ABSTRACT
The radical-based theory proposes that three major classes of bactericidal antibiotics, i.e., β-lactams, quinolones, and aminoglycosides, have in common the downstream formation of lethal levels of reactive oxygen species (ROS) as part of the killing mechanism. If bactericidal antibiotics exhibit a common mechanism, then it is to be expected that the acquisition of resistance against these drugs would have some shared traits as well. Indeed, cells made resistant to one bactericidal antibiotic more rapidly became resistant to another. This effect was absent after induced resistance to a bacteriostatic drug. De novo acquisition of resistance to one bactericidal antibiotic provided partial protection to killing by bactericidal antibiotics from a different class. This protective effect was observed in short-term experiments. No protective effect was detected during 24-h exposures, suggesting that cross-resistance did not occur. In the wild-type strain, exposure to bactericidal antibiotics increased intracellular ROS levels. This increase in ROS levels was not observed when strains resistant to these drugs were exposed to the same concentrations. These results indicate that de novo acquisition of resistance to the bactericidal drugs tested involves a common cellular response that provides protection against ROS accumulation upon exposure to this type of antibiotics. A central mechanism or at least a few common elements within the separate mechanisms possibly play a role during the acquisition of antibiotic resistance.
KEYWORDS: bactericidal antibiotics, de novo resistance, reactive oxygen species
INTRODUCTION
Development of resistance to antibiotics is often caused by sublethal levels of antibiotics in the environment (1). To design interventions that prevent or slow the induction of resistance, the molecular mechanisms of the development of resistance must be understood.
The difference between bactericidal and bacteriostatic antibiotics is not always well defined. Three classes are usually considered bactericidal, i.e., β-lactams that inhibit cell wall synthesis (2), fluoroquinolones that interfere with DNA synthesis (3), and aminoglycosides that target ribosomal function (4). The radical-based theory was proposed to explain the observation that cells exposed to bactericidal antibiotics show increased levels of reactive oxygen species (ROS) (5). This theory includes an important role for secondary ROS formation as part of the killing mechanism of all bactericidal antibiotics. Interaction of the drug with its target results in an altered cellular metabolic state, in which hyperstimulation of the electron transport chain results in increased production of superoxide, a normal by-product of aerobic respiration (5, 6). Destabilization of iron-sulfur clusters results in the formation of hydroxyl radicals through the Fenton reaction. Although not the sole factor deciding antibiotic lethality, the radicals produced after antibiotic stimulation were shown to contribute to the killing activity of all bactericidal antibiotics (7). Although the role of ROS in antibiotic action has been heavily debated (8–10), evidence accumulated over the past few years seems to support the importance of ROS in bactericidal action (7, 11). The original theory has been expanded to fit a range of antibacterial compounds (12, 13), as well as different microorganisms (14–17). In Escherichia coli, the addition of antioxidants (18) or hydroxyl radical scavengers (19, 20) partially protects against killing by bactericidal antibiotics. Similarly, different species of bacteria that are deficient in scavenging enzyme activity show increased susceptibility to bactericidal antibiotics (20, 21).
Although specific target mutations are known to occur during adaptation to individual antibiotics (22–24), the existence of a shared downstream mechanism of action for all bactericidal antibiotics raises the question of whether such shared elements also contribute during the acquisition of resistance. Mechanisms reducing the levels of ROS induced during antibiotic treatment would thus increase survival rates and possibly could also reduce mortality rates for cells exposed to a second bactericidal antibiotic. Therefore, two questions are raised. (i) Do cells made resistant to one bactericidal antibiotic adapt faster than susceptible cells to a second, similar drug? (ii) Are killing rates affected? If ROS indeed play a central role in killing by bactericidal antimicrobials, then it is to be expected that cells made resistant to one of those antibiotics would not show increases in intracellular ROS levels upon exposure to a second antimicrobial; this should result in moderate reductions in the rates of killing by a second bactericidal antibiotic.
RESULTS
Acquisition of resistance.
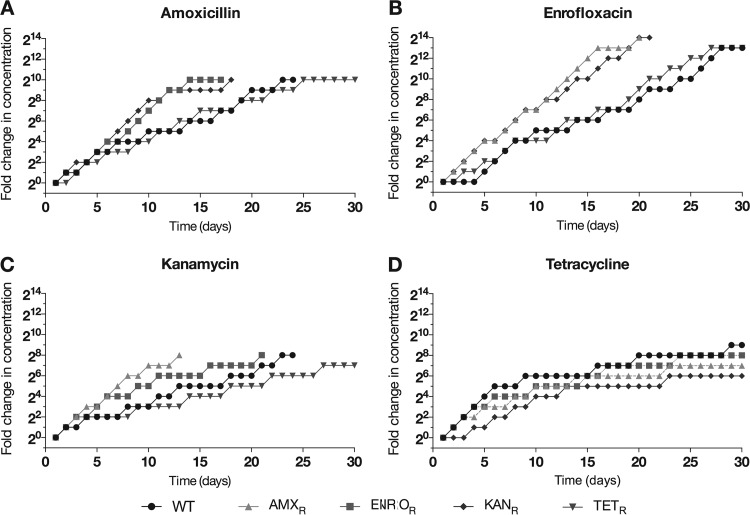
Wild-type cells were adapted to a first antibiotic in duplicate, and each of two duplicate replicates was adapted to a second antibiotic. Therefore, each data point is an average of four replicates. Cells that were exposed to increasing concentrations of antibiotics acquired resistance to a bactericidal antibiotic faster if they had previously been made resistant to another bactericidal antibiotic (Fig. 1A, B, and C). In contrast, cells with previously induced resistance to the bacteriostatic antibiotic tetracycline did not acquire resistance to a bactericidal antibiotic faster than wild-type E. coli cells. Resistance to tetracycline never exceeded concentrations of 64 μg/ml, except for adaptation of a single replicate of an enrofloxacin-resistant strain to tetracycline at levels of 128 μg/ml (data not shown). The rate of secondary adaptation to tetracycline for strains resistant to a bactericidal antibiotic did not differ from that for the fully susceptible control (Fig. 1D).
FIG 1.
Acquisition of resistance to amoxicillin (A), enrofloxacin (B), kanamycin (C), and tetracycline (D) by wild-type (WT) E. coli and strains resistant to amoxicillin (AMXR), enrofloxacin (ENROR), kanamycin (KANR), or tetracycline (TETR). Cells were adapted by doubling the antibiotic concentration when cultures with the higher concentration of antibiotic reached an OD600 similar to that of already adapted cultures. Results shown are the averages of four independent experiments.
Cross-resistance and cross-protection.
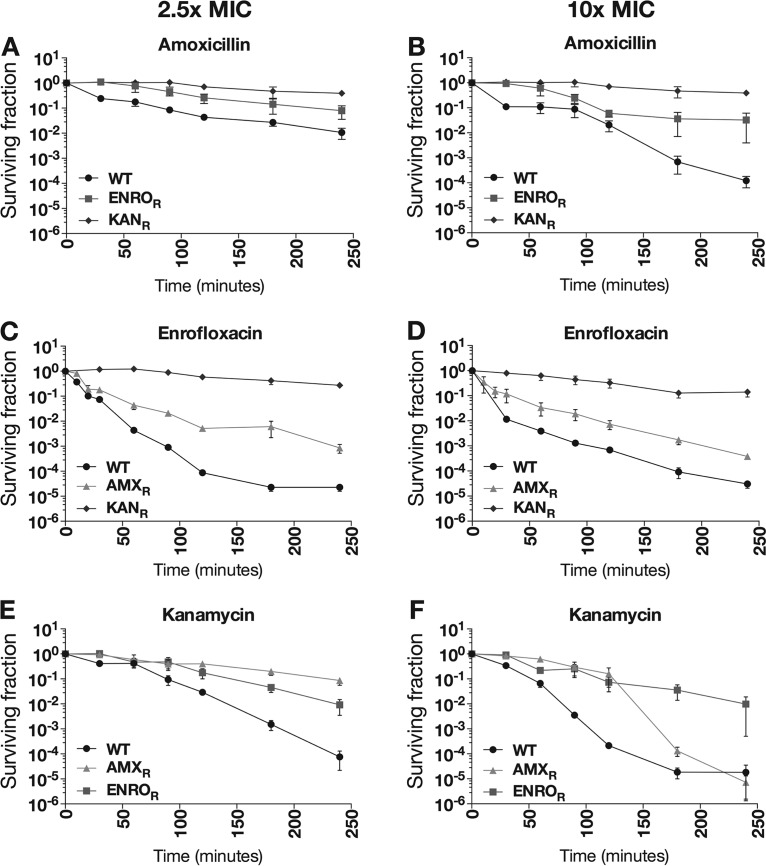
Cells made resistant by exposure to increasing levels of a single bactericidal antibiotic (either amoxicillin, enrofloxacin, or kanamycin) survived longer than fully susceptible wild-type cells in the presence of another bactericidal antibiotic in short-term (up to 4-h) killing experiments (Fig. 2). To determine whether resistance to one antibiotic provided protection against a different bactericidal antibiotic, the demise of wild-type cells was compared to that of cells with previously induced resistance to the first antibiotic. Although actual killing rates upon addition of 2.5 or 10 times the MIC of the second antibiotic differed, wild-type E. coli was always eliminated faster than the strains with induced resistance to the first drug. Resistance to kanamycin offered the most protection against killing by other antibiotics. In all cases, induced resistance to one bactericidal antibiotic reduced the killing efficiency of the others.
FIG 2.
Killing efficiency of amoxicillin, enrofloxacin, and kanamycin in resistant strains. Wild-type (WT) E. coli and strains resistant to amoxicillin (AMXR), enrofloxacin (ENROR), or kanamycin (KANR) were treated for 4 h with 2.5 times (A, C, and E) or 10 times (B, D, and F) the MIC level of amoxicillin (A and B), enrofloxacin (C and D), or kanamycin (E and F). Results shown are the averages of at least three independent experiments. Error bars indicate deviations as standard errors of the mean.
During longer-term (24-h) measurements to determine the MICs and minimal bactericidal concentrations (MBCs), no cross-resistance was observed in the strains with induced resistance to the three bactericidal antibiotics mentioned above and to tetracycline (Table 1). This outcome might seem to contradict the protection against other antibiotics observed in short-term measurements, but the kinetics of the killing process over 4 h are such that death after 24 h for almost all cells is to be expected. The exception is kanamycin-resistant cells exposed to amoxicillin, partially because of the large difference between MIC and MBC values for amoxicillin, compared to the other two bactericidal drugs. However, because neither the MIC nor the MBC of amoxicillin for kanamycin-resistant cells differed from that for wild-type cells (Table 1), this cannot be considered cross-resistance.
TABLE 1.
MICs and MBCs of amoxicillin, enrofloxacin, kanamycin, and tetracycline for wild-type E. coli and strains resistant to all four antibiotics
| Strain | MIC (μg/ml)a |
MBC (μg/ml) |
||||||
|---|---|---|---|---|---|---|---|---|
| AMX | ENRO | KAN | TET | AMX | ENRO | KAN | TET | |
| Wild-type | 4 | 0.25 | 8 | 0.5 | 64 | 1 | 64 | NA |
| AMX-resistant | 4,096 | 0.25 | 16 | 0.5 | 4,096 | 1 | 32 | NA |
| ENRO-resistant | 4 | >4,096 | 8 | 0.5 | 64 | >4,096 | 64 | NA |
| KAN-resistant | 4 | 0.25 | 1,024 | 0.5 | 64 | 1 | >1,024 | NA |
| TET-resistant | 4 | 0.5 | 8 | 64 | 64 | 1 | 64 | NA |
AMX, amoxicillin; ENRO, enrofloxacin; KAN, kanamycin; TET, tetracycline; NA, not applicable.
ROS concentrations after antibiotic stimulation.
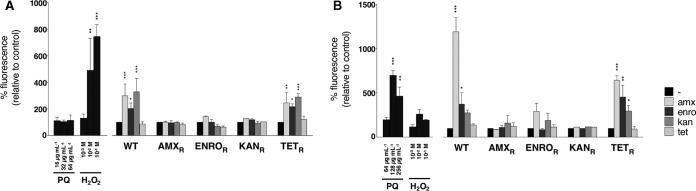
Wild-type cells exposed to MIC levels of the three bactericidal antibiotics during a 2-h treatment in minimal medium showed 2- to 3-fold increases in ROS levels, compared to the untreated condition (Fig. 3A). These increases in ROS levels were not induced by tetracycline. A similar pattern could be observed when tetracycline-resistant cells were treated with the same concentrations of the four antibiotics. In contrast, when strains resistant to amoxicillin, enrofloxacin, or kanamycin were treated with any of these antibiotics, no significant increases in fluorescence could be detected, indicating that ROS levels were not affected. Treatment with paraquat did not result in a significant increase in fluorescence, but treatment with 1 to 10 mM H2O2 resulted in increased ROS levels, indicating the physiological relevance of the ROS induced by antibiotic treatment.
FIG 3.
Antibiotic-induced ROS production in strains resistant to antibiotics. Wild-type (WT) E. coli and strains resistant to amoxicillin (AMXR), enrofloxacin (ENROR), kanamycin (KANR), or tetracycline (TETR) were grown to an OD600 of 0.2 and treated for 2 h with MIC levels of amoxicillin (amx), enrofloxacin (enro), kanamycin (kan), or tetracycline (tet), in M9 medium (A) or LB medium (B). For physiological controls, wild-type E. coli was treated with 0.5 times (16 μg/ml in M9 medium or 64 μg/ml in LB medium), 1 times (32 μg/ml in M9 medium or 128 μg/ml in LB medium), or 2 times (64 μg/ml in M9 medium or 256 μg/ml in LB medium) the MIC level of paraquat (PQ) or 1, 10, or 100 mM H2O2. Results shown are the averages of at least three independent experiments (two independent experiments for physiological controls). Error bars indicate deviations as standard errors of the mean. One-way ANOVA was used to determine statistical significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The growth rate and thus the metabolic activity of E. coli were higher in rich medium than in minimal medium (μ = 1.03 in LB medium versus μ = 0.49 in M9 medium; data not shown). The same experimental setup was used to study the responses of E. coli to antibiotic treatment in rich LB medium (Fig. 3B). Again, wild-type and tetracycline-resistant cells responded in similar manners to treatment with any of the four antibiotics. However, whereas treatment of cells resistant to either amoxicillin or kanamycin did not result in significant increases in ROS levels, treatment of enrofloxacin-resistant cells with amoxicillin did. This effect was much smaller than that in wild-type or tetracycline-resistant cells. An even smaller effect could be observed when the enrofloxacin-resistant strain was treated with kanamycin, but this increase was not significant (P = 0.33). Stimulation with antibiotics resulted in a greater increase in fluorescence than did treatment with either MIC levels of paraquat or up to 100 mM H2O2. Overall, accumulation of ROS in resistant strains upon stimulation with bactericidal antibiotics was, if not abolished, at least strongly reduced.
DISCUSSION
Reactive oxygen species have been proposed to play an important role in the killing mechanisms of bactericidal antibiotics (5–7). Here, we expand on this radical-based theory by investigating the role of ROS in antibiotic resistance acquired de novo. The partial protection of strains with acquired resistance to a bactericidal antibiotic against a second antibiotic with an unrelated killing mechanism suggests that at least one common factor is in play. A possible candidate for that role would be ROS. The following mechanisms could be imagined. Previously acquired resistance against a first bactericidal drug induces ROS protection mechanisms. The amounts of ROS produced upon exposure to MIC levels of antibiotics are reduced in these cells, in comparison to susceptible cells, which accelerates adaptation against a second bactericidal antimicrobial. The decreased ROS concentrations in adapted cells could also promote the development of resistance through ROS-induced mutagenesis (25), as ROS induced by bactericidal antibiotics have been shown to result in oxidation of the nucleotide pool (26, 27). Similar results were obtained in a Streptococcus pneumoniae strain resistant to either penicillin, ciprofloxacin, or kanamycin (28). In contrast to wild-type S. pneumoniae, cells resistant to one of those three bactericidal antibiotics did not respond by producing ROS upon exposure to another.
The mechanism causing reduced accumulation of ROS in antibiotic-resistant E. coli has not yet been determined. Protection against ROS induced by DNA mutations could be expected to have long-term effects and thus increase both MICs and MBCs. This was not observed; therefore, the protective effect of resistance acquired de novo against one bactericidal antibiotic toward a second cannot be explained by target-related mutations.
To reduce the amounts of ROS produced upon antibiotic stimulation, several points in the pathway activated upon exposure to an antibiotic could be targeted. For a S. pneumoniae strain resistant to penicillin, the reduced ROS levels upon stimulation with other bactericidal antibiotics were shown to be attributable to a nonsense mutation in a putative iron permease (29), but no generalized mechanism that explains the ROS-mediated tolerance in S. pneumoniae has yet been identified.
Antibiotic-induced ROS originate from a metabolic response involving hyperstimulation of the tricarboxylic acid (TCA) cycle (5, 7). By decreasing the flux through the TCA cycle, antibiotic-induced ROS levels could be reduced. Indeed, in Staphylococcus epidermidis clinical strains, TCA cycle defects provide protection to killing by β-lactam antibiotics and prevent ROS production upon stimulation with oxacillin (30). Similarly, in Salmonella enterica serovar Typhimurium, reduced flux through the TCA cycle increases the cells' ability to resist oxidative stress (31).
In addition to limiting the production of ROS upon antibiotic stimulation, ROS levels could be reduced by increasing the cells' response to oxidative stress. In E. coli, ROS concentrations that exceed normal physiological concentrations result in activation of the OxyRS (32) and SoxRS (33) regulons. Using gfp reporters for the promoters of OxyS and SoxS (7), we attempted to measure upregulation of the oxidative stress response in wild-type and resistant cells exposed to antibiotics. However, even though gfp reporters for OxyS and SoxS seemed to show the expected increases in expression, an unexpected and inconsistent increase in gfp fluorescence under control conditions (data not shown) made the data uninterpretable.
An alternative pathway of interest could be the glyoxylate cycle, an anaplerotic metabolic pathway that bypasses the TCA cycle (34). Increased flux through this pathway decreases the amount of NADH that is produced, thereby limiting the amounts of ROS that can be produced. The glyoxylate shunt was found to be upregulated upon exposure to oxidative stress in E. coli (35), Pseudomonas aeruginosa (36), and Burkholderia cenocepacia (16). Additionally, Mycobacterium tuberculosis strains with knockout mutations of isocitrate lyase, the first enzyme in the glyoxylate cycle, showed higher oxidative stress levels upon antibiotic treatment, as well as increased susceptibility to bactericidal antibiotics (37).
In conclusion, we hypothesize that a common protective mechanism is activated upon the acquisition of de novo antibiotic resistance to bactericidal antibiotics. Although the exact nature of this protective effect is unclear, this mechanism partially protects against the increased levels of ROS that are normally associated with exposure to bactericidal antibiotics. Because bactericidal antibiotics still exert their killing effects in the absence of ROS (7), the reduced ROS concentrations protect only against short-term bactericidal effects. In accordance with this line of reasoning, no cross-resistance was observed when MICs or MBCs were measured.
MATERIALS AND METHODS
Bacterial strains, growth media, and antibiotics.
The antibiotic-susceptible E. coli strain MG1655 was used as the wild-type strain throughout this study. Cells were routinely grown at 37°C in M9 medium supplemented with 55 mM glucose or in LB medium. For the experiments documenting the evolution of resistance, cells were grown in phosphate-buffered defined minimal Evans medium supplemented with 55 mM glucose (pH 6.9) (38). Cells were grown in flasks, shaken at 200 rpm, or in tubes, continuously shaken at 300 rpm. Stock solutions of antibiotics contained 10 mg/ml of the respective antibiotic and were kept at 4°C for up to 1 week. Growth rates were determined using round-bottom 96-well plates, with continuous shaking at 600 rpm, in a Thermo Scientific Multiskan FC photometer. The optical density at 600 nm (OD600) was measured.
Evolution of resistance.
Evolution experiments following the development of resistance were performed according to the protocol described previously (39). Based on MIC measurements, different starting concentrations were chosen to induce resistance to each antibiotic (amoxicillin, 1.25 μg/ml; enrofloxacin, 0.0625 μg/ml; kanamycin, 4 μg/ml; tetracycline, 0.25 μg/ml). Cells were transferred for 30 days or until they were able to grow at previously determined concentrations (1,024 μg/ml for enrofloxacin, kanamycin, and tetracycline and 1,280 μg/ml for amoxicillin). Strains isolated at the end of each evolution experiment were used as resistant strains for other experiments as needed, including adaptation to a second antibiotic.
Each adaptation of the wild-type E. coli strain to an antibiotic was independently performed twice. The resulting resistant strains from these first rounds were also used to acquire resistance to the second antibiotic in another two independent experiments, resulting in four secondary rounds of adaptation for each antibiotic. For adaptation to tetracycline, the resistant strain obtained from the first evolution experiment was used for all four experiments to induce resistance to a second antibiotic. To normalize the data, the starting concentration of each antibiotic was set to 1. All four independent experiments are displayed as one average by taking the mean of all four experiments and rounding the number to the nearest whole number.
MIC and MBC measurements.
MICs were determined by measuring the OD600, as described previously (38). To determine MBCs, 10 μl from each well of the 96-well plate used to measure MICs was spotted on LB agar plates, and the LB agar plates were incubated overnight at 37°C. The lowest concentration at which no growth could be visibly detected was considered the MBC.
Killing curves.
To determine the killing efficiency of bactericidal antibiotics, cells from an overnight culture were inoculated to an OD600 of 0.1, in a total volume of 2.5 ml, and directly treated with MIC levels of antibiotic. A sample was taken before the addition of antibiotics, to determine the CFU at time zero. Cells were incubated at 37°C, with shaking at 300 rpm. At selected time points, 25 μl of sample was used to make 10-fold dilutions with 1× phosphate-buffered saline (PBS) (pH 7.4) in a 96-well plate; 10 μl of each dilution was spotted on LB agar plates, and colonies were counted after overnight incubation at 37°C. For each time point, the counts of three technical repeats were averaged. At least three independent experiments were performed for each experimental condition.
ROS measurements.
The levels of ROS were measured using the fluorescent dye 2′,7′-dichlorofluorescein diacetate (H2DCFDA) (7). For experiments performed in M9 medium, cells from an overnight culture were inoculated in fresh medium to an OD600 of 0.1. After the culture reached an OD600 of ∼0.3 to 0.4, cells were rediluted to an OD600 of 0.1 and dye was added to a final concentration of 10 μM. A culture without H2DCFDA was included as a control. Cells were incubated with the dye for 1 h at 37°C, in the dark. Aliquots of 3 ml were used for antibiotic treatment. Cells were incubated for 2 h at 37°C. For analysis, cells were diluted 10-fold in 1× PBS (pH 7.4). Fluorescence was measured in 96-well plates in a Fortessa flow cytometer (Becton Dickinson), with excitation at 488 nm and emission at 510 nm.
For LB measurements, cells from an overnight culture were diluted 1:1,000 in fresh medium containing 10 μM H2DCFDA and were incubated at 37°C. For control measurements, cells were grown in the absence of the dye. When the culture reached an OD600 of ∼0.2, 3 ml of culture was used for each antibiotic treatment. Cells were incubated for 2 h at 37°C. Samples containing the dye were kept in the dark at all times. For analysis, cells were diluted 100-fold in 1× PBS (pH 7.4) and fluorescence was measured in a Gallios flow cytometer (Beckman Coulter), with excitation at 488 nm and emission at 505 to 545 nm.
To correct for autofluorescence induced by antibiotic treatment (40), all data points were normalized to controls from cells treated with antibiotics in the absence of H2DCFDA. At least three independent experiments were performed for each experimental condition. Data were analyzed using FlowJo. Statistical significance was determined by one-way analysis of variance (ANOVA) with Bonferroni correction.
ACKNOWLEDGMENTS
We thank James J. Collins for providing laboratory space and technical assistance to carry out part of this study, as well as for critically reading the manuscript.
This study was financed by the Netherlands Food and Consumer Product Safety Authority.
REFERENCES
- 1.Ter Kuile BH, Kraupner N, Brul S. 2016. The risk of low concentrations of antibiotics in agriculture for resistance in human health care. FEMS Microbiol Lett 363:fnw210. doi: 10.1093/femsle/fnw210. [DOI] [PubMed] [Google Scholar]
- 2.Tomasz A. 1979. The mechanism of the irreversible antimicrobial effects of penicillins: how the beta-lactam antibiotics kill and lyse bacteria. Annu Rev Microbiol 33:113–137. doi: 10.1146/annurev.mi.33.100179.000553. [DOI] [PubMed] [Google Scholar]
- 3.Levine C, Hiasa H, Marians KJ. 1998. DNA gyrase and topoisomerase IV: biochemical activities, physiological roles during chromosome replication, and drug sensitivities. Biochim Biophys Acta 1400:29–43. doi: 10.1016/S0167-4781(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 4.Jana S, Deb JK. 2006. Molecular understanding of aminoglycoside action and resistance. Appl Microbiol Biotechnol 70:140–150. doi: 10.1007/s00253-005-0279-0. [DOI] [PubMed] [Google Scholar]
- 5.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 6.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. 2007. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol 3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, Takahashi N, Chan CT, Lobritz MA, Braff D, Schwarz EG, Ye JD, Pati M, Vercruysse M, Ralifo PS, Allison KR, Khalil AS, Ting AY, Walker GC, Collins JJ. 2014. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A 111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keren I, Wu Y, Inocencio J, Mulcahy LR, Lewis K. 2013. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 339:1213–1216. doi: 10.1126/science.1232688. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Imlay JA. 2013. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. doi: 10.1126/science.1232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezraty B, Vergnes A, Banzhaf M, Duverger Y, Huguenot A, Brochado AR, Su SY, Espinosa L, Loiseau L, Py B, Typas A, Barras F. 2013. Fe-S cluster biosynthesis controls uptake of aminoglycosides in a ROS-less death pathway. Science 340:1583–1587. doi: 10.1126/science.1238328. [DOI] [PubMed] [Google Scholar]
- 11.Van Acker H, Coenye T. 2017. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol 25:456–466. doi: 10.1016/j.tim.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Sampson TR, Liu X, Schroeder MR, Kraft CS, Burd EM, Weiss DS. 2012. Rapid killing of Acinetobacter baumannii by polymyxins is mediated by a hydroxyl radical death pathway. Antimicrob Agents Chemother 56:5642–5649. doi: 10.1128/AAC.00756-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi H, Yang Z, Weisshaar JC. 2015. Single-cell, real-time detection of oxidative stress induced in Escherichia coli by the antimicrobial peptide CM15. Proc Natl Acad Sci U S A 112:E303–E310. doi: 10.1073/pnas.1417703112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeom J, Imlay JA, Park W. 2010. Iron homeostasis affects antibiotic-mediated cell death in Pseudomonas species. J Biol Chem 285:22689–22695. doi: 10.1074/jbc.M110.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calhoun LN, Kwon YM. 2011. The ferritin-like protein Dps protects Salmonella enterica serotype Enteritidis from the Fenton-mediated killing mechanism of bactericidal antibiotics. Int J Antimicrob Agents 37:261–265. doi: 10.1016/j.ijantimicag.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 16.Van Acker H, Sass A, Bazzini S, De Roy K, Udine C, Messiaen T, Riccardi G, Boon N, Nelis HJ, Mahenthiralingam E, Coenye T. 2013. Biofilm-grown Burkholderia cepacia complex cells survive antibiotic treatment by avoiding production of reactive oxygen species. PLoS One 8:e58943. doi: 10.1371/journal.pone.0058943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lupien A, Billal DS, Fani F, Soualhine H, Zhanel GG, Leprohon P, Ouellette M. 2013. Genomic characterization of ciprofloxacin resistance in a laboratory-derived mutant and a clinical isolate of Streptococcus pneumoniae. Antimicrob Agents Chemother 57:4911–4919. doi: 10.1128/AAC.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goswami M, Mangoli SH, Jawali N. 2006. Involvement of reactive oxygen species in the action of ciprofloxacin against Escherichia coli. Antimicrob Agents Chemother 50:949–954. doi: 10.1128/AAC.50.3.949-954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. 2012. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci U S A 109:12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Zhao X, Malik M, Drlica K. 2010. Contribution of reactive oxygen species to pathways of quinolone-mediated bacterial cell death. J Antimicrob Chemother 65:520–524. doi: 10.1093/jac/dkp486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bizzini A, Zhao C, Auffray Y, Hartke A. 2009. The Enterococcus faecalis superoxide dismutase is essential for its tolerance to vancomycin and penicillin. J Antimicrob Chemother 64:1196–1202. doi: 10.1093/jac/dkp369. [DOI] [PubMed] [Google Scholar]
- 22.Corvec S, Caroff N, Espaze E, Marraillac J, Reynaud A. 2002. −11 Mutation in the ampC promoter increasing resistance to β-lactams in a clinical Escherichia coli strain. Antimicrob Agents Chemother 46:3265–3267. doi: 10.1128/AAC.46.10.3265-3267.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drlica K, Hiasa H, Kerns R, Malik M, Mustaev A, Zhao X. 2009. Quinolones: action and resistance updated. Curr Top Med Chem 9:981–998. doi: 10.2174/156802609789630947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handel N, Schuurmans JM, Feng Y, Brul S, ter Kuile BH. 2014. Interaction between mutations and regulation of gene expression during development of de novo antibiotic resistance. Antimicrob Agents Chemother 58:4371–4379. doi: 10.1128/AAC.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohanski MA, DePristo MA, Collins JJ. 2010. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi N, Gruber CC, Yang JH, Liu X, Braff D, Yashaswini CN, Bhubhanil S, Furuta Y, Andreescu S, Collins JJ, Walker GC. 2017. Lethality of MalE-LacZ hybrid protein shares mechanistic attributes with oxidative component of antibiotic lethality. Proc Natl Acad Sci U S A 114:9164–9169. doi: 10.1073/pnas.1707466114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giroux X, Su WL, Bredeche MF, Matic I. 2017. Maladaptive DNA repair is the ultimate contributor to the death of trimethoprim-treated cells under aerobic and anaerobic conditions. Proc Natl Acad Sci U S A 114:11512–11517. doi: 10.1073/pnas.1706236114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dridi B, Lupien A, Bergeron MG, Leprohon P, Ouellette M. 2015. Differences in antibiotic-induced oxidative stress responses between laboratory and clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 59:5420–5426. doi: 10.1128/AAC.00316-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fani F, Leprohon P, Legare D, Ouellette M. 2011. Whole genome sequencing of penicillin-resistant Streptococcus pneumoniae reveals mutations in penicillin-binding proteins and in a putative iron permease. Genome Biol 12:R115. doi: 10.1186/gb-2011-12-11-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas VC, Kinkead LC, Janssen A, Schaeffer CR, Woods KM, Lindgren JK, Peaster JM, Chaudhari SS, Sadykov M, Jones J, AbdelGhani SM, Zimmerman MC, Bayles KW, Somerville GA, Fey PD. 2013. A dysfunctional tricarboxylic acid cycle enhances fitness of Staphylococcus epidermidis during β-lactam stress. mBio 4(4):e00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. 2013. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci U S A 110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng M, Aslund F, Storz G. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 33.Nunoshiba T, Hidalgo E, Amabile Cuevas CF, Demple B. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J Bacteriol 174:6054–6060. doi: 10.1128/jb.174.19.6054-6060.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornberg HL. 1966. The role and control of the glyoxylate cycle in Escherichia coli. Biochem J 99:1–11. doi: 10.1042/bj0990001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rui B, Shen T, Zhou H, Liu J, Chen J, Pan X, Liu H, Wu J, Zheng H, Shi Y. 2010. A systematic investigation of Escherichia coli central carbon metabolism in response to superoxide stress. BMC Syst Biol 4:122. doi: 10.1186/1752-0509-4-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn S, Jung J, Jang IA, Madsen EL, Park W. 2016. Role of glyoxylate shunt in oxidative stress response. J Biol Chem 291:11928–11938. doi: 10.1074/jbc.M115.708149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nandakumar M, Nathan C, Rhee KY. 2014. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun 5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 38.Schuurmans JM, Nuri Hayali AS, Koenders BB, ter Kuile BH. 2009. Variations in MIC value caused by differences in experimental protocol. J Microbiol Methods 79:44–47. doi: 10.1016/j.mimet.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 39.van der Horst MA, Schuurmans JM, Smid MC, Koenders BB, ter Kuile BH. 2011. De novo acquisition of resistance to three antibiotics by Escherichia coli. Microb Drug Resist 17:141–147. doi: 10.1089/mdr.2010.0101. [DOI] [PubMed] [Google Scholar]
- 40.Renggli S, Keck W, Jenal U, Ritz D. 2013. Role of autofluorescence in flow cytometric analysis of Escherichia coli treated with bactericidal antibiotics. J Bacteriol 195:4067–4073. doi: 10.1128/JB.00393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]