ABSTRACT
We evaluated the diversity of group B Streptococcus (GBS) vaginal carriage populations in pregnant women. For this purpose, we studied each isolate present in a primary culture of a vaginal swab using a new approach based on clustered regularly interspaced short palindromic repeats (CRISPR) locus analysis. To evaluate the CRISPR array composition rapidly, a restriction fragment length polymorphism (RFLP) analysis was performed. For each different pattern observed, the CRISPR array was sequenced and capsular typing and multilocus sequence typing (MLST) were performed. A total of 970 isolates from 10 women were analyzed by CRISPR-RFLP. Each woman carrying GBS isolates presented one to five specific “personal” patterns. Five women showed similar isolates with specific and unique restriction patterns, suggesting the carriage of a single GBS clone. Different patterns were observed among isolates from the other five women. For three of these, CRISPR locus sequencing highlighted low levels of internal modifications in the locus backbone, whereas there were high levels of modifications for the last two women, suggesting the carriage of two different clones. These two clones were closely related, having the same ancestral spacer(s), the same capsular type and, in one case, the same ST, but showed different antibiotic resistance patterns in pairs. Eight of 10 women were colonized by a single GBS clone, while two of them were colonized by two strains, leading to a risk of selection of more-virulent and/or more-resistant clones during antibiotic prophylaxis. This CRISPR analysis made it possible to separate isolates belonging to a single capsular type and sequence type, highlighting the greater discriminating power of this approach.
KEYWORDS: CRISPR. Streptococcus agalactiae, carriage, genotyping, diversity
INTRODUCTION
Streptococcus agalactiae, or group B Streptococcus (GBS), is a major pathogen in human medicine, particularly in neonatology, where this bacterium is still the leading cause of invasive infections in industrialized countries (1). Prevention of neonatal infection associated with GBS requires the detection of vaginal carriage in pregnant women, followed by intrapartum antibiotic prophylaxis for those colonized (2, 3). Indeed, GBS colonizes the gastrointestinal and genital tracts of 10 to 30% of healthy women (4–6).
GBS isolates were initially distinguished on the basis of differences in capsular polysaccharides (CPS), leading to the definition of 10 different serotypes (Ia, Ib, and II to IX) (7, 8). While this typing method highlighted pathogenicity differences among serotypes (serotype III GBS strains are strongly associated with neonatal invasive infections [9, 10]), it was suboptimal in its discriminatory capacity for studying the epidemiology of GBS infection. Thus, several higher-throughput and higher-resolution subtyping methods were developed, such as multilocus sequence typing (MLST) and multilocus variable-number tandem repeat (VNTR) analysis (MLVA) (11–13). Based on the combination of alleles for seven housekeeping genes, MLST defines sequence types (ST), which can be clustered in clonal complexes (CC) when six of the seven alleles are common. Based on the number of repeats at each of the 6 VNTR loci, MLVA defines a profile consisting of a series of allele numbers.
Clustered regularly interspaced short palindromic repeats (CRISPR) loci consist of short (25- to 40-bp) direct repeats (DR) that are highly conserved within a given CRISPR array, interspaced by various sequences of similar size, called spacers, which are derived from foreign genetic elements. CRISPR and CRISPR-associated (Cas) proteins form the CRISPR-Cas system, which provides defense against mobile genetic elements (MGE) such as phages or plasmids (14). This immune system is adaptive and confers specific and acquired immunity based on short nucleic acid sequence integration from the invaders. A core feature of the CRISPR-Cas system is the ability to acquire a novel spacer in a specific manner at the leader end of the locus, indicating exposure to invasive elements over time. Spacers at the leader end were recently acquired and provide insight into recent MGE exposure. In contrast, ancestral spacers at the trailer end of the CRISPR array provide information about the phylogenetic lineage and are correlated with phylogenetic groups defined by other typing methods. For bacterial phylogenetic analysis, CRISPR-based typing appears to be a practical and powerful alternative (15–17). Subtyping using CRISPR has been employed for other bacterial species such as Yersinia and Salmonella (18, 19). In GBS, a type II-A system, associated with the CRISPR1 locus, is ubiquitous and functional. The similarity between CRISPR1 spacer sequences and MGE sequences has been previously emphasized (20, 21). Comparative sequence analysis across numerous isolates has revealed that this locus is extremely diverse and evolves in vivo, reflecting the dynamics of the system (20–22).
The aim of this study was to determine the homogeneity of GBS populations in pregnant women at the time of their GBS screening. For this purpose, we used an original approach based on a CRISPR array study to analyze all the isolates present in a primary culture from a vaginal swab. To rapidly evaluate the composition of the spacers, we screened isolates by restriction fragment length polymorphism (RFLP) of the CRISPR1 array and, when different restriction patterns were obtained, CRISPR1 loci were sequenced and isolates typed using the most common GBS typing methods (capsular typing and MLST). This method enabled us to compare numerous isolates and to target potentially different clones with the high discriminatory power of CRISPR analysis.
MATERIALS AND METHODS
Bacterial isolates.
Isolates were collected between July and September 2016 from the department of obstetrics, gynecology, and fetal medicine of the Tours University Hospital, Tours, France. We randomly selected vaginal swabs from 10 pregnant women. GBS specimen collection was conducted during the late third trimester (35 to 37 weeks' gestation), observing the French recommendations on screening for GBS carriage (2). The protocol was reviewed and approved by the Ethics Committee in Human Research of the University Hospital Center of Tours (Tours, France). We selected plates with at least 20 and up to 200 isolates for practical reasons. Each swab was spread on a selective agar plate (group B Streptococcus differential agar, Granada medium; BD) and incubated for 18 to 48 h at 35 ± 2°C under anaerobic conditions according to the manufacturer's recommendations. Bacterial identification was based on growth and orange pigment formation on Granada agar and confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Microflex; Bruker Daltonics). Each isolate present on the agar plate was reisolated and stored at −80°C for the duration of the study and grown secondarily on TSH agar (Trypticase soy agar supplemented with 5% horse blood; bioMérieux).
DNA extraction.
Genomic DNA was extracted following enzymatic lysis with mutanolysin (Sigma). A bacterial suspension of 1.5 McFarland standard was prepared in 250 μl water containing 25 U of mutanolysin. The suspension was incubated at 56°C for 1 h, followed by 10 min at 100°C, leading to cell lysis. Lysates were centrifuged at 1,500 × g for 3 min, and the DNA-containing supernatants were collected.
CRISPR1 locus amplification.
CRISPR1 locus amplification was performed in a T3000 thermocycler (Biometra) using CRISPR1 SEQ-F and CRISPR1 SEQ-R primers targeting the CRISPR1-flanking regions, as previously described (20). PCR amplification was performed in a total volume of 25 μl, containing 0.5 μM each primer, 0.2 μM deoxynucleoside triphosphate (dNTP), 1.5 mM MgCl2, 0.02 U/μl Go Taq polymerase (Promega), 1× PCR buffer, and 1 μl extracted DNA. The PCR mixtures were heated to 94°C for 5 min, followed by 40 cycles of a denaturation step at 94°C for 30 s, an annealing step at 58°C for 30 s, and an elongation step at 72°C for 3 min, ending with a final extension step at 72°C for 10 min. PCR amplification was verified by electrophoretic migration in a 1% agarose gel. If amplification did not enable us to restrict the PCR product due to low signal intensity, isolates were amplified using internal primers designed after sequencing and covering the entire CRISPR1 locus (Table 1). Indeed, for these CRISPR arrays, two amplifications were necessary, one targeting the 5′ end and one targeting the 3′ end of the locus.
TABLE 1.
Primers used for amplification and sequencing
| Oligonucleotide name | Used for: (patient IDa/isolate) | Sequence (5′→3′) |
|---|---|---|
| CRISPR1 seq-F | All patients and isolates | GAAGACTCTATGATTTACCGC |
| CRISPR1 seq-R | All patients and isolates | CAGCAATCACTAAAAGAACCAAC |
| Sp803F | W1/All isolates | TCTAATGTATACTGTAACTGAGGCAG |
| Sp504F | W4/All isolates | TTTATCCCTAGTAGGTT |
| Sp539F | W8/A2 | TCTAAGTGCTCGACCATC |
| Sp8R | W8/A2 | TATCCATCTCGGTGAGATGAGAATTAGCTT |
| Sp31R | W1/All isolates | ATCAGTACCAACAAATGATTTTGTACCATC |
| Sp36R | W3/All isolates | ACATATCCTTTTGTTAGGTCAAAAGAAGAT |
| Sp177R | W9/All isolates | AGTTACTGTTGAGGGTAGTCC |
| 275F | W3/All isolates | ACAGACAAAGAAGATGGCAAG |
Patients are identified by “W” followed by a number, i.e., W1 is woman 1.
RFLP.
Restriction enzymes were selected using NEBcutter V2.0 (http://nc2.neb.com/NEBcutter2/) based on previously sequenced CRISPR1 arrays. We selected two enzymes with three or four cutters for most of the CRISPR1 arrays tested: DdeI and MnlI. Restriction was performed separately for each enzyme in a T3000 thermocycler (Biometra) according to the manufacturer's recommendations. Restriction products were analyzed by electrophoretic migration in a 1.5% agarose gel. For isolates with CRISPR1 arrays showing different sizes and/or different patterns by RFLP, the CRISPR1 locus was sequenced; MLST and capsular typing were performed.
CRISPR1 locus sequencing and analysis.
CRISPR1 locus sequencing was performed as previously described (20). Primers targeting internal spacers were required to complete the sequencing of the PCR products for CRISPR1 regions exceeding 1.3 kb (Table 1).
DNA sequences were analyzed and assembled using the BioEdit sequence alignment editor version 7.2.5 (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Spacers, repeats, and flanking regions for each sequence were determined using a macro-enabled Excel tool (kindly provided by P. Horvath, DuPont). This application was used to identify and extract CRISPR features from nucleotide sequences and provided graphic representations of spacers as colored cells in Excel spreadsheets. Spacer sequences were compared to the previously established dictionary of spacers (20). New spacers identified in this study expanded the dictionary and were numbered incrementally. The spacer dictionary is available online (http://crispr.i2bc.paris-saclay.fr/CRISPRcompar/Dict/Dict.php).
MLST and capsular typing.
MLST was carried out as previously described (12). Allelic profiles and ST were assigned using the international MLST database (http://pubmlst.org/sagalactiae/). Clonal complexes were defined using eBurst analysis (http://eburst.mlst.net/v3/mlst_datasets/) (23). Capsular type identification was performed using PCR multiplex assays as described previously (24).
Antibiotic susceptibility testing.
Susceptibility to antibiotics was tested and interpreted according to the 2016 EUCAST guidelines using a standardized disk diffusion method (http://www.eucast.org/clinical_breakpoints/). We tested the following 14 antibiotics in an MHF medium (Mueller-Hinton agar, 5% horse blood, 20 mg/liter β-NAD): benzylpenicillin (1 unit), ampicillin (2 μg), norfloxacin (10 μg), levofloxacin (5 μg), erythromycin (15 μg), lincomycin (15 μg), pristinamycin (15 μg), gentamicin (500 μg), tetracycline (30 μg), rifampin (5 μg), trimethoprim-sulfamethoxazole (1.25 to 23.75 μg), vancomycin (5 μg), teicoplanin (30 μg), nitrofurantoin (100 μg). To confirm the differences in antibiotic susceptibility observed by disk diffusion, MICs were determined for some isolates by Etest (bioMérieux) for erythromycin and clindamycin.
RESULTS
Population studied.
A total of 970 isolates were recovered from 10 GBS-colonized women during their last month of pregnancy. The average age of the cohort was 31 years at sampling time (median, 31 years; minimum, 25 years; maximum, 38 years). Vaginal swabs were taken from each woman and rapidly seeded to agar plates. All GBS isolates present on these plates were then analyzed. An average of 97 isolates per woman was considered (median, 108; minimum, 20; maximum, 164) (Table 2), illustrating differences in bacterial carriage density.
TABLE 2.
Characteristics of the cohort study and results of CRISPR-RFLP using DdeIa
| Woman ID (age in yr) | No. of isolates | No. of isolates (%) with pattern no.: |
||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| W1 (35) | 164 | 162 (99) | 2 (1) | |||
| W2 (34) | 130 | 130 (100) | ||||
| W3 (32) | 48 | 47 (98) | 1 (2) | |||
| W4 (27) | 64 | 64 (100) | ||||
| W5 (25) | 144 | 144 (100) | ||||
| W6 (26) | 121 | 121 (100) | ||||
| W7 (28) | 60 | 60 (100) | ||||
| W8 (32) | 20 | 13 (65) | 7 (35) | |||
| W9 (38) | 97 | 92 (95) | 2 (2) | 1 (1) | 1 (1) | 1 (1) |
| W10 (29) | 125 | 92 (74) | 33 (26) | |||
A total of 970 isolates were studied. Each pattern was specific for a given woman. Five women carried GBS isolates with a single pattern. The other five carried GBS isolates with several patterns (up to five different patterns). The same distribution was observed with MnlI.
Culture morphology.
No phenotypic differences were observed between isolates from any given woman except for one (woman 10), from whom 125 isolates were present on the plate. Among them, 33 isolates were more pigmented on Granada selective agar and more hemolytic on blood agar than the other 92 isolates. These observed colony pigmentation differences may be explained by the level of production of the orange carotenoid pigment, which is tightly regulated.
CRISPR1 amplification and RFLP analysis.
For 922 isolates, amplification was performed using CRISPR1 seq-F and CRISPR1 seq-R primers. For 48 isolates from one woman (woman 3), DNA was amplified using internal primers due to low signal intensity. Indeed, two amplifications were necessary for these isolates, and restriction was performed for the two amplicons (96 PCR products, 192 restrictions). CRISPR1 locus sizes were evaluated after PCR amplification and compared for each isolate from the same woman. For 5 of the 10 women, all isolates presented a CRISPR1 locus of the same size (women 2, 4, 5, 6, and 7). For the other five women, a difference in CRISPR1 locus size was observed. Differences were observed for 1% (two isolates, woman 1) to 35% (seven isolates, woman 8) of isolates.
For each isolate, we performed a CRISPR-RFLP with the restriction enzymes DdeI and MnlI. The two restrictions proceeded and were analyzed separately. The resulting restriction fragments were separated according to length by gel electrophoresis. Among all the GBS isolates, a total of 18 different gel band patterns were detected by CRISPR-RFLP. All women studied carried one to five GBS isolates with specific gel band patterns. Each gel band pattern was specific to the isolates from a given woman.
For five women, no differences were observed after CRISPR enzymatic restriction (Table 2). In contrast, different restriction patterns were observed for isolates from the other five women: minor patterns represented 1% (2/164 isolates, woman 1) to 35% (7/20 isolates, woman 8) of all isolates. The two enzymes used detected the same differences. For four of these five women, only two patterns were observed, suggesting the presence of two different clones. For the last woman (woman 9), five patterns were observed, suggesting the presence of five different clones. Nonetheless, for all women, one clone was predominant, representing at least 65% of isolates.
Sequencing.
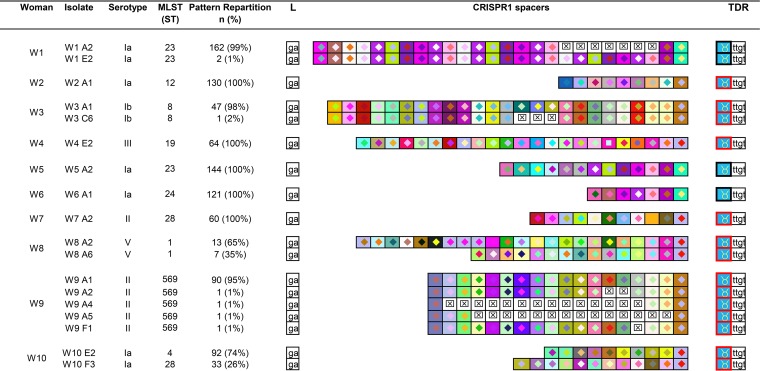
All CRISPR1 loci showing different restriction patterns were sequenced and analyzed (18 isolates, 10 women) (Fig. 1). Each woman carried GBS, characterized by a specific CRISPR array. For three of the five women whose isolates showed CRISPR variations by CRISPR-RFLP (women 1, 3, and 9), variations concerned a single CRISPR type. Indeed, spacers were acquired or deleted in the middle part of the locus, whereas ancestral and more recently acquired spacers were conserved. For woman 1, isolates presented a CRISPR1 locus with one repetition of seven spacers in 99% of isolates (162/164) and two repetitions in 1% of isolates (2/164, e.g., isolates W1 E2). For woman 3, the differences in restriction patterns were due to a middle locus deletion of three spacers for one isolate (isolate W3 C6). For woman 9, four isolates presented a middle locus deletion of one (isolate W9 F1), two (isolates W9 A2), or 14 (isolates W9 A4 and W9 A5) spacers (Fig. 1). For two of the five women with different CRISPR-RFLP, isolates had two different CRISPR types (women 8 and 10). The CRISPR1 locus, however, presented common ancestral spacer(s) among all isolates (four [woman 8] and one [woman 10]).
FIG 1.
Spacer composition of CRISPR1 arrays for GBS isolates from 10 women. Only one array is represented by different patterns observed after RFLP. The CRISPR1 arrays are represented using a macro-enabled Excel tool whereby spacers are converted into two-color symbols based on the spacer sequence. Gaps (indicating missing spacers) are shown with a boxed “×” symbol after alignment of identical spacers between strains of the same group. Terminal direct repeats (TDRs) are represented by different color borders according to their sequence. The leader sequence (L) is represented by its last two nucleotides. For each woman, the main CRISPR array was represented first. Each woman presented a specific CRISPR array. Three of the 10 women carried isolates belonging to a single CRISPR type with internal modification of the CRISPR1 array (women 1, 3, and 9). Two of the 10 women carried isolates with different CRISPR1 arrays, indicating the presence of two clones (women 8 and 10).
Antibiotic susceptibility testing.
Antibiotic susceptibility profiles were determined for the two women with different CRISPR type isolates as a supplementary phenotypic character. Various susceptibilities were observed for the two clones. Isolates from woman 8 were resistant to erythromycin and susceptible to lincomycin (inhibition zone diameters, 17 mm and 29 mm, respectively; erythromycin MIC, 1 mg/liter; clindamycin MIC, 0.125 mg/liter) for the major clone, whereas the minor clone was resistant to both erythromycin and lincomycin (inhibition zone diameter, 7 mm for both; erythromycin MIC, >256 mg/liter; clindamycin MIC, >256 mg/liter). Concerning woman 10, isolates were resistant to both erythromycin and lincomycin (inhibition zone diameters, 17 mm and 15 mm, respectively; erythromycin MIC, 4 mg/liter; clindamycin MIC, 0.125 mg/liter) for the major clone and susceptible to both erythromycin and lincomycin (inhibition zone diameters, 28 mm and 30 mm, respectively; erythromycin MIC, 0.064 mg/liter; clindamycin MIC, 0.064 mg/liter) for the minor clone.
Capsular typing and MLST.
Capsular typing and MLST were performed for each isolate showing a different CRISPR-RFLP pattern (Fig. 1). Among isolates belonging to the 12 different CRISPR types, 10 could not be distinguished by capsular typing (isolates from women 1, 2, 5, and 6 and the two CRISPR types isolated from woman 10 [type Ia]; from women 7 and 9 [type II] and the two CRISPR types isolated from woman 8 [type V]). Six could not be distinguished by MLST (isolates from women 1 and 5, which shared the same ST, ST23; isolates from woman 7 and from the minor clone from woman 10, sharing ST28, and the two CRISPR types isolated from woman 8 with the same ST, ST1). All isolates from a given woman presented the same capsular type and the same ST, except those from woman 10, which presented two different STs among all isolates tested (Fig. 1).
Most isolates belonged to capsular types Ia (five women, corresponding to 684 isolates) and II (two women, corresponding to 154 isolates). Capsular types Ib, III, and V, respectively, were isolated from the three other women, corresponding to 132 isolates. Concerning MLST analysis, isolates belonged to CC23 for three women (women 1 and 5 [ST23] and woman 6 [ST24], corresponding to 429 isolates), to CC10 for three women (women 2 [ST12], 3 [ST8], and 9 [ST569], corresponding to 272 isolates), to CC19 for three women (woman 7 and a minor clone from woman 10 [ST28] and woman 4 [ST19], corresponding to 157 isolates), and CC1 for two women (woman 8 [ST1] and a major clone from woman 10 [ST4], corresponding to 112 isolates).
DISCUSSION
The objective of our study was to explore GBS phylogenetic diversity in a vaginal population taken from pregnant women at the time of their screening, specifically at the end of pregnancy. Given the previous data on CRISPR typing analysis (21, 22), the strategy used was based on CRISPR array analysis. We developed an original approach to compare CRISPR composition based on a CRISPR-RFLP analysis to screen GBS isolates, followed by sequencing and analysis of the CRISPR array.
In our study, GBS carriage in pregnant women was divided into three categories based on CRISPR1 locus diversity. For one-half of the women (women 2, 4, 5, 6, and 7), all isolates shared the same CRISPR1 locus, demonstrating homogenous carriage.
In the second category, isolates shared a CRISPR1 locus with the same backbone (women 1, 3, and 9), but some isolates had a CRISPR array with some minor variations. These internal CRISPR1 locus modifications were spacer deletions (women 3 and 9) or duplications (woman 1). This phenomenon appeared rarely and was probably the result of slipped misalignment during DNA replication or homologous recombination (20, 22, 25). This phenomenon concerned up to 5% of isolates (woman 9), with various numbers of spacers (1 to 14 spacers) but never the spacers located at the CRISPR1 array extremities. Thus, the CRISPR locus appears to evolve in vivo mainly through the loss and acquisition of internal spacers. These first two categories consisted of the same type of CRISPR isolates, which probably belonged to the same clone. The same capsular type and the same ST also point to the fact that these were a single clone.
In the latter category, GBS isolates for women 8 and 10 displayed two different CRISPR types. Two clones coexisted with proportions of 65% to 35% (woman 8) and 74% to 26% (woman 10). In this group, the two clones from each woman shared similarities among ancestral spacers: four for woman 8 and one for woman 10. Moreover, for woman 8, the two clones detected shared the same serotype (serotype V) and the same ST (ST1), suggesting a common ancestor. For woman 10, the two clones belonged to two different STs (ST4 and ST28), though both were from serotype Ia. Although we analyzed isolates from a small number of women, we observed the favoring presence of a phylogenetic lineage between GBS clones carried simultaneously, as previously suggested by minor differences in pulsed-field gel electrophoresis (PFGE) patterns (≤2 bands) (26). We can hypothesize that a specific vaginal environment promoted the development of these lineages. To our knowledge, no link between phylogenetic group and vaginal environment has been described. However, the “cocarriage” of several clones might represent an advantage for improving adaptation and virulence by genetic exchange, even more so with closely related clones, which could promote horizontal gene transfer (27–30). Moreover, bacteriophages may modify the equilibrium between several clones, eventually leading to the elimination of some of them according to the S. agalactiae CRISPR-Cas system present in the phage-infected isolate. For women 8 and 10, the two clones presented phenotypic differences in hemolytic activity and antimicrobial susceptibilities (GBS isolates from woman 10) and in antimicrobial susceptibilities (GBS isolates from woman 8). We note that genetic diversity was not linked to richness of carriage. For woman 8, who carried two clones simultaneously, only 20 isolates were obtained on the agar plate, whereas 125 isolates were present for woman 10.
Although clonal diversity was limited for each individual person, it was high for the entire cohort. Each woman presented a “personal” profile of GBS carriage strains. Nevertheless, the study was conducted on just 10 women. In contrast, the previously described PFGE and randomly amplified polymorphic DNA (RAPD) analyses failed to distinguish profiles for some isolates from different women (26, 31, 32). In our study, the CRISPR-RFLP approach appears to be more discriminant than serotyping and MLST. Among the 12 different CRISPR types, 10 and 6 isolates could not be distinguished by capsular typing and MLST, respectively (Fig. 1).
The isolates studied belonged to five different capsular types (Ia, Ib, II, III, V) and to four CCs (CC23, CC10, CC19, and CC1). These serotypes are widely represented and account for more than 90% of GBS isolates worldwide (12, 33–36). Serotype Ia was particularly frequent (present in one-half of the women), whereas serotype III, associated with CC17, which is one of the most prevalent serotypes in carriage among pregnant women, was isolated for just one woman (37–39). This could be explained by the limited number of women studied and the selection of plates with intermediate to heavy carriage (at least 20 isolates), while serotype III isolates appear to be involved in “light” vaginal carriage mainly (37). Concerning MLST analysis and CC clustering, the isolates belonged to four CCs (CC23, CC10, CC19, and CC1), which are the predominant CCs among colonizing strains during pregnancy (35, 36). No ST17 strains (associated with serotype III) were isolated (12, 33, 34).
The three typing methods used in our study (capsular typing, MLST, and CRISPR typing) showed that the GBS population was relatively uniform in all the women examined, with the same capsular type for each carrier, the same ST for all but one, the specific CRISPR type for 8 of the 10 carriers, and no more than two clones per carrier. Previous typing approaches performed to evaluate GBS diversity in carriage have given similar results, with more than two-thirds of GBS carriers colonized by a single clone (same serotype [96%] and the same PFGE profile [86 to 90% in the same sample] with no more than three clones per carrier [26, 31, 40, 41]). Using RAPD analysis, a higher genotypic diversity per individual was reported (same genotype for 39% of carriers with up to four clones per carrier) (32). While the use of broth could explain part of this diversity, RAPD is now notoriously technically dependent and difficult to reproduce, which might explain part of this variability. These studies compared only a few selected isolates on agar plates from vaginal and rectal swabs (5 to 26 isolates) (26, 31, 40, 41). Our study reinforces previous results on the diversity of GBS carriage with the analysis of a large number of isolates per sample. Here, we considered all the isolates present on the agar plates; thus, we analyzed a higher number of colonies per woman (about 100 on average) than did previous studies. To more precisely characterize the genetic links between isolates, however, the whole-genome approach appears to be suitable. Although this approach remains expensive for large studies, it could be helpful in studying clones, especially those from women 8 and 10.
One of the difficulties of our approach was the choice of restriction enzymes because of the high polymorphism of the spacer sequences. The enzymes selected had to provide sufficient restriction to separate isolates but not so much as to be detectable. Here, the two enzymes selected separated the considered CRISPR array into three to six fragments. However, two restriction sites were located in conserved CRISPR flanking regions and did not provide supplemental information about locus composition. The use of two different enzymes improved the discriminatory power of this approach while remaining useful and affordable.
In routine clinical laboratory practice, bacteriological analyses such as antibiotic sensitivity testing or typing are often performed on a single or a very small number of isolates. The isolates selected, however, may not represent the entire population, as several clones may coexist. For two women studied, two clones coexisted and shared paired differences in antibiotic susceptibility. Although the two clones remained susceptible to beta-lactam antibiotics, which are the first-line antibiotics, they had different susceptibilities to the macrolides and lincosamides tested, which are used as alternative drugs for patients allergic to penicillin.
Enzymatic restriction makes it possible to perform rapid comparisons of locus CRISPR compositions for large-scale studies. Applied to CRISPR loci, this is a good screening method before CRISPR sequencing for comparing isolates and can be used to separate isolates belonging to the same ST. Vaginal carriage from pregnant women is relatively homogenous, with a single clone for 8 of the 10 women and at least two closely related clones simultaneously. Along with possibly being a source of selection for resistant strains, cocarriage of GBS clones may contribute to increasing bacterial virulence by horizontal transfer. Further studies are needed to expand our results to a larger number of pregnant women. Moreover, long-term follow-up of GBS carriage and the interaction between different clones or between very close strains is also an interesting question for understanding the coexistence of bacterial populations in vivo. Monitoring based on CRISPR array analysis may be a useful tool for deciphering these interactions.
ACKNOWLEDGMENTS
We are most grateful to P. Horvath for providing the macro-enabled Excel tool for CRISPR analysis and for fruitful discussions. We are most grateful to P. Glaser for providing the initial spacer dictionary. We also thank the Institut de Biologie Intégrative de la Cellule (I2BC) for maintaining the CRISPR's web server (http://crispr.i2bc.paris-saclay.fr) and especially Christine Pourcel.
REFERENCES
- 1.Edmond KM, Kortsalioudaki C, Scott S, Schrag SJ, Zaidi AK, Cousens S, Heath PT. 2012. Group B streptococcal disease in infants aged younger than 3 months: systematic review and meta-analysis. Lancet 379:547–556. doi: 10.1016/S0140-6736(11)61651-6. [DOI] [PubMed] [Google Scholar]
- 2.Agence Nationale d'Accréditation et d'Évaluation en Santé. 2001. Antenatal prevention of early neonatal bacterial infection (September 2001). https://www.has-sante.fr/portail/upload/docs/application/pdf/Antenatal_prevention.pdf. [PubMed]
- 3.Schrag S, Gorwitz R, Fultz-Butts K, Schuchat A. 2002. Prevention of perinatal group B streptococcal disease. Revised guidelines from CDC. MMWR Recomm Rep 51(RR-11):1–22. [PubMed] [Google Scholar]
- 4.Regan JA, Klebanoff MA, Nugent RP. 1991. The epidemiology of group B streptococcal colonization in pregnancy. Vaginal Infections and Prematurity Study Group. Obstet Gynecol 77:604–610. [PubMed] [Google Scholar]
- 5.Stoll BJ, Schuchat A. 1998. Maternal carriage of group B Streptococci in developing countries. Pediatr Infect Dis J 17:499–503. doi: 10.1097/00006454-199806000-00013. [DOI] [PubMed] [Google Scholar]
- 6.Campbell JR, Hillier SL, Krohn MA, Ferrieri P, Zaleznik DF, Baker CJ. 2000. Group B streptococcal colonization and serotype-specific immunity in pregnant women at delivery. Obstet Gynecol 96:498–503. [DOI] [PubMed] [Google Scholar]
- 7.Cieslewicz MJ, Chaffin D, Glusman G, Kasper D, Madan A, Rodrigues S, Fahey J, Wessels MR, Rubens CE. 2005. Structural and genetic diversity of group B Streptococcus capsular polysaccharides. Infect Immun 73:3096–3103. doi: 10.1128/IAI.73.5.3096-3103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slotved H-C, Kong F, Lambertsen L, Sauer S, Gilbert GL. 2007. Serotype IX, a proposed new Streptococcus agalactiae serotype. J Clin Microbiol 45:2929–2936. doi: 10.1128/JCM.00117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuchat A. 1998. Epidemiology of group B streptococcal disease in the United States: shifting paradigms. Clin Microbiol Rev 11:497–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poyart C, Réglier-Poupet H, Tazi A, Billoët A, Dmytruk N, Bidet P, Bingen E, Raymond J, Trieu-Cuot P. 2008. Invasive group B streptococcal infections in infants, France. Emerg Infect Dis 14:1647–1649. doi: 10.3201/eid1410.080185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haguenoer E, Baty G, Pourcel C, Lartigue M-F, Domelier A-S, Rosenau A, Quentin R, Mereghetti L, Lanotte P. 2011. A multi locus variable number of tandem repeat analysis (MLVA) scheme for Streptococcus agalactiae genotyping. BMC Microbiol 11:171. doi: 10.1186/1471-2180-11-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones N, Bohnsack JF, Takahashi S, Oliver KA, Chan M-S, Kunst F, Glaser P, Rusniok C, Crook DWM, Harding RM, Bisharat N, Spratt BG. 2003. Multilocus sequence typing system for group B Streptococcus. J Clin Microbiol 41:2530–2536. doi: 10.1128/JCM.41.6.2530-2536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radtke A, Lindstedt B-A, Afset JE, Bergh K. 2010. Rapid multiple-locus variant-repeat assay (MLVA) for genotyping of Streptococcus agalactiae. J Clin Microbiol 48:2502–2508. doi: 10.1128/JCM.00234-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 15.Grissa I, Vergnaud G, Pourcel C. 2009. Clustered regularly interspaced short palindromic repeats (CRISPRs) for the genotyping of bacterial pathogens. Methods Mol Biol 551:105–116. doi: 10.1007/978-1-60327-999-4_9. [DOI] [PubMed] [Google Scholar]
- 16.Shariat N, Dudley EG. 2014. CRISPRs: molecular signatures used for pathogen subtyping. Appl Environ Microbiol 80:430–439. doi: 10.1128/AEM.02790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou R, Dudley EG. 2016. CRISPR-based typing and next-generation tracking technologies. Annu Rev Food Sci Technol 7:395–411. doi: 10.1146/annurev-food-022814-015729. [DOI] [PubMed] [Google Scholar]
- 18.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 19.Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, de Romans S, Lim C, Roux C, Passet V, Diancourt L, Guibourdenche M, Issenhuth-Jeanjean S, Achtman M, Brisse S, Sola C, Weill F-X. 2012. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7:e36995. doi: 10.1371/journal.pone.0036995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Sanchez M-J, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P. 2012. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol 85:1057–1071. doi: 10.1111/j.1365-2958.2012.08172.x. [DOI] [PubMed] [Google Scholar]
- 21.Lier C, Baticle E, Horvath P, Haguenoer E, Valentin A-S, Glaser P, Mereghetti L, Lanotte P. 2015. Analysis of the type II-A CRISPR-Cas system of Streptococcus agalactiae reveals distinctive features according to genetic lineages. Front Genet 6:214. doi: 10.3389/fgene.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beauruelle C, Pastuszka A, Horvath P, Perrotin F, Mereghetti L, Lanotte P. 2017. CRISPR: a useful genetic feature to follow vaginal carriage of group B Streptococcus. Front Microbiol 8:1981. doi: 10.3389/fmicb.2017.01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imperi M, Pataracchia M, Alfarone G, Baldassarri L, Orefici G, Creti R. 2010. A multiplex PCR assay for the direct identification of the capsular type (Ia to IX) of Streptococcus agalactiae. J Microbiol Methods 80:212–214. doi: 10.1016/j.mimet.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Brochet M, Rusniok C, Couvé E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc Natl Acad Sci U S A 105:15961–15966. doi: 10.1073/pnas.0803654105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Ruiz M, Rodríguez-Granger JM, Bautista-Marín MF, Romero-Noguera J, Rosa-Fraile M. 2004. Genetic diversity of Streptococcus agalactiae strains colonizing the same pregnant woman. Epidemiol Infect 132:375–378. doi: 10.1017/S0950268803001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen P, Huang HV. 1986. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics 112:441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majewski J, Zawadzki P, Pickerill P, Cohan FM, Dowson CG. 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J Bacteriol 182:1016–1023. doi: 10.1128/JB.182.4.1016-1023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salloum M, van der Mee-Marquet N, Valentin-Domelier A-S, Quentin R. 2011. Diversity of prophage DNA regions of Streptococcus agalactiae clonal lineages from adults and neonates with invasive infectious disease. PLoS One 6:e20256. doi: 10.1371/journal.pone.0020256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Domelier A-S, van der Mee-Marquet N, Sizaret P-Y, Hery-Arnaud G, Lartigue M-F, Mereghetti L, Quentin R. 2009. Molecular characterization and lytic activities of Streptococcus agalactiae bacteriophages and determination of lysogenic-strain features. J Bacteriol 191:4776–4785. doi: 10.1128/JB.00426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen SM, Uldbjerg N, Kilian M, Sorensen UBS. 2004. Dynamics of Streptococcus agalactiae colonization in women during and after pregnancy and in their infants. J Clin Microbiol 42:83–89. doi: 10.1128/JCM.42.1.83-89.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El Aila N, Tency I, Claeys G, Saerens B, De Backer E, Temmerman M, Verhelst R, Vaneechoutte M. 2009. Genotyping of Streptococcus agalactiae (group B Streptococci) isolated from vaginal and rectal swabs of women at 35-37 weeks of pregnancy. BMC Infect Dis 9:153. doi: 10.1186/1471-2334-9-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohnsack JF, Whiting A, Gottschalk M, Dunn DM, Weiss R, Azimi PH, Philips JB, Weisman LE, Rhoads GG, Lin F-YC. 2008. Population structure of invasive and colonizing strains of Streptococcus agalactiae from neonates of six U.S. academic centers from 1995 to 1999. J Clin Microbiol 46:1285–1291. doi: 10.1128/JCM.02105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber CA, McOdimba F, Pflueger V, Daubenberger CA, Revathi G. 2011. Characterization of invasive and colonizing isolates of Streptococcus agalactiae in East African adults. J Clin Microbiol 49:3652–3655. doi: 10.1128/JCM.01288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manning SD, Lewis MA, Springman AC, Lehotzky E, Whittam TS, Davies HD. 2008. Genotypic diversity and serotype distribution of group B Streptococcus isolated from women before and after delivery. Clin Infect Dis 46:1829–1837. doi: 10.1086/588296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones N, Oliver KA, Barry J, Harding RM, Bisharat N, Spratt BG, Peto T, Crook DW, Consortium OGBS. 2006. Enhanced invasiveness of bovine-derived neonatal sequence type 17 group B Streptococcus is independent of capsular serotype. Clin Infect Dis 42:915–924. doi: 10.1086/500324. [DOI] [PubMed] [Google Scholar]
- 37.van der Mee-Marquet N, Jouannet C, Domelier A-S, Arnault L, Lartigue M-F, Quentin R. 2009. Genetic diversity of Streptococcus agalactiae strains and density of vaginal carriage. J Med Microbiol 58:169–173. doi: 10.1099/jmm.0.005827-0. [DOI] [PubMed] [Google Scholar]
- 38.Jones N. 2006. Carriage of group B Streptococcus in pregnant women from Oxford, UK. J Clin Pathol 59:363–366. doi: 10.1136/jcp.2005.029058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunze M, Ziegler A, Fluegge K, Hentschel R, Proempeler H, Berner R. 2011. Colonization, serotypes and transmission rates of group B Streptococci in pregnant women and their infants born at a single University Center in Germany. J Perinat Med 39:417–422. doi: 10.1515/JPM.2011.037. [DOI] [PubMed] [Google Scholar]
- 40.Anthony BF, Eisenstadt R, Carter J, Kim KS, Hobel CJ. 1981. Genital and intestinal carriage of group B streptococci during pregnancy. J Infect Dis 143:761–766. doi: 10.1093/infdis/143.6.761. [DOI] [PubMed] [Google Scholar]
- 41.Brzychczy-Włoch M, Pabian W, Majewska E, Zuk MG, Kielbik J, Gosiewski T, Bulanda MG. 2014. Dynamics of colonization with group B streptococci in relation to normal flora in women during subsequent trimesters of pregnancy. New Microbiol 37:307–319. [PubMed] [Google Scholar]