ABSTRACT
Ten IMP-8-producing Escherichia coli isolates were recovered from surveillance cultures of a neonatal intensive care unit; eight of the isolates were clonally related. A 168.2-kb blaIMP-8 plasmid was fully sequenced, and it corresponded to the recently described IncA/C1-ST13 plasmid. This plasmid was detected in all isolates, even in those that were not clonally related. One unrelated isolate was also resistant to colistin and positive for mcr-1. This marker was located in a 62.7-kb IncI2 plasmid, which was also fully sequenced.
KEYWORDS: Escherichia coli, IMP-8 metallo-beta-lactamase, IncA/C1 plasmid, IncI2 plasmid, mcr-1
TEXT
The emergence and spread of carbapenemase-producing bacteria are major concerns for public health systems worldwide. IMP-type metallo-β-lactamases (MBLs) were first identified in the early 1990s in Pseudomonas aeruginosa in Japan and since then have been globally reported, mostly in P. aeruginosa and in other nonfermenting Gram-negative bacilli (1). Studies performed in Argentina reported the presence of IMP-13 in P. aeruginosa and IMP-8 in Enterobacter cloacae (2–4). IMP-8 was initially described in Klebsiella pneumoniae in Taiwan, where it became the dominant MBL among Enterobacteriaceae (5). The presence of the blaIMP-8 gene was reported in nosocomial and environmental E. coli isolates in association with conjugative plasmids belonging to IncA/C and IncFIB, respectively. As in other MBL-coding genes, blaIMP-8 was found to be located in a class 1 integron (4–6).
The rising frequency of carbapenem-resistant Enterobacteriaceae infections prompted the use of colistin as a last therapeutic option. However, the scenario became more complex as a consequence of the silent spread of plasmid-carried mcr-1 in the past decade (7, 8).
The aim of this study was to characterize carbapenem-resistant E. coli isolates recovered from the active surveillance cultures of a neonatal intensive care unit (NICU) at a hospital in Buenos Aires, Argentina. Surveillance cultures are routinely conducted on patients admitted to the NICU in this hospital. During November to December 2016, 10 carbapenem-resistant E. coli isolates were recovered from seven patients on CHROMagar KPC (K. pneumoniae carbapenemase). Antibiotic susceptibility was assessed by microdilution tests according to the CLSI guidelines and the use of automated systems (Vitek 2, bioMérieux, France). All isolates were found to be resistant to trimethoprim-sulfamethoxazole and cephalosporins, with intermediate susceptibility or resistance to imipenem and/or meropenem (9). Nine of the 10 isolates were resistant to amikacin, gentamicin, and ciprofloxacin, and they displayed a wild-type phenotype with respect to colistin (i.e., they were susceptible according the EUCAST guidelines). The one remaining isolate was categorized as non-wild type for colistin (i.e., resistant according the EUCAST guidelines), and it was intermediate for aminoglycosides and ciprofloxacin (Table 1) (9). All isolates showed a positive synergy test result with EDTA (1 μmol), suggesting the presence of an MBL (10, 11). PCR amplifications for the most common MBL-coding genes were conducted with specific primers and plasmid DNA as the template (11–13). Nucleotidic sequences of the amplified fragments showed 100% identity with blaIMP-8 for all samples. This marker was located on a conjugative plasmid, which was successfully transferred to E. coli CAG 12177. According to a PCR-based replicon typing method proposed previously, the plasmid corresponded to the IncA/C group, similarly to those seen in isolates previously reported in Singapore (5, 14).
TABLE 1.
Clinical data and microbiological characteristics of blaIMP-8-harboring isolatesa
| Newborn | Bacterial isolate/day of isolationb/no. of days of NICU stay before isolation | Treatment/no. of days of treatment | Risk factor/GA/wt of newborn (g) | Antimicrobial susceptibility (MIC [μg/ml])c |
Pulsotype | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | AMS | PTZ | FEP | CAZ | CTX | IMP | MEM | GEN | AKN | CIP | COL | TMS | |||||
| 1 | G2116/1/89 | MEM/21, COL/21, VAN/21 | Preterm birth/30 wks/870 | >32 | >32 | >128 | >64 | >64 | >64 | 8 | 16 | >16 | >64 | 1 | 2 | >320 | I |
| 2 | G1116/13/40 | AM/10, GEN/7, CTX/5, AKN/5, COL/8 | Preterm birth/28 wks/1,260 | >32 | >32 | >128 | 16 | >64 | >64 | 4 | 8 | >16 | >64 | 1 | 1 | >320 | I |
| G1216/28/56 | >32 | >32 | >128 | >64 | >64 | >64 | 8 | 8 | >16 | >64 | 1 | 1 | >320 | II | |||
| G1316/41/69 | >32 | >32 | >128 | >64 | >64 | >64 | 2 | 8 | >16 | >64 | 1 | 1 | >320 | I | |||
| 3 | G4116/28/41 | AM/10, GEN/7 | Term birth/40 wks/3,370 | >32 | >32 | >128 | >64 | >64 | >64 | 8 | 16 | >16 | >64 | 1 | 1 | >320 | I |
| 4 | G3116/28/8 | CTX/4, AKN/4 | Term birth/38 wks/3,210 | >32 | >32 | >128 | >64 | >64 | >64 | 8 | 8 | >16 | >64 | 1 | 2 | >320 | I |
| G3216/41/21 | >32 | >32 | >128 | >64 | >64 | >64 | 4 | 8 | 8 | 32 | <0.25 | 8 | >320 | III | |||
| 5 | G6116/43/16 | AM/5, GEN/5 | Preterm birth/34 wks/2,060 | >32 | >32 | >128 | >64 | >64 | >64 | 2 | 8 | >16 | >64 | 1 | 1 | >320 | I |
| 6 | G7116/43/28 | VAN/12, MEM/12 | Preterm birth/32 wks/1,610 | >32 | >32 | 128 | >64 | >64 | >64 | 2 | 8 | >16 | >64 | 1 | 1 | >320 | I |
| 7 | G5116/49/41 | Preterm birth/37 wks/2,500 | >32 | >32 | >128 | >64 | >64 | >64 | 2 | 8 | >16 | >64 | 1 | 1 | >320 | I | |
AM, ampicillin; AMS, ampicillin-sulbactam; PTZ, piperacillin-tazobactam; FEP, cefepime; CAZ, ceftazidime; CTX, cefotaxime; IMP, imipenem; MEM, meropenem; GEN, gentamicin; AKN, amikacin; CIP, ciprofloxacin; COL, colistin; TMS, trimethoprim-sulfamethoxazole; VAN, vancomycin; GA, gestational age.
For the data representing the day of isolation, day 1 corresponds to the index case; the other days of isolation were determined with respect to that of the index case.
MICs for IMP, MEM, and COL were assessed by manual procedures, whereas those of the others were assessed by automated methods (Vitek 2). Data corresponding to the mcr-1-harboring isolate are indicated in bold.
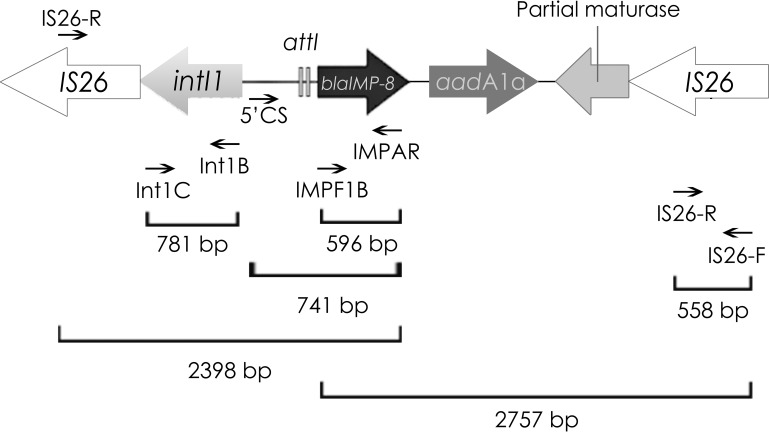
blaIMP-8 was located in a 168.2-kb plasmid, which was purified by the standard plasmid DNA phenol-chloroform purification protocol initially proposed by Kado and Liu, adding two extraction steps performed with chloroform to remove any remaining phenol (15). Purified plasmids were fully sequenced by the use of a MiSeq sequencer (Illumina). The sequencing reads were assembled using SPAdes V3.9 with the following statistical parameters: largest contig, 153,183 bp; N50, 90,320 bp; L50, 2. The plasmid presented 227 open reading frames and had an average of 50.3% G+C content. Genes were predicted and annotated using the RAST tool and PROKKA software and were also manually curated (GenBank accession no. [AN] MG550958) (16). Using the ResFinder tool, other resistance markers such as blaTEM-1B, aadA1, aph(3′)VIa, and sul1 were detected in the same plasmid. blaIMP-8 was associated with a class 1 integron flanked by two IS26 elements. The blaIMP-8-carrying integron lacked the typical 3′ conserved sequence (qacEΔ1 and sul1) but instead harbored a truncated sequence of a retron-type RNA-directed DNA polymerase (maturase), which has been reported to be likely involved in cassette gene generation (17, 18) (Fig. 1). The genetic platform for blaIMP-8 was confirmed in all isolates by PCR mapping and sequencing performed with the primers shown in Fig. 1. Plasmid multilocus sequence typing (pMLST) was performed to determine the IncA/C replicon type (https://pubmlst.org/plasmid/). The IncA/C1 replicon, detected in this plasmid, was coincident with data corresponding to the recently deposited sequence type 13 (ST13) plasmid from Citrobacter freundii, which was isolated 20 years ago in Argentina. These plasmids are closely related to the ST11 IncA/C1 RA1 plasmid and to the recently published ST12 (19, 20).
FIG 1.
Genetic context of plasmid-borne blaIMP-8 detected in this study (MG550958). The primers used for PCR mapping were as follows: IMP-F1B (GTTTTGTAGCATTGCTACCGCAG) and IMP-AR (GTTTTGCCTTACCATATTTGGA), IS26-F (TCACTCCACGATTTACCGCT) and IS26-R (CTTACCAGGCGCATTTCGCC), Int1B (GCGTTCGGTCAAGGTTCTGG) and Int1C (CGTGATGCCTGCTTGTTCTA), and 5′CS (GCTTGCTGCTTGGATGCC).
As previously mentioned, one isolate (E. coli G3216) was also colistin resistant and rendered a positive PCR result for mcr-1; this gene was located in a conjugative plasmid that was successfully transferred to E. coli CAG 12177. In accordance with previous reports, the full sequence of the mcr-1-harboring plasmid (pG3216) showed that mcr-1 was flanked upstream by pap2 (type 2 phosphatidic acid phosphatase) and downstream by the relaxase NikB-coding gene (GenBank AN MF693349). This plasmid was 62.7 kb in size and presented 88 open reading frames with an average of 42.5% G+C content. It did not harbor any further resistance gene and was almost identical (99% identity) to that previously reported in our country (GenBank AN KY471314) (21). Using the PlasmidFinder tool, pG3216 was found to be associated with the IncI2 group. Hence, hicA and hicB, which are related to the IncI2 incompatibility group and involved in type II toxin-antitoxin (TA) systems, were identified (22). The coding genes for the toxin RelE and the antitoxin StbE, which belong to the RelE/ParE TA system superfamily, were located together (23). Similarly to other IncI2 plasmids, pG3216 displayed a typical backbone responsible for plasmid replication, maintenance, and self-transfer by conjugation (24).
The E. coli phylogenetic group was determined according to the method previously described by Clermont et al. (25), and all of the isolates corresponded to phylogroup D (25). The clonal relationship was investigated by the use of XbaI pulsed-field gel electrophoresis (XbaI-PFGE) and MLST (http://enterobase.warwick.ac.uk/species/ecoli/allele_st_search). Eight of the 10 isolates were clustered in pulsotype I and did not correspond to any of the assigned sequence types (adk 332, fumC 594, gyrB 428, icd 517, mdh 292, purA 373, recA 262). E. coli G1216 presented a different pulsotype (pulsotype II), which corresponded to, among others, a single-locus variant (SLV) of ST69 (CC69) (adk 21, fumC 35, gyrB 27, icd 6, mdh 286, purA 5, recA 4). The mcr-1-positive E. coli G3216 isolate represented pulsotype III, which corresponds to a SLV of ST5377 and ST7395 (adk 35, fumC 37, gyrB 29, icd 25, mdh 416, purA 564, recA 73). These results indicate that the isolates included in this study do not belong to the STs in which blaIMP-8 was previously reported (ST131, ST359, ST457, and ST410) (5, 26).
Despite the fact that IMP-8-producing Enterobacteriaceae are frequently detected in Asia, in our country (Argentina), they have been encountered only sporadically (4). Both CHROMagar KPC and EDTA-based synergy tests were useful to perform early and accurate detection of colonized neonates with MBL-producing E. coli, thereby contributing to the reduction of the spread of such microorganisms and probably of the subsequent infections. E. coli phylogroup D includes extraintestinal pathogens and multidrug-resistant isolates (27). Although these strains are reported to be responsible for severe diseases, none of the seven neonates included in this study developed IMP-8 E. coli infections. Moreover, none of them received antibiotic treatment for this colonization and all of them were medically discharged. Dissemination of E. coli which produced IMP-8 MBL and belonged to pulsotype I occurred in this neonatal ward until hygiene measures and contact isolation were strengthened. It can be speculated that horizontal transmission of the blaIMP-8 Inc A/C1 plasmid may have occurred, as this marker was also detected in the unrelated isolates; E. coli G1216 (pulsotype II) recovered from a patient also colonized with pulsotype I E. coli and E. coli G3216 (pulsotype III), which also harbored the IncI2 plasmid containing mcr-1 (Table 1). Furthermore, the class 1 integron platform containing blaIMP-8 could be self-mobilized as it was flanked by two identical IS26 copies, which could act as composite transposons (28).
Plasmid-mediated colistin resistance, commonly found in carbapenem-susceptible strains, has also been reported in Enterobacteriaceae producing different carbapenemases such as VIM-1, VIM-2, NDM-5, NDM-9, IMP-4, KPC-2, and OXA-48 (29–35). In our country, mcr-1 was previously described both in clinical isolates and in poultry farms (8, 36). Here, we have described the presence of mcr-1 in a blaIMP-8-producing E. coli isolate, which adds an extra level of plasticity in the evolving epidemiology of carbapenem and colistin resistance.
Our results highlight the importance of establishing screening schemes and rapid laboratory diagnostic tests to ensure the implementation of efficient infection control measures.
Accession number(s).
The complete sequences of the plasmids analyzed in this study have been deposited at DDBJ/EMBL/GenBank under accession numbers MG550958 and MF693349.
ACKNOWLEDGMENTS
We thank V. Pebe for his collaboration and for providing the clinical data.
This study was supported by UBACyT grants to M.R. and G.G. (20020150100174BA and 20020130100432BA), by PICT grants to M.R., G.G., and D.C. (2013-0858, 2015-1925, and 2015-2844), and by PIP grant 11220120100400CO to G.G. and M.R.
REFERENCES
- 1.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. 1991. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 35:147–151. doi: 10.1128/AAC.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomez S, Rapoport M, Togneri A, Viegas-Caetano J, Faccone D, Corso A, Petroni A, Pasteran F. 2011. Emergence of metallo-beta-lactamases in Enterobacteriaceae from Argentina. Diagn Microbiol Infect Dis 69:94–97. doi: 10.1016/j.diagmicrobio.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 3.Santella G, Cuirolo A, Almuzara M, Palombarani S, Sly G, Radice M, Gutkind G. 2010. Full resistance and decreased susceptibility to carbapenems in IMP-13-producing Pseudomonas aeruginosa isolates from an outbreak. Antimicrob Agents Chemother 54:1381–1382. doi: 10.1128/AAC.00399-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Togneri AM, Gomez SA, Podesta LB, Perez MP, Faccone DF, Rios LE, Ganetea MA, Anchordoqui MS, Pasteran FG, Corso AC. 2013. Dissemination of blaIMP-8 among Enterobacteriaceae isolates from a Buenos Aires hospital. Rev Argent Microbiol 45:104–109. (In Spanish.) [DOI] [PubMed] [Google Scholar]
- 5.Koh TH, Cao D, Tee NW, Teo JW. 2014. Escherichia coli with bla(IMP-8) in Singapore. Antimicrob Agents Chemother 58:617. doi: 10.1128/AAC.01754-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kieffer N, Poirel L, Bessa LJ, Barbosa-Vasconcelos A, da Costa PM, Nordmann P. 2016. VIM-1, VIM-34, and IMP-8 carbapenemase-producing Escherichia coli strains recovered from a Portuguese river. Antimicrob Agents Chemother 60:2585–2586. doi: 10.1128/AAC.02632-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez JE, Figueroa Espinosa RA, Redondo LM, Cejas D, Gutkind GO, Chacana PA, Di Conza JA, Fernandez-Miyakawa ME. 2017. Plasmid-mediated colistin resistance in Escherichia coli recovered from healthy poultry. Rev Argent Microbiol 49:297–298. doi: 10.1016/j.ram.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 9.CLSI. 2017. Performance standards for antimicrobial susceptibility testing. vol 37, approved standard M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 10.Arakawa Y, Shibata N, Shibayama K, Kurokawa H, Yagi T, Fujiwara H, Goto M. 2000. Convenient test for screening metallo-beta-lactamase-producing gram-negative bacteria by using thiol compounds. J Clin Microbiol 38:40–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagniez G, Radice M, Cuirolo A, Rodriguez H, Vay C, Famiglietti A, Gutkind G. 2006. Prevalence of metallo-beta-lactamase in carbapenem resistant Pseudomonas aeruginosa at a university hospital of Buenos Aires City. Rev Argent Microbiol 38:33–37. [PubMed] [Google Scholar]
- 12.Cejas D, Almuzara M, Santella G, Tuduri A, Palombarani S, Figueroa S, Gutkind G, Radice M. 2008. Phenotypic and genotypic characterization of imipenem-resistant Pseudomonas aeruginosa isolated in a Buenos Aires hospital. Rev Argent Microbiol 40:238–245. (In Spanish.) [PubMed] [Google Scholar]
- 13.Poirel L, Dortet L, Bernabeu S, Nordmann P. 2011. Genetic features of blaNDM-1-positive Enterobacteriaceae. Antimicrob Agents Chemother 55:5403–5407. doi: 10.1128/AAC.00585-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 15.Kado CI, Liu ST. 1981. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol 145:1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 17.Centrón D, Roy PH. 2002. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob Agents Chemother 46:1402–1409. doi: 10.1128/AAC.46.5.1402-1409.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sunde M. 2005. Class I integron with a group II intron detected in an Escherichia coli strain from a free-range reindeer. Antimicrob Agents Chemother 49:2512–2514. doi: 10.1128/AAC.49.6.2512-2514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock SJ, Phan MD, Peters KM, Forde BM, Chong TM, Yin WF, Chan KG, Paterson DL, Walsh TR, Beatson SA, Schembri MA. 24 January 2017. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother doi: 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esposito EP, Gaiarsa S, Del Franco M, Crivaro V, Bernardo M, Cuccurullo S, Pennino F, Triassi M, Marone P, Sassera D, Zarrilli R. 2017. A novel IncA/C1 group conjugative plasmid, encoding VIM-1 metallo-beta-lactamase, mediates the acquisition of carbapenem resistance in ST104 Klebsiella pneumoniae isolates from neonates in the intensive care unit of V. Monaldi hospital in Naples. Front Microbiol 8:2135. doi: 10.3389/fmicb.2017.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tijet N, Faccone D, Rapoport M, Seah C, Pasteran F, Ceriana P, Albornoz E, Corso A, Petroni A, Melano RG. 2017. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS One 12:e0180347. doi: 10.1371/journal.pone.0180347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li G, Shen M, Lu S, Le S, Tan Y, Wang J, Zhao X, Shen W, Guo K, Yang Y, Zhu H, Rao X, Hu F, Li M. 2016. Identification and characterization of the HicAB toxin-antitoxin system in the opportunistic pathogen Pseudomonas aeruginosa. Toxins (Basel) 8:113. doi: 10.3390/toxins8040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unterholzner SJ, Poppenberger B, Rozhon W. 2013. Toxin-antitoxin systems: biology, identification, and application. Mob Genet Elements 3:e26219. doi: 10.4161/mge.26219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun J, Li XP, Yang RS, Fang LX, Huo W, Li SM, Jiang P, Liao XP, Liu YH. 2016. Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob Agents Chemother 60:5014–5017. doi: 10.1128/AAC.00774-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan JJ, Tsai LH, Wu JJ. 2012. Emergence of the IMP-8 metallo-β-lactamase in the Escherichia coli ST131 clone in Taiwan. Int J Antimicrob Agents 40:281–282. doi: 10.1016/j.ijantimicag.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 28.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du H, Chen L, Tang YW, Kreiswirth BN. 2016. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect Dis 16:287–288. doi: 10.1016/S1473-3099(16)00056-6. [DOI] [PubMed] [Google Scholar]
- 30.Mulvey MR, Mataseje LF, Robertson J, Nash JH, Boerlin P, Toye B, Irwin R, Melano RG. 2016. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 16:289–290. doi: 10.1016/S1473-3099(16)00067-0. [DOI] [PubMed] [Google Scholar]
- 31.Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. 2016. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 16:281. doi: 10.1016/S1473-3099(16)00006-2. [DOI] [PubMed] [Google Scholar]
- 32.Yao X, Doi Y, Zeng L, Lv L, Liu JH. 2016. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis 16:288–289. doi: 10.1016/S1473-3099(16)00057-8. [DOI] [PubMed] [Google Scholar]
- 33.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Kasbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T. 2016. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis 16:282–283. doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhang XF, Doi Y, Huang X, Li HY, Zhong LL, Zeng KJ, Zhang YF, Patil S, Tian GB. 2016. Possible transmission of mcr-1-harboring Escherichia coli between companion animals and human. Emerg Infect Dis 22:1679–1681. doi: 10.3201/eid2209.160464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon M, Melzl H, Hiergeist A, Richert K, Falgenhauer L, Pfeifer Y, Gerlach RG, Fuchs K, Reischl U, Gessner A, Jantsch J. 2017. Colistin- and carbapenem-resistant Klebsiella oxytoca harboring blaVIM-2 and an insertion in the mgrB gene isolated from blood culture. Int J Med Microbiol 307:113–115. doi: 10.1016/j.ijmm.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, Corso A. 2016. First description of mcr-1-mediated colistin resistance in human infections caused by Escherichia coli in Latin America. Antimicrob Agents Chemother 60:4412–4413. doi: 10.1128/AAC.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]