ABSTRACT
Carbapenemase-producing Pseudomonadaceae have increasingly been reported worldwide, with an ever-increasing heterogeneity of carbapenem resistance mechanisms, depending on the bacterial species and the geographical location. OXA-198 is a plasmid-encoded class D β-lactamase involved in carbapenem resistance in one Pseudomonas aeruginosa isolate from Belgium. In the setting of a multicenter survey of carbapenem resistance in P. aeruginosa strains in Belgian hospitals in 2013, three additional OXA-198-producing P. aeruginosa isolates originating from patients hospitalized in one hospital were detected. To reveal the molecular mechanism underlying the reduced susceptibility to carbapenems, MIC determinations, whole-genome sequencing, and PCR analyses to confirm the genetic organization were performed. The plasmid harboring the blaOXA-198 gene was characterized, along with the genetic relatedness of the four P. aeruginosa isolates. The blaOXA-198 gene was harbored on a class 1 integron carried by an ∼49-kb IncP-type plasmid proposed as IncP-11. The same plasmid was present in all four P. aeruginosa isolates. Multilocus sequence typing revealed that the isolates all belonged to sequence type 446, and single-nucleotide polymorphism analysis revealed only a few differences between the isolates. This report describes the structure of a 49-kb plasmid harboring the blaOXA-198 gene and presents the first description of OXA-198-producing P. aeruginosa isolates associated with a hospital-associated cluster episode.
KEYWORDS: IncP-11, carbapenemase, integron, outbreak, plasmid
INTRODUCTION
Resistance to carbapenems is increasing worldwide, leading to ever more restricted therapeutic choices (1). In Pseudomonas aeruginosa, resistance to β-lactams is mostly due to overproduction of the chromosome-encoded cephalosporinase, to cell wall impermeability, to alteration of the outer membrane protein OprD, or to overexpression of efflux pumps, often in association with the aforementioned mechanisms or with the acquisition of β-lactamases through horizontal gene transfer (HGT) (2). Several class B carbapenemases (i.e., VIM, IMP, NDM, and SPM) and to a lesser extent certain class A carbapenemases (i.e., GES and KPC) have been involved in the carbapenem resistance of P. aeruginosa (2–5). Since these carbapenemases are acquired transferable resistance determinants, they have the greatest clinical impact in hospitals worldwide (1, 4). Although they have been frequently observed in Enterobacteriaceae with OXA-48 and in Acinetobacter baumannii with OXA-23, OXA-40, OXA-58, and OXA-143, carbapenem-hydrolyzing class D β-lactamases (CHDLs) have been observed very rarely in P. aeruginosa, To the best of our knowledge, OXA-40 was detected in two clonally unrelated, imipenem-resistant, clinical isolates in Spain, OXA-48- and OXA-181-producing P. aeruginosa strains were isolated in India and in the United Kingdom (from a patient repatriated from India), respectively, and a novel CHDL, OXA-198, that belongs to a new group of class D β-lactamases was observed in a clinical isolate of P. aeruginosa in Belgium (6–10). In this work, we have characterized three additional OXA-198-producing P. aeruginosa isolates, along with their plasmids harboring the blaOXA-198 gene.
RESULTS AND DISCUSSION
Clinical characteristics of the four OXA-198-producing P. aeruginosa cases.
Clinical histories of the four cases are summarized in Table 1. Hospital A (first case in 2010) and hospital B (three later cases in 2012 and 2013) are located 120 km from each other, with no histories of patient transfer between the two institutions. Remarkably, none of the patients had traveled abroad. Isolates 5301-1 and 5301-2 were recovered in the same intensive care unit (ICU) and during the same time period, suggesting the occurrence of cross-transmission between the two patients. In contrast, the third patient (isolate 5301-4) was transferred from another Belgian hospital more than 6 weeks after the death of patient 5301-2, thus excluding the occurrence of direct patient-to-patient cross-transmission. The origin of colonization for the third patient remains unknown; it could have been due to a contaminated respiratory device or other possible environmental reservoirs.
TABLE 1.
Clinical features of the four OXA-198-producing P. aeruginosa cases
| Patient no. | Age (yr) | Sexa | Hospital | Date of hospitalization | Ward | Date of isolation | Site (colonization or infection) | Isolate | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 79 | M | A | April 2010 | ICU | April 2010 | Tracheal aspirates; VAP/sepsis (infection) | PA41437 | Meropenem (1 g t.i.d.) then ceftazidime (2 g t.i.d.) | Deceased (related to infection); cardiac insufficiency and acute renal failure |
| 2 | 67 | M | B | December 2012 | Medicine/ICU | December 2012 | Bronchial aspirate (colonization) | 5301-1 | None | Deceased (not directly related to infection) |
| 3 | 65 | F | B | February 2013 | Surgery/ICU | February 2013 | Tracheal aspirates, blood, and peritoneal fluid; postcolectomy, peritonitis/sepsis (infection) | 5301-2 | Amoxicillin-clavulanic acid then piperacillin-tazobactam then meropenem plus amikacin | Deceased (related to infection) |
| 4 | 65 | M | B | May 2013 | ICU | May 2013 | Tracheal aspirate; VAP/sepsis (infection) | 5301-4 | Piperacillin-tazobactam then ceftazidime | Deceased (related to infection) |
M, male; F, female; VAP, ventilator-associated pneumonia; t.i.d., three times a day.
Antimicrobial susceptibilities and clonal relationship.
All four P. aeruginosa isolates were resistant to penicillins and penicillin-inhibitor combinations and displayed decreased susceptibility to carbapenems (Table 2). They were also resistant to aminoglycosides and fluoroquinolones (highlighting their phenotype of multidrug resistance), but they remained susceptible to expanded-spectrum cephalosporins (ceftazidime [except 5301-4] and cefepime [PA41437]), to the ceftazidime-avibactam combination, and to colistin. The isolates 5301-4 and PA41437, which were already more resistant to expanded-spectrum cephalosporins, were also resistant to ceftolozane-tazobactam. All four strains belonged to serotype O11. Multilocus sequence typing (MLST) analysis of the four isolates revealed an allelic profile for acs, aro, gua, mut, nuo, pps, and trp of alleles 18, 4, 5, 3, 1, 17, and 13, respectively, which corresponded to sequence type 446 (Table 3), a type already reported for patients from different continents and clinical origins, as deposited in the PUBMLST database.
TABLE 2.
MICs of antimicrobial agents for the four OXA-198-producing P. aeruginosa isolates (broth microdilution method using Sensititre plates)
| Isolate | MIC (μg/ml)a |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TIC | TCC | PTZ | CAZ | CZA | C/T | FEP | IPM | MEM | AMK | TM | GM | CIP | CST | |
| 5301-1 | >128 | >128 | 32 | 2 | 2 | 4 | 1 | 16 | 2 | >64 | >64 | 8 | >32 | 2 |
| 5301-2 | >128 | >128 | 16 | 2 | 2 | 4 | 1 | 16 | 2 | >64 | >64 | 8 | >32 | 2 |
| 5301-4 | >128 | >128 | 64 | 16 | 4 | 8 | 4 | 32 | 4 | >64 | >64 | 8 | >32 | 2 |
| PA41437 | >128 | >128 | 16 | 2 | 2 | 8 | 16 | >32 | 16 | >64 | >64 | 16 | >32 | 1 |
TIC, ticarcillin; TCC, ticarcillin-clavulanic acid; PTZ, piperacillin-tazobactam; CAZ, ceftazidime; CZA, ceftazidime-avibactam; C/T, ceftolozane-tazobactam; FEP, cefepime; IPM, imipenem; MEM, meropenem; AMK, amikacin; TM, tobramycin; GM, gentamicin; CIP, ciprofloxacin; CST, colistin.
TABLE 3.
Genetic features of plasmid pOXA-198 and OXA-198-producing P. aeruginosa isolates
| Plasmid or strain | Total no. of nucleotides (contigs) | GC proportion (%) | Sequence typea | Resistome |
|---|---|---|---|---|
| pOXA-198 | 66,550 | 58.1 | ND | blaOXA-198, aacA7, catB7-like, QacEΔ1, sul1 |
| 5301-1 | 7,005,891 | 65.9 | 446 | blaOXA-198, blaOXA-50-like, blaPAO-like, fosA-like, aph(3′)-IIb, aacA7, catB7-like, cmlA1, QacEΔ1, sul1 |
| 5301-2 | 7,042,955 | 66.0 | 446 | blaOXA-198, blaOXA-50-like, blaPAO-like, fosA-like, aph(3′)-IIb, aacA7, catB7-like, cmlA1, QacEΔ1, sul1 |
| 5301-4 | 7,050,840 | 66.0 | 446 | blaOXA-198, blaOXA-50-like, blaPAO-like, fosA-like, aph(3′)-IIb, aacA7, catB7-like, cmlA1, QacEΔ1, sul1 |
| PA41437 | 7,100,158 | 66.0 | 446 | blaOXA-198, blaOXA-50-like, blaPAO-like, fosA-like, aph(3′)-IIb, aacA7, catB7-like, cmlA1, QacEΔ1, sul1 |
ND, not determined.
Together, these findings indicated that all four isolates were clonally related. Single-nucleotide polymorphism (SNP) analysis revealed that isolates 5301-1 and 5301-2 were identical, whereas 1 SNP was identified in 5301-4 and 31 SNPs were identified in PA41437, in comparison to 5301-1. These 31 SNPs could be explained by the fact that PA41437 was isolated in 2010 and the three other isolates were isolated in 2013. Interhospital spread of a single OXA-198-producing P. aeruginosa strain could not be evidenced by epidemiological data, since there was a time interval of 3 years, a distance of 120 km between the two hospitals, and an absence of any patient exchange or transfer between the two hospitals. It should be highlighted that, besides the three isolates reported here, no other OXA-198-producing P. aeruginosa isolates were identified in a multicenter survey conducted in 67 Belgian hospitals in 2013 (196 isolates intermediate/resistant to carbapenems and resistant to at least three classes of antibiotics) (Y. Glupczynski, unpublished data). Furthermore, in a similar and more recent Belgium multicenter survey performed in 2016 (163 multidrug-resistant, carbapenem-intermediate/resistant P. aeruginosa isolates obtained from patients from 56 different hospitals), no single OXA-198-producing P. aeruginosa isolate could be evidenced.
Besides the presence of the blaOXA-198 gene, the resistome of the four tested isolates revealed the presence of several resistance genes, including a catB7-like gene (98.4% nucleotide identity with catB7), qacEΔ1, and sul1 (conferring resistance to quaternary ammonium and sulfonamides, respectively) (Table 3). These resistance traits were carried by the plasmid pOXA-198 (Fig. 1A). Immediately downstream of the blaOXA-198 gene, a copy of aacA7 coding for AAC(6′)-I was detected. Other resistance determinants were identified from the whole-genome annotation, including a novel variant of the naturally occurring oxacillinase gene blaOXA-50-like (3 amino acid changes) and the chromosomal cephalosporinase gene blaPAO, as well as a fosA-like gene (99.0% identity with fosA), cmlA1, and a aph(3′)-IIb gene. Substitutions in topoisomerases were also investigated, to identify potential mutations that could explain the fluoroquinolone resistance. Typical substitutions in the quinolone-resistance-determining region (QRDR), i.e., T83I in GyrA and S87L and A587T in ParC, were identified. To explain the high MICs for imipenem, mutations in the OprD2-porin-coding genes causing alterations or modifications were investigated. A deletion of 88 nucleotides, encompassing the 5′ end of the oprD2 gene and the ribosomal binding site (RBS), was identified; the deletion resulted in loss of the RBS and the first 9 amino acids of OprD2, suggesting the absence of the OprD2 porin.
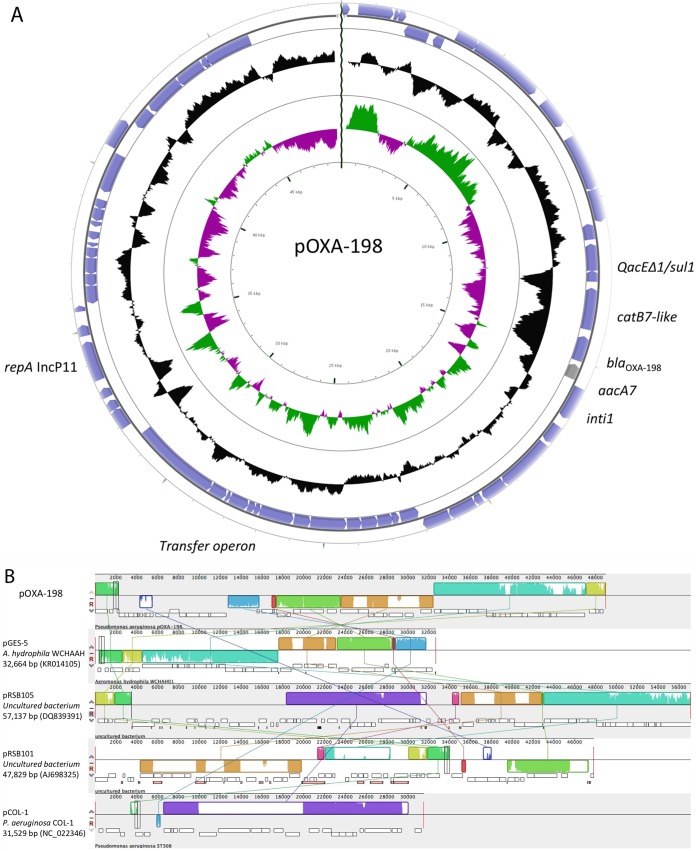
FIG 1.
Structure of pOXA-198. (A) Circular representation of pOXA-198, determined using CGview (http://wishart.biology.ualberta.ca/cgview). Coding sequences are represented by arrows. The two inner circles display CG content and CG skew. (B) Comparison with other plasmid backbones, using Mauve software (http://darlinglab.org/mauve/mauve.html). Multiple plasmid alignment was performed with the related plasmid pGES-5 (GenBank accession number KR014105), pRSB105 (GenBank accession number DQ839391), pRSB101 (GenBank accession number AJ698325), and pCOL-1 (GenBank accession number NC_022346). Conserved synteny is represented by boxes, and rearrangements are indicated by lines.
Structure of plasmid pOXA-198.
Sequencing yielded 20,490 high-quality reads, giving rise to 4,395,424 bp, which were derived from the pOXA-198 library. De novo assembly, performed using CLC Workbench 10.0.1 (Qiagen, Courtaboeuf, France), yielded 31 contigs, of which only 17 were derived from pOXA-198. After gap closures and verification of the position of each contig by PCR, the overall structure of pOXA-198 included a replication, a partition, partial conjugation, and accessory modules (Fig. 1A). This plasmid was untypeable using PCR-based replicon typing (PBRT) targeting enterobacterial plasmids and did not correspond to any of the 10 known IncP-type plasmids commonly identified in Pseudomonas species (11); therefore, we propose to name it IncP-11. Research on synteny was performed using Mauve software (Fig. 1B). This plasmid shared common features with pRSB101 (GenBank accession number AJ698325) and pRSB105 (GenBank accession number DQ839391), including the replication module. These two plasmids were identified from uncultured bacteria in activated sludge (12, 13). BLAST searches of GenBank revealed that IncP-11 was found on plasmid pRSB105 along with IncP-6 and on plasmid pRSB101, which harbored only IncP-11. This IncP-11 replicon confers the ability to replicate in P. aeruginosa, in Escherichia coli, and in Xanthomonas campestris (12). The partitioning system associated with the replicase showed high levels of identity with partitioning systems discovered in environmental bacteria such as Xanthomonas, Aeromonas, and Gulbenkiania species, thus suggesting a likely environmental origin for these IncP-11-bearing plasmids. Several genes showing homology with transfer operons have been identified, but a complete transfer operon is absent from this plasmid. This could explain why this plasmid could not be transferred by conjugation, as described previously (10).
Concerning the accessory genes identified within the plasmid sequence, the blaOXA-198 gene is embedded within a class 1 integron, together with the catB7-like gene (10). Several remnant structures of transposons, i.e., Tn402-like and TnAS3-like, both belonging to the Tn3 family of transposons, have also been identified within the pOXA198 sequence. These two transposons are likely to be inactive due to partial deletions. As for partitioning and replication systems, these not yet fully characterized transposons shared homology with transposons identified in soil bacteria. Another mobile element, similar to ISPa38, which was identified previously in P. aeruginosa, was also identified. The element present on plasmid pOXA-198 shares with ISPa38 only the presence of tnpA, the likely transposase gene, and it lacks the seven other open reading frames present in ISPa38 (14). The remaining part of the plasmid contains mainly hypothetical proteins with unknown function, which need to be characterized in more detail.
Comparison of pOXA-198 with pOXA-198 from three other P. aeruginosa isolates.
Mapping the reads obtained from whole-genome sequencing (WGS) of the four OXA-198-producing P. aeruginosa isolates against pOXA-198 revealed that the plasmids from the four isolates displayed 100% nucleotide sequence identity.
Conclusions.
We described here the molecular features of the blaOXA-198-harboring plasmid. This plasmid belonged to a novel IncP type that we named IncP-11, based on sequence homology. The transfer operon was partly deleted, explaining why the transfer by conjugation failed. This could limit the spread of this plasmid and favor clonal diffusion.
Genome analysis of the four OXA-198-producing P. aeruginosa isolates revealed that the isolates were clonally related. However, neither a reservoir nor any common source of transmission between the patients harboring P. aeruginosa OXA-198 isolates 5301-1 and 5301-2 and the last isolate (isolate 5301-4) could be detected. These isolates could be considered to belong to a cluster episode (rather than an outbreak) and could possibly reflect interregional spread of this clone, although this could not be demonstrated. Remarkably, only 31 SNPs were identified between the first isolate and the three later isolates, which were collected from patients in two different and distantly located hospitals without any obvious link or context. Overall, this study suggests that the spread of the blaOXA-198 gene is linked to a single P. aeruginosa clone rather than dispersion of a plasmid, which is not self-transferable.
MATERIALS AND METHODS
Bacterial strains, antimicrobial susceptibility, and PCR.
P. aeruginosa isolate PA41437 was originally isolated in 2010 from several lower respiratory tract specimens from a patient with acute exacerbation of chronic bronchitis who had been admitted to hospital A, as described previously (10). Three additional P. aeruginosa clinical isolates (5301-1, 5301-2, and 5301-4) were recovered from clinical specimens from patients hospitalized at hospital B in Belgium in 2013. The three OXA-198-positive P. aeruginosa strains were collected in the setting of a multicenter survey on antimicrobial resistance in P. aeruginosa that was organized by the Belgian national reference center in early 2014. The P. aeruginosa reference strain PAO1, P. aeruginosa PU21, and Escherichia coli TOP10 (Invitrogen, Merelbeke, Belgium), harboring the natural plasmid pOXA-198, were used (10, 15, 16).
Bacterial species identification was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with a Microflex LT mass spectrometer (Bruker Daltonik GmbH, Leipzig, Germany). In vitro determination of MICs for 12 antimicrobial agents was performed with the microdilution method, using customized Sensititre plates (BEGN3B; Trek Diagnostic Systems), and results were interpreted according to CLSI breakpoints (17).
Specific primers were used to detect the most common carbapenemase genes, as described previously (10). Detection of the blaOXA-198 gene was performed using primers OXA-41437FW (5′-CTCGAATTCATGCATAAACACATGAGTAAG-3′) and OXA-41437RV (5′-CTCAAGCTTTTATTCGATCCCCCTTT-3′) (10).
O serotyping of P. aeruginosa isolates.
O serotyping was performed for all four isolates with the slide agglutination test, using polyvalent antisera and 16 monovalent antisera (designated O1 to O16), according to the manufacturer's instructions (Bio-Rad, Marnes-La-Coquette, France).
DNA extractions.
Plasmid DNA from P. aeruginosa PA41437 was extracted using a plasmid maxiprep kit (Qiagen), according to the manufacturer's instructions. Total DNA was extracted from colonies using an UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Ozyme, Saint-Quentin, France), according to the manufacturer's instructions. The DNA concentrations and purity were controlled with a Qubit 2.0 fluorometer, using double-stranded DNA high-sensitivity and/or broad-range assay kits (Life Technologies) (18).
Whole-genome sequencing.
The DNA library for the plasmid extracted from E. coli strain TOP10 (pOXA-198) (2 by 250-bp paired-end reads) and those for the whole genomes of the four P. aeruginosa isolates (2 by 150-bp paired-end reads) were prepared using Nextera XT kits v2 (18). The libraries were run on a HiSeq automated system (Illumina, Paris, France). Reads were trimmed to remove poor-quality sequences (Q values of ≥20, with a word size of 34). De novo assembly was performed using CLC Workbench 9.0.1 (Qiagen). The acquired antimicrobial resistance genes were identified using the Resfinder server v2.1 (http://cge.cbs.dtu.dk/services/ResFinder-2.1) (19). MLST was performed in silico using the MLST server (https://cge.cbs.dtu.dk/services/MLST/), and SNP tree construction was performed using CSI phylogeny (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) (20, 21). The plasmid and genomes were annotated using the RAST server (22).
Contigs from plasmid pOXA-198 were considered DNA contamination if they showed at least 90% similarity with E. coli K-12 substrain MG1655 genomic DNA sequences (GenBank accession number NC_000913.2), using the BLASTn algorithm. Gap closures and verification of the position of each contig were performed by PCRs. Circular representation was obtained using DNA plotter (http://www.sanger.ac.uk/science/tools/dnaplotter) (23). Alignment to observe the synteny of pOXA-198 was performed using Mauve software (http://darlinglab.org/mauve/mauve.html) (24).
Electroporation.
Hypothetical natural plasmids were extracted and analyzed by electrophoresis on a 0.7% agarose gel, as described previously (18, 25). They were introduced into E. coli TOP10 by electroporation, using a Gene Pulser II (Bio-Rad). Recombinant clones were selected on tryptic soy agar supplemented with 50 μg/ml ticarcillin (Sigma, St. Quentin Fallavier, France).
Accession number(s).
The nucleotide sequence data reported in this paper have been deposited in the EMBL/GenBank nucleotide sequence database under accession numbers MG958650 for pOXA-198 and NMPW00000000, NMPV00000000, NMPU00000000, and NMPT00000000 for P. aeruginosa PA41437, 5301-1, 5301-2, and 5301-4, respectively.
ACKNOWLEDGMENTS
We are very grateful to Dilek Imanci from the sequencing platform of Hôpital Bicêtre and to Linda Tlili for technical assistance in plasmid reconstruction.
This work was supported by the Assistance Publique-Hôpitaux de Paris, by a grant from Université Paris Sud (grant EA 7361), and by the LabEx LERMIT, supported by a grant from the French National Research Agency (grant ANR-10-LABX-33). This work was also funded in part by a grant from the Joint Program Initiative on Antimicrobial Resistance (grant ANR-14-JAMR-0002). The Belgian national reference center is partially supported by the Belgian Ministry of Social Affairs, through a fund within the health insurance system.
We have no conflicts of interest to declare.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland: http://www.who.int/drugresistance/documents/surveillancereport/en. [Google Scholar]
- 2.Nordmann P, Naas T. 2009. P. aeruginosa and β-lactams, p 157–174. In Courvalin P, LeClercq R, Rice LB (ed), Antibiogram. ASM Press, Washington, DC. [Google Scholar]
- 3.Naas T, Dortet L, Iorga BI. 2016. Structural and functional aspects of class A carbapenemases. Curr Drug Targets 17:1006–1028. doi: 10.2174/1389450117666160310144501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potron A, Poirel L, Nordmann P. 2015. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 45:568–585. doi: 10.1016/j.ijantimicag.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob Agents Chemother 54:24–38. doi: 10.1128/AAC.01512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sevillano E, Gallego L, García-Lobo JM. 2009. First detection of the OXA-40 carbapenemase in P. aeruginosa isolates, located on a plasmid also found in A. baumannii. Pathol Biol (Paris) 57:493–495. doi: 10.1016/j.patbio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Borah VV, Saikia KK, Hazarika NK. 2016. First report on the detection of OXA-48 β-lactamase gene in Escherichia coli and Pseudomonas aeruginosa co-infection isolated from a patient in a tertiary care hospital in Assam. Indian J Med Microbiol 34:252–253. doi: 10.4103/0255-0857.176842. [DOI] [PubMed] [Google Scholar]
- 9.Meunier D, Doumith M, Findlay J, Mustafa N, Mallard K, Anson J, Panagea S, Pike R, Wright L, Woodford N, Hopkins KL. 2016. Carbapenem resistance mediated by blaOXA-181 in Pseudomonas aeruginosa. J Antimicrob Chemother 71:2056–2057. doi: 10.1093/jac/dkw087. [DOI] [PubMed] [Google Scholar]
- 10.El Garch F, Bogaerts P, Bebrone C, Galleni M, Glupczynski Y. 2011. OXA-198, an acquired carbapenem-hydrolyzing class D β-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother 55:4828–4833. doi: 10.1128/AAC.00522-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shintani M, Takahashi Y, Yamane H, Nojiri H. 2010. The behavior and significance of degradative plasmids belonging to Inc groups in Pseudomonas within natural environments and microcosms. Microbes Environ 25:253–265. doi: 10.1264/jsme2.ME10155. [DOI] [PubMed] [Google Scholar]
- 12.Szczepanowski R, Krahn I, Linke B, Goesmann A, Pühler A, Schlüter A. 2004. Antibiotic multiresistance plasmid pRSB101 isolated from a wastewater treatment plant is related to plasmids residing in phytopathogenic bacteria and carries eight different resistance determinants including a multidrug transport system. Microbiology 150:3613–3630. doi: 10.1099/mic.0.27317-0. [DOI] [PubMed] [Google Scholar]
- 13.Schlüter A, Szczepanowski R, Kurz N, Schneiker S, Krahn I, Pühler A. 2007. Erythromycin resistance-conferring plasmid pRSB105, isolated from a sewage treatment plant, harbors a new macrolide resistance determinant, an integron-containing Tn402-like element, and a large region of unknown function. Appl Environ Microbiol 73:1952–1960. doi: 10.1128/AEM.02159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pragasam AK, Yesurajan F, Priya Doss G, George B, Ragupathi NKD, Walia K, Veeraraghavan B. 2016. Draft genome sequence of extremely drug-resistant Pseudomonas aeruginosa (ST357) strain CMC_VB_PA_B22862 isolated from a community-acquired bloodstream infection. Genome Announc 4:e01092-16. doi: 10.1128/genomeA.01092-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girlich D, Bonnin RA, Jousset A, Naas T. 2017. Promoter characterization and expression of the blaKPC-2 gene in Escherichia coli, Pseudomonas aeruginosa and Acinetobacter baumannii. J Antimicrob Chemother 72:1597–1601. doi: 10.1093/jac/dkx044. [DOI] [PubMed] [Google Scholar]
- 16.Dortet L, Flonta M, Boudehen YM, Creton E, Bernabeu S, Vogel A, Naas T. 2015. Dissemination of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa in Romania. Antimicrob Agents Chemother 59:7100–7103. doi: 10.1128/AAC.01512-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 18.Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, Dortet L, Glaser P, Glupczynski Y, Naas T. 2017. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 61:e01697-16. doi: 10.1128/AAC.01697-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. 2014. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9(8):e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. doi: 10.1016/0147-619X(84)90063-5. [DOI] [PubMed] [Google Scholar]