ABSTRACT
Candida auris has simultaneously emerged on five continents as a fungal pathogen causing nosocomial outbreaks. The challenges in the treatment of C. auris infections are the variable antifungal susceptibility profiles among clinical isolates and the development of resistance to single or multiple classes of available antifungal drugs. Here, the in vitro susceptibility to echinocandin antifungal drugs was determined and FKS1 sequencing was performed on 106 C. auris clinical isolates. Four isolates were identified to be resistant to all tested echinocandins (MIC ≥ 4 mg/liter) and harbored an S639F mutation in FKS1 hot spot region 1. All remaining isolates were FKS1 wild type (WT) and echinocandin susceptible, with micafungin being the most potent echinocandin (MIC50 = 0.125 mg/liter). Antifungal susceptibility testing with caspofungin was challenging due to the fact that all FKS1 WT isolates exhibited an Eagle effect (also known as the paradoxical growth effect), which occurred at various intensities. To assess whether the Eagle effect resulted in pharmacodynamic resistance, 8 representative isolates were evaluated for their in vivo drug response in a murine model of invasive candidiasis. All isolates were susceptible to caspofungin at a human therapeutic dose, except for those harboring the S639F mutation. The data suggest that only isolates carrying mutations in FKS1 are echinocandin resistant and that routine in vitro testing of C. auris isolates for susceptibility to caspofungin by the broth microdilution method should be viewed cautiously or avoided.
KEYWORDS: Candida, Candida auris, anidulafungin, antifungal resistance, antifungal susceptibility testing, caspofungin, echinocandins, micafungin, susceptibility
INTRODUCTION
Candida auris is a novel Candida species that was first reported in 2009 after being isolated from the external ear canal discharge of a patient in Japan (1). Subsequently, various types of C. auris infections have been reported from many countries located on five continents (2–18). Bloodstream infection has been the most frequently reported type of invasive infection, with the rate of mortality ranging from 30 to 60% (19). C. auris isolates have been recovered from various types of clinical specimens, including typically sterile body fluids, respiratory sections, urine, bile, tissues, wounds, and mucocutaneous swabs (2, 14–16, 20, 21). Patients infected with C. auris are usually critically ill with serious underlying medical conditions and significant health care exposure (2, 15, 16, 20). Moreover, it was reported that C. auris can extensively contaminate health care environments. Lockhart et al. concluded that its behavior is quite unique for a fungal pathogen and speculated that it is a skin commensal rather than part of the gastrointestinal microbiota (22).
Notably reduced susceptibility to azoles, polyenes, and echinocandins is often observed among C. auris isolates. Like other Candida species, antifungal susceptibility testing (AFST) for C. auris has been performed using various standardized tests, including Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution methods, the Etest gradient diffusion method, and the Vitek 2 antifungal susceptibility system. Yet analysis of the susceptibility results obtained by these tests is not straightforward and can be problematic since no antifungal breakpoints for C. auris have been defined by CLSI or EUCAST. C. auris was initially suspected to be intrinsically resistant to fluconazole, since reports dealing with Indian isolates indicated their nearly universal resistance to this drug (6, 23, 24). A recent study on 350 isolates from India reported that 3% (11 of 350 isolates) had low fluconazole MICs (2 to 8 mg/liter). Furthermore, the ERG11 sequences of isolates with low fluconazole MICs (1 to 2 mg/liter) exhibited the wild-type genotype, whereas the amino acid substitutions Y132 and K143, responsible for azole resistance in C. albicans, were present in fluconazole-resistant (32 to ≥64 mg/liter) C. auris isolates (25). It was also reported that C. auris can exhibit reduced susceptibility to other triazole antifungal drugs, including voriconazole, posaconazole, itraconazole, and isavuconazole (5, 6, 8, 14, 16, 24). This is troubling, given that azoles are a mainstay in the treatment of Candida infections and access to antifungals other than fluconazole is often lacking in resource-limited countries. However, as more isolates from around the world have been tested, the number of isolates with fluconazole MIC values similar to those of C. glabrata (in the 2 to 8 mg/liter range) has increased, and fluconazole resistance is now believed to be acquired (22). Furthermore, C. auris isolates exhibit variable susceptibility to the polyene antifungal drug amphotericin B (5, 9, 15, 16, 21, 26–28). The concern about resistance to azoles and amphotericin B has led to the recommendation for the use of echinocandins as a first-line therapy, pending susceptibility testing (29, 30). However, C. auris isolates with a reduced susceptibility to one or more drugs in the echinocandin class (MICs, ≥4 mg/liter for anidulafungin [ANF] and micafungin [MCF] and ≥2 mg/liter for caspofungin [CAS]) have been detected (5, 6, 16, 24, 27). An alarming 37% rate of resistance to CAS, measured by broth microdilution, was reported on the basis of an analysis of 102 C. auris isolates from 4 hospitals in India (23). Moreover, a number of C. auris isolates have demonstrated elevated MICs to multiple classes of antifungal agents, raising the possibility of pan-drug resistance (5, 23).
The emergence of multidrug-resistant C. auris is a multifactorial phenomenon that raises many serious public health concerns. A thorough understanding of the molecular insights into this organism is crucial for improved management. In this paper, we present the results of a comprehensive study (in vitro AFST, in vivo drug efficacy studies, and molecular resistance determinant analysis) on the activity of the echinocandin class of antifungal drugs against C. auris isolates.
RESULTS
Antifungal susceptibility testing.
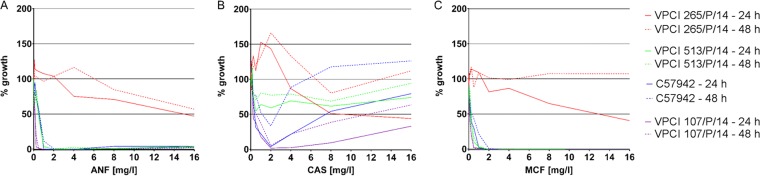
AFST was performed for a collection of 106 C. auris isolates from Colombia (n = 56), India (n = 40), and the Antimicrobial Resistance (AR) Isolate Bank (n = 10). The MIC distribution and the MICs of ANF, CAS, and MCF for individual isolates at 24 h and 48 h are shown in Table 1 and Table S1 in the supplemental material, respectively. Only 4 Indian isolates from a total of 106 isolates (3.8%) exhibited highly elevated MICs of ≥4 mg/liter at 24 h and were considered presumptively resistant to all tested echinocandins (ANF, CAS, MCF). These isolates were recovered from the blood of four different patients treated in intensive care units. Three of the patients were from the same hospital, and their echinocandin exposure history was unknown. One patient from a different hospital was given CAS for 10 days, but the specimen was drawn on the 4th day of therapy. MCF was the most potent echinocandin, with an MIC50 of 0.125 mg/liter and an MIC90 of 0.5 mg/liter at 24 h. Reading of the CAS MICs was difficult in many cases, since all tested isolates exhibited an Eagle effect (also known as the paradoxical growth effect), the intensity of which differed among the isolates. The growth of isolates exhibiting the most prominent Eagle effect was not significantly inhibited (≥50%) at 24 h. At 48 h, these isolates grew over the entire range of CAS concentrations. In order to better investigate this phenomenon, absorbance measurements (595 nm) were performed and growth intensities were calculated for four isolates (isolate VPCI 265/P/14, which was resistant to all echinocandins; isolate VPCI 513/P/14, which was susceptible to all echinocandins with a high CAS Eagle effect; isolate C57942, which was susceptible to all echinocandins with a medium CAS Eagle effect; and isolate VPCI 107/P/14, which was susceptible to all echinocandins with a low CAS Eagle effect) in the presence of ANF, CAS, and MCF at 24 h and 48 h (Fig. 1). The results of this experiment confirmed the observations that C. auris isolates do not exhibit a typical inhibition pattern in the presence of an increasing CAS concentration. In the case of ANF, spectrophotometric readings also revealed a very low intensity of the Eagle effect that was not visible to the naked eye. Moreover, in order to exclude the possibility of resistance, 100 μl of the cells of isolates VPCI 513/P/14 and C57942 growing at 1 mg/liter and 16 mg/liter was recovered from their respective wells, cultured overnight on yeast extract peptone dextrose (YPD) medium, and subjected to a repeat susceptibility test. All recovered isolates exhibited the same pattern of growth (Eagle effect) as the parental isolates, thus demonstrating that no stable genetic mechanism of resistance was acquired.
TABLE 1.
Distribution of Candida auris isolates MICs at 24 h and 48 h
| Echinocandin | Time of testing (h) | No. of isolates for which the MIC (mg/liter) was as followsa: |
MIC (mg/liter) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.03 | 0.06 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | >16 | GMb | MIC50 | MIC90 | ||
| ANF | 24 | 1 | 6 | 23 | 18 | 45 | 9 | 0 | 1 | 0 | 1 | 2 | 0.34 | 0.5 | 1 |
| 48 | 1 | 6 | 19 | 17 | 31 | 27 | 1 | 0 | 0 | 2 | 2 | 0.42 | 0.5 | 1 | |
| CAS | 24 | 0 | 0 | 6 | 51 | 42 | 3 | 0 | 0 | 3 | 1 | 0 | 0.38 | 0.25 | 0.5 |
| 48 | 0 | 0 | 2 | 14 | 46 | 21 | 1 | 0 | 0 | 1 | 21 | 1.06 | 0.5 | >16 | |
| MCF | 24 | 1 | 10 | 46 | 36 | 9 | 0 | 0 | 0 | 0 | 4 | 0 | 0.20 | 0.125 | 0.5 |
| 48 | 0 | 6 | 29 | 38 | 21 | 8 | 0 | 0 | 0 | 0 | 4 | 0.28 | 0.25 | 1 | |
The underlined values indicate modal (most frequent) MIC.
GM, geometric mean.
FIG 1.
Growth intensity-drug concentration curves for Candida auris isolates which presented different patterns of response to anidulafungin (A), caspofungin (B), and micafungin (C). VPCI 265/P/14 was resistant to all echinocandins, VPCI 513/P/14 was susceptible to all echinocandins with a high CAS Eagle effect, C57942 was susceptible to all echinocandins with a medium CAS Eagle effect, and VPCI 107/P/14 was susceptible to all echinocandins with a low CAS Eagle effect. Growth intensity was calculated as follows: (i) the absorbance of the no-growth control was subtracted from the absorbance of all the wells, and (ii) the values obtained were compared to the value for the growth control (which was not treated with an antifungal drug), which was set at 100%.
FKS1 analysis.
FKS1 sequencing was performed for all C. auris isolates (Table 2), and 102 echinocandin-sensitive isolates presented the wild-type (WT) genotype. The four high MIC isolates, which were presumptively echinocandin resistant, exhibited a serine-to-phenylalanine amino acid substitution (S639F) in the position equivalent to well-characterized position S645 in FKS1 hot spot region 1 (HS1) in Candida albicans. Isolates representing different clades were separated by 20 single nucleotide polymorphisms (SNPs) not resulting in amino acid substitutions.
TABLE 2.
Candia auris FKS1 sequence analysis results
| FKS1 hot spot | Nucleotide sequence (nta positions or mutation) | Amino acid sequence (type) | Isolates |
|---|---|---|---|
| FKS1 HS1 | TTCTTGACTTTGTCCTTGAGAGATCCT (1903–1929) | FLTLSLRDP (WT) | All except 4 (mentioned below) |
| TTCTTGACTTTGTTCTTGAGAGATCCT (C1916T) | FLTLFLRDPb (S639F) | VPCI 1133/P/13, VPCI 265/P/14, VPCI 462/P/14, VPCI 471a/P/14c | |
| FKS1 HS2 | GACTGGATTAGACGTTATACCTTG (4048–4071) | DWIRRYTL (WT) | All |
nt, nucleotide.
The boldface F represents the mutated amino acid.
Isolates VPCI 1133/P/13 - R, VPCI 265/P/14, VPCI 462/P/14, and VPCI 471a/P/14 - R were resistant to all the echinocandins tested.
Pharmacodynamic response of C. auris in a mouse model of invasive candidiasis.
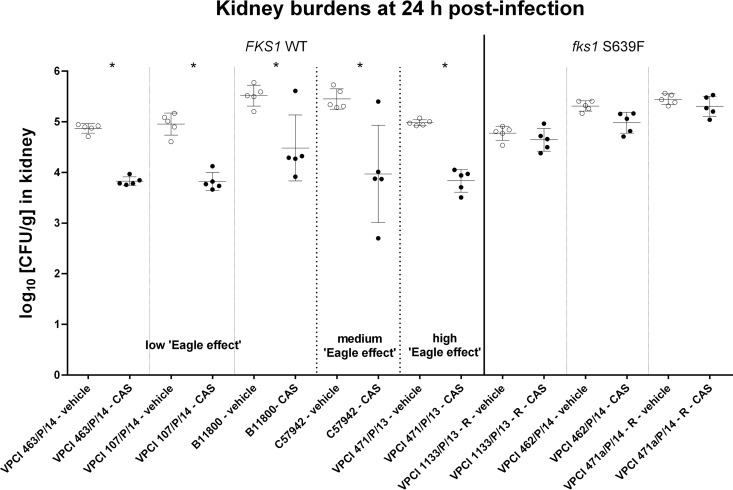
In order to evaluate whether the in vitro Eagle effect had any impact on the in vivo drug response of the C. auris isolates to echinocandins, mice were challenged with 8 C. auris isolates representing different susceptibility profiles and FKS1 genotypes and treated with a single humanized therapeutic dose of CAS. At 24 h postinfection, mice challenged with fks1 S639F mutants failed to respond to the drug, as there was no significant difference in the fungal kidney burdens between the CAS- and vehicle-treated groups (Fig. 2). In contrast, mice infected with C. auris FKS1 WT isolates, irrespective of the MIC and the intensity of the Eagle effect observed in vitro, had a significant (P < 0.05) ∼1 log10 CFU/g kidney burden reduction upon CAS treatment relative to the kidney burden in the vehicle-treated controls. Two individual mouse outliers, one from the isolate B11800-infected and CAS-treated group and the other from the isolate C57942-infected and CAS-treated group, in which the kidney burdens were not decreased upon CAS treatment, were observed. However, the presence of these two outliers did not affect the statistical significance of the general kidney burden differences between CAS-treated and untreated mice infected with the WT isolates.
FIG 2.
Comparison of the kidney burdens among the different treatment groups of mice infected with C. auris FKS1 WT isolates which exhibited Eagle effects of different intensities in vitro (isolates VPCI 463/P/14, VPCI 107/P/14, and B11800 exhibited a low Eagle effects; isolate C57942 exhibited a medium Eagle effect; and isolate VPCI 471/P/13 exhibited a high Eagle effect) and C. auris fks1 mutant isolates with the S639F substitution (isolates VPCI 1133/P/13 - R, VPCI 462/P/14, and VPCI 471a/P/14 - R, which were resistant to all echinocandins tested) at 24 h postinfection.
DISCUSSION
Even though Candida albicans remains the most frequently isolated Candida species in the clinical setting, an apparent trend toward an increasing prevalence of infections caused by non-albicans Candida spp. has been observed in recent years. One of them is C. auris, which has recently emerged as a nosocomial pathogen in many locations all over the world. A concerning attribute of C. auris is the fact that isolates are often characterized by reduced susceptibility to azoles, polyenes, and echinocandins, thus limiting antifungal treatment options and making the management of patients infected with C. auris more challenging (19).
The development of echinocandin resistance has been documented in C. auris isolates recovered from patients treated with these drugs in multiple geographic areas. This is particularly worrisome, given the recommendation for the use of echinocandins as an empirical treatment for C. auris infection prior to the availability of specific susceptibility testing results (29, 30). In the present study, AFST according to the CLSI guidelines was performed, and four Indian isolates were recognized to be resistant to all tested echinocandins (ANF, CAS, MCF), since they exhibited MICs of ≥4 mg/liter at 24 h (31). These isolates were further identified to harbor an S639F mutation in FKS1. A different amino acid substitution in the same position (S639P) was recently reported by Berkow and Lockhart in echinocandin-resistant C. auris isolates (32). FKS1 encodes glucan synthase, which is the target for echinocandin class antifungal drugs. The major mechanism of echinocandin resistance in other Candida spp. is amino acid substitutions in Fks subunits of glucan synthase, which decrease the sensitivity of the enzyme to the drugs (33). Amino acid changes associated with resistance occur in two highly conserved hot spot (HS) regions of Fks (34–36) in all Candida species, and mutations in codons S645 and F641 (C. albicans) cause the most pronounced resistance phenotypes (34, 37, 38). Correspondingly, the two equivalent HS regions in C. auris FKS1 are present at positions F635 to P643 and D1350 to L1357 of FKS1, and the amino acid substitution S639F is equivalent to S645F in C. albicans (Table 3). Resistance caused by fks1 S639F in C. auris was further confirmed in vivo in the mouse model of invasive candidiasis. Mice infected with the fks1 S639F mutant isolates did not respond to CAS treatment, with the mice having high kidney burdens comparable to those in the kidneys of the vehicle-administered controls.
TABLE 3.
Fks1 HS1 equivalents in Candida spp.
| Species | Fks1 HS1 sequence |
|---|---|
| C. auris | F635LTLSLRDP |
| C. albicans | F641LTLSLRDP |
| C. glabrata | F625LILSIRDP |
| C. parapsilosis | F652LTLSLRDA |
| C. tropicalis | F650LTLSLRDP |
MCF was the most potent echinocandin in the MIC testing, which is consistent with the AFST results of other groups (6, 27) Moreover, micafungin demonstrated the most prominent pharmacodynamic response in comparison to those of fluconazole and amphotericin B in a C. auris murine candidemia pharmacokinetic/pharmacodynamic study (39).
A significant challenge with obtaining an accurate AFST MIC readout for CAS was encountered, since all tested isolates exhibited an Eagle effect (also known as the paradoxical growth effect), with the intensities of the Eagle effect varying among the isolates. This phenomenon refers to reduced fungicidal activity at high concentrations of antifungals, mainly in in vitro circumstances (40). It was widely reported that paradoxical growth varies in terms of the medium, species, strain, and type of echinocandin. Among the currently approved echinocandins, CAS is most notably associated with the Eagle effect. It was first described in 2002, when it was noticed that higher concentrations of CAS seemed to be less effective against C. albicans than lower concentrations of the drug (41). The Eagle effect is not associated with a genetically stable resistant fungal subpopulation, where fungal growth would be expected at all concentrations above the MIC, even at the lower concentration range (42). Moreover, the effect is not due to mutations in FKS and is not related to the upregulated activity of glucan synthase in the presence of the drug. It was shown that echinocandins are effective in vivo in the treatment of invasive murine candidiasis caused by C. dubliniensis strains, despite the presence of the Eagle effect in vitro (43). Similarly, our study demonstrated that the observed Eagle effect did not affect the in vivo drug response of C. auris isolates, of which the only determinant impacting the pharmacodynamic response was the FKS1 genotype.
The echinocandin resistance in our C. auris collection was characterized by 3.8% prevalence and the same underlying mechanism of FKS1 HS1 mutation. Comparable levels of resistance (2% and 7%, respectively) were reported for C. auris isolates collected in India (10 hospitals) and by the CDC (from hospitals in Pakistan, India, South Africa, and Venezuela) (5, 25). We speculate that the previously reported 37% CAS resistance rate, without supporting molecular data (23), may possibly be due to the fact that isolates exhibiting a high Eagle effect were categorized as resistant, thereby overestimating the rate of echinocandin resistance in C. auris. Thus, as with many Candida species, CAS testing appears to provide an unreliable measure of resistance prevalence (44). The most solid information can be obtained by performing MCF AFST and/or FKS1 sequencing, where resistance is reflected by an MIC of ≥2 mg/liter and the presence of the characteristic mutation, respectively.
In any event, susceptibility testing for an emerging fungal species like C. auris is complicated by the lack of breakpoints enabling AFST result interpretation and very little knowledge of the correlation between MICs and clinical outcomes. Currently, guidance for C. auris MIC interpretation, based on information gathered for related Candida species and expert opinion, is provided by the CDC (31). Presumably, new guidance documents will soon be developed by EUCAST and CLSI.
In summary, our results demonstrate for a global collection of C. auris isolates that the prevalence of resistance was 3.8%, which is on the order of that for C. glabrata. As with many Candida species, standardized CLSI susceptibility testing with CAS should be viewed cautiously or avoided since an Eagle effect results in an overestimation of the resistant population. These “pseudoresistant” strains responded well to the drug in a murine infection model; only clinical isolates with an fks1 S639F mutation failed to respond in this model, and these isolates were best identified by AFST with MCF or by FKS1 sequence analysis.
MATERIALS AND METHODS
Fungal isolates and culture conditions.
A total of 106 C. auris isolates were investigated in this study. Forty C. auris isolates were obtained from Vallabhbhai Patel Chest Institute, University of Delhi (Delhi, India), 48 isolates were obtained from Clinica General del Norte (Barranquilla, Colombia), 8 isolates were obtained from a clinic of high complexity (Santa Marta, Colombia), and 10 isolates were from the Candida auris Panel from the Antimicrobial Resistance (AR) Isolate Bank (Centers for Disease Control and Prevention in collaboration with the Food and Drug Administration). Isolates were grown at 37°C on yeast extract peptone dextrose (YPD) agar plates prior to testing. Species identification of all Candida isolates was performed by sequencing of the rDNA region, amplified with primers Fun-rDNAF (5′-GGTCATTTAGAGGAAGTAAAAGTCG-3′) and Fun-rDNAR (5′-YGATATGCTTAAGTTCAGCGGGTA-3′) (S. Katiyar, personal communication), and further identified by nucleotide BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) analysis, as well as real-time PCR (45).
AFST.
Antifungal susceptibility testing was performed at least in duplicate for each strain in accordance with the guidelines described in CLSI document M27-A3 (46). C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 were used as quality control strains. ANF (Pfizer, New York, NY), CAS (Merck & Co. Inc., Rahway, NJ), and MCF (Astellas Pharma, USA, Inc., Deerfield, IL) were obtained as standard powders from their manufacturers, and stock solutions were prepared by dissolving the compounds in water (CAS, MCF) or 100% dimethyl sulfoxide (DMSO) (ANF). The MIC endpoints were defined as the lowest drug concentration that caused a prominent decrease (≥50%) in visual growth in relation to the growth of the controls. Additionally, an absorbance measurement (595 nm) was performed at 24 and 48 h for selected strains which exhibited an Eagle effect of different intensities.
DNA extraction.
DNA from fungal isolates was prepared by a 10-min incubation of a single colony in 100 μl of extraction buffer (60 mM sodium bicarbonate [NaHCO3], 250 mM potassium chloride [KCl], 50 mM Tris, pH 9.5) at 95°C and subsequent addition of 100 μl anti-inhibition buffer (2% bovine serum albumin). After vortex mixing, this DNA-containing solution was used for PCR (47).
FKS1 gene analysis.
On the basis of analysis of the sequence with GenBank accession number XM_018312389, primers for C. auris FKS1 amplification and sequencing were designed (Table 4). The primers were then synthesized by Integrated DNA Technologies. PCR mixtures were prepared in a total volume of 30 μl, consisting of 15 μl of 2× EmeraldAmp Max PCR master mix (TaKaRa Bio Inc.), 1 μl of each primer (CauFKS1_F and CauFKS1_R) at 10 μM, and 2 μl of DNA. PCR was performed in a T100 thermal cycler (Bio-Rad Laboratories, Inc.). The thermal profile included initial denaturation for 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 40°C, and 4 min at 72°C. The presence of amplicons was examined electrophoretically on 1% agarose gels stained with GelStar (Lonza). Amplicons were purified by using a ZR DNA sequencing cleanup kit (Zymo Research) and sequenced by Genewiz. Sequencing results were analyzed by SeqMan Pro (version 14) software (Lasergene; DNAStar).
TABLE 4.
Primers used in the study
| Primer | Sequence |
|---|---|
| CauFKS1_F | ATGTCTTACGATAACAATC |
| CauFKS1_R | TTAGAATGCCTTTGTAGTATAG |
| CauFKS1_R178-87 | AGT TCT GGT AAC CGT CCA TG |
| CauFKS1_F653-71 | GAA GTG GTT TTT CGC CTC G |
| CauFKS1_F1256-77 | AGA GAT ACA TGA GAT TGG GTG |
| CauFKS1_F1863-82 | TCT TTG GGT CAC TGT GTT TG |
| CauFKS1_F2446-66 | CGT ATC AGT TTC TTT GCT CAG |
| CauFKS1_F3048-67 | CGC CGA GTT TTT GTT GAG AG |
| CauFKS1_F3674-94 | GTA TGA CTG CCA TGT TGA GAG |
| CauFKS1_F4241-61 | CAG ACT TGA CCG TTG GTG GTG |
| CauFKS1_F4878-98 | CTG TCT TGG AAT GGC TTG TTG |
In vivo drug response assessment in a murine model of invasive candidiasis.
Six-week-old female BALB/c mice (Charles River Laboratories) weighing 16 to 18 g were used for all animal experiments. Mice were housed in presterilized filter-top cages and maintained in accordance with American Association for Accreditation of Laboratory Care criteria. The animal study was approved by the Rutgers Institutional Animal Care and Use Committee.
A well-established murine neutropenic disseminated candidiasis model was used for this study (48). A total of 80 mice were randomized into 16 different infection/antifungal therapy arms. The sample size of this animal experiment was considered adequate on the basis of the resource equation method (49). Mice were rendered neutropenic by receiving 150 mg/kg and 100 mg/kg of cyclophosphamide via intraperitoneal (i.p.) injection on day −4 and day −1 prior to infection, respectively. Five C. auris WT isolates which exhibited Eagle effects of different intensities in vitro (isolates VPCI 463/P/14, VPCI 107/P/14, and B11800 had a low Eagle effect, isolate C57942 had a medium Eagle effect, and isolate VPCI 471/P/13 had a high Eagle effect) and three C. auris S639F mutant isolates (echinocandin-resistant isolates VPCI 1133/P/13 - R, VPCI 462/P/14, and VPCI 471a/P/14 - R) were used in the study. The organisms were subcultured in liquid YPD medium at 37°C with shaking overnight. Cells were collected by centrifugation, washed twice with sterile phosphate-buffered saline (PBS), and counted with a hemocytometer. The inoculum was adjusted to 2 × 107 CFU/ml, and 50 μl was used to infect each mouse. The actual infection dose was verified by determination of viable counts on YPD plates spread with proper dilutions of the inoculum and incubated at 37°C for 24 h. On day 0, mice were infected with 1 × 106 CFU of the respective C. auris strain via retro-orbital injection. Groups of 5 mice were given a single dose of vehicle (sterile saline) or CAS at 4 mg/kg at 1 h postinfection via i.p. injection. At the experiment endpoint (24 h postinoculation), mice from each group were euthanized via CO2 inhalation, and the kidneys were aseptically removed for enumeration of the fungal burdens. All graphical data are expressed as scattered data points with means and standard deviations (bars) and were statistically analyzed by analysis of variance (ANOVA) using Prism computer software (version 6; GraphPad Software, Inc., San Diego, CA). The differences in the burdens between the test and the control groups were assessed by post hoc analyses, using Dunnett's or Dunn's multiple-comparison test (when group values did not fit a Gaussian distribution). A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the U.S. National Institutes of Health (AI109025) and Astellas Pharma (Echinocandin Reference Center) to D.S.P.
We thank Cristina Jiménez-Ortigosa for her expert assistance in experimental design and analysis of the results. We also thank Min-Hee Lee, Xin Hou, Irina Kolesnikova, Enriko Dolgov, Yevgeniy Senin, and George Rasic for their great help with the animal experiments.
D.S.P. receives funding from the U.S. National Institutes of Health and contracts from Astellas, Scynexis, Cidara, and Amplyx. He serves on advisory boards for Astellas, Cidara, Amplyx, Scynexis, and Matinas. In addition, D.S.P. has an issued U.S. patent concerning echinocandin resistance. I.B. is a speaker for Merck Sharp & Dohme, Pfizer SA, and Stendhal Pharma and has received research grants from Merck Sharp & Dohme. The remaining authors have no potential conflicts of interest.
We alone are responsible for the content and writing of the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00238-18.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MN, Shin JH, Sung H, Lee K, Kim EC, Ryoo N, Lee JS, Jung SI, Park KH, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. 2009. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis 48:e57–e61. doi: 10.1086/597108. [DOI] [PubMed] [Google Scholar]
- 4.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2017. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis 64:134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhary A, Anil Kumar V, Sharma C, Prakash A, Agarwal K, Babu R, Dinesh KR, Karim S, Singh SK, Hagen F, Meis JF. 2014. Multidrug-resistant endemic clonal strain of Candida auris in India. Eur J Clin Microbiol Infect Dis 33:919–926. doi: 10.1007/s10096-013-2027-1. [DOI] [PubMed] [Google Scholar]
- 7.Emara M, Ahmad S, Khan Z, Joseph L, Al-Obaid I, Purohit P, Bafna R. 2015. Candida auris candidemia in Kuwait, 2014. Emerg Infect Dis 21:1091–1092. doi: 10.3201/eid2106.150270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A, Finn T. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23:195–203. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magobo RE, Corcoran C, Seetharam S, Govender NP. 2014. Candida auris-associated candidemia, South Africa. Emerg Infect Dis 20:1250–1251. doi: 10.3201/eid2007.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borman AM, Szekely A, Johnson EM. 2017. Isolates of the emerging pathogen Candida auris present in the UK have several geographic origins. Med Mycol 55:563–567. doi: 10.1093/mmy/myw147. [DOI] [PubMed] [Google Scholar]
- 11.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF, Armstrong-James D, Fisher MC. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz Gaitan AC, Moret A, Lopez Hontangas JL, Molina JM, Aleixandre Lopez AI, Cabezas AH, Mollar Maseres J, Arcas RC, Gomez Ruiz MD, Chiveli MA, Canton E, Peman J. 2017. Nosocomial fungemia by Candida auris: first four reported cases in continental Europe. Rev Iberoam Micol 34:23–27. doi: 10.1016/j.riam.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 13.McCarthy M. 2016. Hospital transmitted Candida auris infections confirmed in the US. BMJ 355:i5978. doi: 10.1136/bmj.i5978. [DOI] [PubMed] [Google Scholar]
- 14.Morales-Lopez SE, Parra-Giraldo CM, Ceballos-Garzon A, Martinez HP, Rodriguez GJ, Alvarez-Moreno CA, Rodriguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2017. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013-August 2016. Am J Transplant 17:296–299. doi: 10.1111/ajt.14121. [DOI] [PubMed] [Google Scholar]
- 17.Mohsin J, Hagen F, Al-Balushi ZAM, de Hoog GS, Chowdhary A, Meis JF, Al-Hatmi AMS. 2017. The first cases of Candida auris candidaemia in Oman. Mycoses 60:569–575. doi: 10.1111/myc.12647. [DOI] [PubMed] [Google Scholar]
- 18.Arauz AB, Caceres DH, Santiago E, Armstrong P, Arosemena S, Ramos C, Espinosa-Bode A, Borace J, Hayer L, Cedeno I, Jackson BR, Sosa N, Berkow EL, Lockhart SR, Rodriguez-French A, Chiller T. 2018. Isolation of Candida auris from 9 patients in Central America: importance of accurate diagnosis and susceptibility testing. Mycoses 61:44–47. doi: 10.1111/myc.12709. [DOI] [PubMed] [Google Scholar]
- 19.Chowdhary A, Sharma C, Meis JF. 2017. Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog 13:e1006290. doi: 10.1371/journal.ppat.1006290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 21.Sarma S, Kumar N, Sharma S, Govil D, Ali T, Mehta Y, Rattan A. 2013. Candidemia caused by amphotericin B and fluconazole resistant Candida auris. Indian J Med Microbiol 31:90–91. doi: 10.4103/0255-0857.108746. [DOI] [PubMed] [Google Scholar]
- 22.Lockhart SR, Berkow EL, Chow N, Welsh RM. 2017. Candida auris for the clinical microbiology laboratory: not your grandfather's Candida species. Clin Microbiol News 39:99–103. doi: 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arendrup MC, Prakash A, Meletiadis J, Sharma C, Chowdhary A. 2017. Comparison of EUCAST and CLSI reference microdilution MICs of eight antifungal compounds for Candida auris and associated tentative epidemiological cutoff values. Antimicrob Agents Chemother 61:e00485-17. doi: 10.1128/AAC.00485-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009-17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 26.Prakash A, Sharma C, Singh A, Kumar Singh P, Kumar A, Hagen F, Govender NP, Colombo AL, Meis JF, Chowdhary A. 2016. Evidence of genotypic diversity among Candida auris isolates by multilocus sequence typing, matrix-assisted laser desorption ionization time-of-flight mass spectrometry and amplified fragment length polymorphism. Clin Microbiol Infect 22:277.e1–e9. doi: 10.1016/j.cmi.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 27.Sharma C, Kumar N, Pandey R, Meis JF, Chowdhary A. 2016. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microbes New Infect 13:77–82. doi: 10.1016/j.nmni.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shin JH, Kim MN, Jang SJ, Ju MY, Kim SH, Shin MG, Suh SP, Ryang DW. 2012. Detection of amphotericin B resistance in Candida haemulonii and closely related species by use of the Etest, Vitek-2 yeast susceptibility system, and CLSI and EUCAST broth microdilution methods. J Clin Microbiol 50:1852–1855. doi: 10.1128/JCM.06440-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chowdhary A, Voss A, Meis JF. 2016. Multidrug-resistant Candida auris: ‘new kid on the block’ in hospital-associated infections? J Hosp Infect 94:209–212. doi: 10.1016/j.jhin.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Public Health England. 2017. Guidance for the laboratory investigation, management and infection prevention and control for cases of Candida auris. Last updated 11 August 2017 Public Health England, London, United Kingdom. [Google Scholar]
- 31.Centers for Disease Control and Prevention. 2017. Recommendations for identification of Candida auris. Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/fungal/diseases/candidiasis/recommendations.html Accessed 5 December 2017. [Google Scholar]
- 32.Berkow EL, Lockhart SR. 2018. Activity of CD101, a long-acting echinocandin, against clinical isolates of Candida auris. Diagn Microbiol Infect Dis 90:196–197. doi: 10.1016/j.diagmicrobio.2017.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Perlin DS. 2015. Mechanisms of echinocandin antifungal drug resistance. Ann N Y Acad Sci 1354:1–11. doi: 10.1111/nyas.12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Effron G, Park S, Perlin DS. 2009. Correlating echinocandin MIC and kinetic inhibition of fks1 mutant glucan synthases for Candida albicans: implications for interpretive breakpoints. Antimicrob Agents Chemother 53:112–122. doi: 10.1128/AAC.01162-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katiyar SK, Edlind TD. 2009. Role for Fks1 in the intrinsic echinocandin resistance of Fusarium solani as evidenced by hybrid expression in Saccharomyces cerevisiae. Antimicrob Agents Chemother 53:1772–1778. doi: 10.1128/AAC.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson ME, Katiyar SK, Edlind TD. 2011. New Fks hot spot for acquired echinocandin resistance in Saccharomyces cerevisiae and its contribution to intrinsic resistance of Scedosporium species. Antimicrob Agents Chemother 55:3774–3781. doi: 10.1128/AAC.01811-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perlin DS. 2011. Current perspectives on echinocandin class drugs. Future Microbiol 6:441–457. doi: 10.2217/fmb.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Effron G, Lee S, Park S, Cleary JD, Perlin DS. 2009. Effect of Candida glabrata FKS1 and FKS2 mutations on echinocandin sensitivity and kinetics of 1,3-beta-d-glucan synthase: implication for the existing susceptibility breakpoint. Antimicrob Agents Chemother 53:3690–3699. doi: 10.1128/AAC.00443-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lepak AJ, Zhao M, Berkow EL, Lockhart SR, Andes DR. 2017. Pharmacodynamic optimization for treatment of invasive Candida auris infection. Antimicrob Agents Chemother 61:e00791-17. doi: 10.1128/AAC.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vanstraelen K, Lagrou K, Maertens J, Wauters J, Willems L, Spriet I. 2013. The Eagle-like effect of echinocandins: what's in a name? Expert Rev Anti Infect Ther 11:1179–1191. doi: 10.1586/14787210.2013.841543. [DOI] [PubMed] [Google Scholar]
- 41.Ramage G, VandeWalle K, Bachmann SP, Wickes BL, Lopez-Ribot JL. 2002. In vitro pharmacodynamic properties of three antifungal agents against preformed Candida albicans biofilms determined by time-kill studies. Antimicrob Agents Chemother 46:3634–3636. doi: 10.1128/AAC.46.11.3634-3636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens DA, Espiritu M, Parmar R. 2004. Paradoxical effect of caspofungin: reduced activity against Candida albicans at high drug concentrations. Antimicrob Agents Chemother 48:3407–3411. doi: 10.1128/AAC.48.9.3407-3411.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marine M, Pastor FJ, Sahand IH, Ponton J, Quindos G, Guarro J. 2009. Paradoxical growth of Candida dubliniensis does not preclude in vivo response to echinocandin therapy. Antimicrob Agents Chemother 53:5297–5299. doi: 10.1128/AAC.00980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Florl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas SR, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother 57:5836–5842. doi: 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kordalewska M, Zhao Y, Lockhart SR, Chowdhary A, Berrio I, Perlin DS. 2017. Rapid and accurate molecular identification of the emerging multidrug-resistant pathogen Candida auris. J Clin Microbiol 55:2445–2452. doi: 10.1128/JCM.00630-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 47.Brillowska-Dabrowska A, Nielsen SS, Nielsen HV, Arendrup MC. 2010. Optimized 5-hour multiplex PCR test for the detection of Tinea unguium: performance in a routine PCR laboratory. Med Mycol 48:828–831. doi: 10.3109/13693780903531579. [DOI] [PubMed] [Google Scholar]
- 48.Andes D. 2005. Use of an animal model of disseminated candidiasis in the evaluation of antifungal therapy. Methods Mol Med 118:111–128. [DOI] [PubMed] [Google Scholar]
- 49.Charan J, Kantharia ND. 2013. How to calculate sample size in animal studies? J Pharmacol Pharmacother 4:303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.