ABSTRACT
Co-trimoxazole is one of the antimicrobials of choice for treating Stenotrophomonas maltophilia infections. Most works on the molecular epidemiology of the resistance to this drug combination are based on the analysis of sul genes. Nevertheless, the existence of clinical co-trimoxazole-resistant S. maltophilia isolates that do not harbor sul genes has been reported. To investigate potential mutations that can reduce the susceptibility of S. maltophilia to co-trimoxazole, spontaneous S. maltophilia co-trimoxazole-resistant mutants isolated under different co-trimoxazole concentrations were studied. All mutants presented phenotypes compatible with the overexpression of either SmeVWX (94.6%) or SmeDEF (5.4%). Indeed, the analysis of a selected set of strains showed that the overexpression of either of these efflux pumps, which was due to mutations in their regulators smeRv and smeT, respectively, was the cause of co-trimoxazole resistance. No other efflux pump was overexpressed in any of the studied mutants, indicating that they do not participate in the observed resistance phenotype. The analysis of mutants overexpressing or lacking SmeDEF or SmeVWX shows that SmeDEF contributes to the intrinsic and acquired resistance to co-trimoxazole in S. maltophilia, whereas SmeVWX only contributes to acquired resistance. It is important to highlight that all mutants were less susceptible to other antibiotics, including chloramphenicol and quinolones. Since both SmeVWX and SmeDEF are major determinants of quinolone resistance, the potential cross-selection of resistance to co-trimoxazole and quinolones, when either of the antimicrobials is used, is of particular concern for the treatment of S. maltophilia infections.
KEYWORDS: co-trimoxazole, SmeDEF, SmeVWX, Stenotrophomonas maltophilia
INTRODUCTION
Stenotrophomonas maltophilia is an opportunistic pathogen that is responsible for nosocomial infections, mainly in patients with underlying diseases (1), with particular relevance in the case of cystic fibrosis patients (2–4). In addition, S. maltophilia is considered a prototype of intrinsically resistant organisms due to its low susceptibility to several of the antimicrobials used in clinical practice, a phenotype that is at least partly due to the presence of several genes encoding antibiotic resistance determinants in its genome (5–7). In addition, in vitro studies and analyses of clinical isolates have shown that strains presenting even higher levels of resistance due to mutation or to the acquisition of resistance genes are not infrequent (6–12). One of the antimicrobials used for the treatment of S. maltophilia is co-trimoxazole, a combination of the antibiotics trimethoprim and sulfamethoxazole, which target different steps of the folic acid biosynthesis pathway. Unfortunately, the percentage of S. maltophilia co-trimoxazole-resistant isolates has increased in recent years.
The presence of the sul genes (sul1 and sul2), which encode variants of the dihydropteroate synthase that are not inhibited by sulfonamides, has been proposed to be a major cause of co-trimoxazole resistance in S. maltophilia (13–15). These elements, which are involved in sulfonamide resistance, are frequently present in integrons and in mobile elements, a situation that helps their dissemination among several different microorganisms, including S. maltophilia. Nevertheless, despite the presence of sul genes in some S. maltophilia co-trimoxazole-resistant isolates, resistant strains that do not carry sul genes have been isolated as well (16, 17). This situation indicates that S. maltophilia may possess mechanisms of co-trimoxazole resistance besides the presence of sul genes in its genome that remain to be established.
Indeed, different studies have shown that the activity of some efflux pumps might affect S. maltophilia co-trimoxazole susceptibility. Among these studies, it has been shown that the overexpression of SmeDEF reduces the susceptibility to co-trimoxazole of S. maltophilia, while the inactivation of this efflux pump increases the bacterial susceptibility to this antimicrobial (18). Other studies have shown that the deletion of the efflux pump SmeYZ or of the outer membrane protein TolC, the latter required for the activity of the efflux pump SmeOP, increases the susceptibility of S. maltophilia to co-trimoxazole (19–21). These findings reinforce the idea that, in addition to the acquisition of sul genes, there are other mechanisms, such as via efflux pumps, which could contribute to the acquisition of resistance to co-trimoxazole in S. maltophilia.
To identify these potential mechanisms, we have selected S. maltophilia co-trimoxazole-resistant mutants at different concentrations of co-trimoxazole and have determined the underlying mechanisms of resistance in the selected mutants. The results shed light on the ignored aspects of S. maltophilia co-trimoxazole resistance and show that the main mechanism of mutation-driven co-trimoxazole resistance is the overexpression of efflux pumps, mainly of SmeVWX but also, in minor proportions, of SmeDEF. In both cases, the overexpression is associated with mutations in the regulator genes of these efflux pumps, smeRv and smeT, respectively.
RESULTS
Isolation and phenotypic characterization of spontaneous co-trimoxazole-resistant strains.
Spontaneous co-trimoxazole-resistant mutants of S. maltophilia strain D457 were selected in Mueller-Hinton agar plates containing 2, 4, 8, 16, or 32 mg/liter of co-trimoxazole. Here, we use the operational definition of resistance by which a strain is considered resistant if “it has a higher MIC value for the studied compound than its parental wild-type strain” (22). Ninety-four mutants isolated from plates containing different amounts of co-trimoxazole were chosen for further analysis. After isolation, the putative mutants were regrown two sequential times in plates without antibiotics and afterwards reseeded in antibiotic-containing plates. All of them, with one exception, grew in the presence of co-trimoxazole, indicating that in most of the mutants, the observed phenotype of resistance was not due to the transient adaptation of S. maltophilia to the presence of the antibiotic.
To analyze the susceptibility of the 93 mutants, these strains, as well as the parental strain D457 and its derivative D457R that overexpresses SmeDEF (Table 1), were grown in a 96-well microtiter plate and seeded in antibiotic-containing plates with the aid of a 96-pin replicator as described in Material and Methods. As Fig. 1 and Table S1 in the supplemental material show, co-trimoxazole-resistant mutants display different phenotypes of susceptibility to the tested antibiotics. These differential phenotypes enabled the classification of S. maltophilia co-trimoxazole-resistant mutants into four groups: chloramphenicol and quinolone resistant (84 strains); quinolone resistant (4 strains); chloramphenicol, quinolone, and erythromycin resistant (3 strains), and quinolone and erythromycin resistant (2 strains). The types of mutants isolated varied according to the amount antibiotic used for the selection. As expected, the variability of the phenotypes is higher at the lowest concentration of the selective agent. The fact that resistance to co-trimoxazole is associated with a reduced susceptibility to antibiotics belonging to different structural families suggests that the overexpression of efflux pumps, able to extrude different substrates, might be the basis for the observed resistance.
TABLE 1.
Bacteria and plasmid used in this work
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| S. maltophilia | ||
| D457 | Clinical strain, wild type | 29 |
| MBS411 | D457 ΔsmeE | 11, 46 |
| PBT02 | D457 with pPBT04 plasmid | 31 |
| PBT06 | MBS287 with pPBT04 plasmid | 31 |
| D457R | D457 mutant overexpressing SmeDEF efflux pump | 29 |
| MBS287 | D457 mutant overexpressing SmeVWX efflux pump | 11 |
| MBS704 | D457 ΔsmeW | 31 |
| MBS509-MBS602 | Mutants isolated in different amounts of co-trimoxazole | This work |
| MBS708 | MBS509 with pPBT04 plasmid | This work |
| MBS709 | MBS512 with pPBT04 plasmid | This work |
| MBS713 | MBS520 with pPBT04 plasmid | This work |
| MBS716 | MBS534 with pPBT04 plasmid | This work |
| MBS718 | MBS560 with pPBT04 plasmid | This work |
| E. coli | ||
| CC118λpir | λpir lysogen from CC118, Tetr, Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1 | 45 |
| 1047(pRK2013) | Strain containing the pRK2013 helper plasmid, Kanr | 47 |
| Plasmid | ||
| pPBT04 | Cloning vector pSEVA237Y containing smeVWX promoter region | 31 |
FIG 1.
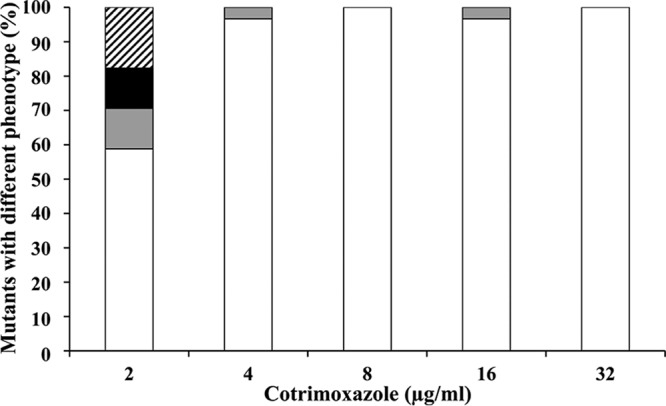
Phenotypes of mutants selected at different co-trimoxazole concentrations. The percentages of each category of mutants selected at each co-trimoxazole concentration are shown. White bars, mutants with chloramphenicol and quinolone resistance; gray bars, mutants with quinolone resistance; black bars, mutants with erythromycin and quinolone resistance; striped bars, mutants with erythromycin, chloramphenicol, and quinolone resistance.
The acquisition of antibiotic resistance might result in fitness costs that might be reflected in a lower growth rate (23–25). To address this possibility, the growth rates of a set of mutants presenting different phenotypes of resistance were analyzed in comparison to that of the wild-type parental strain D457. As shown in Fig. S1, the acquisition of co-trimoxazole resistance was always associated with a growth impairment.
Acquired co-trimoxazole resistance is associated with overexpression of the efflux pumps SmeDEF and SmeVWX.
As stated above, the observed multidrug-resistant phenotypes matched with the potential overexpression of efflux pumps. In particular, the phenotype of the larger group of mutants, which are chloramphenicol and quinolone resistant, is consistent with SmeVWX overexpression (11, 26). Other less-abundant mutants, presenting reduced susceptibility to chloramphenicol and quinolones and erythromycin resistance, display a phenotype consistent with the overexpression of SmeDEF (27, 28).
Two mutants presenting each of the phenotypes were chosen to analyze the possibility that the overexpression of any of the eight efflux pumps described in S. maltophilia (5, 6) could be responsible of these phenotypes. The expression of the efflux pumps encoded by smeABC, smeDEF, smeGH, smeIJK, smeMN, smeOP, smeVWX, and smeYZ was analyzed by real-time reverse transcriptase PCR (RT-PCR). As shown in Table 2, only the efflux pumps SmeDEF and SmeVWX were overexpressed in the selected mutants (data not shown for the other analyzed efflux pumps). Mutants presenting either chloramphenicol and quinolone or just quinolone resistance phenotypes (MBS509, MBS534, MBS560, and MBS571) overexpressed the efflux pump SmeVWX, while mutants presenting either chloramphenicol, quinolone, and erythromycin resistance or quinolone and erythromycin resistance (MBS510, MBS517, MBS518, and MBS522) overexpressed the SmeDEF efflux pump. Taking into consideration the distribution of phenotypes shown in Fig. 1, these results indicate that mutants overexpressing SmeDEF are selected mainly at low co-trimoxazole concentrations, while SmeVWX mutants are selected in the full range of concentrations of the antimicrobial.
TABLE 2.
Expression levels of efflux pump genes smeV and smeD
| Strain | Phenotypea | Fold change ± SEMb |
|
|---|---|---|---|
| smeV | smeD | ||
| D457 | 1.00 ± 0.0 | 1.00 ± 0.0 | |
| D457R | 0.33 ± 0.03 | 6.99 ± 3.61 | |
| MBS509 | Q | 55.03 ± 9.40 | 0.80 ± 0.19 |
| MBS510 | Q/CHL/ERY | 0.87 ± 0.11 | 8.59 ± 1.55 |
| MBS517 | Q/ERY | 0.71 ± 0.17 | 7.54 ± 1.14 |
| MBS518 | Q/ERY | 0.64 ± 0.28 | 7.99 ± 1.58 |
| MBS522 | Q/CHL/ERY | 0.74 ± 0.26 | 5.46 ± 1.01 |
| MBS534 | Q | 30.41 ± 4.82 | 0.75 ± 0.10 |
| MBS560 | Q/CHL | 48.05 ± 19.48 | 0.58 ± 0.05 |
| MBS571 | Q/CHL | 91.20 ± 41.68 | 0.71 ± 0.09 |
Antibiotic resistance phenotype. Q, quinolone resistance; Q/CHL/ERY, quinolone, chloramphenicol, and erythromycin resistance; Q/CHL, quinolone and chloramphenicol resistance; Q/ERY, quinolone and erythromycin resistance.
Expression levels are fold change of each mutant versus the wild-type strain, D457. Data are the means from three independent experiments. Fold changes larger than five are highlighted in bold.
To further confirm that the selected mutants overexpressed these efflux pumps, two different approaches were utilized. The overexpression of the efflux pump SmeDEF was confirmed by Western blotting (27) as described in Materials and Methods. As shown in Fig. 2, MBS510, MBS517, MBS518, and MBS522 presented levels of SmeF similar to those of the D457R mutant, which is known to overexpress SmeDEF (11, 27, 29, 30), while the expression levels of SmeF in MBS509, MBS534, MBS560, and MBS571 were similar to those found in the parental wild-type D457. These results are in agreement with the data obtained by real-time RT-PCR.
FIG 2.
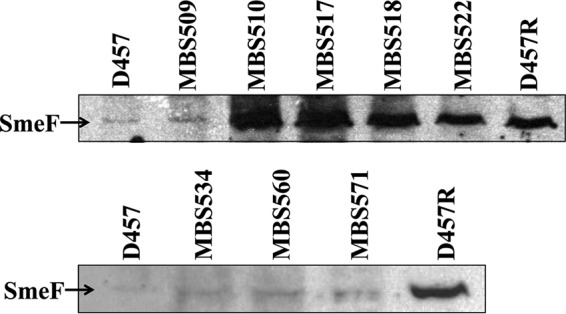
Expression of SmeF by co-trimoxazole-resistant mutants. The level of expression of SmeDEF was measured by Western blotting using an anti-SmeF antibody. D457, wild-type S. maltophilia strain; D457R, SmeDEF-overexpressing mutant. In agreement with the real-time RT-PCR data, the mutants MBS510, MBS517, MBS518, and MBS522 overexpress SmeDEF.
Concerning smeVWX, its increased expression was confirmed using a fluorescent reporter under the control of the smeV promoter as described previously (31). As Fig. 3 shows, MBS708, MBS716, and MBS718 presented higher fluorescence levels than PBT02 (a derivative of the wild-type strain D457) and MBS713 (derived from the MBS520 mutant that overexpresses SmeDEF) and similar levels to those observed in PBT06 (a derivative of MBS287, which overexpresses SmeVWX).
FIG 3.
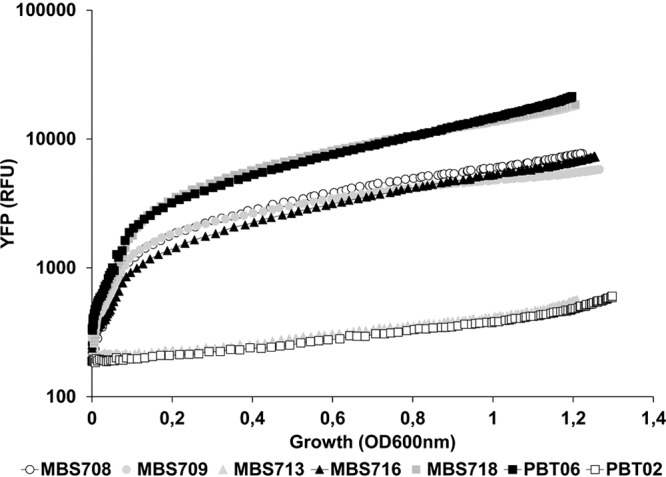
Analysis of smeVWX expression by co-trimoxazole-resistant mutants. The level of expression of smeVWX was analyzed using a fluorescent reporter as described in Materials and Methods. MBS708, MBS709, MBS716, MBS718, and PBT06 were derived from MBS509, MBS512, MBS534, MBS560, and MBS287, respectively. In agreement with real-time RT-PCR data, the expression of the fluorescent reporter YFP was higher in these mutants than in the control strains PBT02 (a derivative of the wild-type strain D457) and MBS713 (derived from MBS520, a mutant that overexpresses SmeDEF but not SmeVWX).
Overexpression of the efflux pumps SmeDEF and SmeVWX are associated with mutations in the genes smeT and smeRv encoding their respective local regulators.
It has been described that the overexpression of SmeDEF and SmeVWX is associated with mutations in their regulators genes, smeT and smeRv, respectively (9, 11, 30).
The smeT and smeRv genes of 24 co-trimoxazole-resistant mutants (Table 3), including those analyzed above, were amplified by PCR, and the resulting amplicons were sequenced. All five mutants overexpressing SmeDEF presented mutations in smeT. Three of the studied mutants presented the amino acid change Leu166Gln, which has been previously described in other SmeDEF-overproducing mutants (30). The other two mutants presented a Lys insertion in position 161 (Lys160_Ile161insLys) (Table 3).
TABLE 3.
Mutations identified in co-trimoxazole-resistant S maltophilia strains
| Co-trimoxazole conc. (mg/liter)a | Strain | Phenotypeb | Mutationc |
|
|---|---|---|---|---|
| SmeRv | SmeT | |||
| 2 | MBS509d | Q | GCC→ACC (Ala265Thr) | |
| 2 | MBS510e | Q/CHL/ERY | CTG→CAG (Leu166Gln) | |
| 2 | MBS512 | Q/CHL | IS3 insertion | |
| 2 | MBS513 | Q/CHL | GAC→GGC (Asp302Gly) | |
| 2 | MBS514 | Q/CHL | TGC→TGG (Cys310Trp) | |
| 2 | MBS515 | Q/CHL | GGC→AGC (Gly266Ser) | |
| 2 | MBS516 | Q/CHL | GAC→AAC (Asp302Asn) | |
| 2 | MBS517e | Q/ERY | CTG→CAG (Leu166Gln) | |
| 2 | MBS518e | Q/ERY | CTG→CAG (Leu166Gln) | |
| 2 | MBS520 | Q/CHL/ERY | Lys160_Ile161insLys | |
| 2 | MBS522e | Q/CHL/ERY | Lys160_Ile161insLys | |
| 2 | MBS523 | Q | TGC→TTC (Cys310Phe) | |
| 4 | MBS530 | Q/CHL | GGC→GAC (Gly266Asp) | |
| 4 | MBS534d | Q | GGC→GAC (Gly266Cys) | |
| 4 | MBS540 | Q/CHL | GCG→CCG (Ala308Pro) | |
| 8 | MBS560d | Q/CHL | GGC→AGC (Gly266Ser) | |
| 8 | MBS563 | Q/CHL | GGC→AGC (Gly266Ser) | |
| 8 | MBS567 | Q/CHL | GGC→GAC (Gly266Asp) | |
| 16 | MBS571d | Q/CHL | GGC→GAC (Gly266Asp) | |
| 16 | MBS572 | Q | GCC→GAC (Ala265Asp) | |
| 16 | MBS580 | Q/CHL | GGC→AGC (Gly266Ser) | |
| 32 | MBS583 | Q/CHL | GGC→GAC (Gly266Asp) | |
| 32 | MBS584 | Q/CHL | GGC→GAC (Gly266Asp) | |
| 16 | MBS593 | Q/CHL | GGC→AGC (Gly266Ser) | |
Co-trimoxazole concentration used to isolate the mutant (data refer to trimethoprim concentration, as the ratio of trimethoprim to sulfamethoxazole is 1:5).
Antibiotic phenotype. Q, quinolone resistance; Q/CHL/ERY, quinolone, chloramphenicol, and erythromycin resistance; Q/CH, quinolone and chloramphenicol resistance; Q/ERY, quinolone and erythromycin resistance.
Mutations identified in smeRv or smeT gene in each mutant.
Overexpression of smeV confirmed by real time RT-PCR (see Table 2).
Overexpression of smeD confirmed by real time RT-PCR (see Table 2).
Concerning the 19 analyzed mutants presenting phenotypes compatible with SmeVWX overexpression, 11 of them presented mutations previously associated with the overexpression of this efflux pump (11), namely, Gly266Asp (5 strains), Gly266Ser (5 strains), and Cys310Phe (1 strain) (Table 3), while seven presented novel mutations, Ala265Thr, Ala265Asp, Gly266Cys, Asp302Gly, Cys310Trp, Ala308Pro, and Asp302Asn. In one of the mutants, MBS512, the inactivation of smeRv was due to the insertion of an IS3 family insertion sequence, which is present at six different positions in the genome of S. maltophilia D457 (accession number NC_017671.1).
SmeVWX is not involved in the intrinsic co-trimoxazole resistance of S. maltophilia.
We have previously described that SmeDEF contributes to intrinsic and acquired co-trimoxazole resistance in S. maltophilia (18). The results presented in the current work indicate that SmeVWX, when overexpressed, is a relevant element in the acquisition of S. maltophilia co-trimoxazole resistance as well. Nevertheless, information on the role of SmeVWX in intrinsic resistance is absent. To address this issue, the susceptibility to co-trimoxazole of the MBS704 strain, which presents a deletion of the smeW gene (31), was determined. The susceptibility of the wild-type D457 strain, as well as mutants overexpressing either SmeDEF or SmeVWX or were defective in SmeE, was measured. As show in Table 4, the inactivation of SmeW did not increase the susceptibility of S. maltophilia to co-trimoxazole, indicating that in contrast to SmeDEF, the efflux pump SmeVWX is involved in just acquired resistance, without any role in the intrinsic resistance to co-trimoxazole of S. maltophilia.
TABLE 4.
MICs of mutants without or overexpressing the efflux pump SmeDEF or SmeVWX
| Strain | Phenotype | SXTa MIC (mg/liter) |
|---|---|---|
| D457 | Wild type | 0.25 |
| MBS411 | Defective in SmeE | 0.19 |
| D457R | SmeDEF overexpression | 2 |
| MBS704 | Defective in SmeW | 0.25 |
| MBS287 | SmeVWX overexpression | 2 |
SXT, co-trimoxazole (trimethoprim-sulfamethoxazole, 1:19) test trips.
DISCUSSION
S. maltophilia is an opportunistic pathogen of increasing relevance at hospitals (1) and is characterized by its low susceptibility to several antibiotics (5, 6). Co-trimoxazole is one of the antimicrobials of choice for treating S. maltophilia infections, but unfortunately, S. maltophilia resistant strains are increasingly isolated from patients (15, 32) Most epidemiological surveys concentrate their analyses on the presence of sul genes in S. maltophilia co-trimoxazole-resistant clinical isolates (13–15). Nevertheless, it has been reported that some co-trimoxazole-resistant S. maltophilia strains do not contain sul genes (16, 17). This indicates that there must exist mechanisms other than the presence of sul with relevance for S. maltophilia co-trimoxazole resistance. For example, using in vitro-constructed mutants, we previously described that the efflux pump SmeDEF may contribute to the intrinsic and acquired (when overexpressed) resistance to co-trimoxazole in S. maltophilia (18). Nevertheless, this study does not provide evidence on the mechanisms that are actually selected when S. maltophilia is challenged with co-trimoxazole.
Our results show that the mutations selected in the presence of co-trimoxazole are found in smeT and smeRv, which encode the transcriptional regulators of smeDEF and smeVWX, respectively. The regulator SmeT belongs to TetR family of transcriptional repressors, while SmeRv belongs to the LysR family of transcriptional regulators, which can be activators and repressors. In both cases, the selected mutations were localized in the C-terminal region of each regulatory protein, outside the DNA binding domain. In the case of SmeT, the C-terminal domain is mainly involved in ligand binding and dimerization, and mutations were localized in the dimerization region (33). For SmeRv, this part of the protein is predicted to be the effector binding motif, and mutations in this region, as those selected in the current work, will most likely modify the activity of this regulator. Indeed, the analyzed mutations correlate with an increased expression of the corresponding efflux pump and with changes in the susceptibility to antibiotics belonging to different structural families, a result that fits with previous results showing that the overexpression either SmeDEF or SmeVWX is associated with multidrug resistance (MDR) to several antibiotics (9, 11). In addition, our results indicate that none of the other efflux pumps described so far are overexpressed in the studied co-trimoxazole-resistant mutants.
We previously showed that under quinolone selective pressure, SmeDEF overexpression is the main mechanism of resistance among the selected mutants, while the role of SmeVWX overexpression is less relevant (11). However, when co-trimoxazole is used as the selective pressure, we observe the opposite; most mutants present a phenotype compatible with SmeVWX overexpression and just a few mutants, isolated in the presence of low co-trimoxazole concentrations, overexpress SmeDEF. These data strongly suggest that the overexpression of either SmeVWX or SmeDEF may imply a differential fitness cost when S. maltophilia is growing in the presence of either of the antibiotics, despite the fact that SmeDEF and SmeVWX can extrude both quinolones and co-trimoxazole. We also found that the acquisition of co-trimoxazole resistance is associated with impaired growth when resistant mutants are compared with the wild-type strain. This fitness cost may reduce the chances for the maintenance and the spread of these mutants after their selection (34). Nevertheless, it is important to recall that the overexpression of either SmeDEF or SmeVWX has been shown to be a relevant cause of S. maltophilia antibiotic resistance in clinical resistant isolates (9, 35–37).
It is worth mentioning that one of the risk factors for the acquisition of co-trimoxazole resistance by S. maltophilia is previous treatment with fluoroquinolones (38). Our results provide one explanation for these findings: SmeDEF is the major determinant of quinolone resistance in S. maltophilia (11, 37, 39) and its expression confers resistance to co-trimoxazole, whereas SmeVWX is also able of extruding both antibiotics when overexpressed, although its contribution to intrinsic resistance is minor.
The findings that co-trimoxazole selects multidrug-resistant mutants and that resistance to this antibiotic is not specific or confined to the presence of sul genes are of particular concern, given the ample use of co-trimoxazole in the treatment of S. maltophilia infections. Although the present results are based on the analysis of in vitro-selected mutants, it is important to highlight that S. maltophilia quinolone-resistant isolates overexpressing either SmeDEF or SmeVWX have been isolated from infected patients (9, 37, 40), providing evidence of the relevance of our in vitro results for understanding the selection of antibiotic-resistant mutants at hospitals.
MATERIALS AND METHODS
Strains, plasmids, and bacterial growth.
The strains and plasmid used in this work are described in Table 1. To select resistant mutants, S. maltophilia was seeded on Mueller-Hinton (MH) agar plates (41) containing different concentrations of co-trimoxazole (trimethoprim-sulfamethoxazole [1:5], Soltrim, injectable; Laboratorios Almofarma SL, Barcelona, Spain). The concentrations of co-trimoxazole described in this article refer to the amount of trimethoprim, being the trimethoprim-sulfamethoxazole ratio is 1:5 unless otherwise stated. Escherichia coli strains were grown in LB (41) with kanamycin (25 mg/liter for strain 1047 carrying pRK2013 and 50 μg/ml for strains carrying pPBT04). S. maltophilia strains containing the plasmid pPBT04 were grown in LB with 500 μg/ml kanamycin. All strains were grown at 37°C.
Isolation of co-trimoxazole S. maltophilia spontaneous mutants.
The concentration of co-trimoxazole most suitable for selection was determined by applying 5 μl of an overnight culture of S. maltophilia, as well as serial dilutions (10−1 to 10−3) of the culture, to MH agar plates containing different amounts of the antimicrobial. The results were recorded after 24 h at 37°C.
To isolate spontaneous co-trimoxazole mutants, 100 μl of dilutions 10−1 (109 CFU) and 10−2 (108 CFU) of S. maltophilia overnight cultures were spread on MH agar plates containing the concentration of co-trimoxazole that inhibits growth under these conditions or concentrations above this value (2×, 4×, 8×, and 16×). Colonies were selected after 48 to 72 h of incubation at 37°C. Each chosen colony was replicated two times on MH plates without antibiotic and then again in plates with antibiotic to discard false-positive or transient adaptations to the presence of the antimicrobial.
Antibiotic susceptibility assay.
MICs of erythromycin, chloramphenicol, norfloxacin, ofloxacin, nalidixic acid, and ciprofloxacin were determined by 2-fold agar dilution in MH agar plates, using a 96-pin replicator. The plates were incubated for 24 h at 37°C, and the data were the results from at least three independent assays. The co-trimoxazole MICs of the analyzed strains were determined on MH agar plates by using MIC test trips (Liofilchem).
RNA extraction and estimation of gene expression.
Overnight cultures were used to inoculate 40 ml of LB at an optical density at 600 nm (OD600) of 0.05. These new cultures were grown until an OD600 of 0.6 to 0.7, and 30 ml of each culture was recovered and centrifuged, and the RNA was extracted using an RNase kit (Qiagen) according to the manufacturer's instructions. The RNA was treated with DNase Turbo (Ambion) to remove any DNA present and cleaned using the RNase kit according to the manufacturer's instructions. Afterwards, the expression of the best-known S. maltophilia efflux pumps (smeABC, smeDEF, smeGH, smeIJK, smeMN, smeOP, smeVWX, and smeYZ) was analyzed by real-time RT-PCR. For this, 400 ng of RNA was retrotranscribed using the High-Capacity cDNA reverse transcription kit (Applied Biosystems). The expression of the first gene of each of the operons (smeA, smeD, smeG, smeI, smeM, smeY, smeO, and smeV) was analyzed using the primers and conditions of amplification described in reference 42. The primers for ftsZ1/2, which is used as a reference for normalization, are described in reference 11. The data obtained from three independent experiments were analyzed using the 2−ΔΔCT method (43).
Western blotting.
It was shown that the level of SmeF expression is a good marker of SmeDEF expression (27). Hence, to compare the levels of expression between the selected mutants and the wild-type strain, whole-cell extracts from overnight cultures were loaded in 12% SDS-PAGE gels (44). After electrophoresis, the proteins were transferred to an Immobilon-P membrane (Millipore, Billerica, MA, USA), and the Western blot analysis was performed as described previously (27). SmeF was detected with a polyclonal anti-SmeF antibody (27) at a 1:5,000 dilution and with a secondary goat anti-rabbit-horseradish peroxidase (HRP) conjugate (Bio-Rad) at 1:40,000. The bands were detected by chemiluminescence using the Immobilon Western kit (Millipore).
Analysis of smeVWX expression.
The plasmid pPBT04 (Table 1) that contains a gene encoding yellow fluorescent protein (YFP), whose expression is under the control of the promoter of smeVWX, was introduced into different co-trimoxazole-resistant mutants by triple conjugation as described previously (45). Transconjugants containing the plasmid were selected in LB with 500 mg/liter kanamycin and 20 mg/liter imipenem to remove E. coli.
Strains containing the plasmid were grown at 37°C for 22 h, and the OD600 of the cultures and their fluorescence (excitation at 508 nm and emission at 540 nm) were measured in a Tecan Infinite 200 plate reader.
Identification of mutations in smeT and smeRv.
The complete smeT and smeRv genes were amplified using a PCR master mix (Promega) and the primers SmeRvR/SmeRvF (11) for smeRv and 27/43 (30) for smeT. The PCR products were purified using a Qiagen kit according to the manufacturer's instructions and were sequenced by Macrogen (Amsterdam, The Netherlands) using the same primers.
Supplementary Material
ACKNOWLEDGMENTS
Work in our laboratory is supported by grants from the Instituto de Salud Carlos III (Spanish Network for Research on Infectious Diseases [RD16/0016/0011]), the Spanish Ministry of Economy and Competitiveness (BIO2017-83128-R)CNB, and the Autonomous Community of Madrid (B2017/BMD-3691).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00301-18.
REFERENCES
- 1.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esposito A, Pompilio A, Bettua C, Crocetta V, Giacobazzi E, Fiscarelli E, Jousson O, Di Bonaventura G. 2017. Evolution of Stenotrophomonas maltophilia in cystic fibrosis lung over chronic infection: a genomic and phenotypic population study. Front Microbiol 8:1590. doi: 10.3389/fmicb.2017.01590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spicuzza L, Sciuto C, Vitaliti G, Di Dio G, Leonardi S, La Rosa M. 2009. Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients. Eur J Clin Microbiol Infect Dis 28:191–195. doi: 10.1007/s10096-008-0605-4. [DOI] [PubMed] [Google Scholar]
- 4.Gibson RL, Burns JL, Ramsey BW. 2003. Pathophysiology and management of pulmonary infections in cystic fibrosis. Am J Respir Crit Care Med 168:918–951. doi: 10.1164/rccm.200304-505SO. [DOI] [PubMed] [Google Scholar]
- 5.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez MB. 2015. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia. Front Microbiol 6:658. doi: 10.3389/fmicb.2015.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez MB, Hernandez A, Martinez JL. 2009. Stenotrophomonas maltophilia drug resistance. Future Microbiol 4:655–660. doi: 10.2217/fmb.09.45. [DOI] [PubMed] [Google Scholar]
- 8.Cha MK, Kang CI, Kim SH, Cho SY, Ha YE, Chung DR, Peck KR, Song JH. 2016. Emergence of fluoroquinolone-resistant Stenotrophomonas maltophilia in blood isolates causing bacteremia: molecular epidemiology and microbiologic characteristics. Diagn Microbiol Infect Dis 85:210–212. doi: 10.1016/j.diagmicrobio.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 9.García-León G, Ruiz de Alegria Puig C, Garcia de la Fuente C, Martinez-Martinez L, Martinez JL, Sanchez MB. 2015. High-level quinolone resistance is associated with the overexpression of smeVWX in Stenotrophomonas maltophilia clinical isolates. Clin Microbiol Infect 21:464–467. doi: 10.1016/j.cmi.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Bernardini A, Corona F, Dias R, Sanchez MB, Martinez JL. 2015. The inactivation of RNase G reduces the Stenotrophomonas maltophilia susceptibility to quinolones by triggering the heat shock response. Front Microbiol 6:1068. doi: 10.3389/fmicb.2015.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-León G, Salgado F, Oliveros JC, Sanchez MB, Martinez JL. 2014. Interplay between intrinsic and acquired resistance to quinolones in Stenotrophomonas maltophilia. Environ Microbiol 16:1282–1296. doi: 10.1111/1462-2920.12408. [DOI] [PubMed] [Google Scholar]
- 12.Liaw SJ, Lee YL, Hsueh PR. 2010. Multidrug resistance in clinical isolates of Stenotrophomonas maltophilia: roles of integrons, efflux pumps, phosphoglucomutase (SpgM), and melanin and biofilm formation. Int J Antimicrob Agents 35:126–130. doi: 10.1016/j.ijantimicag.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Barbolla R, Catalano M, Orman BE, Famiglietti A, Vay C, Smayevsky J, Centron D, Pineiro SA. 2004. Class 1 integrons increase trimethoprim-sulfamethoxazole MICs against epidemiologically unrelated Stenotrophomonas maltophilia isolates. Antimicrob Agents Chemother 48:666–669. doi: 10.1128/AAC.48.2.666-669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung HS, Kim K, Hong SS, Hong SG, Lee K, Chong Y. 2015. The sul1 gene in Stenotrophomonas maltophilia with high-level resistance to trimethoprim/sulfamethoxazole. Ann Lab Med 35:246–249. doi: 10.3343/alm.2015.35.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. 2007. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis 13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu LF, Chen GS, Kong QX, Gao LP, Chen X, Ye Y, Li JB. 2016. Increase in the prevalence of resistance determinants to trimethoprim/sulfamethoxazole in clinical Stenotrophomonas maltophilia isolates in China. PLoS One 11:e0157693. doi: 10.1371/journal.pone.0157693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrera-Heredia SA, Pezina-Cantu C, Garza-Gonzalez E, Bocanegra-Ibarias P, Mendoza-Olazaran S, Morfin-Otero R, Camacho-Ortiz A, Villarreal-Trevino L, Rodriguez-Noriega E, Palau-Davila L, Maldonado-Garza HJ, Flores-Trevino S. 2017. Risk factors and molecular mechanisms associated with trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia in Mexico. J Med Microbiol 66:1102–1109. doi: 10.1099/jmm.0.000550. [DOI] [PubMed] [Google Scholar]
- 18.Sánchez MB, Martinez JL. 2015. The efflux pump SmeDEF contributes to trimethoprim-sulfamethoxazole resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 59:4347–4348. doi: 10.1128/AAC.00714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YW, Hu RM, Yang TC. 2013. Role of the pcm-tolCsm operon in the multidrug resistance of Stenotrophomonas maltophilia. J Antimicrob Chemother 68:1987–1993. doi: 10.1093/jac/dkt148. [DOI] [PubMed] [Google Scholar]
- 20.Lin YT, Huang YW, Chen SJ, Chang CW, Yang TC. 2015. The SmeYZ efflux pump of Stenotrophomonas maltophilia contributes to drug resistance, virulence-related characteristics, and virulence in mice. Antimicrob Agents Chemother 59:4067–4073. doi: 10.1128/AAC.00372-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin CW, Huang YW, Hu RM, Yang TC. 2014. SmeOP-TolCSm efflux pump contributes to the multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 58:2405–2408. doi: 10.1128/AAC.01974-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martínez JL, Coque TM, Baquero F. 2015. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol 13:116–123. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 23.Hernando-Amado S, Sanz-Garcia F, Blanco P, Martinez JL. 2017. Fitness costs associated with the acquisition of antibiotic resistance. Essays Biochem 61:37–48. doi: 10.1042/EBC20160057. [DOI] [PubMed] [Google Scholar]
- 24.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol Appl 8:273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes D, Andersson DI. 2017. Evolutionary trajectories to antibiotic resistance. Annu Rev Microbiol 71:579–596. doi: 10.1146/annurev-micro-090816-093813. [DOI] [PubMed] [Google Scholar]
- 26.Chen CH, Huang CC, Chung TC, Hu RM, Huang YW, Yang TC. 2011. Contribution of resistance-nodulation-division efflux pump operon smeU1-V-W-U2-X to multidrug resistance of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 55:5826–5833. doi: 10.1128/AAC.00317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alonso A, Martinez JL. 2000. Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 44:3079–3086. doi: 10.1128/AAC.44.11.3079-3086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Li XZ, Poole K. 2001. SmeDEF multidrug efflux pump contributes to intrinsic multidrug resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:3497–3503. doi: 10.1128/AAC.45.12.3497-3503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alonso A, Martinez JL. 1997. Multiple antibiotic resistance in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 41:1140–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez P, Alonso A, Martinez JL. 2002. Cloning and characterization of SmeT, a repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. Antimicrob Agents Chemother 46:3386–3393. doi: 10.1128/AAC.46.11.3386-3393.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blanco P, Corona F, Sanchez MB, Martinez JL. 2017. Vitamin K3 induces the expression of the Stenotrophomonas maltophilia SmeVWX multidrug efflux pump. Antimicrob Agents Chemother 61:e02453-. doi: 10.1128/AAC.02453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang CH, Lin JC, Lin HA, Chang FY, Wang NC, Chiu SK, Lin TY, Yang YS, Kan LP, Yang CH, Chan MC, Yeh KM. 2016. Comparisons between patients with trimethoprim-sulfamethoxazole-susceptible and trimethoprim-sulfamethoxazole-resistant Stenotrophomonas maltophilia monomicrobial bacteremia: a 10-year retrospective study. J Microbiol Immunol Infect 49:378–386. doi: 10.1016/j.jmii.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Hernández A, Ruiz FM, Romero A, Martinez JL. 2011. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog 7:e1002103. doi: 10.1371/journal.ppat.1002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonso A, Morales G, Escalante R, Campanario E, Sastre L, Martinez JL. 2004. Overexpression of the multidrug efflux pump SmeDEF impairs Stenotrophomonas maltophilia physiology. J Antimicrob Chemother 53:432–434. doi: 10.1093/jac/dkh074. [DOI] [PubMed] [Google Scholar]
- 35.Gould VC, Okazaki A, Howe RA, Avison MB. 2004. Analysis of sequence variation among smeDEF multi drug efflux pump genes and flanking DNA from defined 16S rRNA subgroups of clinical Stenotrophomonas maltophilia isolates. J Antimicrob Chemother 54:348–353. doi: 10.1093/jac/dkh367. [DOI] [PubMed] [Google Scholar]
- 36.Chang LL, Chen HF, Chang CY, Lee TM, Wu WJ. 2004. Contribution of integrons, and SmeABC and SmeDEF efflux pumps to multidrug resistance in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother 53:518–521. doi: 10.1093/jac/dkh094. [DOI] [PubMed] [Google Scholar]
- 37.Alonso A, Martinez JL. 2001. Expression of multidrug efflux pump SmeDEF by clinical isolates of Stenotrophomonas maltophilia. Antimicrob Agents Chemother 45:1879–1881. doi: 10.1128/AAC.45.6.1879-1881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang CH, Lin JC, Chang FY, Yu CM, Lin WS, Yeh KM. 2017. Risk factors for hospital acquisition of trimethoprim-sulfamethoxazole resistant Stenotrophomonas maltophilia in adults: a matched case-control study. J Microbiol Immunol Infect 50:646–652. doi: 10.1016/j.jmii.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez P, Alonso A, Martinez JL. 2004. Regulatory regions of smeDEF in Stenotrophomonas maltophilia strains expressing different amounts of the multidrug efflux pump SmeDEF. Antimicrob Agents Chemother 48:2274–2276. doi: 10.1128/AAC.48.6.2274-2276.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gould VC, Avison MB. 2006. SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J Antimicrob Chemother 57:1070–1076. doi: 10.1093/jac/dkl106. [DOI] [PubMed] [Google Scholar]
- 41.Atlas R. 1993. Handbook of microbiological media. CRC Press, Boca Raton, FL. [Google Scholar]
- 42.García-León G, Hernández A, Hernando-Amado S, Alavi P, Berg G, Martínez JL. 2014. A function of SmeDEF, the major quinolone resistance determinant of Stenotrophomonas maltophilia, is the colonization of plant roots. Appl Environ Microbiol 80:4559–4565. doi: 10.1128/AEM.01058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.de Lorenzo V, Timmis KN. 1994. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol 235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 46.García-León G, Sánchez MB, Martínez JL. 2012. The inactivation of intrinsic antibiotic resistance determinants widens the mutant selection window for quinolones in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 56:6397–6399. doi: 10.1128/AAC.01558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


