ABSTRACT
Rift Valley fever virus (RVFV) is a mosquito-borne, zoonotic virus that infects ruminants, including cattle, sheep, goats, camels, and buffalo. Multiplexing diagnostic assays that can simultaneously detect antibodies against multiple RVFV antigens offer a high-throughput test for disease surveillance and vaccine evaluations. We describe the improvement and evaluation of a previously developed fluorescence microsphere immunoassay (FMIA) for the detection of IgG and IgM antibodies against the RVFV glycoprotein (Gn) and the immunogenic nucleocapsid protein (Np). Well-characterized vaccinated and experimentally infected ruminant sera were used for the evaluation of the assay. Recombinant viral proteins were produced and then coupled to polystyrene magnetic beads for analysis using the Luminex MAGPIX system with xMAP technology. The FMIA was performed in parallel with virus neutralization tests. Our results revealed the highest median fluorescence intensity (MFI) values for the detection of IgG antibodies against RVFV Np, indicating that this antigen would be a good candidate for a screening assay. The Np and Gn targets could differentiate infected animals from animals vaccinated with a candidate subunit vaccine formulation based on the RVFV Gn and Gc proteins. The results presented in this report demonstrate that FMIA provides a rapid and robust serological diagnostic tool for the detection of antibodies against RVFV. The targets developed in this assay provide the basis for the development of a companion diagnostic test for an RVFV Gn/Gc subunit vaccine that is capable of differentiating infected from vaccinated animals (DIVA), as well as a multiplex serodiagnostic assay that can simultaneously screen for several ruminant diseases.
KEYWORDS: fluorescence microsphere immnoassay, Rift Valley fever virus, serology
INTRODUCTION
Rift Valley fever virus (RVFV) is an arthropod-borne, zoonotic virus (genus Phlebovirus, family Phenuiviridae) that is a significant threat to domestic ruminants and humans. The virus was first discovered in 1930 in Kenya's Great Rift Valley (1), and since then, outbreaks have been documented in other countries within Africa (2). In 2000, RVFV spread beyond Africa when outbreaks occurred in the Arabian Peninsula (3). Because of global climate changes that are expanding its vector distribution and growing international trade, the risk of RVFV introduction into countries where RVFV is not endemic is increasing (4). Thus, there is renewed demand for the development of safe, rapid, and accurate diagnostic assays.
Traditional serological methods for RVFV include complement fixation, hemagglutination inhibition, immunodiffusion, virus neutralization tests (VNTs), and enzyme-linked immunosorbent assays (ELISAs) (5). Currently, VNTs and ELISAs are the more commonly used tests for disease outbreaks and surveillance studies. VNTs are highly specific and are the current gold standard serological method (6). However, VNTs are labor- and time-intensive and thus rarely used as a screening tool. Additionally, unless a nonvirulent strain is used (7, 8), the assay must be performed in appropriate biocontainment. ELISAs offer a rapid and safe means to detect antibodies to RVFV in animals and humans. Several formats using whole-cell lysate or purified viral antigen have been developed and validated (9–13). However, the production of these reagents must be done within biocontainment facilities, resulting in a high production cost and the risk of incomplete virus inactivation. Recombinant proteins have been explored as a safe and easy antigen production method for detecting antibodies with ELISAs (14–20). RVFV recombinant nucleocapsid and glycoproteins are immunogenic and have been demonstrated to be strong targets for the serological detection of RVFV. Although ELISAs are sensitive and reliable screening tests, they can only test one analyte at a time.
While ELISAs detect only a single viral antigen, fluorescence microsphere immunoassays (FMIA) allow for simultaneous detection of antibodies to multiple pathogen targets with one sample. Additionally, antibodies to several antigens of a single virus can be detected, consequently increasing the specificity of the assay. The use of FMIA enables high-throughput analysis and uses a smaller sample volume (21, 22). The Luminex technology uses colored magnetic polystyrene beads that are covalently coupled to capture antigens that bind to analytes in liquid suspension. A charge-coupled device (CCD) camera identifies the color-coded beads and detects a secondary fluorescent conjugate that is attached to the target antibody in an indirect format (Fig. 1). Because of its multiplexing capability, the FMIA has been used for the differentiation of infected animals from vaccinated animals (DIVA) for diseases like avian influenza, foot-and-mouth disease, and West Nile virus (23–25). An RVFV bead-based suspension assay has been previously developed to differentiate vaccinated animals from nonvaccinated animals (26). However, there are no validated tests for the detection of RVFV antibodies that differentiate vaccinated from infected animals. The versatility and potentially improved diagnostic accuracy of the FMIA make it a promising alternative to traditional serological methods.
FIG 1.
The FMIA utilizes color-coded polystyrene microspheres that can be coated with protein. (A and B) The Luminex MAGPIX system has up to 50 spectrally distinct bead sets to allow for the simultaneous detection of multiple biological targets in a single sample. (C) Once the target of interest is captured, each bead is individually read using a CCD camera. (D) RVFV recombinant proteins are covalently coupled to the beads and incubated with sera for the detection of primary antibodies. A biotin-labeled antispecies secondary antibody followed by a fluorescent conjugate (streptavidin-phycoerythrin) is added to detect the presence of primary antibody.
Highly sensitive yet specific tests like the FMIA are critical for the rapid detection of emerging and zoonotic animal diseases. In this study, we optimized and evaluated the FMIA for the detection of RVFV antibodies against several antigen targets in sheep and cattle. The targets included the immunogenic nucleocapsid (Np), a truncated glycoprotein Gn (minus the transmembrane domain), the main virulence factor nonstructural protein NSs, and nonstructural protein NSm. The assay was compared side by side to the VNT. We also demonstrated the use of the FMIA as a DIVA-compliant platform for a candidate RVFV Gn/Gc subunit vaccine.
MATERIALS AND METHODS
Serum samples.
A total of 154 cattle and 272 sheep sera from prior experimental RVFV animal studies were used for the evaluation. These included sheep and cattle inoculated with vaccine strain MP12 (27) and wild-type strains Saudi Arabia 2001 (SA01) and Kenya 2006 (Ken06) (28, 29) and in the presence or absence of a candidate Gn/Gc subunit vaccine (30, 31). These sample sources are summarized in Table S1 in the supplemental material. All studies were conducted under biosafety level 3-enhanced conditions at the Biosecurity Research Institute (Manhattan, KS). Sera were heat inactivated and safety tested for further diagnostic evaluation at biosafety level 2 by using a modified method that successfully inactivated high-titer samples. Briefly, all sera were inactivated by adding 0.25% Tween 20 at a dilution of 1:10 to the serum and heating samples to 60°C in a water bath for 2 h as previously described (29).
Additional sera from an experimental study of an RVFV MP-12-NSm deletion vaccine candidate in sheep were tested. Briefly, a reverse genetics system using the MP12 strain produced a vaccine virus in which a portion of the NSm gene had been removed (arMP-12ΔNSm21/384) (32). A total of 119 sheep samples were tested for the evaluation.
RVFV challenge sera were obtained from an experimental challenge with wild-type Egyptian isolate ZH501 (33). Sheep and cattle were inoculated and kept up to 35 days postinoculation (dpi) to assess antibody production. A total of 4 sheep samples and 7 cattle samples were tested. These samples served as positive-control sera for the FMIA.
Sera from animals never exposed to RVFV were used as the negative serum set, which included 165 sheep and 325 cattle serum samples. All samples were obtained from animals born and raised in the United States. The sera were heat inactivated by following the protocol described above.
Production of recombinant proteins.
RVFV Gn was produced from the coding sequence taken from the RVFV MP12 strain (GenBank DQ380208) and modified to remove the transmembrane domain. The sequence was synthesized (IDT, Coralville, IA) and cloned into a pHUE expression vector (34) and then transformed into BL-21 Escherichia coli cells (New England Labs, Coralville, IA). Gene expression was induced with 1 mM isopropyl-β-thiogalactoside in LB culture medium with constant shaking for 4 h at 37°C. Bacterial cells were harvested by centrifugation and then purified using the PrepEase histidine-tagged protein purification high-yield midi kit (Affymetrix, USB Corp., Cleveland, OH) under denaturing conditions. Purified protein was then precipitated to remove urea and exchanged with 1× phosphate-buffered saline (PBS) and 0.1% SDS, as previously described (35). The precipitated protein was concentrated using Amicon Ultra 2-ml centrifugal filters (EMD Millipore Corporation, Billerica, MA) with a molecular size cutoff of 30 kDa. The protein concentration was measured by the Bradford protein assay (Bio-Rad, Hercules, CA). Purified protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10%) and stained with Coomassie brilliant blue (Bio-Rad, Hercules, CA).
Purified recombinant RVFV Np, NSm, and NSs proteins were produced in an E. coli expression system as previously described (36). Before use, proteins were analyzed by SDS-PAGE (10%) and stained with Coomassie brilliant blue (Bio-Rad, Hercules, CA) to visualize purity and integrity.
Non-RVFV recombinant proteins were used as negative-control coupled beads to account for nonspecific binding of antibodies. PCV2 was selected to represent a non-RVFV viral nucleocapsid protein that was expressed in an E. coli expression system using the pHUE vector and purified using the PrepEase histidine-tagged protein purification high-yield midi kit as described above under denaturing conditions. Purified protein was then analyzed by SDS-PAGE.
All proteins were analyzed by Western blot analysis to assess recognition of antigens by monoclonal and polyclonal antibodies. Briefly, 10 ng of each recombinant protein was separated by SDS gel electrophoresis and then transferred onto a polyvinylidene difluoride (PVDF) membrane by electroblotting using the TransBlot Turbo transfer pack on the TransBlot Turbo transfer system (Bio-Rad, Hercules, CA). The membranes were blocked with a protein-free blocking buffer (G-Biosciences, St. Louis, MO) at 4°C overnight with rocking. The membranes were washed three times using PBS with 0.1% Tween 20. To visualize the reactivity of proteins against ruminant sera, membranes were incubated with a strong positive sheep serum experimentally infected with the RVFV ZH501 strain (33) at a dilution of 1:100 for 1 h at room temperature. The cross-reactivity of proteins was also tested using negative lamb serum at a dilution of 1:100. After washing, rabbit anti-sheep IgG (H+L)–horseradish peroxidase (HRP) conjugate (Bio-Rad, Hercules, CA) at a dilution of 1:20,000 was applied for 1 h at room temperature. To visualize the reactivity of RVFV recombinant proteins against monoclonal antibodies, primary antibodies against Np and Gn were commercially acquired from Maine Biotechnology Services (MAB240P) (Portland, ME) and BEI Resources (NR43190) (Manassas, VA), respectively. All monoclonal antibodies were diluted 1:1,000 and incubated for 1 h at room temperature. After washing, a peroxidase AffiniPure donkey anti-mouse IgG (Jackson ImmunoResearch, Inc., West Grove, PA) secondary antibody was added at a 1:100,000 dilution for 1 h at room temperature. After a final wash, Clarity ECL blotting substrate (Bio-Rad, Hercules, CA) was added for 5 min for the detection of proteins. The membrane was visualized using the Chemidoc MP imaging system (Bio-Rad, Hercules, CA).
Conjugation of antigens to Luminex carboxylated beads.
Recombinant proteins were covalently coupled to MagPlex microspheres (Luminex Corp., Austin, TX) by using a two-step carbodiimide reaction as previously described (36). Briefly, 6.25 × 106 microsphere beads per target were activated and then incubated with their respective recombinant protein for 3 h at room temperature with rotation. Coupled bead sets were stored in blocking buffer (1% fish gelatin in PBS with 0.05% Tween 20 and 0.05% sodium azide, pH 7.4) overnight to reduce nonspecific binding of antibodies to the beads. The efficiency of the protein coupling reaction was confirmed using a mouse penta-His antibody, bovine serum albumin (BSA)-free (Qiagen, Germantown, MD), according to the bead manufacturer's instructions. Bead sets were stored at 4°C and used within 4 weeks to minimize signal loss due to protein degradation. To minimize photobleaching effects on the beads, all coupling reactions, storage, and use of beads were done in reduced lighting. The final concentration of each coupled bead set was manually counted using a hemacytometer.
Optimization.
The amount of RVFV protein coupled to the beads was optimized to identify the strongest signal against positive sera for each coupled bead set. The beads were coated and tested with various protein concentrations, including 12.5, 25, 50, and 100 μg/ml per antigen. The antigen concentration that provided the strongest signal-to-noise ratio for all targets was used for testing samples.
The FMIA platform was optimized to determine the ideal concentrations of sera and reagents. A well-characterized strong positive-control serum and a negative-control serum were serially diluted in a checkerboard fashion to optimize the signal-to-noise ratios. Serum, antibody, and fluorescent reporter dilutions were selected based on an ideal signal-to-noise ratio for both the Np and Gn bead targets.
A set of beads coupled to a non-RVFV viral recombinant antigen (PCV2 nucleocapsid) was included to account for nonspecific binding of antibodies to the recombinant antigens coupled to the beads. A bead set with no coupled antigen was included to account for background due to antibodies nonspecifically binding to the beads. Each bead set was tested in singleplex format and then in multiplex format to identify any cross-reactivity between multiplexing bead sets and detection antibodies.
FMIA.
Assays were run on the Luminex MAGPIX system (Luminex Corporation, Austin, TX). All sheep serum samples were diluted 1:400 in assay buffer (1% fish gelatin in PBS with 0.05% Tween 20 and 0.05% sodium azide, pH 7.4). All cattle serum samples were diluted 1:200 in assay buffer. Samples were transferred to a 96-well round-bottom polystyrene plate (Corning, Inc., Corning, NY) and plated in triplicate at 50 μl per well. Control sera, which included a strong positive, a medium positive, a low positive, and a negative serum sample, were added to each plate to serve as an internal quality control. An additional well with only the bead sets and assay buffer was added to each plate to calculate background signal. Each well received 50 μl assay buffer containing each bead set at a concentration of 2,500 total beads per antigen. Plates were covered with foil to minimize light exposure and then incubated for 30 min at room temperature on a plate shaker set at 800 rpm. Using a magnetic bead separator, the plates were washed three times using 190 μl of assay buffer per well. The antispecies secondary antibody was diluted in assay buffer and added at a volume of 50 μl per well, and the wells were incubated for 30 min. Ovine antibodies were detected with rabbit anti-sheep biotinylated IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted to 1 μg/ml and rabbit anti-sheep biotinylated IgM (MyBioSource, San Diego, CA) diluted to 1 μg/ml. Bovine antibodies were detected with goat anti-bovine biotinylated IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) diluted to 1 μg/ml. The plates were washed as described previously, and then 50 μl of the fluorescent conjugate streptavidin-phycoerythrin (SAPE) (Moss, Inc., Pasadena, MD) was diluted to 1 μg/ml and added to each well. The plates were incubated for another 30 min and then washed. The beads in each well were resuspended in 100 μl assay buffer, and the plate was analyzed on the MAPGIX system using xPONENT version 4.2 software (Luminex Corporation, Austin, TX). Analysis of fluorescence was set to measure 100 beads per bead set per well. Results were recorded as median fluorescence intensity (MFI). Background signal detected from the blank well containing only beads and assay buffer was subtracted from each sample well. The average MFI was calculated for each triplicate set and then converted to a sample/positive control (S/P) ratio to normalize the results across the plates. The S/P ratio was calculated by using the following formula: S/P = (mean MFI of test sample − mean MFI of negative control)/(mean MFI of strong positive control − mean MFI of negative control).
Virus neutralization test.
For validation of the FMIA, a plaque reduction neutralization test was performed as described previously (30). Briefly, RVFV MP12 virus was diluted to approximately 50 PFU per 250 μl in minimum essential medium (MEM) (Thermo Fisher Scientific, Grand Island, NY) containing 2% BSA (Sigma-Aldrich, St. Louis, MO). Sera were serially diluted 2-fold from 1:10 to 1:1,280 in MEM with 2% fetal bovine serum (FBS) and 1% penicillin-streptomycin-amphotericin B (Thermo Fisher Scientific, Grand Island, NY). The diluted sera were mixed at a 1:1 volume with diluted MP12 virus on a 96-well plate and then incubated for 1 h at 37°C. Virus-serum mixtures were then used to inoculate a confluent monolayer of Vero cells (ATCC CCL-81) on 12-well plates. The plates were incubated for 1 h at 37°C with gentle rocking every 15 min, an overlay with 1% methylcellulose (Sigma-Aldrich, St. Louis, MO) and MEM was then added to all wells, and the plates were incubated for 5 days at 37°C. The formation of plaques was visualized, and the plaques were counted after staining them with crystal violet fixative stain (0.5% crystal violet, 1% formaldehyde, ethanol, and glacial acetic acid in water) for 1 h at room temperature. Neutralizing antibody titers were determined using an 80% neutralization cutoff, which corresponds to the reciprocal titer of the highest serum dilution at which the number of plaques is reduced by ≥80% compared to that of the MP12 strain virus control. A sample was considered positive if there was a detectable titer at or above the dilution of 1:10 to indicate exposure to the virus.
Statistics.
Pearson's correlation coefficient was calculated to compare the singleplex and multiplex FMIA platforms and to compare the FMIA and VNT. Two-graph receiver operating characteristic (TG-ROC) analysis was used for determining the FMIA cutoff values for each target. Youden's index was used to calculate the diagnostic accuracy for each target by the following formula: J = [Sn + (Sp − 1)], where Sn is sensitivity and Sp is specificity. The Np (1) and Gn (2) targets were tested in series and in parallel using the following formulas: Sn (series) = Sn1 × Sn2, Sn (parallel) = 1 − [(1 − Sn1) × (1 − Sn2)], Sp (series) = 1 − [(1 − Sp1) × (1 − SP2)], Sp (parallel) = Sp1 × Sp2. A t test was used to analyze differences in antibody detection between three strains of RVFV used for experimental challenge. Analysis was done using Graph Pad Prism software (version 7.0) (Graphpad Software, Inc., La Jolla, CA).
RESULTS
Expression and immunogenicity of protein targets.
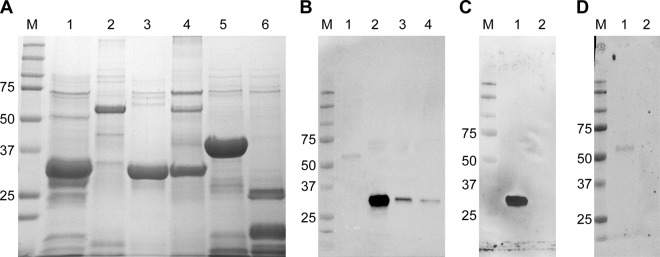
All FMIA panel proteins were produced, purified, and evaluated for antibody reactivity (Fig. 2). RVFV proteins were reactive against positive sheep serum at 28 days postinoculation (dpi) with the RVFV ZH501 strain. Np showed the strongest reactivity, followed by NSs, Gn, and, finally, NSm (Fig. 2B). Negative-control proteins were nonreactive. Negative lamb serum, tested against all proteins, was also nonreactive (data not shown). Further testing of Np and Gn using anti-Np and anti-Gn monoclonal antibodies revealed that anti-Np reacted strongly with the Np protein but not with Gn (Fig. 2C), while anti-Gn reacted with the Gn protein, and not with Np (Fig. 2D), albeit more weakly than anti-Np and Np. A similar examination of the NSs and NSm proteins was not possible due to the lack of working antibodies. Overall, these results demonstrate the antibody reactivity of target proteins to be used for the FMIA.
FIG 2.
RVFV and negative-control recombinant proteins used in FMIA. (A) All proteins were analyzed by SDS-PAGE to visualize their integrity. Lane M, molecular size marker (in kilodaltons); lane 1, RVFV Np (32 kDa); lane 2, RVFV Gn (54 kDa); lane 3, RVFV NSs (34 kDa); lane 4, RVFV NSm (30 kDa); lane 5, GFP (37 kDa); lane 6, PCV2 (22 kDa). (B) Western blot analyses were done to confirm the presence of the RVFV proteins. All four RVFV targets were probed with a positive sheep serum. Lane M, molecular size marker (in kilodaltons); lane 1, RVFV Gn; lane 2, RVFV Np; lane 3, RVFV NSs; lane 4, RVFV NSm. (C and D) RVFV proteins Np and Gn were further tested against monoclonal antibodies. (C) Monoclonal antibody MAB240P was used against Np. Lane 1, RVFV Np; lane 2, RVFV Gn. (D) Monoclonal antibody NR43190 was used against Gn. Lane 1, RVFV Gn; lane 2, RVFV Np.
FMIA optimization.
Optimization of concentrations of the RVFV Np and Gn proteins revealed that 25 μg/ml of each offered the strongest signal-to-noise ratio for both bead targets. When seeking the optimal serum dilution for detecting IgG and IgM antibodies, serial dilution of the sheep and cattle sera revealed a concentration-dependent MFI signal pattern (data not shown).
Initially, IgG antibody detection by FMIA in both sheep and cattle sera had high background signals. Therefore, subtraction of the porcine circovirus type 2 nucleocapsid (PCV2) and the blank (negative-control) beads' MFI signals for each sample accounted for the nonspecific binding of primary antibodies to the bead-coupled recombinant antigen and to the beads, respectively. These two adjustments afforded a better interpretation of the RVFV antigen targets detecting antibodies in the sera.
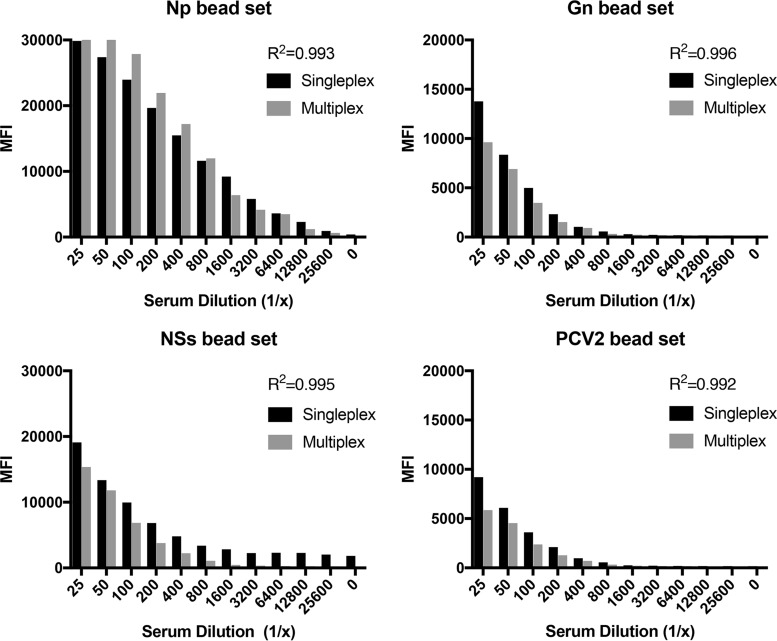
Each bead set was tested in singleplex and then in multiplex format against control sera to identify any cross-reactivity between bead sets (Fig. 3). The correlation coefficient for each bead set was high (R2 = 0.99), demonstrating minimal cross-reactivity between bead sets. There was no statistical difference between the singleplex and multiplex formats (P value > 0.05).
FIG 3.
Comparison of singleplex and multiplex formats of FMIA. Each bead set was tested in singleplex format and then in multiplex format against positive sheep sera. The correlation coefficient (R2) between the singleplex and multiplex formats is provided for each bead set.
The MFI signals for the NSs and NSm bead sets against ruminant sera were weaker than the MFI signals from the Np and Gn bead sets. Detection of antibodies against the NSs target was variable in known-positive samples, making NSs a poor target for a screening assay. MFI signals against the NSm target were at or below the assay cutoff in known-positive samples and by Western blotting. Additionally, the NSm recombinant protein was only weakly reactive against positive sera (Fig. 2B). Due to these issues, which require additional optimization, the NSs and NSm bead sets were excluded from further analysis, and evaluation of the FMIA focused on Np and Gn targets.
Humoral immune response detection by FMIA.
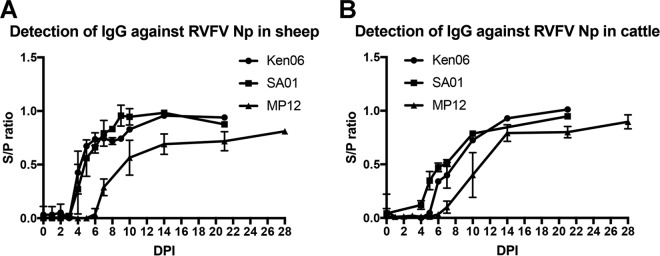
IgG antibody detection over time for the Np target was demonstrated using samples from experimental challenge studies with three RVFV strains: SA01, Ken06, and MP12. There were no detectable IgG responses for any of the targets to mock-challenged animal sera (data not shown). Sheep IgG antibodies were detected as early as 4 dpi for SA01 and Ken06 and 7 dpi for MP12 (Fig. 4A). Peak responses occurred by 10 dpi for SA01 and Ken06 and by 14 dpi for MP12. Cattle IgG antibodies were detected by 4 dpi for SA01, 5 dpi for Ken06, and 8 dpi for MP12 (Fig. 4B). Peak responses occurred at 21 dpi for SA01 and Ken06 and at 14 dpi for MP12. Regardless of species, the FMIA results for the Np target were similar for SA01 and Ken06 samples. While the MP12 vaccine strain samples consistently produced a weaker MFI signal, statistically the finding was not significant.
FIG 4.
Detection of IgG antibodies in serum with an RVFV Np bead set during RVFV infections with SA01, Ken06, and MP12 strains. Total IgG detection of all sheep and cattle are plotted by days postinoculation (DPI). (A) Antibody detection in sheep; (B) antibody detection in cattle.
As demonstrated with the Np target, the Gn target was also detected by IgG subtype antibodies in the challenge studies. In comparison to Np, the Gn target had an overall lower MFI signal in both species (data not shown). In sheep samples, detection of IgG antibody production occurred at 8 dpi for all strains. Ken06 had the strongest signal, with a peak response at 14 dpi. For SA01, the signal increased until 10 dpi and then decreased throughout the rest of the study. As seen with the Np target, MP12-induced antibodies overall showed the weakest signal of all the strains. In cattle samples, detection of IgG antibodies by the Gn target was variable and the background signal was higher. Overall, the Np target was more sensitive than the Gn target in the detection of IgG antibodies earlier in an infection time course (P value < 0.05).
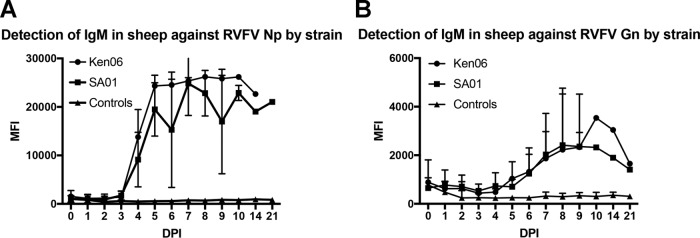
The detection of IgM antibodies against RVFV Np and Gn targets was similarly tested in sheep sera. IgM antibodies were detected against the Np target as early as 4 dpi for the SA01 and Ken06 RVFV strains (Fig. 5A). The peak MFI signal occurred by 7 dpi and then declined starting at 8 dpi for SA01 and 14 dpi for Ken06. As with the IgG detection, Ken06 had a higher MFI signal than SA01 at most time points, although the differences were not statistically significant. For the Gn target, a rise in IgM antibodies was detected at 6 dpi, with a peak signal by 9 dpi and then a decrease in signal by 21 dpi (Fig. 5B). Similar to IgG antibody detection, the detection of IgM by the Gn revealed an overall lower signal than that of the Np target. Cattle sera were not evaluated for IgM detection due to the high background noted in the IgG FMIA against the Gn target.
FIG 5.
Detection of IgM antibodies in sheep serum over the course of experimental RVFV infections with SA01 and Ken06. Total IgM detection of all sheep against the RVFV Np target (A) and Gn target (B) are plotted by days postinoculation (DPI). The controls are serum samples from sheep that were mock inoculated.
Comparison of FMIA with VNT.
A total of 518 sheep and 447 cattle samples were tested by the gold standard assay, VNT. A statistical comparison of the FMIA and VNT showed the two assays to be highly correlated (R2 = 0.89) when a comparison was made between the FMIA panel detecting IgG antibodies against the Np target for sheep inoculated with wild-type strains (Ken06, SA01) (Fig. 6A). Also, a good correlation (R2 = 0.71) was seen when the VNT was compared to the FMIA for detection of IgG antibodies against Gn (Fig. 6B). There was a good correlation (R2 = 0.91) when a comparison was made against the Np target with sheep inoculated with MP12, but the correlation was weaker in a comparison against the Gn target (R2 = 0.55).
FIG 6.
Comparison of virus neutralization test (VNT) to FMIA. FMIA results are represented on the y axis as an S/P ratio; VNT results are represented on the x axis as log 2 of serum dilutions. Correlation coefficients (R2 values) between VNT and FMIA were determined from the wild-type RVFV strains (Ken06, SA01). (A) Correlation between the FMIA Np target with VNT (P value < 0.05); (B) correlation between the FMIA Gn target with VNT (P value < 0.05).
Diagnostic accuracy.
Cutoff MFI values for the Np and Gn bead targets were determined by two-graph receiver operating characteristic (TG-ROC) analysis. All samples were classified positive or negative by VNT results. The Youden's index (YI) was used to determine the “optimal” cutoff value for which sensitivity (Sn) + [specificity (Sp) − 1] is maximized (37). Table 1 summarizes the cutoffs, Sn, Sp, YI, and area under the curve (AUC) by each bead target in sheep and cattle. Table 1 also includes the Sn and Sp for the two targets (Np and Gn) when tested in series and in parallel using sheep samples.
TABLE 1.
Diagnostic accuracy of RVFV FMIA for sheep and cattle serum samplesa
| Serum samples | Bead set | Cutoff (MFI) determined by TG-ROC analysis | % Sensitivity (95% CI) | % Specificity (95% CI) | YI (%) | AUC (%) |
|---|---|---|---|---|---|---|
| Cattle | Np | 2,500 | 94 (89–97) | 92 (88–95) | 85 | 95 |
| Gn | 3,800 | 89 (85–92) | 67 (59–75) | 56 | 83 | |
| Sheep | Np | 13,000 | 98 (97–99) | 97 (83–100) | 95 | 99 |
| Gn | 9,400 | 99 (97–99) | 51 (37–65) | 50 | 74 | |
| Series (sheep) | 83 | 98 | ||||
| Parallel (sheep) | 99 | 62 |
Disease status was categorized by VNT results. The cutoff values were determined by two-graph receiver operating characteristic (TG-ROC) analysis. CI, confidence interval; YI, Youden's index; AUC, area under the curve.
DIVA compatibility with candidate RVFV vaccines.
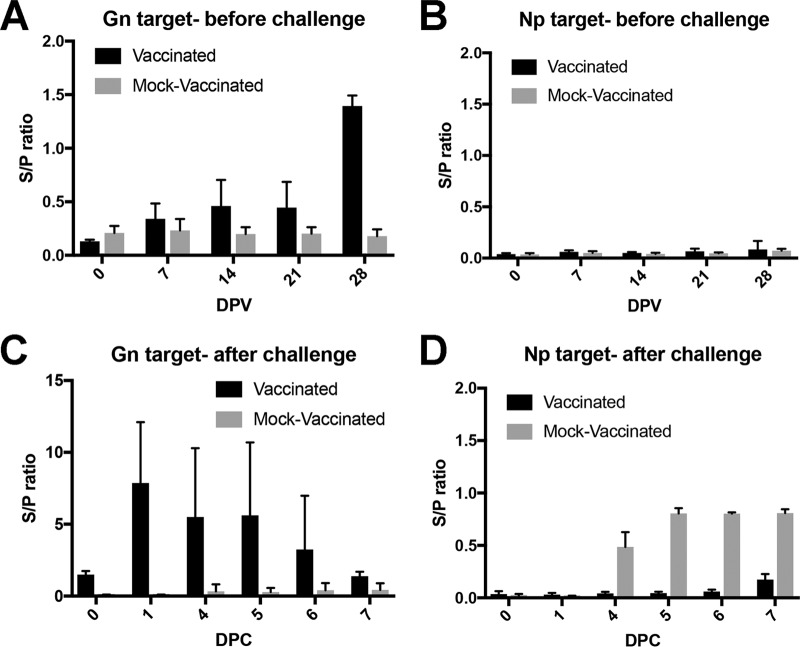
Sheep sera from a candidate Gn/Gc subunit vaccine study demonstrated the DIVA capability of the FMIA. The DIVA capability of the FMIA was not evaluated in cattle due to the lack of available Gn/Gc subunit vaccine cattle sera. In the sheep study, vaccinated and mock-vaccinated animals were readily separable (Fig. 7). The FMIA detected a rise in antibodies against Gn by 7 dpi in sera collected after the initial vaccination (Fig. 7A). After a booster vaccination at 21 dpi, there was a stronger MFI signal against the Gn target. No antibodies against the Np target were detected, as expected (Fig. 7B). Mock-inoculated animals had no rise in IgG antibodies against either the Gn or Np target. All animals were then challenged with a wild-type RVFV strain and monitored for 7 days. Sera from animals that were vaccinated had a higher MFI signal against the Gn target than did the mock-vaccinated animals (Fig. 7C) (P value < 0.05). IgG antibodies were detected against the Np target as early as 4 dpi in the mock-vaccinated animal sera, and the MFI values in these mock-vaccinated animals were higher than those in the vaccinated animals, which remained unchanged after the challenge (Fig. 7D) (P value < 0.05). This difference in observed MFI demonstrated the FMIA's DIVA capability.
FIG 7.
RVFV Np/Gn FMIA enables differentiation of vaccinated from infected sheep. Sheep were initially vaccinated with a Gn/Gc subunit vaccine and then challenged with Ken06 and maintained for 7 days. The subunit vaccine was given at 0 days postvaccination (DPV) and then boosted at 21 DPV. Detection of IgG antibodies in sera against the Gn target (A) and the Np target (B) is shown. At 35 DPV, all sheep were challenged (0 DPC). IgG production in sera was monitored using the Gn target (C) and the Np target (D). Np served as the DIVA-compatible marker.
DISCUSSION
FMIA is a growing technology that offers a versatile and rapid multiplexing platform for diagnostic testing and epidemiological studies. Yet there are limited reports about the use of FMIA in the detection of transboundary animal diseases (23, 26, 38–41). In this study, we demonstrate the ability of the FMIA to detect antibodies against RVFV recombinant antigens. We have previously developed the FMIA for RVFV Np, Gn, NSs, and NSm to detect antibodies in serum samples from experimental infection trials (36). However, during field trials, high background signal was noted in this assay. Subsequently, we modified and evaluated a new FMIA that focused on recombinant RVFV Np and Gn proteins as antigenic targets for the detection of IgG and IgM antibodies in experimentally infected ruminant sera.
Recombinant RVFV Np is an ideal diagnostic target for serological tests since it is considered the immunogenic protein that induces early production of antibodies during infection (16, 42). Our study found RVFV Np to be highly immunogenic, with strong MFI signals for detecting both IgM and IgG antibodies. Diagnostic accuracy was high with sheep serum samples and even higher with cattle serum samples. There was a stronger correlation (R2 = 0.89) between the FMIA Np target and the gold standard VNT assay than between Np-based ELISAs and VNTs (17). Lastly, the timing at which antibodies against Np were detected by FMIA supports what has been seen with recombinant Np-based ELISAs (15–17). Furthermore, the Np target detected antibodies earlier during infection than the secondary Gn target in the FMIA. Therefore, the Np target is ideal for early disease screening of samples by FMIA.
RVFV glycoproteins are also important antigen targets, as they are the components of RVFV that are most exposed to the immune system during infection (43). The FMIA results for the Gn target when using sheep sera were comparable to the results of a Gn-based IgG ELISA (19). The FMIA Gn target correlation to VNT was weaker than that for Np but still reasonable. This difference was due to the VNT detecting neutralizing antibodies earlier in the infection than were detected by the FMIA Gn target, resulting in a higher false-negative rate. It should be noted that the VNT titers for most of the Gn FMIA-negative samples were borderline positive (<1:40). The Gn bead set could not be fully evaluated for cattle samples due to a high background signal, which has been previously noted in other serological assays when using cattle sera (D. S. McVey, personal communication). Future development and optimization of smaller Gn peptides with critical epitopes for antibody binding may reduce cross-reactivity of antibodies and offer a simpler way to produce antigen targets versus whole recombinant proteins (44, 45). Additionally, expression of Gn in a nonbacterial system may reduce cross-reactivity from bacterial contaminants as well as support posttranslational protein modification that may improve antigenicity, as was seen in the report of van der Wal et al., in which the Gn was more antigenic than the Np (26, 46).
When the Np target and the Gn target were tested in singleplex, the Np target had high Sn and Sp, while the Gn target had high Sn but poor Sp. The YI and AUC values also demonstrate the higher accuracy of the Np target than of the Gn. Most serum samples tested were collected at times of early antibody production, when antibodies against Np are present but it may be too early for antibodies against Gn. Additional sera from later time points need to be tested to better evaluate the Gn target. Overall, the cattle FMIA panel had higher MFI signals than the sheep FMIA panel, which resulted in higher assay cutoff values. Addressing the background issues with cattle sera may reduce the cutoff values to a level comparable to that of the sheep assay.
To evaluate the FMIA in multiplex, the Np and Gn targets were tested in parallel and in a series. The analysis was done only with sheep sera due to the high background signal observed with cattle sera. When testing targets in a series, the Sp improved to 98%, in comparison to the Sp of singleplex testing (92% for Np, 67% for Gn). When testing targets in parallel, the Sn improved to 99%, in comparison to that of singleplex testing (94% for Np, 89% for Gn). However, the Sp is greatly reduced in parallel testing. When testing the targets in multiplex, testing in a series will offer a highly specific assay that is ideal for confirmatory testing.
RVFV Np and Gn are important as DIVA-compatible targets for a candidate RVFV glycoprotein-based vaccine. Recently, a candidate Newcastle disease virus vaccine expressing RVFV glycoproteins was used to evaluate a bead-based suspension array for differentiating vaccinated and nonvaccinated animals (26). Our study demonstrated that the FMIA is DIVA compatible with a candidate RVFV Gn/Gc subunit vaccine. Additionally, we were able to differentiate vaccinated and nonvaccinated animals prechallenge. DIVA vaccines and companion tests are important tools in control and eradication programs of high-consequence animal diseases. Further evaluation of our FMIA with other DIVA-compatible RVFV vaccines such as NSm deletion vaccine candidates is under way (32).
With several candidate NSm and NSs deletion RVFV vaccines being evaluated, RVFV NSm and NSs are valuable diagnostic targets for companion DIVA assays. NSm serves as a DIVA marker in some recombinant RVFV vaccine candidates (47–50). However, NSm was a poor antigenic target in the FMIA due to a weak antibody response compared to that of Np and Gn. Additional work may further optimize a recombinant NSm that could be used as an effective antigenic target on the FMIA panel; however, the host response to NSm may be too limited to be useful. Several NSs deletion vaccines are being developed; thus, this protein could be used as a DIVA-compatible marker (47, 50–52). Recombinant NSs was used as a target in the FMIA, and strong MFI signals were detected to antibodies against NSs. However, the antibody response was variable between infected animals, similar to what has been reported with NSs-based ELISAs (53). Therefore, NSs could be used as a confirmatory RVFV target but not a reliable DIVA marker, except on the herd level. Further investigation is needed to understand the mechanism of the observed variable antibody responses of less immunogenic RVFV antigens to develop a diagnostic assay that can be applied for testing on the individual animal level.
The FMIA is expected to be more sensitive than traditional solid-phase assays like ELISAs because of the freedom of the antibodies to bind to epitopes in suspension (21). However, current antigen targets demonstrate slightly reduced sensitivity. This may be due to the coupling of antigen to beads interfering with the antibodies that are accessing epitopes. Another factor may be the potential inactivation of epitopes during the carbodiimide coupling reaction (54). Introduction of spacers near epitopes of interest would enhance the ability of the protein targets to detect antibodies and improve antigen orientation. Evaluation of shorter peptides for RVFV should be done to identify key epitopes that can be used for the FMIA.
In this study, we evaluated a multiplexing serological assay using recombinant RVFV Np and Gn. The initial background signal seen during field trials has since been resolved, and future evaluation of the assay will include additional field samples. The results from this study demonstrate that the FMIA offers a high-throughput and versatile multiplexing tool for animal disease surveillance. This assay can be used for routine serological testing in countries where RVFV is endemic or can be incorporated into a foreign animal disease early warning strategy in countries where RVFV is not endemic. The RVFV Np and Gn antigen targets could be combined into a larger multiplexing panel for screening several ruminant diseases, such as bluetongue disease, Wesselsbron disease, peste des petits ruminants, and diseases caused by other abortive pathogens. Furthermore, the FMIA can be used as a DIVA-compatible assay for candidate RVFV subunit and recombinant vaccines.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Kansas State NBAF Transition Funds, Kansas State University College of Veterinary Medicine, the USDA Agricultural Research Service under project number 3020-32000-009-00D, and the U.S. Department of Homeland Security under grant award number DHS-2010-ST-061-AG0001.
RVFV MP-12-NSm deletion vaccine study sera were kindly provided by John Morrill, University of Texas Medical Branch, through Doug Watts, University of Texas at El Paso. RVFV-positive sera were provided by Hana Weingartl, National Centre for Foreign Animal Disease, Canadian Food Inspection Agency, Winnipeg, Manitoba, Canada. We thank Kathleen Yeater, USDA, ARS Plains Area, for assistance with the statistics. We thank Maryka Smith (KSU) for technical support and Mal Rooks Hoover (KSU) for assistance with medical illustration.
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Department of Homeland Security or U.S. Department of Agriculture.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01626-17.
REFERENCES
- 1.Daubney R, Hudson JR. 1931. Enzootic hepatitis or Rift Valley fever. An undescribed virus disease in sheep cattle and man from East Africa. J Pathol Bacteriol 34:545–579. [Google Scholar]
- 2.Nanyingi MO, Munyua P, Kiama SG, Muchemi GM, Thumbi SM, Bitek AO, Bett B, Muriithi RM, Njenga MK. 2015. A systematic review of Rift Valley Fever epidemiology 1931-2014. Infect Ecol Epidemiol 5:28024. doi: 10.3402/iee.v5.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balkhy HH, Memish ZA. 2003. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int J Antimicrob Agents 21:153–157. doi: 10.1016/S0924-8579(02)00295-9. [DOI] [PubMed] [Google Scholar]
- 4.Rolin AI, Berrang-Ford L, Kulkarni MA. 2013. The risk of Rift Valley fever virus introduction and establishment in the United States and European Union. Emerg Microbes Infect 2:e81. doi: 10.1038/emi.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swanepoel R, Struthers JK, Erasmus MJ, Shepherd SP, McGillivray GM, Erasmus BJ, Barnard BJ. 1986. Comparison of techniques for demonstrating antibodies to Rift Valley fever virus. J Hyg (Lond) 97:317–329. doi: 10.1017/S0022172400065414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.OIE. 2016. OIE terrestrial manual, chapter 2.1.18. OIE, Paris, France: http://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/2.01.18_RVF.pdf. [Google Scholar]
- 7.Wichgers Schreur PJ, van Keulen L, Kant J, Kortekaas J. 2017. Four-segmented Rift Valley fever virus-based vaccines can be applied safely in ewes during pregnancy. Vaccine 35:3123–3128. doi: 10.1016/j.vaccine.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 8.Caplen H, Peters CJ, Bishop DH. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol 66:2271–2277. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- 9.Niklasson B, Peters CJ, Grandien M, Wood O. 1984. Detection of human immunoglobulins G and M antibodies to Rift Valley fever virus by enzyme-linked immunosorbent assay. J Clin Microbiol 19:225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paweska JT, Mortimer E, Leman PA, Swanepoel R. 2005. An inhibition enzyme-linked immunosorbent assay for the detection of antibody to Rift Valley fever virus in humans, domestic and wild ruminants. J Virol Methods 127:10–18. doi: 10.1016/j.jviromet.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 11.Paweska JT, Burt FJ, Swanepoel R. 2005. Validation of IgG-sandwich and IgM-capture ELISA for the detection of antibody to Rift Valley fever virus in humans. J Virol Methods 124:173–181. doi: 10.1016/j.jviromet.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Paweska JT, Burt FJ, Anthony F, Smith SJ, Grobbelaar AA, Croft JE, Ksiazek TG, Swanepoel R. 2003. IgG-sandwich and IgM-capture enzyme-linked immunosorbent assay for the detection of antibody to Rift Valley fever virus in domestic ruminants. J Virol Methods 113:103–112. doi: 10.1016/S0166-0934(03)00228-3. [DOI] [PubMed] [Google Scholar]
- 13.Paweska JT, Smith SJ, Wright IM, Williams R, Cohen AS, Van Dijk AA, Grobbelaar AA, Croft JE, Swanepoel R, Gerdes GH. 2003. Indirect enzyme-linked immunosorbent assay for the detection of antibody against Rift Valley fever virus in domestic and wild ruminant sera. Onderstepoort J Vet Res 70:49–64. [PubMed] [Google Scholar]
- 14.McElroy AK, Albariño CG, Nichol ST. 2009. Development of a RVFV ELISA that can distinguish infected from vaccinated animals. Virol J 6:125. doi: 10.1186/1743-422X-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jansen van Vuren P, Potgieter AC, Paweska JT, van Dijk AA. 2007. Preparation and evaluation of a recombinant Rift Valley fever virus N protein for the detection of IgG and IgM antibodies in humans and animals by indirect ELISA. J Virol Methods 140:106–114. doi: 10.1016/j.jviromet.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Fafetine JM, Tijhaar E, Paweska JT, Neves LC, Hendriks J, Swanepoel R, Coetzer JA, Egberink HF, Rutten VP. 2007. Cloning and expression of Rift Valley fever virus nucleocapsid (N) protein and evaluation of a N-protein based indirect ELISA for the detection of specific IgG and IgM antibodies in domestic ruminants. Vet Microbiol 121:29–38. doi: 10.1016/j.vetmic.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 17.Paweska JT, van Vuren PJ, Kemp A, Buss P, Bengis RG, Gakuya F, Breiman RF, Njenga MK, Swanepoel R. 2008. Recombinant nucleocapsid-based ELISA for detection of IgG antibody to Rift Valley fever virus in African buffalo. Vet Microbiol 127:21–28. doi: 10.1016/j.vetmic.2007.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Williams R, Ellis CE, Smith SJ, Potgieter CA, Wallace D, Mareledwane VE, Majiwa PA. 2011. Validation of an IgM antibody capture ELISA based on a recombinant nucleoprotein for identification of domestic ruminants infected with Rift Valley fever virus. J Virol Methods 177:140–146. doi: 10.1016/j.jviromet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Jäckel S, Eiden M, Balkema-Buschmann A, Ziller M, van Vuren PJ, Paweska JT, Groschup MH. 2013. A novel indirect ELISA based on glycoprotein Gn for the detection of IgG antibodies against Rift Valley fever virus in small ruminants. Res Vet Sci 95:725–730. doi: 10.1016/j.rvsc.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 20.Jäckel S, Eiden M, Dauber M, Balkema-Buschmann A, Brun A, Groschup MH. 2014. Generation and application of monoclonal antibodies against Rift Valley fever virus nucleocapsid protein NP and glycoproteins Gn and Gc. Arch Virol 159:535–546. doi: 10.1007/s00705-013-1867-4. [DOI] [PubMed] [Google Scholar]
- 21.Christopher-Hennings J, Araujo KP, Souza CJ, Fang Y, Lawson S, Nelson EA, Clement T, Dunn M, Lunney JK. 2013. Opportunities for bead-based multiplex assays in veterinary diagnostic laboratories. J Vet Diagn Invest 25:671–691. doi: 10.1177/1040638713507256. [DOI] [PubMed] [Google Scholar]
- 22.Elshal MF, McCoy JP. 2006. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods 38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clavijo A, Hole K, Li M, Collignon B. 2006. Simultaneous detection of antibodies to foot-and-mouth disease non-structural proteins 3ABC, 3D, 3A and 3B by a multiplexed Luminex assay to differentiate infected from vaccinated cattle. Vaccine 24:1693–1704. doi: 10.1016/j.vaccine.2005.09.057. [DOI] [PubMed] [Google Scholar]
- 24.Balasuriya UB, Shi PY, Wong SJ, Demarest VL, Gardner IA, Hullinger PJ, Ferraro GL, Boone JD, De Cino CL, Glaser AL, Renshaw RW, Ledizet M, Koski RA, MacLachlan NJ. 2006. Detection of antibodies to West Nile virus in equine sera using microsphere immunoassay. J Vet Diagn Invest 18:392–395. doi: 10.1177/104063870601800413. [DOI] [PubMed] [Google Scholar]
- 25.Watson DS, Reddy SM, Brahmakshatriya V, Lupiani B. 2009. A multiplexed immunoassay for detection of antibodies against avian influenza virus. J Immunol Methods 340:123–131. doi: 10.1016/j.jim.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 26.van der Wal FJ, Achterberg RP, de Boer SM, Boshra H, Brun A, Maassen CB, Kortekaas J. 2012. Bead-based suspension array for simultaneous detection of antibodies against the Rift Valley fever virus nucleocapsid and Gn glycoprotein. J Virol Methods 183:99–105. doi: 10.1016/j.jviromet.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Wilson WC, Bawa B, Drolet BS, Lehiy C, Faburay B, Jasperson DC, Reister L, Gaudreault NN, Carlson J, Ma W, Morozov I, McVey DS, Richt JA. 2014. Evaluation of lamb and calf responses to Rift Valley fever MP-12 vaccination. Vet Microbiol 172:44–50. doi: 10.1016/j.vetmic.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Faburay B, Gaudreault NN, Liu Q, Davis AS, Shivanna V, Sunwoo SY, Lang Y, Morozov I, Ruder M, Drolet B, Scott McVey D, Ma W, Wilson W, Richt JA. 2016. Development of a sheep challenge model for Rift Valley fever. Virology 489:128–140. doi: 10.1016/j.virol.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 29.Wilson WC, Davis AS, Gaudreault NN, Faburay B, Trujillo JD, Shivanna V, Sunwoo SY, Balogh A, Endalew A, Ma W, Drolet BS, Ruder MG, Morozov I, McVey DS, Richt JA. 2016. Experimental infection of calves by two genetically-distinct strains of Rift Valley fever virus. Viruses 8:E145. doi: 10.3390/v8050145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faburay B, Lebedev M, McVey DS, Wilson W, Morozov I, Young A, Richt JA. 2014. A glycoprotein subunit vaccine elicits a strong Rift Valley fever virus neutralizing antibody response in sheep. Vector Borne Zoonotic Dis 14:746–756. doi: 10.1089/vbz.2014.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faburay B, Wilson WC, Gaudreault NN, Davis AS, Shivanna V, Bawa B, Sunwoo SY, Ma W, Drolet BS, Morozov I, McVey DS, Richt JA. 2016. A recombinant Rift Valley fever virus glycoprotein subunit vaccine confers full protection against Rift Valley fever challenge in sheep. Sci Rep 6:27719. doi: 10.1038/srep27719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrill JC, Laughlin RC, Lokugamage N, Pugh R, Sbrana E, Weise WJ, Adams LG, Makino S, Peters CJ. 2013. Safety and immunogenicity of recombinant Rift Valley fever MP-12 vaccine candidates in sheep. Vaccine 31:559–565. doi: 10.1016/j.vaccine.2012.10.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weingartl HM, Miller M, Nfon C, Wilson WC. 2014. Development of a Rift Valley fever virus viremia challenge model in sheep and goats. Vaccine 32:2337–2344. doi: 10.1016/j.vaccine.2014.02.066. [DOI] [PubMed] [Google Scholar]
- 34.Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. 2004. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci 13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giménez-Lirola LG, Mur L, Rivera B, Mogler M, Sun Y, Lizano S, Goodell C, Harris DL, Rowland RR, Gallardo C, Sánchez-Vizcaíno JM, Zimmerman J. 2016. Detection of African swine fever virus antibodies in serum and oral fluid specimens using a recombinant protein 30 (p30) dual matrix indirect ELISA. PLoS One 11:e0161230. doi: 10.1371/journal.pone.0161230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hossain MM, Wilson WC, Faburay B, Richt J, McVey DS, Rowland RR. 2016. Multiplex detection of IgG and IgM to Rift Valley fever virus nucleoprotein, nonstructural proteins, and glycoprotein in ovine and bovine. Vector Borne Zoonotic Dis 16:550–557. doi: 10.1089/vbz.2014.1721. [DOI] [PubMed] [Google Scholar]
- 37.Greiner M, Pfeiffer D, Smith RD. 2000. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev Vet Med 45:23–41. doi: 10.1016/S0167-5877(00)00115-X. [DOI] [PubMed] [Google Scholar]
- 38.Pinette MM, Rodriguez-Lecompte JC, Pasick J, Ojkic D, Leith M, Suderman M, Berhane Y. 2014. Development of a duplex Fluorescent Microsphere Immunoassay (FMIA) for the detection of antibody responses to influenza A and Newcastle disease viruses. J Immunol Methods 405:167–177. doi: 10.1016/j.jim.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Lupiani B, Mozisek B, Mason PW, Lamichhane C, Reddy SM. 2010. Simultaneous detection of avian influenza virus NP and H5 antibodies in chicken sera using a fluorescence microsphere immunoassay. Avian Dis 54:668–672. doi: 10.1637/8818-040209-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 40.Chen TH, Lee F, Lin YL, Pan CH, Shih CN, Lee MC, Tsai HJ. 2013. Development of a Luminex assay for the detection of swine antibodies to non-structural proteins of foot-and-mouth disease virus. J Immunol Methods 396:87–95. doi: 10.1016/j.jim.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deregt D, Furukawa-Stoffer TL, Tokaryk KL, Pasick J, Hughes KMB, Hooper-McGrevy K, Baxi S, Baxi MK. 2006. A microsphere immunoassay for detection of antibodies to avian influenza virus. J Virol Methods 137:88–94. doi: 10.1016/j.jviromet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Jansen van Vuren P, Paweska JT. 2009. Laboratory safe detection of nucleocapsid protein of Rift Valley fever virus in human and animal specimens by a sandwich ELISA. J Virol Methods 157:15–24. doi: 10.1016/j.jviromet.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet Res 41:61. doi: 10.1051/vetres/2010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gori A, Peri C, Quilici G, Nithichanon A, Gaudesi D, Longhi R, Gourlay L, Bolognesi M, Lertmemongkolchai G, Musco G, Colombo G. 2016. Flexible vs rigid epitope conformations for diagnostic- and vaccine-oriented applications: novel insights from the Burkholderia pseudomallei BPSL2765 Pal3 epitope. ACS Infect Dis 2:221–230. doi: 10.1021/acsinfecdis.5b00118. [DOI] [PubMed] [Google Scholar]
- 45.Rizwan M, Ronnberg B, Cistjakovs M, Lundkvist A, Pipkorn R, Blomberg J. 2016. Serology in the digital age: using long synthetic peptides created from nucleic acid sequences as antigens in microarrays. Microarrays (Basel) 5:E22. doi: 10.3390/microarrays5030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faburay B, Wilson W, McVey DS, Drolet BS, Weingartl H, Madden D, Young A, Ma W, Richt JA. 2013. Rift Valley fever virus structural and nonstructural proteins: recombinant protein expression and immunoreactivity against antisera from sheep. Vector Borne Zoonotic Dis 13:619–629. doi: 10.1089/vbz.2012.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bird BH, Albariño CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. 2008. Rift Valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J Virol 82:2681–2691. doi: 10.1128/JVI.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrill JC, Laughlin RC, Lokugamage N, Wu J, Pugh R, Kanani P, Adams LG, Makino S, Peters CJ. 2013. Immunogenicity of a recombinant Rift Valley fever MP-12-NSm deletion vaccine candidate in calves. Vaccine 31:4988–4994. doi: 10.1016/j.vaccine.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weingartl HM, Nfon CK, Zhang S, Marszal P, Wilson WC, Morrill JC, Bettinger GE, Peters CJ. 2014. Efficacy of a recombinant Rift Valley fever virus MP-12 with NSm deletion as a vaccine candidate in sheep. Vaccine 32:2345–2349. doi: 10.1016/j.vaccine.2013.12.064. [DOI] [PubMed] [Google Scholar]
- 50.Gowen BB, Bailey KW, Scharton D, Vest Z, Westover JB, Skirpstunas R, Ikegami T. 2013. Post-exposure vaccination with MP-12 lacking NSs protects mice against lethal Rift Valley fever virus challenge. Antiviral Res 98:135–143. doi: 10.1016/j.antiviral.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller R, Saluzzo JF, Lopez N, Dreier T, Turell M, Smith J, Bouloy M. 1995. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg 53:405–411. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- 52.Lihoradova OA, Indran SV, Kalveram B, Lokugamage N, Head JA, Gong B, Tigabu B, Juelich TL, Freiberg AN, Ikegami T. 2013. Characterization of Rift Valley fever virus MP-12 strain encoding NSs of Punta Toro virus or sandfly fever Sicilian virus. PLoS Negl Trop Dis 7:e2181. doi: 10.1371/journal.pntd.0002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez JC, Billecocq A, Durand JP, Cêtre-Sossah C, Cardinale E, Marianneau P, Pépin M, Tordo N, Bouloy M. 2012. The nonstructural protein NSs induces a variable antibody response in domestic ruminants naturally infected with Rift Valley fever virus. Clin Vaccine Immunol 19:5–10. doi: 10.1128/CVI.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van der Wal FJ, Jelsma T, Fijten H, Achterberg RP, Loeffen WL. 2016. Towards a peptide-based suspension array for the detection of pestivirus antibodies in swine. J Virol Methods 235:15–20. doi: 10.1016/j.jviromet.2016.04.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.