ABSTRACT
Streptococcus mutans, the organism most frequently associated with the development of dental caries, is able to utilize a diverse array of carbohydrates for energy metabolism. One such molecule is trehalose, a disaccharide common in human foods, which has been recently implicated in enhancing the virulence of epidemic strains of the pathogen Clostridium difficile. In this study, mutants with deletions of all three genes in the putative S. mutans trehalose utilization operon were characterized, and the genes were shown to be required for wild-type levels of growth when trehalose was the only carbohydrate source provided. Interestingly, the TreR transcriptional regulator appeared to be critical for responding to oxidative stress and for mounting a protective stress tolerance response following growth at moderately acidic pH. mRNA sequencing (RNA-seq) of a treR deletion mutant suggested that in S. mutans, TreR acts as a trehalose-sensing activator of transcription of the tre operon, rather than as a repressor, as described in other species. In addition, deletion of treR caused the downregulation of a number of genes involved in genetic competence and bacteriocin production, supporting the results of a recent study linking trehalose and the S. mutans competence pathways. Finally, deletion of treR compromised the ability of S. mutans to inhibit the growth of the competing species Streptococcus gordonii and Lactococcus lactis. Taking the results together, this study solidifies the role of the S. mutans tre operon in trehalose utilization and suggests novel functions for the TreR regulator, including roles in the stress response and competitive fitness.
IMPORTANCE S. mutans is the primary etiologic agent of dental caries, which globally is the most common chronic disease. S. mutans must be able to outcompete commensal organisms in its dental plaque niche in order to establish persistence and pathogenesis. To that end, S. mutans metabolizes a diverse array of carbohydrates to generate acid and impede its acid-sensitive neighbors. Additionally, S. mutans utilizes quorum signaling through genetic competence-associated pathways to induce production of toxins to kill its rivals. This study definitively shows that the S. mutans trehalose utilization operon is required for growth in trehalose. Furthermore, this study suggests that the S. mutans TreR transcriptional regulator has a novel role in virulence through regulation of genes involved in genetic competence and toxin production.
KEYWORDS: Streptococcus mutans, gene regulation, mutacin, oxidative stress, trehalose
INTRODUCTION
Streptococcus mutans, the oral pathogen most frequently associated with the development of dental caries, relies chiefly on human host foodstuffs as an energy source (1). As the omnivorous human diet is quite diverse, it is to the benefit of S. mutans to be able to utilize as many host dietary macromolecules as possible. S. mutans is truly a master of carbohydrate metabolism, capable of exploiting a greater assortment of carbohydrate types than other sequenced Gram-positive organisms (2, 3). S. mutans encodes at least 14 phosphoenolpyruvate:phosphotransferase systems (PTS) and 2 ABC transporters for sugar transport (2, 3). After import, sugars are broken down by appropriate hydrolases and enter the Emden-Meyerhof-Parnas (EMP) glycolytic pathway, which is the primary method of ATP generation in S. mutans. The resulting pyruvate is then fermented to a number of mainly acidic end products, a process that regenerates the NAD+ needed for a continuation of flux through the EMP pathway (S. mutans lacks a canonical electron transport chain). It is these acidic end products of fermentation that lower the local pH in dental plaque such that demineralization of the adjacent tooth surface occurs and the process of caries formation is initiated (4).
Trehalose is a disaccharide consisting of two α-glucose units joined by an α,α-1,1-α-glucosidic bond. Foods common in the human diet that naturally contain trehalose include shellfish, bread, beer, mushrooms, and seaweed. Following the discovery of a novel trehalose synthesis method in 2000, trehalose has seen increased use as a food additive and is used in a diverse array of foods, including pasta, ground beef, and ice cream. Trehalose is both an established solute in regulation of osmotic pressure/heat shock and a cryoprotectant in a variety of prokaryotic and eukaryotic species, including the Gram-positive organisms Bacillus subtilis and Lactobacillus acidophilus (5, 6). A tre operon, encoding the machinery to import and utilize trehalose as a carbon source, is widely conserved throughout Gram-positive bacteria (6). In the majority of cases, the operon consists of genes encoding a trehalose-specific PTS, a hydrolase capable of hydrolyzing trehalose to glucose and glucose-6-phosphate, and a transcriptional regulator that senses trehalose levels (6). In Lactococcus lactis, the tre operon also encodes a trehalose phosphate phosphorylase and a β-phosphoglucomutase (6). Regulation of this operon is most well understood in B. subtilis, where the multimeric TreR regulator binds to a tre operator in the promoter region of the operon and represses transcription of the operon when trehalose is absent. Trehalose-6-phosphate binds to TreR and inhibits multimerization of the protein and thus its ability to repress transcription. The B. subtilis tre operon is also regulated by carbon catabolite repression (CCR), through the global regulator carbon catabolite control protein (CcpA). Canonically, CCR represses use of nonpreferred sugars, such as trehalose, in the presence of glucose. In this manner, B. subtilis senses the presence of trehalose and accordingly upregulates the production of proteins needed to utilize it as an energy source (7–9). More recently, trehalose was shown to enhance virulence of the significant intestinal pathogen Clostridium difficile, likely through increased toxin production (10).
S. mutans UA159 is able to grow on trehalose as a sole carbon source, and its genome contains a putative tre operon encoding a trehalose-6-phosphate hydrolase (treA [alternatively annotated treC], SMU.2037), a trehalose-specific EIIABC PTS component (treB [alternatively annotated treP or pttB], SMU.2038), and a GntR family regulator of the trehalose operon (treR, SMU.2040) (2, 3). A previous study used cDNA microarray analysis to examine global gene expression in batch cultures of S. mutans UA159 grown in trehalose versus glucose, and it was observed that treA and treB were highly upregulated during growth in trehalose (3). The results suggested that the S. mutans tre operon is important for trehalose metabolism and is likely regulated via a trehalose-sensing mechanism, similar to the TreR-mediated repression observed in B. subtilis (3). Another study indicated that although S. mutans could grow on trehalose, acid production was significantly reduced compared to that with growth on glucose, leading the authors to suggest the use of trehalose as a less-cariogenic sugar substitute (11). More recent analyses have shown that the expression of S. mutans treA and treB was elevated during growth in galactose compared to glucose and upon deletion of ccpA, indicating that CcpA may regulate tre expression through CCR in S. mutans (12). Further, it has been shown that growth in trehalose significantly increased the transformation efficiency of S. mutans by inducing expression of comX, comS, and comYA (13), indicating that trehalose may serve as an important signaling molecule in S. mutans.
To persist in plaque biofilm and cause disease, S. mutans must outcompete commensal streptococci that occupy the same niche. These commensals include peroxigenic streptococcal species such as S. sanguinis and S. gordonii, which produce bactericidal levels of H2O2 to kill rivals, such as S. mutans (14, 15). Therefore, the ability of S. mutans to contend with oxidative stress generated by H2O2 is paramount to its ability to prevail as a significant player in the dental plaque community (16–18). In addition to reducing the local environmental pH to impair less-acid-tolerant competitors, S. mutans utilizes toxins, including the nonlantibiotic mutacins IV, V, and VI, to kill adversaries (19). In S. mutans, production of these toxins is intertwined with competence activation pathways through the competence-stimulating peptide (CSP) quorum-sensing peptide (20–22).
In this study, the regulation and function of the tre operon in S. mutans were investigated and characterized. All three genes in the operon were required for wild-type levels of growth when trehalose was the sole carbon source provided. In S. mutans, TreR appeared to function as an activator of transcription of the tre operon, rather than a repressor, under the conditions tested. Surprisingly, deletion of treR severely impacted the ability of S. mutans to tolerate oxidative stress and mount an effective stress tolerance response. Further investigation via mRNA sequencing (RNA-seq) of a treR deletion mutant confirmed that loss of treR affected transcription of a number of genes outside the tre operon, particularly several involved in competence and toxin production. Additionally, loss of treR compromised the ability of S. mutans to inhibit growth of the competing organisms Streptococcus gordonii and Lactococcus lactis. Taking the results together, this study confirms the role of the S. mutans tre operon in trehalose utilization and suggests that the S. mutans TreR transcriptional regulator acts in a disparate manner, and has an expanded role, compared to its previously characterized counterpart in B. subtilis.
RESULTS
The S. mutans tre operon is required for trehalose utilization.
As a resident of the oral cavity, S. mutans derives a significant portion of its energy from carbohydrates harvested during transient contact with food being consumed by the human host. S. mutans is well adapted to the diverse human diet and is able to utilize numerous carbon sources present in human food, including trehalose. In S. mutans, the predicted trehalose utilization (tre) operon consists of three genes: a 1,629-bp putative trehalose-6-phosphate hydrolase gene (treA, SMU.2037), a 1,968-bp trehalose-specific EIIABC PTS component gene (treB, SMU.2038), and a 714-bp gene for a putative GntR family repressor of the trehalose operon (treR, SMU.2040) (2).
Although previous work determined that growth in trehalose resulted in upregulation of treA and treB (3), it remained unclear whether the tre operon was required for the growth of S. mutans in trehalose. To elucidate the role of the tre operon genes in sugar metabolism, a treR deletion mutant (ΔtreR strain) was generated as described in Materials and Methods. treA and treB deletion mutants (ΔtreA and ΔtreB strains) have been previously described (23). Cultures of S. mutans UA159 and the ΔtreA, ΔtreB, and ΔtreR mutants were grown in a Bioscreen C in either tryptone-yeast extract (TY) plus 1% glucose or TY plus 1% trehalose. During growth in glucose-containing medium, all four strains had nearly identical growth rates (Fig. 1A). However, when cultures were grown in TY medium containing trehalose, compared to UA159, the ΔtreB strain was significantly impaired in its ability to grow, the ΔtreR strain was even more severely impaired in its ability to grow, and the ΔtreA strain was completely unable to grow (Fig. 1B). Meanwhile, UA159 exhibited nearly identical growth kinetics during growth in trehalose versus glucose. These assays were also performed under anaerobic conditions, which generated similar results (data not shown). Taken together, these data show that treA is absolutely required for growth in medium containing trehalose and that disruption of treB or treR significantly impairs the ability of S. mutans to grow in trehalose. To confirm specifically that the loss of these genes was indeed preventing growth of the strains in trehalose, complemented strains of the ΔtreA and ΔtreR strains were generated as described in Materials and Methods and designated treA-C and treR-C strains, respectively. Complementation of both treR and treA faithfully restored the phenotype of the parent strain during growth in trehalose (Fig. 1C). Construction of a treB complement strain was recalcitrant to our methods, likely due to toxicity of the transmembrane treB gene in Escherichia coli.
FIG 1.
The tre operon is required for wild-type growth on trehalose. (A and B) Growth of the parent strain S. mutans UA159 and its ΔtreR, ΔtreB, and ΔtreA derivatives in TY plus 1% (vol/vol) glucose (A) or TY plus 1% (vol/vol) trehalose (B). (C) Growth of the parent strain S. mutans UA159 and the treR and treA complement strains (treR-C and treA-C strains) in TY plus 1% (vol/vol) trehalose. n = 10 for each condition. Insets in each panel provide the mean doubling time (D.T.) ± standard deviation for each strain, if applicable.
In S. mutans, treR is critical for tolerance of oxidative stress.
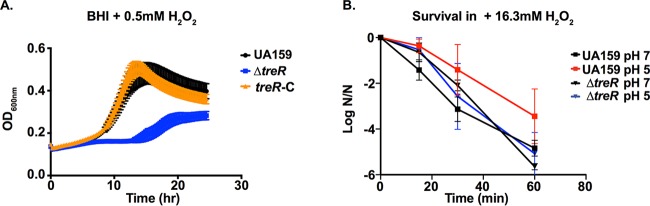
Previous screening of an S. mutans mutant library indicated that deletion of treA or treB modestly impaired the growth of S. mutans under several conditions of acid and oxidative stress (23). In this study, growth of the ΔtreR strain was characterized under acid and oxidative stress. Surprisingly, compared to that of UA159, growth of ΔtreR strain was severely impaired in the presence of 0.5 mM H2O2 (Fig. 2A), while growth of the treR-C strain was nearly identical to that of the parent strain. Growth of the ΔtreR strain was similar to that of UA159 in medium buffered to pH 5.4 or pH 5.6 (data not shown). Typically, growth of S. mutans at the moderately acidic pH of 5 induces the acid tolerance response (ATR), which protects the cells from further insult, including extreme acid and oxidative stress. Compared to UA159, after steady-state growth at pH 5 to induce the ATR, the ΔtreR strain was able to survive an acid challenge (pH 2.5) at a similar rate (data not shown); however, the ΔtreR strain was significantly compromised in its ability to survive a peroxide challenge (16.3 mM H2O2), indicating a defect in the ATR in the absence of treR (Fig. 2B). These results were remarkable, as TreR is predicted to be simply a local regulator with a role strictly in trehalose utilization.
FIG 2.
The ΔtreR strain is impaired in its ability to grow in the presence of oxidative stress. (A) The parent strain, S. mutans UA159, the ΔtreR strain, and the treR-C (treR complement) strain were grown in triplicate in BHI plus 0.5 mM H2O2 (n = 10, biological replicates). A representative graph is shown. (B) Sensitivity of S. mutans to H2O2 challenge following growth at steady-state pH 7 or steady-state pH 5 as indicated. Results are expressed as the log10 (n/n0) where n is the viable cell count at each time point and n0 is the viable cell count at time zero. Error bars represent the standard deviation.
Loss of treR impacts the S. mutans transcriptome during growth on several carbon sources, and TreR is likely an activator of transcription of treA and treB.
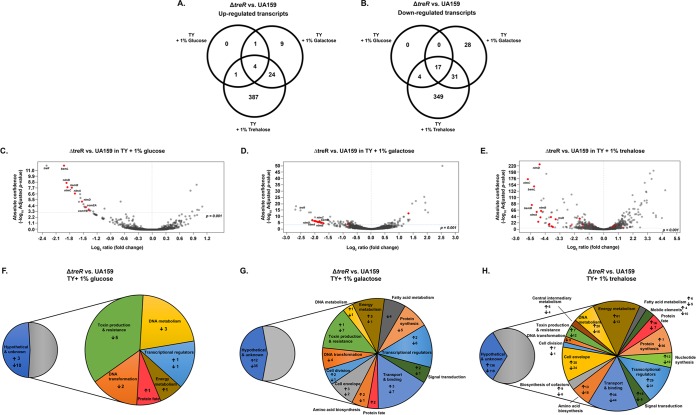
Since the ΔtreR strain was, unexpectedly, defective in its ability to tolerate oxidative stress, we hypothesized that in S. mutans, TreR may be involved in the regulation of genes outside its operon. Although S. mutans does not appear to accumulate trehalose in response to stress, the disaccharide may serve as an important signaling molecule (13). To examine the effects of the loss of TreR-mediated regulation on the S. mutans transcriptome, RNA-seq was performed on S. mutans UA159 or ΔtreR strains grown in TY plus 1% glucose, 1% galactose, or 1% trehalose. In the ΔtreR strain, compared to UA159, at a Benjamini-Hochberg adjusted P value cutoff of <0.001, there were 6 genes upregulated and 21 genes downregulated during growth in glucose (see Table S1 in the supplemental material), 136 genes upregulated and 80 genes downregulated during growth in galactose (see Table S2 in the supplemental material), and 426 genes upregulated and 401 genes downregulated during growth in trehalose (see Table S3 in the supplemental material). A summary of the number of genes and functional classes differentially regulated is provided in Fig. 3A, B, and F to H, and a list of genes of interest that were differentially regulated in the ΔtreR strain compared to UA159 is provided in Table 1. As an internal control, treR was significantly downregulated under all 3 conditions, congruent with the fact that the ΔtreR strain is a deletion mutant and no treR transcript should be present (Table 1). Interestingly, treA and treB were significantly downregulated in the ΔtreR strain compared to UA159 during growth in trehalose and did not have significantly altered expression in the ΔtreR strain compared to UA159 during growth in glucose or galactose (Table 1). These observations suggest that under the conditions tested, TreR acts as an activator of transcription of the tre operon in the presence of trehalose, rather than as a repressor that is derepressed in the presence of trehalose (as in B. subtilis [9]). If S. mutans TreR is indeed a trehalose-sensitive activator of tre transcription, a likely explanation for the large number of genes differentially regulated in the ΔtreR strain compared to UA159 during growth in trehalose is general stress of the organism due to an inability to efficiently import and utilize the only available carbon source. This is supported by the impaired growth phenotype (Fig. 1B), as well as the vast number of genes involved in energy metabolism and protein synthesis that are differentially regulated in the ΔtreR strain during growth in trehalose (Fig. 3H).
FIG 3.
Differentially regulated genes in the ΔtreR strain compared to S. mutans UA159. (A and B) Venn diagrams illustrating the number of genes significantly (P < 0.001; Benjamini-Hochberg procedure) upregulated (A) or downregulated (B) in the ΔtreR strain, compared to UA159, during growth in TY plus 1% glucose, galactose, or trehalose, based on results from RNA-seq. (C to E) Volcano plots illustrating the transcriptome of the ΔtreR strain versus UA159 during growth in TY plus 1% glucose (C), 1% galactose (D), or 1% trehalose (E). The vertical dashed line indicates 0-fold change in expression, and the horizontal dashed line indicates a Benjamini-Hochberg adjusted P value of 0.001, which was used as the cutoff in our analyses. Data points representing transcripts assigned to the functional classes “toxin production and resistance” or “DNA transformation” are colored red for visualization. Specific data points representing genes of interest are individually labeled. (F to H) Pie charts of functional classes with altered expression during growth of the ΔtreR strain, compared to UA159, in TY plus 1% glucose (F), TY plus 1% galactose (G), or TY plus 1% trehalose (H). The numbers of genes upregulated are indicated next to the up arrows, and the numbers of downregulated genes are indicated next to the down arrows. Genes encoding hypothetical or unknown proteins are separated out in the smaller pie charts to allow for better visualization of genes with known functions.
TABLE 1.
Selected genes of interest differentially regulated in the ΔtreR strain compared to UA159 during growth in the indicated media
| Locus tag | Product | Functional class | Gene(s) | Differential expression, ΔtreR/UA159 (log2; P < 0.001) in: |
||
|---|---|---|---|---|---|---|
| TY + 1% glucose | TY + 1% galactose | TY + 1% trehalose | ||||
| SMU_1906c | Hypothetical protein | Cellular processes: toxin production and resistance | bsmB | −1.74 | −1.96 | −5.41 |
| SMU_1905c | Putative bacteriocin secretion protein | Cellular processes: toxin production and resistance | bsmL | −1.91 | −1.70 | −5.24 |
| SMU_1915 | Competence-stimulating peptide, precursor | Hypothetical proteins: conserved | comC | NSa | NS | −2.43 |
| SMU_1917 | ComE response regulator | Signal transduction: two-component systems | comE | NS | NS | −1.09 |
| SMU_625 | Putative competence protein | Cellular processes: DNA transformation | comEA | −1.44 | −1.73 | −3.83 |
| SMU_498 | Putative late competence protein | Unknown function: general | comF | NS | NS | −3.62 |
| SMU_1987 | Putative ABC transporter, ATP-binding protein ComYA; late competence protein | Cellular processes: DNA transformation | comYA | NS | NS | −4.76 |
| SMU_1985 | Putative ABC transporter ComYB; probably part of the DNA transport machinery | Cellular processes: DNA transformation | comYB | −1.37 | −1.53 | −3.91 |
| SMU_1984 | Putative competence protein ComYC | Cellular processes: DNA transformation | comYC | NS | NS | −4.12 |
| SMU_1983 | Putative competence protein ComYD | Cellular processes: DNA transformation | comYD | NS | NS | −4.06 |
| SMU_1913c | Putative immunity protein, BLpL-like | Hypothetical proteins | immA | NS | NS | −3.97 |
| SMU_150 | Hypothetical protein | Cellular processes: toxin production and resistance | nlmA | −1.68 | −2.02 | −5.04 |
| SMU_151 | Hypothetical protein | Hypothetical proteins: conserved | nlmB | −1.86 | −1.87 | −4.72 |
| SMU_1914c | Hypothetical protein | Cellular processes: toxin production and resistance | nlmC | −1.84 | −1.91 | −5.71 |
| SMU_423 | Hypothetical protein | Cellular processes: toxin production and resistance | nlmD | −1.52 | −2.09 | −4.83 |
| SMU_2037 | Putative trehalose-6-phosphate hydrolase TreA | Energy metabolism: biosynthesis and degradation of polysaccharides | treA | NS | NS | −2.98 |
| SMU_2038 | Putative PTS, trehalose-specific IIABC component | Signal transduction: PTS | treB | NS | NS | −3.07 |
| SMU_2040 | Putative transcriptional regulator; repressor of the trehalose operon | Regulatory functions: DNA interactions | treR | −2.30 | −2.52 | −3.47 |
NS, not significant.
In the ΔtreR strain, compared to UA159, all of the genes differentially regulated during growth in medium containing glucose, and the vast majority of genes differentially regulated during growth in medium containing galactose, were also differentially regulated during growth in at least one of the other two sugars (Fig. 3A and B). During growth in glucose, several genes involved in competence as well as toxin production resistance and DNA metabolism were downregulated in the ΔtreR stain compared to UA159 (Fig. 3C and F; Tables 1 and S1). These included comEA and comYB, as well as the genes encoding the nonlantibiotic toxins mutacin IV (nlmA and nlmB), mutacin V (nlmC), and mutacin VI (nlmD), which are known to kill other oral Lactobacillales (19). These trends continued, and were even more striking, in the ΔtreR strain during growth in medium containing galactose (Fig. 3D and G; Tables 1 and S2). In addition, a number of transcriptional regulators and genes involved in signal transduction, including spxA2 (formerly spxB), ciaH, and cpsY, were differentially regulated during growth in galactose (Fig. 3G; Table S2). Although differential expression of nearly half of the organism's transcriptome was observed in the ΔtreR stain grown in medium containing trehalose, compared to UA159, major trends included a downregulation of genes involved in protein synthesis and competence and an upregulation of genes involved in protein fate and degradation, as well as energy metabolism (Fig. 3H; Table S3). Taken together, the results from RNA-seq indicate that the S. mutans TreR regulator appears to function differently than described in other species: as an activator of tre transcription, rather than a repressor. In addition, the genes differentially regulated in the ΔtreR strain, compared to UA159, during growth in glucose and galactose suggest that TreR acts, either directly or indirectly, as a more global regulator in S. mutans than previously thought and is involved in competence and toxin production.
Deletion of treR significantly reduces the ability of S. mutans to kill competing species.
To determine if the reduced expression of the genes encoding mutacins IV, V, and VI in the ΔtreR strain actually impaired the ability of S. mutans to kill and compete with neighboring species, a deferred-antagonism assay (24, 25) was performed using rich medium (brain heart infusion [BHI]) or defined medium (TY plus 1% glucose or TY plus 1% trehalose). S. mutans UA159, the ΔtreR strain, or the treR complement strain (treR-C strain) was plated as an initial colonizer, and an overlay of S. gordonii (which is susceptible to mutacins IV and V) or Lactococcus lactis (which is susceptible to mutacins V and VI) was plated as a secondary colonizer (Fig. 4). While UA159 produced a marked zone of inhibition in the S. gordonii and L. lactis overlays, the ΔtreR strain produced a greatly reduced zone of inhibition in both overlays, indicating a defect in the ability of the ΔtreR strain to inhibit the growth of its competitors, most likely through a reduction in mutacin expression. The treR-C strain produced zones of inhibition that were larger than those of the ΔtreR strain but smaller than those of the parent strain, UA159. The difference in apparent mutacin production between UA159 and the treR-C strain may be due to the fact that the complemented treR gene, while present, is expressed from a different genetic locus in the treR-C strain. All strains produced a larger zone of inhibition during growth on BHI compared to TY medium. The ΔtreR strain produced no zone of inhibition during growth on TY plus 1% trehalose and, in fact, allowed the overlaid species to grow across the top of the colony. In all other cases, the primary colony, at least, prevented growth of S. gordonii or L. lactis across its top, even if there was no discernible clearing of the surrounding lawn. It is unclear whether this is due to poor growth of the ΔtreR strain when trehalose is the only sugar provided (i.e., as seen in Fig. 1), a lack of mutacin production, or a combination of the two. In the case of the L. lactis overlay, growth on TY plus 1% trehalose produced larger zones of inhibition than growth on TY plus 1% glucose in the UA159 and treR-C strains, indicating that exposure to trehalose may increase mutacin production in S. mutans, though more detailed studies exploring a titration of trehalose concentrations and growth in multiple sugars are needed to confirm this hypothesis.
FIG 4.
Loss of treR drastically reduces the ability of S. mutans UA159 to inhibit its competitors, S. gordonii and L. lactis. (A and B) Deferred-antagonism assay, performed as described in Materials and Methods, to observe mutacin production. Cultures of S. mutans UA159, ΔtreR, and treR-C strains were spotted on BHI agar, TY plus 1% glucose agar, and TY plus 1% trehalose agar and incubated overnight. Cultures of S. gordonii (A) or L. lactis (B) were added to 5 ml BHI soft agar and used to overlay the plates containing the S. mutans strains. Zones of inhibition were measured 24 h after addition of the overlay. Six replicate assays were performed, and representative images are shown. (C and D) Areas of the zones of inhibition for the indicated strains in the indicated media for S. gordonii (C) and L. lactis (D). Error bars represent standard deviation, and asterisks denote statistical significance between indicated pairs as determined by Tukey's multiple-comparison test following a two-way analysis of variance (ANOVA) (*, P < 0.05; **, P < 0.01; ****, P < 0.0001) (n = 6).
DISCUSSION
The disaccharide trehalose consists of two α-glucose molecules linked by an α,α-1,1-glucosidic bond. A number of bacterial and eukaryotic species utilize trehalose as a carbon source, as well as as a protectant from heat shock, osmotic stress, and cold (5, 6). Among Gram-positive bacteria, the machinery needed to import and consume trehalose as an energy source is most well understood in Bacillus subtilis. B. subtilis is thought to sense the presence of trehalose and upregulate the machinery needed to import and prepare the disaccharide for use as an energy source. The B. subtilis tre operon is also subject to CCR, mediated by the CcpA protein, and the presence of glucose (7).
In the oral pathogen S. mutans, the ability to metabolize a wide variety of carbohydrates is of significant interest, such that full advantage can be taken of the diverse human host diet (1, 2). Trehalose is among the numerous sugars which S. mutans can exploit, and the S. mutans genome is predicted to encode TreA, TreB, and TreR (2). Similar to the case in B. subtilis, the S. mutans tre operon was upregulated during growth in trehalose or galactose and upon deletion of ccpA (3, 12), suggesting that the operon was regulated in a manner similar to that in B. subtilis. A more recent study showed that in S. mutans, exposure to trehalose significantly increased genetic competence, and suggested that in S. mutans, trehalose may function as a signaling molecule, as well as an energy source (13). In this study, the role of the S. mutans tre operon was characterized, and the role of the TreR regulator was further explored.
To confirm that the tre operon was indeed crucial for utilization of trehalose by S. mutans, the ΔtreA, ΔtreB, and ΔtreR strains were all tested for their ability to grow in medium where trehalose was the sole carbon source provided. Although all three genes were required for wild-type levels of growth in trehalose (Fig. 1B), treA appears to be the most essential gene for trehalose utilization, as the ΔtreA strain was unable to grow under these conditions. This finding is logical, as without TreA, it is likely that S. mutans was unable to break trehalose down to glucose for use in energy metabolism. The ΔtreR and ΔtreB mutants were able to grow, but at a greatly reduced rate and with lower final yields (Fig. 1B). The loss of TreB most likely affected the ability of S. mutans to import trehalose in a high-affinity, specific manner. Instead, other nonspecific, lower-affinity (and less efficient) transport systems may be used. It was initially less clear why the ΔtreR strain was significantly impaired in its ability to grow on trehalose, because if it functioned merely as a repressor of the tre operon, the deletion strain should exhibit increased expression of the machinery needed to utilize trehalose and thus would be unlikely to have impaired growth in trehalose.
Further characterization of the ΔtreR strain revealed a surprising phenotype: increased sensitivity to H2O2 (Fig. 2A). This was unexpected, as TreR has thus far only been described as a local regulator governing trehalose utilization. Typically, induction of the S. mutans ATR protects against both acid and reactive oxygen species (ROS) toxicity; however, deletion of treR appeared to hinder the capacity of S. mutans to mount an ATR that protected against oxidative stress, following growth at the moderately acidic pH of 5 (Fig. 2B). The full arsenal of the ATR is crucial in the capacity of S. mutans to cause disease, as this robust stress response gives S. mutans a significant competitive advantage against less aciduric commensal rivals, several of which produce H2O2 to kill S. mutans (26).
To further examine the phenotype observed in the ΔtreR strain, RNA-seq was performed on the UA159 and ΔtreR strains grown in glucose, trehalose, or galactose (a nonpreferred sugar other than trehalose, i.e., alleviating CCR). RNA-seq revealed that treA and treB were not upregulated upon deletion of treR, as was expected. In fact, upon deletion of treR, treA and treB were significantly downregulated during growth in trehalose and were not differentially regulated during growth in glucose or galactose (Table 1). This suggests that in S. mutans, TreR actually serves as a transcriptional activator of the tre operon when trehalose is present. This hypothesis also explains the growth defect of the ΔtreR strain during growth in trehalose: without TreR-mediated activation of treA and treB expression, S. mutans struggles to import and generate energy from the only carbon source provided. The stress generated from this condition would also serve to explain the vast number of genes (>800) that are differentially regulated in the ΔtreR strain compared to UA159 during growth in trehalose, particularly the upregulation of genes involved in energy metabolism and protein degradation and the concurrent downregulation of genes involved in protein synthesis (see Table S3 in the supplemental material). While this theory appears likely, further work is needed to confirm that TreR acts directly as a transcriptional activator. Elucidation of the S. mutans tre promoter, including downstream effects of the binding and stoichiometry of multiple regulators, will also assist in obtaining a more complete understanding of tre regulation. Indeed, putative binding motifs for the CcpA and FabT regulators have been identified in the S. mutans tre promoter, in addition to the predicted TreR motif. In-depth biochemical studies exploring these regulators and tre expression are in progress.
The majority of the most significantly downregulated genes in the RNA-seq data for the ΔtreR strain were involved in toxin production and the competence signaling pathways (Fig. 3C to E; Table 1). This was particularly interesting given that (i) trehalose increases genetic competence in S. mutans (13), (ii) competence coordinates toxin production in S. mutans (27), and (iii) trehalose was also recently shown to increase toxin production in C. difficile (10). This collective evidence suggests that in S. mutans, TreR may indirectly control expression of toxins through involvement in the competence signaling pathway. Several previous studies have reported that S. mutans strains with deficiencies in competence signaling are also defective in the ATR, stringent response, and tolerance of oxidative stress (including H2O2), which likely explains the oxidative-stress-sensitive phenotype of the ΔtreR strain (28–30). Indeed, analysis using virtual footprint regulon prediction (31) revealed putative TreR binding motifs in the promoters of comC and comEA, supporting the possibility that these genes are regulated by trehalose via TreR (data not shown). The comC gene encodes the precursor for the competence-stimulating peptide (CSP), and thus TreR could have far-reaching effects on S. mutans signaling and transcription. Additional experiments illustrating the binding of TreR to promoters outside the tre operon are needed to confirm the direct regulation of noncanonical targets.
Regardless of the mechanism, it appears that the reduction in expression of the genes encoding mutacins IV, V, and VI in the ΔtreR strain is sufficient to drastically reduce inhibition of S. gordonii or L. lactis, as shown in the antagonism assay (Fig. 4). This finding indicates that regulation by TreR is likely to play a critical role in competition with rival species and therefore the persistence and virulence of S. mutans. The apparent involvement of S. mutans TreR in the competence signaling pathways and mutacin expression is reminiscent of a recent study illustrating that the S. mutans Spx global regulators (SpxA1 and SpxA2) are also crucial for proper competence development and, therefore, mutacin expression (32). Indeed, spxA2 had reduced expression in the ΔtreR strain during growth in galactose, indicating possible overlap of the Spx and TreR regulons (see Table S2 in the supplemental material). SpxA2 is involved in regulation of the oxidative stress response, and genes involved in management of ROS were differentially regulated in the ΔtreR strain, including dpr, ahpF, and nox (Table S3). Both this study and the study by Galvao et al. (32) highlight that much work remains to be done to fully understand the highly complex and convoluted regulatory pathways that have evolved in S. mutans as it continues to contribute to disease in the majority of humans.
In summary, the results of the present study confirm that the S. mutans tre operon is required for growth in trehalose. TreR appears to function as an activator of expression for the tre operon, rather than as a repressor (as is the case in B. subtilis). TreR also appears to be intertwined with competence signaling pathways, which has farther-reaching effects on toxin production and survival during oxidative stress and, accordingly, the overall competitive fitness and virulence of S. mutans. Additional biochemical studies, confirming the direct action of TreR as an activator of expression and its novel role in global signaling, stress response, and toxin production, are currently in progress. The link between trehalose and toxin production (and therefore competitive fitness), demonstrated here with S. mutans and in the recent study with C. difficile (10), provides an avenue for discovering similar associations in other Firmicutes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table 2. The Streptococcus mutans genomic type strain UA159 has been described previously (2, 33). The S. mutans ΔtreA and ΔtreB mutant strains are derivatives of UA159 and have been described previously (23). S. mutans was maintained on brain heart infusion (BHI) agar plates (BD/Difco, Franklin Lakes, NJ) at 37°C in a 5% (vol/vol) CO2–95% air environment. Where applicable, antibiotics were added to a final concentration of 5 μg ml−1 for erythromycin and 1 mg ml−1 for kanamycin. Organisms were cultured in TY medium (3% tryptone, 0.1% yeast extract, 0.5% KOH, 1 mM H3PO4) plus 1% (wt/vol) glucose and were grown in liquid culture or in continuous culture in a BioFlo 2000 fermentor (New Brunswick Scientific, Edison, NJ) as described previously (34, 35). Continuous cultures were grown at a dilution rate of 0.24 h−1 under glucose-limiting conditions (2.3 mM) with a continuous impeller speed of 200 rpm. Steady-state growth at pH 7.0 or 5.0 was maintained by the addition of 2 N KOH. The culture pH was continuously monitored throughout the experiment by an indwelling pH probe (Mettler Toledo, Columbus, OH). After continuous cultures had been maintained for a minimum of 10 generations under a given condition, aliquots of the culture were removed and cells were collected by centrifugation. Cell pellets were washed and stored frozen at −80°C.
TABLE 2.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description (reference[s] or source, if not this study) |
|---|---|
| Strains | |
| Streptococcus mutans | |
| UA159 | Genomic type strain (2, 33) |
| ΔtreA strain | treA deletion strain (23) |
| ΔtreB strain | treB deletion strain (23) |
| ΔtreR strain | treR deletion strain |
| treR-C strain | treR complement strain |
| treA-C strain | treA complement strain |
| Escherichia coli | |
| XL-1 Blue | Host for ptreR-C-Topo (Stratagene) |
| XL10-Gold | Host for ptreR-C (Stratagene) |
| Stellar | Host for ptreA-C (Clontech) |
| Streptococcus gordonii DL1 | Laboratory stock |
| Lactococcus lactis ATCC 11454 | Laboratory stock |
| Plasmids | |
| ptreR-KO | Construct to make the ΔtreR strain (GenScript) |
| ptreR-C-TOPO | Construct for subcloning of treR-C strain |
| pBGE | Vector backbone containing erythromycin resistance cassette to make treR-C strain (gift from José Lemos) |
| ptreR-C | Construct to make treR complement strain [treR-C strain] |
| pSUGKBglII | Vector backbone containing kanamycin resistance cassette to make treA-C strain (36) |
| ptreA-C | Construct to make treA complement strain [treA-C] |
| Primers | |
| 5′treR-C-XbaI-F | Forward primer to make ptreR-Topo, TCTAGAAAATTTTTCCTAGGTTAACCTAGTCa |
| 3′treR-C-BsrGI-R | Reverse primer to make ptreR-Topo, TGTACATTCTTTTGTGCTTTCCTTGTTGTCb |
| 3′treA-C | Primer to make ptreA-C, ATTTAAAAATAGATCTGATCAATTATTGTAAGGAGAATAA |
| 5′treA-C | Primer to make ptreA-C, GAGCTCGAATAGATCTTCAGACTTCTGCTAAAATCGC |
The XbaI site is underlined.
The BsrGI site underlined.
Growth curves were recorded using an automated growth-monitoring device, a Bioscreen C (Growth Curves USA, Haverhill, MA), as described previously (36). The optical density at 600 nm (OD600) was measured every hour with a 10-s shaking period before each reading to evenly suspend cultures. Microaerobic cultures were created using a 50-μl mineral oil overlay of the Bioscreen cultures. Growth in glucose versus trehalose or galactose was measured in TY medium plus 1% glucose, 1% trehalose, or 1% galactose. Where applicable, H2O2 was added to 0.5 mM.
Oxidative stress challenge.
Cultures of S. mutans UA159 and the ΔtreR mutant were grown in continuous culture to steady-state conditions of pH 7 and pH 5 as described above. Aliquots of cells were harvested by centrifugation and subjected to an oxidative stress (16.3 mM H2O2), as previously described (36). Briefly, steady-state samples harvested from the chemostat, at culture pH values of 7 and 5, were resuspended in BHI medium, and hydrogen peroxide was added to a final concentration of 16.3 mM. Aliquots were removed at 0, 15, 30, and 60 min, serially diluted, and plated on BHI agar medium. Viable cells from each condition were counted and used to calculate log (n/n0), where n is the number of colonies obtained at a specific time point and n0 is the number of colonies at time zero.
DNA manipulations.
Chromosomal DNA was isolated from S. mutans as previously described (37). Plasmid DNA was isolated from E. coli as previously described using a E.Z.N.A plasmid minikit (Omega Bio-Tek, Norcross, GA). PCR was carried out with Platinum Taq DNA polymerase (Invitrogen, Carlsbad, CA). Amplicons were isolated and purified via gel electrophoresis as previously described (38). S. mutans UA159 was transformed by previously described methods (33, 39).
Generation of recombinant ΔtreR, treR-C, and treA-C strains. (i) ΔtreR strain.
A UA159 derivative was constructed with the treR open reading frame (ORF) replaced by a kanamycin resistance gene (ΔtreR). A construct designated ptreR-KO was synthesized using the pUC57 vector backbone with the EcoRV site in the multiple-cloning site containing the kanamycin resistance gene flanked on the left side by a NotI restriction site followed by 500 bp upstream of treR and flanked on the right side by 500 bp downstream of treR, followed by an XhoI restriction site (GenScript, Piscataway, NJ). The construct was digested with NotI and XhoI, and the resulting insert was gel purified and transformed into S. mutans UA159. Transformants were selected for Kanr, and one isolate was designated the ΔtreR strain.
(ii) treR-C strain.
ΔtreR was complemented using a single-copy genomic insertion of the SMU.2040 (treR) locus, including the intergenic region between treR and treB, into the gtfA (SMU.881) locus. Under the growth conditions tested, no physiological impact from the disruption of the gtfA locus was detected, as reported previously (40). Primers 5′treR-C-XbaI-F and 3′treR-C-BsrGI-R (Table 2) were used to amplify the treR and cognate promoter, as well as to add XbaI (5′) and BsrGI (3′) restriction sites to the amplicon. The resulting amplicon was gel purified and subcloned into pCR2.1 using a TOPO-TA cloning kit (Invitrogen, Carlsbad, CA) to produce ptreR-C-Topo. The TOPO ligation reaction product was transformed into E. coli XL-1 Blue (Stratagene, La Jolla, CA). The streptococcal integration vector pBGE was a gift from José Lemos and has been previously described (41). ptreR-C-Topo and pBGE were linearized using XbaI and BsrGI, and the fragment of ptreR-C-Topo containing the treR ORF was ligated into the vector backbone of pBGE to produce ptreR-C. The ligation reaction product was transformed into E. coli XL-10 Gold (Stratagene, La Jolla, CA), and positive clones were selected on LB agar medium containing erythromycin (500 μg ml−1). The integrity of the construct was confirmed by sequencing. ptreR-C was transformed into the ΔtreR strain and selected on BHI agar medium containing erythromycin. The complemented strain was designated the treR-C strain, and the integrities of both the deletion and complementation loci were confirmed by sequencing.
(iii) treA-C strain.
ΔtreA was complemented using a single-copy genomic insertion of the SMU.2037 (treA) locus, including the intergenic region between treA and treB, into the gtfA (SMU.881) locus. Under the growth conditions tested, no physiological impact from the disruption of the gtfA locus was detected, as reported previously (40). The streptococcal integration vector pSUGKBglII, described previously (36), was linearized with the restriction enzyme BglII. Primers 3′treA-C and 5′treA-C (Table 2) were used to amplify treA and the intergenic region between treA and treB. The resulting amplicon was ligated into the linearized pSUGKBglII using an In-Fusion HD cloning kit (Clontech, Mountain View, CA) to produce ptreA-C. The cloning reaction product was transformed into E. coli Stellar (Clontech, Mountain View, CA), and positive clones were selected on LB agar medium containing kanamycin (50 μg ml−1). The integrity of the construct was confirmed by sequencing. ptreA-C was transformed into the ΔtreA strain and selected on BHI agar medium containing kanamycin. The complemented strain was designated treA-C, and the integrities of both the deletion and complementation loci were confirmed by sequencing.
RNA-seq.
For mRNA sequencing (RNA-seq), triplicate samples of S. mutans UA159 or the ΔtreR strain were grown at 37°C in a 5% (vol/vol) CO2–95% air atmosphere to mid-log phase in TY medium supplemented with 1% glucose (OD600 of ∼0.6), 1% galactose (OD600 of ∼0.5), or 1% trehalose (for UA159, OD600 of ∼0.7; for the ΔtreR strain, OD600 of ∼0.3). Cells were harvested by centrifugation, and cell pellets were stored at −80°C. The protocols for transcriptomic sequencing were modified from those described by Edlund et al. (42). Briefly, total RNA extraction was performed using the Qiagen RNeasy Power Microbiome kit according to the manufacturer's instructions (Qiagen, Chatsworth, CA). rRNA was removed from the total RNA using the Ribo-Zero (Bacteria) kit (Epicentre, Madison, WI). mRNA was purified using the RNA Clean and Concentrator kit (Zymo Research, Irvine, CA). RNA concentration and integrity were monitored before and after rRNA removal using a Qubit 3.0 fluorometer (Life Technologies, Waltham, MA) and the Agilent RNA ScreenTape assay for the 4200 TapeStation system (Agilent Technologies, Santa Clara, CA). cDNA libraries from rRNA-depleted RNA were generated using random-primed cDNA synthesis methods according to the ScriptSeq v2 RNA-seq library preparation protocol (Epicentre, Madison, WI). Prior to second-strand cDNA synthesis, the ditagged cDNA was purified using the Agencourt AMPure XP system (Beckman Coulter, Brea, CA). ScriptSeq index primers were added to the libraries, which were then PCR amplified for 15 cycles. The final libraries were run on the Illumina NextSeq 550 platform using the High Output v2 150-cycle kit (2 × 75-bp paired-end reads) (Illumina, San Diego, CA).
Barcodes were trimmed, and low-quality and short sequences (<100 bp) were filtered using BBDuk (https://sourceforge.net/projects/bbmap) implemented in Geneious version 11 (Biomatters, Auckland, New Zealand) (43). Expression values for each mRNA sample were generated by mapping of paired reads onto the annotated reference genome of S. mutans UA159, resulting in 175 to 275× mean coverage per sample (range of 4.2 to 6.4M reads mapped per sample). Expression level counts and differential expression calculations in triplicate between conditions were performed using Geneious version 11 (Biomatters) (43) with DESeq2 normalization (44). DESeq2 uses a model based on the negative binomial distribution with variance and mean linked by local regression (45).
Deferred-antagonism assay.
The deferred-antagonism assay was performed as previously described (24, 25). Briefly, cultures of Streptococcus mutans UA159, the ΔtreR derivative strain, and the treR-C complement strain were grown in BHI broth, in triplicate, at 37°C in a 5% (vol/vol) CO2–95% air atmosphere. Following overnight incubation, the cultures were diluted in fresh BHI medium to an OD600 of ∼0.3. An aliquot (8 μl) of each culture was spotted on BHI agar, TY agar plus 1% glucose, or TY agar plus 1% trehalose and incubated for 18 h, following which colonies were exposed to UV light in a Stratalinker (Stratagene, San Diego, CA) for 3 min. Stationary-phase cultures (0.5 ml) of Streptococcus gordonii DL1 (mutacin IV and V sensitive) and Lactococcus lactis ATCC 11454 (mutacin V and VI sensitive) were added to 5 ml BHI agar (0.75%) and used to overlay the BHI agar plate containing the primary colonizer (S. mutans UA159, ΔtreR, or treR-C strain). Plates were incubated overnight before photographing to document zones of inhibition. Areas of the zones of inhibition were measured and recorded. Statistical analysis was performed using a two-way analysis of variance (ANOVA) test followed by Tukey's test of multiple comparisons.
Accession number(s).
RNA-seq data have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) with accession number GSE110275.
Supplementary Material
ACKNOWLEDGMENTS
We thank José Lemos for the gift of the pBGE plasmid.
This study was supported by NIH/NIDCR grants T90-DE021985 (J.L.B. and E.L.L.), F32-DE026947 (J.L.B.), R01-DE020102 and R01-DE026186 (J.S.M., X.H., and W.S.), and R01-DE013683 and R01-DE017425 (R.G.Q.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00057-18.
REFERENCES
- 1.Bowden GH, Hamilton IR. 1998. Survival of oral bacteria. Crit Rev Oral Biol Med 9:54–85. doi: 10.1177/10454411980090010401. [DOI] [PubMed] [Google Scholar]
- 2.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajdic D, Pham VT. 2007. Global transcriptional analysis of Streptococcus mutans sugar transporters using microarrays. J Bacteriol 189:5049–5059. doi: 10.1128/JB.00338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi N, Nyvad B. 2011. The role of bacteria in the caries process: ecological perspectives. J Dent Res 90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 5.Kandror O, DeLeon A, Goldberg AL. 2002. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc Natl Acad Sci U S A 99:9727–9732. doi: 10.1073/pnas.142314099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duong T, Barrangou R, Russell MW, Klaenhammer TR. 2006. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 72:1218–1225. doi: 10.1128/AEM.72.2.1218-1225.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burklen L, Schock F, Dahl MK. 1998. Molecular analysis of the interaction between the Bacillus subtilis trehalose repressor TreR and the tre operator. Mol Gen Genet 260:48–55. doi: 10.1007/s004380050869. [DOI] [PubMed] [Google Scholar]
- 8.Rezacova P, Krejcirikova V, Borek D, Moy SF, Joachimiak A, Otwinowski Z. 2007. The crystal structure of the effector-binding domain of the trehalose repressor TreR from Bacilus subtilis 168 reveals a unique quarternary structure. Proteins 69:679–682. doi: 10.1002/prot.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schock F, Dahl MK. 1996. Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J Bacteriol 178:4576–4581. doi: 10.1128/jb.178.15.4576-4581.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins J, Robinson C, Danhof H, Knetsch CW, van Leeuwen HC, Lawley TD, Auchtung JM, Britton RA. 2018. Dietary trehalose enhances virulence of epidemic Clostridium difficile. Nature 553:291–294. doi: 10.1038/nature25178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neta T, Takada K, Hirasawa M. 2000. Low-cariogenicity of trehalose as a substrate. J Dent 28:571–576. doi: 10.1016/S0300-5712(00)00038-5. [DOI] [PubMed] [Google Scholar]
- 12.Zeng L, Choi SC, Danko CG, Siepel A, Stanhope MJ, Burne RA. 2013. Gene regulation by CcpA and catabolite repression explored by RNA-Seq in Streptococcus mutans. PLoS One 8:e60465. doi: 10.1371/journal.pone.0060465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moye ZD, Son M, Rosa-Alberty AE, Zeng L, Ahn SJ, Hagen SJ, Burne RA. 2016. Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl Environ Microbiol 82:4821–4834. doi: 10.1128/AEM.01205-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng LY, Itzek A, Chen ZY, Kreth J. 2011. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. Int J Oral Sci 3:82–89. doi: 10.4248/IJOS11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryan CS, Kleinberg I. 1995. Bacteria in human mouths involved in the production and utilization of hydrogen peroxide. Arch Oral Biol 40:753–763. doi: 10.1016/0003-9969(95)00029-O. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Tong H, Dong X. 2012. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol 78:2120–2127. doi: 10.1128/AEM.07539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol 63:872–880. doi: 10.1111/j.1365-2958.2006.05546.x. [DOI] [PubMed] [Google Scholar]
- 19.Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol 27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreth J, Merritt J, Shi W, Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol 57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufour D, Cordova M, Cvitkovitch DG, Levesque CM. 2011. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol 193:6552–6559. doi: 10.1128/JB.05968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reck M, Tomasch J, Wagner-Dobler I. 2015. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quivey RG Jr, Grayhack EJ, Faustoferri RC, Hubbard CJ, Baldeck JD, Wolf AS, MacGilvray ME, Rosalen PL, Scott-Anne K, Santiago B, Gopal S, Payne JP, Marquis RE. 2015. Functional profiling in Streptococcus mutans: construction and examination of a genomic collection of gene deletion mutants. Mol Oral Microbiol 30:474–495. doi: 10.1111/omi.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hossain MS, Biswas I. 2011. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol 77:2428–2434. doi: 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hale JD, Ting YT, Jack RW, Tagg JR, Heng NC. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl Environ Microbiol 71:7613–7617. doi: 10.1128/AEM.71.11.7613-7617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker JL, Faustoferri RC, Quivey RG Jr. 2017. Acid-adaptive mechanisms of Streptococcus mutans—the more we know, the more we don't. Mol Oral Microbiol 32:107–117. doi: 10.1111/omi.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan R, Rukke HV, Hovik H, Amdal HA, Chen T, Morrison DA, Petersen FC. 2016. Comprehensive transcriptome profiles of Streptococcus mutans UA159 map core streptococcal competence genes. mSystems 1:e00038-15. doi: 10.1128/mSystems.00038-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YH, Hanna MN, Svensater G, Ellen RP, Cvitkovitch DG. 2001. Cell density modulates acid adaptation in Streptococcus mutans: implications for survival in biofilms. J Bacteriol 183:6875–6884. doi: 10.1128/JB.183.23.6875-6884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi F, Merritt J, Lux R, Shi W. 2004. Inactivation of the ciaH gene in Streptococcus mutans diminishes mutacin production and competence development, alters sucrose-dependent biofilm formation, and reduces stress tolerance. Infect Immun 72:4895–4899. doi: 10.1128/IAI.72.8.4895-4899.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seaton K, Ahn SJ, Sagstetter AM, Burne RA. 2011. A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J Bacteriol 193:862–874. doi: 10.1128/JB.01257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 32.Galvao LC, Rosalen PL, Rivera-Ramos I, Franco GC, Kajfasz JK, Abranches J, Bueno-Silva B, Koo H, Lemos JA. 2017. Inactivation of the spxA1 or spxA2 gene of Streptococcus mutans decreases virulence in the rat caries model. Mol Oral Microbiol 32:142–153. doi: 10.1111/omi.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murchison HH, Barrett JF, Cardineau GA, Curtiss R III. 1986. Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun 54:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fozo EM, Quivey RG Jr. 2004. Shifts in the membrane fatty acid profile of Streptococcus mutans enhance survival in acidic environments. Appl Environ Microbiol 70:929–936. doi: 10.1128/AEM.70.2.929-936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quivey RG Jr, Faustoferri RC, Clancy KA, Marquis RE. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol Lett 126:257–261. doi: 10.1111/j.1574-6968.1995.tb07427.x. [DOI] [PubMed] [Google Scholar]
- 36.Derr AM, Faustoferri RC, Betzenhauser MJ, Gonzalez K, Marquis RE, Quivey RG Jr. 2012. Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl Environ Microbiol 78:1215–1227. doi: 10.1128/AEM.06890-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quivey RG Jr, Faustoferri RC, Belli WA, Flores JS. 1991. Polymerase chain reaction amplification, cloning, sequence determination and homologies of streptococcal ATPase-encoding DNAs. Gene 97:63–68. doi: 10.1016/0378-1119(91)90010-9. [DOI] [PubMed] [Google Scholar]
- 38.Kuhnert WL, Quivey RG Jr. 2003. Genetic and biochemical characterization of the F-ATPase operon from Streptococcus sanguis 10904. J Bacteriol 185:1525–1533. doi: 10.1128/JB.185.5.1525-1533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry D, Kuramitsu HK. 1981. Genetic transformation of Streptococcus mutans. Infect Immun 32:1295–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wen ZT, Burne RA. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31–36. doi: 10.1006/plas.2000.1498. [DOI] [PubMed] [Google Scholar]
- 41.Zeng L, Burne RA. 2009. Transcriptional regulation of the cellobiose operon of Streptococcus mutans. J Bacteriol 191:2153–2162. doi: 10.1128/JB.01641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edlund A, Yang Y, Yooseph S, Hall AP, Nguyen DD, Dorrestein PC, Nelson KE, He X, Lux R, Shi W, McLean JS. 2015. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J 9:2605–2619. doi: 10.1038/ismej.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.