are from a single experiment that was performed in triplicate. Asterisks denote a statistically significant difference (P < 0.05; Student's t test) between the ybeY deletion strain and parental strain 2308 for a given condition.
The Biolog Phenotype MicroArray system provides an inexpensive and rapid means of testing microorganisms for the ability to grow under hundreds of sets of conditions. Here, we employed Biolog Phenotype MicroArrays to analyze the growth of B. abortus 2308 and B. abortus 2308::ΔybeY in a wide variety of different nutrient sources and environments and in the presence of various stressors. Each Biolog Phenotype MicroArray plate was inoculated with 108 CFU/well of the appropriate Brucella strains and incubated for 84 h at 37°C. After 84 h of incubation, each individual well was measured at an optical density (OD) of 590 nm and visually monitored for growth, as indicated by metabolic activity (clear to purple) (see Data Set S1 in the supplemental material). Overall, we observed 27 differences in growth between B. abortus 2308 and the ybeY deletion strain (see Table S1 in the supplemental material). The conditions under which the 2308::ΔybeY mutant grew more efficiently than strain 2308 are highlighted in green, and the conditions under which strain 2308 grew more efficiently than the ybeY strain are highlighted in red. With regard to carbon sources, the ybeY deletion strain was better able than parental strain 2308 to utilize malic acid and laminarin (a storage glucan). However, deletion of ybeY led to the inability of B. abortus to utilize butyric acid or caproic acid as a carbon source for growth. Compared to growth of B. abortus 2308, growth of the ybeY deletion strain was more sensitive to dodecyltrimethyl ammonium bromide, promethazine, alexidine, dichlofluanid, chloroxylenol, sodium m-periodate, lidocaine, josamycin, thioridazine, patulin, and tetrazolium violet. Conversely, growth of B. abortus 2308::ΔybeY was more resistant to the presence of fusaric acid, 1-chloro-2,4-dinitrobenzene, 2-phenylphenol, antimony (III) chloride, pentachloro-phenol, azathioprine, phenethicillin, and lawsone. Taken together, the Biolog Phenotype MicroArray results underscored the diverse metabolic abnormalities that result from the deletion of ybeY in B. abortus.
YbeY impacts the levels of mRNA associated with a variety of cellular systems.
The pleiotropic effects of ybeY loss on cellular RNAs have been well documented in other bacteria (5, 9, 10, 12, 15, 25, 26), and as such, we hypothesized that deletion of ybeY would lead to changes in mRNA levels in B. abortus. Therefore, we employed microarray technology to identify mRNAs that are influenced by YbeY. This experiment was performed using RNA from cultures of B. abortus 2308 and B. abortus 2308::ΔybeY grown in brucella broth to the late exponential phase (Data Set S2). Taking the results together, mRNAs from 84 genes exhibited differential expression levels (>3-fold difference) in the ΔybeY mutant; of these, expression levels of 34 mRNAs were elevated in the ybeY deletion strain whereas those of 51 mRNAs were decreased in the ybeY deletion strain compared to the parental strain (Table 1). The mRNAs that displayed differential quantities in the ybeY deletion strain included those encoding membrane proteins and transport systems; proteins involved in DNA replication and transcriptional or translational regulation; proteins related to flagellar processes; proteins linked to metabolism, signaling, and enzymatic processes; and hypothetical proteins.
TABLE 1.
Differential gene expression in B. abortus 2308::ΔybeYa
| Functional category and designation | Description | Fold expression change (ΔybeY mutant vs strain 2308) |
|---|---|---|
| Membrane proteins and transport systems | ||
| BAB1_0114 | Glycosyl transferase | 3.3 |
| BAB1_0372 | TRAP dicarboxylate transporter, DctM subunit | −3.2 |
| BAB1_0373 | TRAP-type mannitol/chloroaromatic compound transport system | −3.9 |
| BAB1_1589 | Major facilitator transporter | 3.2 |
| BAB1_1679 | MotA/TolQ/ExbB proton channel | 3.5 |
| BAB1_1680 | Biopolymer transport protein ExbD/TolR | 3.5 |
| BAB1_1681 | Cell envelope biogenesis protein TonB | 3.8 |
| BAB1_1792 | Leu/Ile/Val-binding family protein | −4.5 |
| BAB2_0242 | Putative sulfite oxidase subunit YedZ | 3.9 |
| BAB2_0277 | Choline dehydrogenase and related flavoproteins | −16.1 |
| BAB2_0278 | ABC transporter, permease | −9.5 |
| BAB2_0279 | Inner membrane translocator | −10.3 |
| BAB2_0280 | Shikimate kinase | −8.2 |
| BAB2_0281 | ABC transporter ATPase | −9.9 |
| BAB2_0282 | Leu/Ile/Val-binding family protein | −7.5 |
| BAB2_0300 | Inner membrane translocator | −3.2 |
| BAB2_0519 | Periplasmic spermidine/putrescine-binding protein | −3.3 |
| BAB2_0547 | Solute-binding family 1 protein | −3.7 |
| BAB2_0548 | Vacuolar H+-transporting two-sector ATPase subunit C | −5.7 |
| BAB2_0583 | Aromatic amino acid permease | −3.2 |
| BAB2_0584 | Binding protein-dependent transport system inner membrane protein | −4.0 |
| BAB2_0585 | Solute-binding family 1 protein | −3.2 |
| BAB2_0593 | Leu/Ile/Val-binding family protein | −3 |
| BAB2_0700 | Solute-binding family 5 protein | −4.1 |
| BAB2_0822 | Leu/Ile/Val-binding family protein | −6.9 |
| BAB2_0827 | ABC transporter ATPase | −4.1 |
| BAB2_0828 | Glutelin | −4.1 |
| BAB2_0829 | Inner membrane translocator | −5.6 |
| BAB2_0830 | Leu/Ile/Val-binding family protein | −5.4 |
| BAB2_1109 | d-Xylose ABC transporter | −4.8 |
| DNA replication, transcription, and translation | ||
| BAB1_0636 | Response regulator receiver/transcriptional regulatory protein, C terminal | 3.1 |
| BAB1_1100 | Phage integrase | 3.0 |
| BAB1_1362 | Periplasmic binding protein/LacI transcriptional regulator | −3.3 |
| BAB1_1588 | MarR family regulatory protein | 4.6 |
| BAB2_0222 | Response regulator receiver/transcriptional regulatory protein, C terminal | 3.3 |
| BAB2_1099 | Response regulator receiver/transcriptional regulatory protein, C terminal | 3.2 |
| Flagellum related | ||
| BAB2_0299 | Flagellar hook-length control protein | −3.1 |
| BAB2_1106 | Flagellin, C terminal; flagellin, N terminal | 3 |
| Metabolism, signaling, and enzymatic processes | ||
| BAB1_0204 | Zinc-containing alcohol dehydrogenase | −4 |
| BAB1_0303 | Urease accessory protein UreG | 3.2 |
| BAB1_0459 | Transglycosylase-associated protein | −3.0 |
| BAB1_0577 | Choline dehydrogenase | −3.5 |
| BAB1_0637 | ATPase-like ATP-binding protein | 3.3 |
| BAB1_0646 | Endonuclease/exonuclease/phosphatase family protein | 3.2 |
| BAB1_0867 | Hlyoxalase/bleomycin resistance protein/dioxygenase | 3.4 |
| BAB1_1070 | NAD[P]H dehydrogenase | −4.0 |
| BAB1_1299 | Sugar fermentation stimulation protein A | 3.1 |
| BAB1_1461 | SLT domain-containing protein | 3.8 |
| BAB1_1578 | Glutathione S-transferase | 3.3 |
| BAB1_1855 | GCN5-related N-acetyltransferase | 3.6 |
| BAB1_2001 | Aquaporin Z | −3.1 |
| BAB1_2052 | Luciferase | 3.5 |
| BAB2_0243 | Putative sulfite oxidase subunit YedY | 3 |
| BAB2_0821 | Zinc-containing alcohol dehydrogenase | −4.7 |
| BAB2_0823 | Aldehyde dehydrogenase | −4.9 |
| BAB2_0824 | Glucose-methanol-choline oxidoreductase; GMC oxidoreductase | −3.6 |
| BAB2_0825 | Shikimate/quinate 5-dehydrogenase | −4.6 |
| BAB2_0826 | 3-Ketoacyl-(acyl carrier protein) reductase | −4.1 |
| BAB2_0831 | Zinc-containing alcohol dehydrogenase superfamily protein | −4.2 |
| BAB2_0890 | Ribonucleotide reductase stimulatory protein | −3.0 |
| BAB2_0905 | Cytochrome c heme-binding site; 4Fe-4S ferredoxin, iron-sulfur binding domain | −3.2 |
| BAB2_0906 | Nitrate reductase, delta subunit | −3.2 |
| BAB2_0907 | Nitrate reductase, gamma subunit | −3.3 |
| BAB2_1073 | Immunoglobulin/major histocompatibility complex | 3.7 |
| Hypothetical | ||
| BAB1_0147 | Hyp | 3.5 |
| BAB1_0265 | Hyp | −7.0 |
| BAB1_0266 | Hyp | −3.6 |
| BAB1_0418 | Hyp | 6.2 |
| BAB1_0419 | Hyp | 3.4 |
| BAB1_0420 | Hyp | 4.6 |
| BAB1_1296 | Hyp | −3.8 |
| BAB1_1302 | Hyp | 4.3 |
| BAB1_1341 | Hyp | 4 |
| BAB1_1347 | Hyp | 3.3 |
| BAB1_1509 | Hyp | 3.3 |
| BAB1_1793 | Hyp | −3.2 |
| BAB1_1893 | Hyp | −6.8 |
| BAB1_2156 (ybeY) | Hyp | −12.1 |
| BAB2_0223 | Hyp | 5.2 |
| BAB2_0224 | Hyp | 4 |
| BAB2_0276 | Hyp | −5.0 |
| BAB2_0732 | Hyp | −3.2 |
| BAB2_0740 | Hyp | 4.3 |
| BAB2_0759 | Hyp | −3.2 |
| BAB2_0847 | Hyp | −3.0 |
Microarray analysis was performed using total cellular RNA from Brucella strains grown in rich media to late exponential phase, and those genes whose expression was shown to have been altered more than 3-fold in the ybeY deletion strain compared to strain 2308 are shown in the list. Boldface indicates data related to genes previously observed to be required for efficient Brucella infection or as being differentially expressed during intracellular trafficking of Brucella. Hyp, hypothetical protein.
Interestingly, several of the mRNAs identified in the ybeY microarray encode proteins that have been previously characterized as being required for efficient Brucella infection or as being differentially expressed in the bacterium during intracellular trafficking of Brucella. Seven genes, bab2_1099 (encoding FtcR, a flagellar transcriptional regulator), bab2_1106 (encoding flagellin), bab1_0303 (encoding UreG1, a urease accessory protein), bab2_0583 (encoding an ABC transporter permease), bab2_0584 (encoding an ABC transporter permease), bab2_0585 (encoding UgpB, an ABC transporter periplasmic binding protein), and bab1_1302 (encoding a hypothetical protein), have been implicated in Brucella virulence (27–32). Seven other genes, bab1_1679 (encoding an ABC transporter ATPase), bab1_1792 (encoding an ABC transporter periplasmic binding protein), bab2_0282 (encoding an ABC transporter permease), bab2_0700 (encoding an ABC transporter periplasmic binding protein), bab1_1681 (encoding cell envelope biogenesis protein TonB), bab2_0547 (encoding an ABC transporter periplasmic binding protein), and bab2_0548 (encoding an ABC transporter permease), were shown previously to be differentially expressed in Brucella during intracellular infection (33, 34).
Contribution of YbeY-associated genes to Brucella abortus virulence.
Given the large number of systems dysregulated in the B. abortus ybeY deletion strain, it is difficult to draw specific conclusions about the linkages between YbeY and individual mRNAs or systems and the observed phenotypes resulting from the deletion of ybeY. Therefore, to begin to define the YbeY-associated mRNAs that are required for virulence, we constructed strains harboring deletions in the nine genes that exhibited the greatest levels of mRNA difference in our microarray experiments (Table 1). Subsequently, peritoneally derived macrophages from BALB/c mice were infected with parental strain B. abortus 2308, as well as with the B. abortus strains with isogenic deletions of bab2_0277 (encoding choline dehydrogenase and related flavoprotein), bab2_0282 (encoding an ABC transporter permease), bab2_0822 (encoding an ABC transporter periplasmic binding protein), bab2_0548 (encoding an ABC transporter permease), bab2_0830 (encoding an ABC transporter periplasmic binding protein), bab2_1109 (encoding an ABC transporter periplasmic binding protein), bab2_0700 (encoding an ABC transporter periplasmic binding protein), bab1_0265 (encoding a hypothetical protein), and bab1_1070 (encoding NADPH dehydrogenase) (Fig. 4). Of the deletion strains tested, mutants Δbab2_0822, Δbab2_1109, and Δbab2_0700 were less able to survive and replicate in the macrophages than parental strain 2308 at 48 h postinfection, while the other deletion strains displayed wild-type levels of infection. Interestingly, bab2_0822, bab2_1109, and bab2_0700 all encode components of putative ABC transport systems, and these genes are discussed in more detail in the next section. Overall, these experiments demonstrated that several YbeY-associated systems are independently required for the full virulence of B. abortus.
FIG 4.
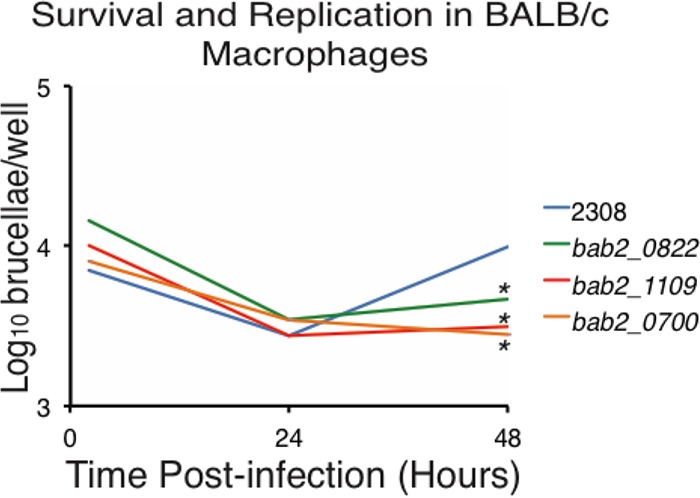
Virulence phenotypes associated with genes differentially expressed in the B. abortus ybeY deletion strain. Cultured peritoneal macrophages from BALB/c mice were infected with B. abortus 2308 or with isogenic deletion strain B. abortus Δbab2_0822, B. abortus Δbab2_1109, or B. abortus Δbab2_0700. At 2, 24, and 48 h postinfection, the macrophages were lysed, and the number of intracellular brucellae present in these phagocytes was determined by serial dilution and plating on agar medium. The asterisk denotes a statistically significant difference (P < 0.05; Student's t test) between the isogenic deletion strains and parental strain 2308 at 48 h postinfection.
DISCUSSION
In this study, we characterized the highly conserved YbeY protein in Brucella abortus. Our findings show that YbeY is necessary for proper cellular morphology, efficient in vitro growth, and full virulence of B. abortus. Moreover, we have defined the repertoire of mRNAs whose levels are connected to YbeY and subsequently determined that several YbeY-controlled genes are independently required for B. abortus virulence.
Generally, there are several similarities between the B. abortus ybeY deletion strain and other well-characterized ybeY deletion strains of other bacterial species. For example, the B. abortus ybeY deletion strain displays a significant growth defect when grown in nutrient-rich media (Fig. 1B), and similarly, V. cholerae, E. coli, Y. enterocolitica, and S. meliloti exhibit various degrees of growth inhibition when ybeY is mutated (9, 14–16). Interestingly, though, the B. abortus ΔybeY strain also has pronounced cellular morphology defects (Fig. 1D) that have not been reported previously in other bacterial ybeY mutants. Finally, the B. abortus ybeY deletion strain is severely compromised in its ability to cope with biologically relevant stresses, such as bile acid, membrane perturbation, and oxidative stresses (Fig. 3), and this too is a phenotype reported for ybeY mutants of V. cholerae and S. meliloti (13, 15). Given the wide array of genes dysregulated in the B. abortus ybeY deletion strain, we cannot conclusively assign a specific YbeY-controlled gene or set of genes to the growth defect, morphological abnormalities, and/or increased sensitivities to external stresses observed in the ybeY deletion strain, but future experiments will be aimed at analyzing specific YbeY-associated genes for links to these phenotypic properties.
Regarding the transcriptomic analysis, we determined that a wide range of mRNAs exhibit significantly altered levels in the B. abortus ybeY deletion strain (Table 1). Due to the large number of mRNAs affected by the deletion of ybeY, it is difficult to ascertain which mRNAs are directly processed by YbeY and which mRNAs YbeY indirectly regulates. Interestingly, our analyses revealed five dysregulated genes in the ybeY deletion strain that encode putative transcriptional regulators, and it is possible that YbeY controls gene expression indirectly through these transcriptional regulatory proteins. Of particular interest is bab2_1099, which encodes the FtcR transcriptional regulator of flagellar genes, as ftcR mRNA levels were elevated >3-fold in the ybeY deletion strain. FtcR is the master transcriptional activator of the flagellar biosynthesis system in B. melitensis, and, importantly, inactivation of FtcR decreases virulence in a mouse model of infection (28). Additionally, expression of fliC (bab2_1106), encoding the major flagellin protein in Brucella, is also significantly elevated in the ybeY deletion strain, and because FtcR is required for FliC production, the observed increase in expression of fliC mRNA in the ΔybeY strain might be due to increased levels of FtcR (27, 28). This is just one example of a possible indirect regulatory link between YbeY and dysregulated mRNAs in B. abortus, and more work is needed to completely characterize the regulatory circuitries associated with YbeY in Brucella strains.
Another prominent element of riboregulation often associated with bacterial YbeY proteins is that of regulatory small RNAs (sRNAs), as demonstrated in S. meliloti, Y. enterocolitica, and V. cholerae (13–15, 35). In these organisms, large variations in sRNA levels have been observed in the corresponding ybeY mutant strains. To date, comparatively few sRNAs have been identified and characterized in Brucella strains (36–40). Given the role of YbeY in bacterial sRNA stability, we assessed the levels of many of the presently known Brucella sRNAs, including AbcR1 and AbcR2, and we did not observe significant differences in sRNA levels between parental B. abortus strain 2308 and the ybeY deletion strain (data not shown). While this was surprising given the well-documented role of YbeY in bacterial sRNA stability and maturation, it is likely that other sRNAs are yet to be identified in Brucella strains, and these sRNAs may well show differences based on the presence of YbeY; however, while unlikely, it is also possible that YbeY in Brucella does not play a major role in sRNA stability and/or maturation. This is an active area of investigation in our laboratory, and future work is aimed at identifying novel Brucella sRNAs, as well as characterizing the effect of YbeY on sRNAs in Brucella.
Overall, it is not surprising that a deletion of ybeY decreases the ability of B. abortus to survive and replicate in macrophages and to colonize the spleens of mice (Fig. 2), as the ΔybeY strain has pronounced growth and morphological defects (Fig. 1). Therefore, we sought to determine if individual YbeY-controlled genes could account for the reduction in virulence independently of the growth aberrations resulting from deletion of ybeY. These experiments identified three genes, bab2_0822, bab2_1109, and bab2_0700, which are required for B. abortus to survive and replicate in murine macrophages (Fig. 4). Importantly, deletion of bab2_0822, bab2_1109, or bab2_0700 did not result in growth inhibition of B. abortus in vitro (see Fig. S1 in the supplemental material). Thus, these genes are linked to YbeY-associated virulence mechanisms in B. abortus but are disconnected from the abnormal growth characteristics of the ΔybeY strain. To date, no empirical information is available describing the function of BAB2_0822, BAB2_1109, and BAB2_0700, but each protein is predicted to act as a periplasmic binding protein likely connected to an ABC-type transport system. Questions remain about the biochemical activity of these proteins and the transport systems with which they function in concert, but our data clearly demonstrate that BAB2_0822, BAB2_1109, and BAB2_0700 are required for the full virulence of B. abortus in macrophages. In the future, it will be interesting to characterize both the regulatory link between YbeY and the mRNAs of bab2_0822, bab2_1109, and bab2_0700 and the roles of BAB2_0822, BAB2_1109, and BAB2_0700 in the biology of B. abortus.
Taking together, the data show that YbeY is a highly conserved bacterial endoribonuclease, and deletion of ybeY in B. abortus results in a pleotropic phenotype characterized by growth abnormalities, increased sensitivities to multiple stresses, and attenuation in cellular and animal models of infection. Additionally, the B. abortus YbeY protein is linked to cellular mRNA levels of genes encoding proteins involved in a variety of processes, including metabolism, flagellar biosynthesis, nutrient transport, and transcriptional regulation. Future work is needed to fully elucidate individual genetic pathways associated with YbeY in the brucellae, as well as to biochemically characterize the endoribonuclease activity of the B. abortus YbeY protein. Moreover, the relationship between YbeY and sRNAs, if one exists, needs to be clearly defined in Brucella. In the end, this work provides important foundational information about YbeY in the brucellae and, furthermore, contributes to better understanding of the diversity of activities controlled by YbeY proteins in bacteria.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
Brucella abortus 2308 and derivative strains were routinely grown on Schaedler blood agar (SBA), which is composed of Schaedler agar (BD, Franklin Lakes, NJ) containing 5% defibrinated bovine blood (Quad Five, Ryegate, MT), or in brucella broth (BD). For cloning, Escherichia coli strain DH5α was grown on tryptic soy agar (BD) or in Luria-Bertani (LB) broth. When appropriate, growth media were supplemented with kanamycin (45 μg/ml) or carbenicillin (100 μg/ml).
Construction of Brucella abortus deletion strains and genetic complementation.
The ybeY gene (bab1_2156; also known as bab_rs26200) in Brucella abortus 2308 was mutated using a nonpolar, unmarked gene excision strategy as described previously (41). Briefly, an approximately 1-kb fragment of the upstream region of each gene extending to the second codon of the coding region was amplified by PCR using primers bab1_2156-Up-For and bab1_2156-Up-Rev and genomic DNA from Brucella abortus 2308 as a template. Similarly, a fragment containing the last two codons of the coding region and extending to approximately 1 kb downstream of the ybeY open reading frame (ORF) was amplified with primers bab1_2156-Down-For and bab1_2156-Down-Rev. The sequences of all oligonucleotide primers used in this study can be found in Table 2, and the plasmids used in the study are listed in Table 3. The upstream fragment was digested with BamHI, the downstream fragment was digested with PstI, and both fragments were treated with polynucleotide kinase in the presence of ATP. Both of the DNA fragments were included in a single ligation mix with BamHI/PstI-digested pNTPS138 (M. R. K. Alley, unpublished data) and T4 DNA ligase (Monserate Biotechnology Group, San Diego, CA). The resulting plasmid (pybeY) was introduced into B. abortus 2308, and merodiploid transformants were obtained by selection on SBA plus kanamycin. A single kanamycin-resistant clone was grown for ∼6 h in brucella broth and then plated onto SBA containing 10% sucrose. Genomic DNA was isolated from sucrose-resistant, kanamycin-sensitive colonies and screened by PCR for loss of the ybeY gene. The method described above was used to construct isogenic mutations of bab2_0277, bab2_0282, bab2_0822, bab2_0548, bab2_0830, bab2_1109, bab2_0700, bab1_1070, and bab1_0265 using the primers specified in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Primer name | Primer sequence (5′→3′)a |
|---|---|
| bab1_2156-Up-For | GCGGATCCTTATGAAACATTGCAAAAGG |
| bab1_2156-Up-Rev | GATCATGATATCAATATGGATCG |
| bab1_2156-Down-For | GATTGACCATGGCTGAACA |
| bab1_2156-Down-Rev | CGCTGCAGTCCAATACGTGGAATTCATAACC |
| ybeY-RC-For | ATGTGGACGGCGCACTGCGCAT |
| ybeY-RC-Rev | GGAATGGCCTGAACCACTTCACC |
| bab2_0548-Up-For | TAGGATCCTTGCAGGAATTTGCCAAATATGA |
| bab2_0548-Up-Rev | CGGCATGCAATTCCGTCGTAAG |
| bab2_0548-Dn-For | CCATGAGCGTCCAATCGCAAGAT |
| bab2_0548-Dn-Rev | TACTGCAGACCAGAAACCCGCCTTCATCAA |
| bab2_0282-Up-For | TAGGATCCATATTTGCTGGCGATGAAATAAG |
| bab2_0282-Up-Rev | TTTCATGAAGTGTTTCCTCCCAG |
| bab2_0282-Dn-For | CAGTAAGAGGCTGGTTTGATGAA |
| bab2_0282-Dn-Rev | TACTGCAGTTTGCGGATAATGCCCATGATG |
| bab2_0277-Up-For | TAGGATCCAAATGCGGCTTACAGCAAGGC |
| bab2_0277-Up-Rev | GGTCATGATTCTATATCCAGTAA |
| bab2_0277-Dn-For | CGGTGAACGGGTTTTCCATCG |
| bab2_0277-Dn-Rev | TACTGCAGAACCAGTGCCTTCACCCAAGG |
| bab1_1070-Up-For | TAGGATCCTAGGACATGACCGATCTCCTTCC |
| bab1_1070-Up-Rev | CATCTGACATCTCCGTTAATCG |
| bab1_1070-Dn-For | ATTACCGCGAAACTGCATGGCT |
| bab1_1070-Dn-Rev | TACTGCAGATATGCGAAAGCTTGACCCG |
| bab2_1109-Up-For | TAGGATCCTTTGAGCGCGGCAGCGATGCA |
| bab2_1109-Up-Rev | TTTCATGCACGTTTCCTCCAA |
| bab2_1109-Dn-For | AAATAAACCTTCTGTTCTGC |
| bab2_1109-Dn-Rev | TACTGCAGAAACATCGTCGACCACCTTGCG |
| bab2_0830-Up-For2 | TAGGATCCGGTCCTGAAGTTCTTGAGCTCGTT |
| bab2_0830-Up-Rev | TCTCATTCTTTTCTCCCTCAA |
| bab2_0830-Dn-For | AAATGATCCTGTGTGGGCG |
| bab2_0830-Dn-Rev | TACTGCAGTTATTCATGCCGGCGCGGTCTAT |
| bab2_0822-Up-For | TAGGATCCTTGGTGCAGGCTGTTCCGTG |
| bab2_0822-Up-Rev | TTCCAATTTTCCCTCCTCTT |
| bab2_0822-Dn-For | CAGTAACAGTCGTCACCGAGGTG |
| bab2_0822-Dn-Rev | TACTGCAGCGAATGGATTTTTCTTCCGCCAC |
| bab1_0265-Up-For | TAGGATCCAAACCAAAAGCCCACAATGAACC |
| bab1_0265-Up-Rev | ACTCAGGTACATAGATTTGTTCC |
| bab1_0265-Dn-For | GAATGAAACCCGACCGTCTTTC |
| bab1_0265-Dn-Rev | TACTGCAGAATTTTCTTCACGACATATGA |
| bab2_0700-Up-For | TAGGATCCTAAGGTCAACTGGATACCTTTCG |
| bab2_0700-Up-Rev | AACCATCGAAAACTCCCATA |
| bab2_0700-Dn-For | AACTAACAAAACGAAACCCCTT |
| bab2_0700-Dn-Rev | TACTGCAGAATGCCGGGAATGCCGAAAAT |
Underlined sequences indicate a restriction endonuclease recognition site.
TABLE 3.
Plasmids used in this study
| Plasmid name | Plasmid descriptiona | Reference or source |
|---|---|---|
| pBBR1MCS-4 | Broad-host-range cloning vector; Ampr | 42 |
| pNPTS138 | Cloning vector; contains sacB gene; Kanr | M. R. K. Alley (unpublished data) |
| pybeY | In-frame deletion of ybeY plus 1 kb of each flanking region in pNPTS138 | This study |
| pybeY-comp | ybeY locus, including the entire promoter region in pBBR1MCS-4 | This study |
| pbab2_0277 | In-frame deletion of bab2_0277 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab2_0282 | In-frame deletion of bab2_0282 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab2_0822 | In-frame deletion of bab2_0822 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab2_0548 | In-frame deletion of bab2_0548 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab2_0830 | In-frame deletion of bab2_0830 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab2_1109 | In-frame deletion of bab2_1109 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab2_0700 | In-frame deletion of bab2_0700 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab1_0265 | In-frame deletion of bab1_0265 plus 1 kb of each flanking region in pNPTS138 | This study |
| pbab1_1070 | In-frame deletion of bab1_0265 plus 1 kb of each flanking region in pNPTS138 | This study |
Ampr, ampicillin resistance; Kanr, kanamycin resistance.
Genetic complementation of the ybeY deletion was achieved by expressing the wild-type ybeY allele from its native promoter in pBBR1MCS-4 (42). The ybeY gene, along with the native ybeY promoter, was amplified by PCR using primers ybeY-RC-For and ybeY-RC-Rev (Table 2) and Pfx polymerase (Invitrogen). The resulting DNA fragment was treated with polynucleotide kinase and then ligated into SmaI-digested pBBR1MCS-4. This construct, pybeY-comp, was introduced into the B. abortus ybeY deletion strain by electroporation, and colonies were selected on SBA plus carbenicillin.
All Brucella strains generated during this study were tested using the crystal violet exclusion assay in order to assess whether a given strain produced a smooth or rough form of lipopolysaccharide (LPS) (43). Briefly, Brucella strains were grown on tryptic soy agar for 72 to 96 h, and the plates were flooded with a dilute (1:1,000) solution of crystal violet for ∼25 s. Cells of parental strain B. abortus 2308 were included as smooth-LPS-producing controls, while B. abortus RB51 served as a rough-LPS-producing control.
Electron microscopy.
Brucella strains were grown to the appropriate phase of growth in brucella broth with constant shaking (200 rpm) at 37°C. When cells reached the exponential and/or stationary phase, cultures were spun down at 16,000 × g for 10 min. Supernatants were discarded, and pellets were washed once with cold H2O followed by vigorous vortex mixing. Cells were spun down for a second time, and supernatants were discarded. The pellets were then fixed in 2.5% to 5% glutaraldehyde, and kill cultures were carried out for 10 days to ensure that no viable bacteria were removed from biosafety level 3 (BSL3) containment. Fixed samples of brucellae were submitted to the Electron Microscopy Services at the Virginia-Maryland College of Veterinary Medicine (VMCVM) for scanning electron microscopy. Samples were then fixed in 0.1 M sodium cacodylate buffer and dehydrated with 15%, 30%, 50%, 70%, 95%, and 100% ethanol. The samples were then mounted on stubs and sputter coated with gold. Cells were then viewed using a Carl Zeiss EVO 40 microscope.
Growth in Biolog Phenotype MicroArray plates.
Phenotype MicroArray plates (Biolog, Inc., Hayward, CA) were utilized to determine phenotypic differences between different B. abortus strains. Strains were grown on SBA plates to produce a lawn of bacteria. Bacteria was collected and suspended in IF-0a GN/GP base (Biolog). The protocol “PM procedures for GN fastidious bacteria” provided by Biolog was followed, and Biolog Phenotype MicroArray plates 1 to 20 were inoculated at a final concentration of 108 CFU/well. Plates were grown without shaking at 37°C for and measured after 84 h of incubation at an OD of 590 nm.
Sensitivity of the B. abortus ΔybeY strain to stressors determined using disk diffusion assays.
Brucella strains were grown on SBA at 37°C under 5% CO2 for 48 to 72 h, and the bacterial cells were harvested into phosphate-buffered saline (PBS) and suspended at a concentration of ∼108 CFU/ml in brucella broth containing 0.6% agar (maintained at 55°C). A 4-ml volume of this suspension was overlaid onto brucella agar plates, and after solidification of the overlay, a sterile 7-mm-diameter Whatman disk was placed in the center of each plate. A 7-μl volume of a deoxycholate (10%), H2O2 (30%), SDS (20%), polymyxin B (10 mg/ml), or paraquat (0.25 M) was applied to each filter disk, and the plates were incubated at 37°C with 5% CO2 for 72 h. Zones of inhibition around each disk were then measured in millimeters.
Microarray analysis.
RNA was isolated from Brucella cultures grown to the late exponential phase in brucella broth (36), and contaminating genomic DNA was removed by treatment with RNase-free DNase I (36). Ten micrograms of each RNA sample from B. abortus 2308 and B. abortus ΔybeY was reverse transcribed, fragmented, and subjected to 3′ biotinylation as previously described (44). The labeled cDNA (1.5 μg) was hybridized to custom-made B. abortus GeneChips (PMD2308a520698F) according to the manufacturer's recommendations for antisense prokaryotic arrays (Affymetrix, Santa Clara, CA). Signal intensities were normalized to the median signal intensity value for each GeneChip, subjected to averaging, and analyzed with GeneSpring X software. RNA species exhibiting a ≥3-fold change in expression between B. abortus 2308 and the ΔyebY strain, as determined by Affymetrix algorithms to be statistically differentially expressed (t test; P < 0.05), were identified. The microarrays used in this study were developed based on B. melitensis biovar abortus 2308 and on all of the B. abortus GenBank entries that were available at the time of design. In total, the predicted open reading frames and intergenic regions were represented on PMD2308a520698F.
Northern blot analysis.
RNA was isolated from Brucella cultures as described previously (36). Ten micrograms of RNA was separated on a denaturing 10% polyacrylamide gel containing 7 M urea and 1× TBE (89 mM Tris base, 89 mM boric acid, and 2 mM EDTA). A low-molecular-weight DNA ladder (New England BioLabs, Ipswich, MA) was labeled with [γ-32P]ATP and polynucleotide kinase, and this radiolabeled ladder was also separated on the polyacrylamide gel. Following electrophoresis in 1× TBE buffer, the ladder and RNA samples were transferred to an Amersham Hybond-N+ membrane (GE Healthcare, Piscataway, NJ) in 1× TBE buffer. The samples were UV cross-linked to the membrane, and the membrane was prehybridized in ULTRAhyb-Oligo buffer (Ambion, Austin, TX) for 45 min at ∼42°C in a rotating hybridization oven. The oligonucleotide probes were end labeled with [γ-32P]ATP and polynucleotide kinase. The radiolabeled probes were incubated with the prehybridized membranes at ∼42°C in a rotating hybridization oven overnight (∼12 h). The membranes were then washed three times for 10 min each time with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 1× SSC, and 0.5× SSC, respectively, at ∼42°C in a rotating hybridization oven. All SSC wash buffers contained 0.1% sodium dodecyl sulfate (SDS). The membranes were then exposed to X-ray film and visualized by autoradiography.
Virulence of Brucella strains in cultured murine macrophages and experimentally infected mice.
Experiments to test the virulence of Brucella strains in primary murine peritoneal macrophages were carried out as described previously (45). Briefly, resident peritoneal macrophages were isolated from BALB/c mice and seeded in 96-well plates in Dulbecco's modified Eagle's medium with 5% fetal bovine serum, and the following day, the macrophages were infected with opsonized brucellae at an MOI of 100:1. After 2 h of infection, extracellular bacteria were killed by treatment with gentamicin (50 μg/ml). For the 2-h time point, the macrophages were then lysed with 0.1% deoxycholate–PBS, and serial dilutions were plated on Schaedler blood agar (SBA). For the 24- and 48-h time points, the cells were washed with PBS following gentamicin treatment, and fresh cell culture medium containing gentamicin (20 μg/ml) was added to the monolayer. At the indicated time point, the macrophages were lysed, and serial dilutions were plated on SBA. Triplicate wells were used for each Brucella strain tested.
Infection and colonization of mice by Brucella strains were performed as described previously by Gee et al. (45). BALB/c mice (5 per Brucella strain) were infected intraperitoneally with ∼5 × 104 CFU of each Brucella strain in sterile PBS. The mice were sacrificed at 4 and 8 weeks postinfection, and serial dilutions of spleen homogenates were plated on SBA to determine CFU counts of brucellae/spleen.
Accession number(s).
The GEO database accession number for the microarray data is GSE113222.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kathy Lowe of the Virginia-Maryland College of Veterinary Medicine for assistance in performing scanning electron microscopy.
This study was supported by grants from the National Institute of Allergy and Infectious Diseases to C.C.C. (AI117648) and R.M.R. (AI48499) and by a grant from the National Institute of General Medical Sciences to G.C.W. (GM31030). G.C.W. is an American Cancer Society Professor.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00105-18.
REFERENCES
- 1.Deutscher MP. 1985. E. coli RNases: making sense of alphabet soup. Cell 40:731–732. doi: 10.1016/0092-8674(85)90330-7. [DOI] [PubMed] [Google Scholar]
- 2.Deutscher MP. 1988. The metabolic role of RNases. Trends Biochem Sci 13:136–139. doi: 10.1016/0968-0004(88)90070-9. [DOI] [PubMed] [Google Scholar]
- 3.Apirion D, Gegenheimer P. 1981. Processing of bacterial RNA. FEBS Lett 125:1–9. doi: 10.1016/0014-5793(81)80984-2. [DOI] [PubMed] [Google Scholar]
- 4.Gegenheimer P, Apirion D. 1981. Processing of procaryotic ribonucleic acid. Microbiol Rev 45:502–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob AI, Kohrer C, Davies BW, RajBhandary UL, Walker GC. 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell 49:427–438. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oganesyan V, Busso D, Brandsen J, Chen S, Jancarik J, Kim R, Kim SH. 2003. Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus. Acta Crystallogr D Biol Crystallogr 59:1219–1223. doi: 10.1107/S0907444903011028. [DOI] [PubMed] [Google Scholar]
- 7.Zhan C, Fedorov EV, Shi W, Ramagopal UA, Thirumuruhan R, Manjasetty BA, Almo SC, Fiser A, Chance MR, Fedorov AA. 2005. The ybeY protein from Escherichia coli is a metalloprotein. Acta Crystallogr Sect F Struct Biol Cryst Commun 61:959–963. doi: 10.1107/S1744309105031131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasouly A, Schonbrun M, Shenhar Y, Ron EZ. 2009. YbeY, a heat shock protein involved in translation in Escherichia coli. J Bacteriol 191:2649–2655. doi: 10.1128/JB.01663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies BW, Kohrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. 2010. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol 78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasouly A, Davidovich C, Ron EZ. 2010. The heat shock protein YbeY is required for optimal activity of the 30S ribosomal subunit. J Bacteriol 192:4592–4596. doi: 10.1128/JB.00448-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grinwald M, Ron EZ. 2013. The Escherichia coli translation-associated heat shock protein YbeY is involved in rRNA transcription antitermination. PLoS One 8:e62297. doi: 10.1371/journal.pone.0062297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandey SP, Winkler JA, Li H, Camacho DM, Collins JJ, Walker GC. 2014. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics 15:121. doi: 10.1186/1471-2164-15-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey SP, Minesinger BK, Kumar J, Walker GC. 2011. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res 39:4691–4708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leskinen K, Varjosalo M, Skurnik M. 2015. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology 161:285–299. doi: 10.1099/mic.0.083097-0. [DOI] [PubMed] [Google Scholar]
- 15.Vercruysse M, Kohrer C, Davies BW, Arnold MF, Mekalanos JJ, RajBhandary UL, Walker GC. 2014. The highly conserved bacterial RNase YbeY is essential in Vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog 10:e1004175. doi: 10.1371/journal.ppat.1004175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies BW, Walker GC. 2008. A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J Bacteriol 190:1118–1123. doi: 10.1128/JB.01521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawal A, Jejelowo O, Chopra AK, Rosenzweig JA. 2011. Ribonucleases and bacterial virulence. Microb Biotechnol 4:558–571. doi: 10.1111/j.1751-7915.2010.00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect Dis 6:91–99. doi: 10.1016/S1473-3099(06)70382-6. [DOI] [PubMed] [Google Scholar]
- 19.Gorvel JP, Moreno E. 2002. Brucella intracellular life: from invasion to intracellular replication. Vet Microbiol 90:281–297. doi: 10.1016/S0378-1135(02)00214-6. [DOI] [PubMed] [Google Scholar]
- 20.Fabrik I, Hartlova A, Rehulka P, Stulik J. 2013. Serving the new masters - dendritic cells as hosts for stealth intracellular bacteria. Cell Microbiol 15:1473–1483. doi: 10.1111/cmi.12160. [DOI] [PubMed] [Google Scholar]
- 21.Roop RM II, Gaines JM, Anderson ES, Caswell CC, Martin DW. 2009. Survival of the fittest: how Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol 198:221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seleem MN, Boyle SM, Sriranganathan N. 2008. Brucella: a pathogen without classic virulence genes. Vet Microbiol 129:1–14. doi: 10.1016/j.vetmic.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Heaton BE, Herrou J, Blackwell AE, Wysocki VH, Crosson S. 2012. Molecular structure and function of the novel BrnT/BrnA toxin-antitoxin system of Brucella abortus. J Biol Chem 287:12098–12110. doi: 10.1074/jbc.M111.332163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu CX, Xu XJ, Zheng K, Liu F, Yang XD, Chen CF, Chen HC, Liu ZF. 2016. Characterization of ribonuclease III from Brucella. Gene 579:183–192. doi: 10.1016/j.gene.2015.12.068. [DOI] [PubMed] [Google Scholar]
- 25.Saramago M, Peregrina A, Robledo M, Matos RG, Hilker R, Serrania J, Becker A, Arraiano CM, Jimenez-Zurdo JI. 2017. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res 45:1371–1391. doi: 10.1093/nar/gkw1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiménez-Zurdo JI, Robledo M. 2017. RNA silencing in plant symbiotic bacteria: insights from a protein-centric view. RNA Biol 14:1672–1677. doi: 10.1080/15476286.2017.1356565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, Danese I, Vandenhaute J, Tibor A, DeBolle X, Letesson JJ. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol 7:687–698. doi: 10.1111/j.1462-5822.2005.00502.x. [DOI] [PubMed] [Google Scholar]
- 28.Léonard S, Ferooz J, Haine V, Danese I, Fretin D, Tibor A, de Walque S, De Bolle X, Letesson JJ. 2007. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J Bacteriol 189:131–141. doi: 10.1128/JB.00712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sangari FJ, Seoane A, Rodriguez MC, Aguero J, Garcia Lobo JM. 2007. Characterization of the urease operon of Brucella abortus and assessment of its role in virulence of the bacterium. Infect Immun 75:774–780. doi: 10.1128/IAI.01244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lestrate P, Delrue RM, Danese I, Didembourg C, Taminiau B, Mertens P, De Bolle X, Tibor A, Tang CM, Letesson JJ. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol Microbiol 38:543–551. doi: 10.1046/j.1365-2958.2000.02150.x. [DOI] [PubMed] [Google Scholar]
- 31.Rossetti CA, Galindo CL, Lawhon SD, Garner HR, Adams LG. 2009. Brucella melitensis global gene expression study provides novel information on growth phase-specific gene regulation with potential insights for understanding Brucella:host initial interactions. BMC Microbiol 9:81. doi: 10.1186/1471-2180-9-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caro-Hernández P, Fernández-Lago L, de Miguel MJ, Martín-Martín AI, Cloeckaert A, Grilló MJ, Vizcaíno N. 2007. Role of the Omp25/Omp31 family in outer membrane properties and virulence of Brucella ovis. Infect Immun 75:4050–4061. doi: 10.1128/IAI.00486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamontagne J, Forest A, Marazzo E, Denis F, Butler H, Michaud JF, Boucher L, Pedro I, Villeneuve A, Sitnikov D, Trudel K, Nassif N, Boudjelti D, Tomaki F, Chaves-Olarte E, Guzman-Verri C, Brunet S, Cote-Martin A, Hunter J, Moreno E, Paramithiotis E. 2009. Intracellular adaptation of Brucella abortus. J Proteome Res 8:1594–1609. doi: 10.1021/pr800978p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roset MS, Alefantis TG, DelVecchio VG, Briones G. 2017. Iron-dependent reconfiguration of the proteome underlies the intracellular lifestyle of Brucella abortus. Sci Rep 7:10637. doi: 10.1038/s41598-017-11283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bobrovskyy M, Vanderpool CK, Richards GR. 2015. Small RNAs regulate primary and secondary metabolism in Gram-negative bacteria. Microbiol Spectr 3(3). doi: 10.1128/microbiolspec.MBP-0009-2014. [DOI] [PubMed] [Google Scholar]
- 36.Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop Ii RM. 2012. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol 85:345–360. doi: 10.1111/j.1365-2958.2012.08117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saadeh B, Caswell CC, Chao Y, Berta P, Wattam AR, Roop RM II, O'Callaghan D. 2016. Transcriptome-wide identification of Hfq-associated RNAs in Brucella suis by deep sequencing. J Bacteriol 198:427–435. doi: 10.1128/JB.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhong Z, Xu X, Li X, Liu S, Lei S, Yang M, Yu J, Yuan J, Ke Y, Du X, Wang Z, Ren Z, Peng G, Wang Y, Chen Z. 2016. Large-scale identification of small noncoding RNA with strand-specific deep sequencing and characterization of a novel virulence-related sRNA in Brucella melitensis. Sci Rep 6:25123. doi: 10.1038/srep25123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y, Ke Y, Xu J, Wang L, Wang T, Liang H, Zhang W, Gong C, Yuan J, Zhuang Y, An C, Lei S, Du X, Wang Z, Li W, Yuan X, Huang L, Yang X, Chen Z. 2015. Identification of a novel small non-coding RNA modulating the intracellular survival of Brucella melitensis. Front Microbiol 6:164. doi: 10.3389/fmicb.2015.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng X, Dong H, Wu Q. 2015. A new cis-encoded sRNA, BsrH, regulating the expression of hemH gene in Brucella abortus 2308. FEMS Microbiol Lett 362:1–7. doi: 10.1093/femsle/fnu017. [DOI] [PubMed] [Google Scholar]
- 41.Caswell CC, Baumgartner JE, Martin DW, Roop RM II. 2012. Characterization of the organic hydroperoxide resistance system of Brucella abortus 2308. J Bacteriol 194:5065–5072. doi: 10.1128/JB.00873-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 43.White PG, Wilson JB. 1951. Differentiation of smooth and nonsmooth colonies of Brucellae. J Bacteriol 61:239–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. 2004. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM II. 2005. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect Immun 73:2873–2880. doi: 10.1128/IAI.73.5.2873-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


