ABSTRACT
The diagnosis of nocardiosis, a severe opportunistic infection, is challenging. We assessed the specificity and sensitivity of a 16S rRNA Nocardia PCR-based assay performed on clinical samples. In this multicenter study (January 2014 to April 2015), patients who were admitted to three hospitals and had an underlying condition favoring nocardiosis, clinical and radiological signs consistent with nocardiosis, and a Nocardia PCR assay result for a clinical sample were included. Patients were classified as negative control (NC) (negative Nocardia culture results and proven alternative diagnosis or improvement at 6 months without anti-Nocardia treatment), positive control (PC) (positive Nocardia culture results), or probable nocardiosis (positive Nocardia PCR results, negative Nocardia culture results, and no alternative diagnosis). Sixty-eight patients were included; 47 were classified as NC, 8 as PC, and 13 as probable nocardiosis. PCR results were negative for 35/47 NC patients (74%). For the 12 NC patients with positive PCR results, the PCR assay had been performed with respiratory samples. These NC patients had chronic bronchopulmonary disease more frequently than did the NC patients with negative PCR results (8/12 patients [67%] versus 11/35 patients [31%]; P = 0.044). PCR results were positive for 7/8 PC patients (88%). There were 13 cases of probable nocardiosis, diagnosed solely using the PCR results; 9 of those patients (69%) had lung involvement (consolidation or nodule). Nocardia PCR testing had a specificity of 74% and a sensitivity of 88% for the diagnosis of nocardiosis. Nocardia PCR testing may be helpful for the diagnosis of nocardiosis in immunocompromised patients but interpretation of PCR results from respiratory samples is difficult, because the PCR assay may also detect colonization.
KEYWORDS: Nocardia, PCR, immunocompromised hosts, nocardiosis, opportunistic infections
INTRODUCTION
Nocardiosis is a rare infection caused by Nocardia spp., environmental Gram-positive bacteria (1). Management of nocardiosis is challenging, with mortality rates ranging from 20 to 30% among patients with invasive infection and up to 50% for patients with central nervous system involvement (1). Clinical and radiological features are diverse and nonspecific and include pneumonia, cutaneous or subcutaneous infections, and brain or other solid organ abscesses. Nocardiosis mainly affects immunocompromised patients, including patients receiving immunosuppressive therapy, post-organ transplantation or post-stem cell transplantation patients, patients with primary immunodeficiencies, and patients with chronic bronchopulmonary disease (1–4). A positive microbiological culture from an infected site remains the diagnostic gold standard. Because of the severity of infection among immunocompromised patients and the many possible differential diagnoses, however, most patients receive antibiotic treatment prior to culture sampling for Nocardia, reducing the sensitivity of microbial cultures in this setting. In addition, although most cultures of Nocardia spp. are positive in 2 to 7 days, culture duration must be extended to 2 to 3 weeks because of slow-growing species. Therefore, alternative techniques are required to improve the diagnosis of nocardiosis.
In 2005, Couble et al. described a 16S rRNA PCR-based assay, with hybridization confirmation, that could be performed directly on human clinical samples for the rapid detection of Nocardia spp. (5). The method was tested with bronchoalveolar lavage (BAL) fluid, sputum, pleural fluid, pus, and biopsy specimens from 18 patients with cultures that were positive for various Nocardia species (Nocardia brasiliensis, Nocardia farcinica, Nocardia nova, Nocardia otitidiscaviarum, and strains formerly classified as Nocardia asteroides). Twenty clinical samples from patients with confirmed tuberculosis were used as negative-control (NC) samples. In that publication, the specificity and sensitivity for detection of Nocardia spp. were both 100%.
A few case reports using the PCR assay have been reported, but the diagnostic performance of Nocardia PCR has never been studied in patients with a wide range of underlying conditions (6, 7). Therefore, our main objective was to assess the specificity and sensitivity of a 16S rRNA PCR-based Nocardia assay performed with clinical samples in a pragmatic setting. A secondary objective was to describe the characteristics of patients with a diagnosis of probable nocardiosis based solely on PCR results, without positive culture findings.
MATERIALS AND METHODS
Study design and setting.
Between 1 January 2014 and 30 April 2015, patients admitted to three French tertiary-care teaching hospitals (Hôpital Necker-Enfants Malades, Hôpital Foch, and Hôpital Avicenne) involved in the management of patients with underlying conditions favoring nocardiosis were prospectively considered for inclusion. During the study period, the three centers used comparable diagnostic evaluation protocols for opportunistic infections in immunocompromised patients, which included Nocardia PCR testing performed on samples from all patients with conditions favoring nocardiosis and clinical or radiological signs consistent with nocardiosis.
Inclusion criteria and classification.
Patients were included if they met all the following criteria: (i) at least one underlying condition favoring nocardiosis, (ii) clinical and/or radiological signs consistent with a diagnosis of nocardiosis, and (iii) a clinical sample sent for Nocardia PCR testing. Underlying conditions considered to favor nocardiosis were solid organ or hematopoietic stem cell transplantation, primary or secondary (human immunodeficiency virus [HIV] infection or immunosuppressive therapy) immunodeficiency, chronic bronchopulmonary disease, and asthma. Clinical and radiological signs considered consistent with nocardiosis were lung consolidation or nodule (with or without cavitation), cerebral abscess, meningitis, muscle abscess, cutaneous or subcutaneous abscess or nodule, dermo-hypodermitis or mycetoma, osteoarthritis, solid organ abscess, and adenitis.
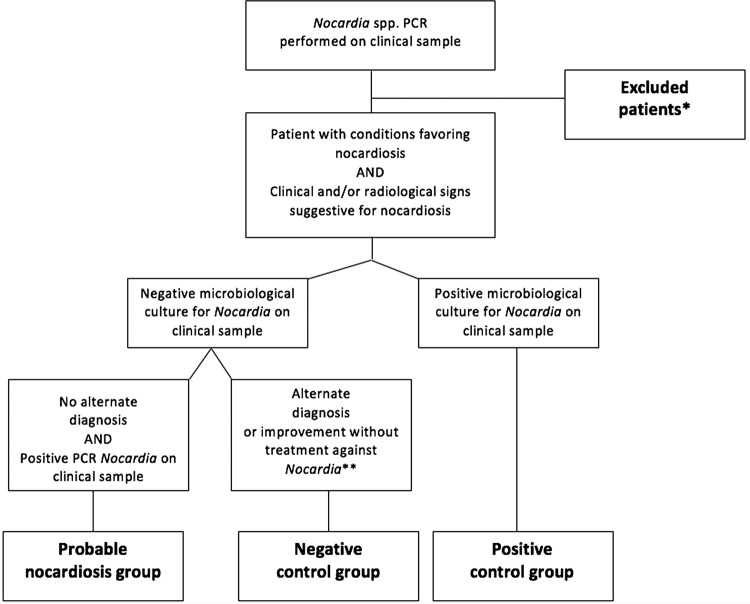
Two senior authors (C.R. and D.L.) reviewed the medical charts of the included patients and retrospectively classified patients into one of the three following groups (Fig. 1). The NC group had negative microscopic examination results, including Gram staining and culture, for Nocardia and a proven alternative diagnosis or improvement within 6 months without anti-Nocardia treatment (≤14 days of antibiotic administration was not considered treatment). The positive-control (PC) group had positive culture results for Nocardia. During the study period, 3 patients were diagnosed as having confirmed nocardiosis based on bacterial culture results and 1 additional patient was considered colonized without clinical and/or radiological signs. Unfortunately, none of these patients had a clinical sample sent for Nocardia PCR testing and so none could be included in our study (see Table S1 in the supplemental material). Therefore, to obtain positive controls, we selected Nocardia culture-positive patients who had had a clinical sample sent to the French Observatory of Nocardiosis (Lyon, France) for both culture and PCR testing between 2010 and 2014. The probable nocardiosis group had positive Nocardia PCR results for a clinical sample, no alternative diagnosis (following a comprehensive microbiological evaluation; see below), and negative culture results for Nocardia. Patients with coinfections were discussed further, to confirm a diagnosis of nocardiosis or to exclude the case. Patients who fit into none of the aforementioned groups were excluded (Table S1).
FIG 1.
Retrospective classification of the study patients. *, patients were excluded (n = 25) on the basis of no underlying condition favoring nocardiosis (n = 8), no proven alternative diagnosis or improvement at 6 months without anti-Nocardia treatment (n = 9), no clinical or radiological signs consistent with nocardiosis (n = 4), or positive microbiological cultures for Nocardia without PCR testing being performed (n = 4). **, antibiotic treatment for ≤14 days was allowed.
Clinical data and definitions.
We recorded demographic data (age and sex), underlying conditions favoring nocardiosis, comorbidities (chronic obstructive pulmonary disease and diabetes mellitus), and prophylaxis with sulfamethoxazole-trimethoprim (SXT) at diagnosis. The type of clinical sample with which the Nocardia PCR assay was performed and the antibiotic regimen prior to sampling were noted. We recorded clinical and radiological signs (involved organ, dissemination, and radiological description), histological and microbiological investigations (bacteriological, mycological, and virological studies) and indirect tests (e.g., antigen- or PCR-based diagnostic tools), and biological findings (neutrophil count and C-reactive protein concentration) at the time of diagnosis.
Microbiology.
Microbiological analyses included prolonged bacteriological cultures (on buffered charcoal-yeast extract [BCYE] agar plates), direct examination (with Gram staining), and cultures for fungi (2 weeks) and mycobacteria (8). Virological investigations (PCR assays for cytomegalovirus and respiratory viruses) were performed with BAL fluid samples in cases with pulmonary involvement. The results of β-d-glucan, galactomannan antigen, and cryptococcal antigen detection, Pneumocystis jirovecii immunofluorescence assays, and PCR assays were recorded if such tests were performed. Cytological or histological analysis was performed for all liquid or tissue samples.
The Nocardia genus PCR was based on amplification and detection of the gene encoding 16S rRNA and was performed at the French Observatory of Nocardiosis. Biological samples were transported at room temperature. Nocardia DNA was extracted with a Mycobacteria tuberculosis respiratory specimen preparation kit (Roche, Meylan, France). NG1 (5′-ACCGACCACAAGGGGG-3′) and NG2 (5′-GGTTGTAAACCTCTTTCGA-3′) primers were used to amplify a Nocardia genus-specific 590-bp fragment of 16S rRNA. In order to amplify a 590-bp fragment of the 16S rRNA gene, amplification was carried out using packaged PCR tubes (Ready-to-Go PCR beads; Amersham Biosciences, Orsay, France). Ten microliters of extracted DNA and primers NG1 (5′-ACCGACCACAAGGGG-3′) and NG2 (5′-GGTTGTAACCTCTTCGA-3′), at a final concentration of 1 μM for each primer, were added for a final volume of 25 μl of PCR mixture. First, we performed an initial denaturation at 94°C for 11 min. After 40 cycles of denaturation at 94°C for 60 s, primer annealing at 55°C for 20 s, and extension at 72°C for 60 s, followed by postamplification extension at 72°C for 10 min, PCR products were purified with the E.Z.N.A. gel extraction kit (Omega Bio-Tek, Vaulx-en-Velin, France). Because the hybridization step is time-consuming and thus difficult to perform in routine practice, we replaced this method with a DNA dilution step, as reported previously (9). Therefore, nondiluted and diluted (1:10, 1:25, 1:100, and 1:250) DNA samples were used to reduce the concentrations of inhibitors. β-Globin PCR was used as a control to monitor specimen processing and DNA extraction. Nocardia species-specific PCR was performed for Nocardia farcinica, targeting the nfa 29510 gene, according to the method described by Hasegawa et al. (10).
Statistical analysis.
The final analysis was conducted after all data had been recorded and verified. Clinical, biological, and radiological data recorded at the time of diagnosis are described. Continuous variables are presented as medians and ranges. Categorical variables are presented as numbers and percentages. The specificity of the PCR for a diagnosis of nocardiosis was calculated as the proportion of negative Nocardia PCR results in the NC group. Nocardia PCR sensitivity was calculated as the proportion of positive Nocardia PCR results in the PC group. Bivariate analyses were performed using Fisher's exact test to compare categorical variables and Student's t test to compare continuous variables.
Ethics.
This study was approved by the Comité de Protection des Personnes Ile-de-France 1 (CCTIRS approval no. 15.1016) and the Commission Nationale de l'Informatique et des Libertés.
RESULTS
Characteristics of the study population and excluded patients.
During the study period, 85 patients were considered for inclusion, and 25 were excluded (see Table S1 in the supplemental material). With the additional 8 PC patients, we therefore studied a total of 68 patients; their characteristics are presented in Table 1. Most of the patients had more than one underlying condition for nocardiosis (43/68 patients [63%]). Twenty-five patients (37%) were receiving prophylaxis with SXT at the time of diagnosis (700 to 5,600 mg of sulfamethoxazole/week). The organs most frequently involved were the lungs (61/68 patients [90%]), and infection was disseminated in 15 of the patients (22%). Nocardia PCR was most often performed with respiratory samples (58/68 patients [85%]).
TABLE 1.
Main characteristics of the study population (n = 68)
| Characteristica | Finding |
|---|---|
| Age at diagnosis (median [range]) (yr) | 54 (5–89) |
| Male (n [%]) | 42 (61.7) |
| Underlying conditions for nocardiosis (n [%])b | |
| Corticosteroid | 27 (39.7) |
| Immunosuppressive treatment | 27 (39.7) |
| Chronic bronchopulmonary disease or asthma | 27 (39.7) |
| Solid organ transplantation | 14 (20.5) |
| Allogeneic HSCT | 14 (20.5) |
| Primary immunodeficiencyc | 9 (13.2) |
| HIV infection | 7 (10.2) |
| Autologous HSCT | 4 (5.8) |
| Comorbidities (n [%]) | |
| Diabetes mellitus (n = 66) | 10 (15.1) |
| Neutropenia (n = 64)d | 2 (3.1) |
| SXT prophylaxis (n [%]) | 25 (36.7) |
| Type of clinical sample for Nocardia PCR testing (n [%]) | |
| Respiratory samples | |
| BAL fluid sample | 40 (58.8) |
| Bronchial sampling specimen | 7 (10.2) |
| Sputum sample | 6 (8.8) |
| Lung biopsy specimen | 2 (2.9) |
| Other specimens | |
| Abscess specimen | 5 (7.3) |
| Cutaneous biopsy specimen | 3 (4.4) |
| Pleural fluid sample | 2 (2.9) |
| CSF sample | 2 (2.9) |
| Lymph node specimen | 1 (1.4) |
| PCR result (n [%]) | |
| Positive | 32 |
| Positive for N. farcinica | 14 (43.7) |
| Clinical/radiological presentation (n [%]) | |
| Pulmonary | 61 (89.7) |
| Disseminated disease | 15 (22) |
| Cutaneous or muscular | 10 (14.7) |
| Cerebral | 6 (8.8) |
| Deep-seated abscess | 4 (5.8) |
| Othere | 8 (11.7) |
| Duration of symptoms before sampling (median [range]) (days) (n = 57) | 30 (1–730) |
| Outcome of death at 6 mo (n [%]) (n = 67) | 10 (14.7) |
BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid; HIV, human immunodeficiency virus; HSCT, hematopoietic stem cell transplantation; SXT, trimethoprim-sulfamethoxazole.
Each patient could have several underlying conditions.
Primary immunodeficiencies included chronic granulomatous disease (n = 3), defect of the interleukin-12 (IL-12) axis (n = 2), severe combined immunodeficiency (n = 2), lymphohistiocytosis (n = 1), and X-linked agammaglobulinemia (n = 1).
Defined as polymorphonuclear cell counts of <500 cells/mm3.
Other includes lymph node (n = 4), bone (n = 2), blood (n = 1), eye (n = 1), and liver (n = 1) involvement.
Nocardia PCR specificity.
Forty-seven patients (30 men and 17 women; median age, 52 years [range, 5 to 83 years]) were classified as NC. PCR was performed with respiratory samples for 41 of those patients (32 BAL fluid samples, 6 bronchial aspiration samples obtained during bronchial endoscopy, 2 sputum samples, and 1 lung biopsy specimen), with pleural fluid samples for 2 patients, with cutaneous biopsy specimens for 2 patients, with a cerebrospinal fluid sample for 1 patient, and with a lymph node tissue specimen for 1 patient. Forty-three patients had another diagnosis (pneumonia with bacterial identification, n = 15; pneumonia without microbial identification, n = 9; noninfectious pneumonia, n = 3; fungal infection, n = 8; mycobacterial infection, n = 3; viral infection, n = 3; hematological malignancy, n = 3; other diseases, n = 3); some patients had several diagnoses. The microbiological evaluations for the NC group are detailed in Table S2 in the supplemental material.
Nocardia PCR test results were negative for 35 of the 47 NC patients and positive for 12, with 6 species-specific PCR assays being positive for N. farcinica. The specificity of the Nocardia PCR assay was thus 74% (35/47 patients). The positive PCR test results for the NC patients were all from respiratory samples; patients in this group were more likely to have chronic bronchopulmonary disease than were patients with negative PCR results (8/12 patients [67%] versus 11/35 patients [31%]; P = 0.044) (Table 2). An alternative diagnosis of fungal, bacterial, or viral infection was identified for one-half of the patients with positive PCR results (6/12 patients).
TABLE 2.
Characteristics of the 47 patients in the negative-control group, according to Nocardia PCR results
| Characteristica | Negative Nocardia PCR result (n = 35) |
Positive Nocardia PCR result (n = 12) |
P (bivariate analysis) |
|---|---|---|---|
| Age at diagnosis (median [range]) (yr) | 53 (5–83) | 49 (17–70) | 0.7 |
| Male (n [%]) | 24 (68.5) | 6 (50) | 0.3 |
| Sample origin (n [%]) | |||
| Pulmonary | 29 (82.8) | 12 (100) | 0.3 |
| Otherb | 6 (17.1) | 0 | |
| Underlying conditions (n [%]) | |||
| Corticosteroid | 13 (37.1) | 4 (33.3) | 1 |
| Immunosuppressive treatment | 13 (37.1) | 7 (58.3) | 0.3 |
| Hematological malignancy | 12 (34.2) | 6 (50) | 0.5 |
| Chronic bronchopulmonary disease | 11 (31.4) | 8 (66.6) | 0.044 |
| HIV | 6 (17.1) | 0 | 0.31 |
| PID | 5 (14.2) | 2 (16.6) | 1 |
| SOT | 9 (25.7) | 3 (25) | 1 |
| SXT prophylaxis (n [%]) | 17 (48.5) | 3 (25) | 0.19 |
| Antibiotic treatment before sampling (n [%]) (n = 46) | 18 (51.4) | 4 (36.3) | 0.33 |
HIV, human immunodeficiency virus; PID, primary immunodeficiency; SOT, solid organ transplantation; SXT, trimethoprim-sulfamethoxazole.
Other includes cutaneous biopsy (n = 2), pleural effusion (n = 2), cerebrospinal fluid (n = 1), and abscess (n = 1).
Nocardia PCR sensitivity.
The 8 PC patients included 3 men and 5 women, with a median age of 54 years (range, 19 to 89 years). One patient was receiving prophylaxis with SXT at diagnosis and 5 patients had received antibiotic treatment before sampling for PCR testing and culture (β-lactams for 4 patients, SXT for 2 patients, and aminoglycosides, fluoroquinolone, and linezolid for 1 patient each). All patients had lung involvement (4 had nodules, 2 had excavations, 1 had nodules and excavations, 1 had lung consolidation, and 1 had interstitial pneumonia), and 2 had disseminated nocardiosis (1 had cerebral and ocular abscesses and 1 had cerebral and other abscesses with lymph node involvement). Bacterial or fungal coinfection was identified for 5 patients. PCR testing was performed with respiratory samples (2 BAL fluid samples, 1 bronchial sampling specimen, and 4 sputum samples) and a muscle abscess sampling specimen (n = 1). The Nocardia species identified were Nocardia abscessus (n = 3), Nocardia cyriacigeorgica (n = 1), N. farcinica (n = 1), N. nova (n = 1), Nocardia paucivorans (n = 1), and Nocardia wallacei (n = 1). After 6 months, 2 patients had died; 1 death was related to nocardiosis (10 days after diagnosis), and 1 death was related to the patient's underlying condition. Outcomes were favorable for the other 6 patients.
Nocardia PCR test results were positive for 7 of the 8 PC patients, giving a sensitivity of 88%. The infection not detected by PCR was caused by N. paucivorans. After identification of this strain in culture, the strain was tested using the Nocardia genus PCR, which yielded positive results. Microscopic examination results were positive for 6/7 PC patients (data were not available for 1 patient), including the aforementioned patient with negative PCR results.
Probable nocardiosis.
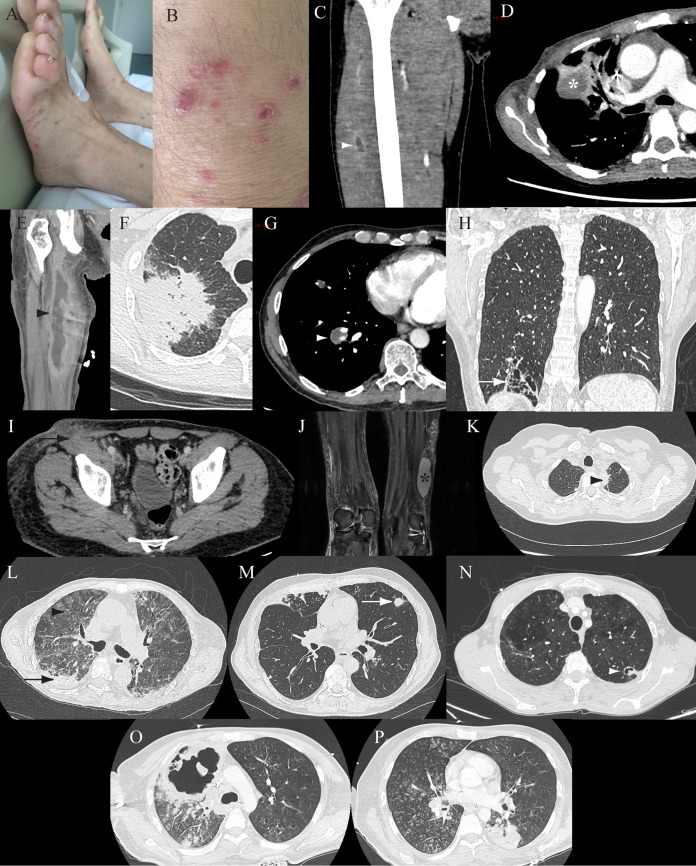
Thirteen cases of probable nocardiosis were diagnosed based on positive Nocardia PCR results despite negative culture and Gram staining results (Table 3). Nine of the patients were men (69.2%), and the median age was 54 years (range, 22 to 82 years). PCR was mostly performed with respiratory samples. Antibiotics were prescribed before microbiological sampling in 5 cases, and 4 patients were receiving prophylaxis with SXT. Coinfections were diagnosed for 5 patients (Table 3). Among the pulmonary infections, we observed lung consolidation in 5 cases, nodules in 5 (1 excavated nodule), and alveolointerstitial pneumonia in 2. Clinical and radiological features are shown in Fig. 2. Two patients required surgical or radiological drainage. Cure was observed for 8 patients, 1 patient experienced relapse, 1 patient was lost to follow-up monitoring after 2 months of antibiotic treatment, and 3 patients died; no deaths were attributed to nocardiosis.
TABLE 3.
Main characteristics of the 13 patients with probable nocardiosis diagnosed solely using PCR results
| Patient no. | Age (yr) | Sexa | Clinical or radiological involvement | Sample(s) for Nocardia PCR | Other tests and findings | Concurrent pathogen | Nocardiosis risk factors | Positive species PCR | SXT prophylaxis (weekly dose of SMT [mg]) | Prior antibiotic treatment | Treatment (overall treatment duration) | Outcome at 6 mo |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 68 | M | Cutaneous | Cutaneous biopsy specimen (PCR positive) | Cutaneous biopsy (negative bacterial, mycological, and mycobacterial cultures, polymorphous inflammatory infiltrate on histological exam, PAS, Ziehl, and Grocott staining negative); Crypto Ag, CMV PCR, and BDG negative | No | Hematological malignancy | No | Yes (2,400) | No | IPM-AMK then CRO-AMK then IPM-CIP then TZP-LZD then CXM-SXT (6 mo) | Cure |
| 2 | 24 | M | Muscular abscess | Muscular abscess specimen (PCR positive) and blood sample (PCR positive) | Abscess puncture (negative Nocardia and mycobacterial cultures, polymorphonuclear cells on histological exam); BDG, GM, A. fumigatus PCR, and Toxoplasma gondii PCR negative | Yes (Staphylococcus aureus) | Hematopoietic stem cell transplantation | Yes (N. farcinica) | No | β-Lactams | LZD then AMC then AMK then AMC (4 mo) | Cure |
| 3 | 63 | F | Disseminated (lung infection and muscular abscess) | Muscular abscess specimen (PCR positive) and BAL fluid sample (PCR positive) | BAL (negative bacterial, mycological, and mycobacterial cultures and GM test); abscess puncture (negative bacterial and mycobacterial cultures and broad-range 16S rRNA PCR); BDG, GM, and A. fumigatus PCR negative | No | Hematopoietic stem cell transplantation and IS | No | No | NA | IPM-AMK then LZD then CRO-AMK then SXT-AMK (7 mo) | Cure |
| 4 | 34 | M | Lung infection | BAL fluid sample (PCR positive) and pulmonary biopsy specimen (PCR negative) | BAL (negative bacterial, mycological, and mycobacterial cultures, histological exam, and GM test); pulmonary biopsy (negative bacterial, mycological, and mycobacterial cultures, epithelioid granuloma on histological exam); BDG positive | No | SOT, CGD, and IS | Yes (N. farcinica) | Yes (5,600) | No | IPM-SXT then SXT (6 mo) | Cure |
| 5 | 54 | M | Lung infection | BAL fluid sample (PCR positive) | BAL (negative bacterial, mycological, and mycobacterial cultures, GM test, and histological exam); pulmonary biopsy (negative bacterial, mycological, and mycobacterial cultures, eosinophilic necrosis); BDG, GM, A. fumigatus PCR, Crypto Ag, and CMV PCR negative | No | Hematological malignancy and chronic bronchopulmonary disease | No | Yes (2,400) | No | IPM, AMK, SXT (NA) | NA |
| 6 | 60 | F | Lung infection | BAL fluid sample (PCR positive) | BAL (negative bacterial, mycological, and mycobacterial cultures and GM test, filamentous form on histological exam, Ziehl staining negative) | No | Chronic bronchopulmonary disease | Yes (N. farcinica) | No | No | AMC (1 mo), LZD (5 mo) | Relapse 7 mo later |
| 7 | 22 | M | Disseminated (subcutaneous abscess and lymph node) | Lymph node biopsy specimen (PCR positive) and cutaneous abscess specimen (PCR positive) | Lymph node biopsy and cutaneous abscess puncture (negative Nocardia, mycological, and mycobacterium cultures); CMV PCR negative | Yes (S. aureus and Staphylococcus epidermidis) | Autologous stem cell transplantation and corticosteroids | Yes (N. farcinica) | Yes (2,400) | β-Lactams, aminoglycosides | IPM-SXT-AMK then SXT then LZD (4 mo) | Death |
| 8 | 82 | F | Muscular abscess | Muscular abscess specimen (PCR positive) | Abscess puncture (negative bacterial, mycological, and mycobacterial cultures, polymorphonuclear cells on histological exam); BDG positive, GM and A. fumigatus PCR negative | No | Corticosteroids and IS | Yes (N. farcinica) | No | No | IPM-AMK (0.2 mo) | Death |
| 9 | 54 | F | Disseminated (lung and bone infection) | BAL fluid sample (PCR positive) | BAL (negative bacterial, mycological, and mycobacterial cultures and histological exam); pulmonary biopsy (negative mycological and mycobacterial cultures, giant cell granuloma with epithelioid cells without necrosis on histological exam) | No | Hematological malignancy | Yes (N. farcinica) | No | No | IPM-SXT then AMK-SXT then SXT (12 mo) | Cure |
| 10 | 64 | M | Lung infection | BAL fluid sample (PCR positive) and lung biopsy specimen (PCR positive) | BAL (negative bacterial, mycological, and mycobacterial cultures, GM test and virus PCR negative); pulmonary biopsy (negative bacterial and mycobacterial cultures); BDG positive, GM negative | Yes (P. jirovecii and A. fumigatus) | SOT, corticosteroids, and IS | No | No | Sulfonamides | SXT then SXT-IPM (NA) | Death |
| 11 | 51 | M | Lung infection | BAL fluid sample (PCR positive) | BAL (negative Nocardia, mycological, and mycobacterial cultures); BDG and GM negative | Yes (Moraxella and Actinomyces naeslundii) | Hematopoietic stem cell transplantation | Yes (N. farcinica) | No | β-Lactams | CXM (6.4 mo) | Cure |
| 12 | 48 | M | Disseminated (meningitis and lung infection) | CSF sample (PCR positive) and bronchial sampling specimen (PCR positive) | BAL and CSF sampling (negative bacterial, mycological, and mycobacterial cultures); Crypto Ag negative | No | HIV | No | No | No | IPM-AMK then IPM-SXT then SXT-MXF (6 mo) | Cure |
| 13 | 40 | M | Lung infection | BAL fluid sample (PCR negative) and lung biopsy specimen (PCR positive) | BAL and pulmonary biopsy (negative Nocardia, mycological, and mycobacterial cultures) | No | Asthma | No | No | β-Lactams, sulfonamides, aminoglycosides | IMP-SXT-AMK then IMP-SXT then SXT (6 mo) | Cure |
M, male; F, female; AMC, amoxicillin-clavulanic acid; AMK, amikacin; BAL, bronchoalveolar lavage; BDG, β-d-glucan; CGD, chronic granulomatous disease; CIP, ciprofloxacin; CMV, cytomegalovirus; CRO, ceftriaxone; Crypto Ag, Cryptococcus antigen; CSF, cerebrospinal fluid; CXM, cefuroxime; GM, galactomannan antigen; HIV, human immunodeficiency virus; IPM, imipenem; IS, immunosuppressive treatment; LZD, linezolid; MXF, moxifloxacin; NA, not available; PAS, periodic acid-Schiff; SMT, sulfamethoxazole; SOT, solid organ transplantation; TZP, piperacillin-tazobactam; SXT, sulfamethoxazole-trimethoprim.
FIG 2.
Clinical and radiological features in a series of 13 patients with probable nocardiosis. (A and B) Patient 1, multiple superficial and purple nodular skin lesions on both legs. (C) Patient 2, coronal enhanced computed tomography (CT) of the right leg, showing a muscle abscess of the thigh (white arrowhead, central low attenuation with peripheral enhancement). (D and E) Patient 3, axial enhanced lung CT (D), showing a right pulmonary mass (white star, central attenuation), and coronal enhanced CT of the right leg (E), showing a muscle abscess of the thigh (black arrowhead, central attenuation with peripheral enhancement). (F) Patient 4, axial nonenhanced lung CT, showing a right spiculated lung consolidation. (G) Patient 5, axial enhanced lung CT, showing a right pulmonary mass (white arrowhead, low density). (H) Patient 6, coronal enhanced lung CT, showing localized bronchiectasis with thin surrounding lung consolidation (white arrow). (I) Patient 7, axial enhanced abdominal CT, showing a subcutaneous abscess and lymph node involvement (black arrow). (J) Patient 8, coronal T2 fat-saturated magnetic resonance imaging of the legs, showing a subcutaneous abscess (black star, hyperintensity). (K) Patient 9, axial nonenhanced lung CT, showing a left apical lung mass (black arrowhead). (L) Patient 10, axial enhanced lung CT, showing a lung consolidation (black arrow) and diffuse ground-glass opacities (black arrowhead). (M) Patient 11, axial nonenhanced lung CT, showing a lung nodule (white arrow). (N) Patient 12, axial nonenhanced lung CT, showing a cavitated lung nodule (white arrowhead). (O and P) Patient 13, axial nonenhanced lung CT, showing bilateral involvement with excavation of 10 cm, micronodules, and lung consolidation.
DISCUSSION
In this first pilot study assessing the 16S rRNA PCR-based Nocardia assay in clinical practice, we observed a specificity of 74% and a sensitivity of 88% for diagnosis of Nocardia infection. In our study population, positive Nocardia PCR results in the NC group were observed only with respiratory samples, suggesting that the PCR may detect airway colonization (1, 11). This hypothesis is supported by the fact that bronchopulmonary disease was more frequent in NC patients with positive Nocardia PCR results (67%) than in those with negative PCR results. A high frequency of upper respiratory tract Nocardia colonization among asymptomatic healthy adults (45/101 subjects [44.5%]) was recently detected using PCR and electrospray ionization-time of flight mass spectrometry (12). Use of a quantitative PCR threshold may help to distinguish airway colonization from infection, as suggested for Pneumocystis (13, 14). To reduce possible misdiagnosis and excess antibiotic treatment, we suggest that Nocardia PCR testing should be used only for patients with a high index of suspicion of nocardiosis, i.e., with an underlying condition favoring nocardiosis, signs consistent with a diagnosis of nocardiosis, and no other obvious diagnosis. Use of antibiotic prophylaxis with SXT in immunocompromised patients with airway colonization may warrant further evaluation.
The sensitivity of the Nocardia PCR assay was not 100%, indicating that a negative Nocardia PCR result does not necessarily exclude a diagnosis of nocardiosis (5). Although the small sample size of our PC group prevents us from drawing any definitive conclusions, our results suggest that no test can completely exclude a diagnosis of nocardiosis in an immunocompromised patient with a clinical picture consistent with this diagnosis.
We described the clinical, biological, and radiological presentations of patients with probable nocardiosis diagnosed solely using positive PCR results, without positive culture results. Because bacteriological cultures are thought to have low sensitivity for detection of Nocardia (although this has never been assessed), especially among antibiotic-treated patients, choosing a diagnostic gold standard is difficult. Our classification criteria, defining probable nocardiosis based on a comprehensive negative evaluation for alternative diagnoses, reflect a real-life approach. For a few patients, improvement with anti-Nocardia treatment was an additional argument in favor of this diagnosis. We decided to include coinfected patients because this situation is frequently (10 to 30%) observed during nocardiosis, mainly among solid organ recipients or HIV-infected patients. The most frequently identified copathogens are Aspergillus fumigatus, Pneumocystis jirovecii, Mycobacterium tuberculosis, Cryptococcus neoformans, Histoplasma capsulatum, and cytomegalovirus (1, 11, 15).
For one-half of our patients with probable nocardiosis, identification at the species level enabled the antibiotic regimen to be adapted. Among the different Nocardia species, N. farcinica has one of the highest degrees of resistance to available antibiotics. Although imipenem and amoxicillin-clavulanic acid are the β-lactam antibiotics that are most frequently active against this pathogen (33 to 100% susceptibility for imipenem and 73 to 100% for amoxicillin-clavulanic acid) (1, 11, 16–19), they should be prescribed in association with amikacin, because of the high frequency of resistant strains (18). In contrast, N. farcinica strains have a low rate of resistance to SXT and no resistance to linezolid. If N. farcinica-specific PCR results are negative, then a broad-spectrum antibiotic, such as SXT, linezolid, or the combination of a β-lactam and amikacin, is required. Among our nocardiosis cases, 4/13 patients with probable nocardiosis and 1/8 patients with proven nocardiosis were receiving SXT prophylaxis, confirming that a diagnosis of nocardiosis should not be ruled out simply because prophylaxis has been given.
During the study period, only 3 patients had a culture-based diagnosis of nocardiosis in our centers, providing a striking contrast with the 13 PCR-based diagnoses. We cannot exclude the possibility that some of our probable cases were the result of colonization rather than true infection; nevertheless, we suspect that the incidence of nocardiosis may be underestimated when diagnosis relies on positive culture results.
In conclusion, Nocardia PCR testing performed directly on a biological sample is a valuable additional assay for the diagnostic evaluation of infections in immunocompromised patients. The interpretation of PCR results may be challenging, however, because of possible detection of airway colonization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Frédéric Imbert (Montélimar) and Patrick Lamarche (Fontenay-le-Comte), Caroline Elie (Unité de Recherche Clinique/Centre d'Investigation Clinique Paris Descartes Necker Cochin, Assistance Publique-Hôpitaux de Paris, Hôpital Necker Enfants Malades, Paris, France), Emilie Cardot (Service de Microbiologie, Hôpital Foch, Suresnes, France), Hervé Lecuyer (Laboratoire de Microbiologie, Hôpital Necker-Enfants Malades, APHP, Paris, France), and Violaine Walewski (Laboratoire de Microbiologie, Hôpital Avicenne, APHP, Bobigny, France) for their help during the study. We thank Karen Pickett for her editorial suggestions.
D.L. was supported by the Bourse Junior 2015-Société de Pathologie Infectieuse de Langue Française.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00002-18.
REFERENCES
- 1.Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ Jr. 2006. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev 19:259–282. doi: 10.1128/CMR.19.2.259-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coussement J, Lebeaux D, van Delden C, Guillot H, Freund R, Marbus S, Melica G, Van Wijngaerden E, Douvry B, Van Laecke S, Vuotto F, Tricot L, Fernández-Ruiz M, Dantal J, Hirzel C, Jais J, Rodriguez-Nava V, Lortholary O, Jacobs F. 2016. Nocardia infection in solid organ transplant recipients: a multicenter European case-control study. Clin Infect Dis 63:338–345. doi: 10.1093/cid/ciw241. [DOI] [PubMed] [Google Scholar]
- 3.Lebeaux D, Morelon E, Suarez F, Lanternier F, Scemla A, Frange P, Mainardi J-L, Lecuit M, Lortholary O. 2014. Nocardiosis in transplant recipients. Eur J Clin Microbiol Infect Dis 33:689–702. doi: 10.1007/s10096-013-2015-5. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Nava V, Durupt S, Chyderiotis S, Freydière A, Karsenty J, de Montclos M, Reix P, Durieu I, Nove-Josserand R, Chiron R, Bremont F, Têtu L, Murris M, Terru D, Godreuil S, Bergeron E, Freney J, Boiron P, Vandenesch F, Marchandin H, Segonds C, Doléans-Jordheim A. 2015. A French multicentric study and review of pulmonary Nocardia spp. in cystic fibrosis patients. Med Microbiol Immunol 204:493–504. doi: 10.1007/s00430-014-0360-3. [DOI] [PubMed] [Google Scholar]
- 5.Couble A, Rodriguez-Nava V, de Montclos MP, Boiron P, Laurent F. 2005. Direct detection of Nocardia spp. in clinical samples by a rapid molecular method. J Clin Microbiol 43:1921–1924. doi: 10.1128/JCM.43.4.1921-1924.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canouï E, Blanc K, Loubinoux J, Valade S, Hamard C, Lefebvre A, Amorim S, Bébéar C, Rodriguez-Nava V, Lebeaux D, Launay O, Alifano M, Rabbat A, Kernéis S. 2017. The value of molecular techniques to diagnose Ureaplasma urealyticum and Nocardia farcinica pleuropneumonia in a patient with diffuse large B-cell lymphoma. Int J Infect Dis 64:93–95. doi: 10.1016/j.ijid.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Marchandin H, Eden A, Jean-Pierre H, Reynes J, Jumas-Bilak E, Boiron P, Laurent F. 2006. Molecular diagnosis of culture-negative cerebral nocardiosis due to Nocardia abscessus. Diagn Microbiol Infect Dis 55:237–240. doi: 10.1016/j.diagmicrobio.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 8.Saubolle MA, Sussland D. 2003. Nocardiosis: review of clinical and laboratory experience. J Clin Microbiol 41:4497–4501. doi: 10.1128/JCM.41.10.4497-4501.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karamon J. 2014. Detection of Echinococcus multilocularis in faeces by nested PCR with the use of diluted DNA samples. Pol J Vet Sci 17:79–83. [DOI] [PubMed] [Google Scholar]
- 10.Hasegawa T, Gonoi T, Ito J, Kogure T, Yazawa K, Mikami Y. 2007. Identification of Nocardia farcinica by a PCR primer amplifying a specific DNA band for the bacterium. Nihon Ishinkin Gakkai Zasshi 48:173–175. doi: 10.3314/jjmm.48.173. [DOI] [PubMed] [Google Scholar]
- 11.Minero MV, Cercenado MME, Rabadan PM, Bouza E, Munoz P. 2009. Nocardiosis at the turn of the century. Medicine (Baltimore) 88:250–261. doi: 10.1097/MD.0b013e3181afa1c8. [DOI] [PubMed] [Google Scholar]
- 12.Vetor R, Murray CK, Mende K, Melton-Kreft R, Akers KS, Wenke J, Spirk T, Guymon C, Zera W, Beckius ML, Schnaubelt ER, Ehrlich G, Vento TJ. 2016. The use of PCR/electrospray ionization-time-of-flight-mass spectrometry (PCR/ESI-TOF-MS) to detect bacterial and fungal colonization in healthy military service members. BMC Infect Dis 16:338. doi: 10.1186/s12879-016-1651-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alanio A, Desoubeaux G, Sarfati C, Hamane S, Bergeron A, Azoulay E, Molina J, Derouin F, Menotti J. 2011. Real-time PCR assay-based strategy for differentiation between active Pneumocystis jirovecii pneumonia and colonization in immunocompromised patients. Clin Microbiol Infect 17:1531–1537. doi: 10.1111/j.1469-0691.2010.03400.x. [DOI] [PubMed] [Google Scholar]
- 14.Huggett J, Taylor M, Kocjan G, Evans H, Morris-Jones S, Gant V, Novak T, Costello A, Zumla A, Miller R. 2008. Development and evaluation of a real-time PCR assay for detection of Pneumocystis jirovecii DNA in bronchoalveolar lavage fluid of HIV-infected patients. Thorax 63:154–159. doi: 10.1136/thx.2007.081687. [DOI] [PubMed] [Google Scholar]
- 15.Ambrosioni J, Lew D, Garbino J. 2010. Nocardiosis: updated clinical review and experience at a tertiary center. Infection 38:89–97. doi: 10.1007/s15010-009-9193-9. [DOI] [PubMed] [Google Scholar]
- 16.Glupczynski Y, Berhin C, Janssens M, Wauters G. 2006. Determination of antimicrobial susceptibility patterns of Nocardia spp. from clinical specimens by Etest. Clin Microbiol Infect 12:905–912. doi: 10.1111/j.1469-0691.2006.01460.x. [DOI] [PubMed] [Google Scholar]
- 17.Munoz J, Mirelis B, Aragon LM, Gutierrez N, Sanchez F, Espanol M, Esparcia O, Gurgui M, Domingo P, Coll P. 2007. Clinical and microbiological features of nocardiosis 1997–2003. J Med Microbiol 56:545–550. doi: 10.1099/jmm.0.46774-0. [DOI] [PubMed] [Google Scholar]
- 18.Schlaberg R, Fisher MA, Hanson KE. 2014. Susceptibility profiles of Nocardia isolates based on current taxonomy. Antimicrob Agents Chemother 58:795–800. doi: 10.1128/AAC.01531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace RJ Jr, Steele LC, Sumter G, Smith JM. 1988. Antimicrobial susceptibility patterns of Nocardia asteroides. Antimicrob Agents Chemother 32:1776–1779. doi: 10.1128/AAC.32.12.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.