ABSTRACT
The conjugation of siderophores to antimicrobial molecules is an attractive strategy to overcome the low outer membrane permeability of Gram-negative bacteria. In this Trojan horse approach, the transport of drug conjugates is redirected via TonB-dependent receptors (TBDR), which are involved in the uptake of essential nutrients, including iron. Previous reports have demonstrated the involvement of the TBDRs PiuA and PirA from Pseudomonas aeruginosa and their orthologues in Acinetobacter baumannii in the uptake of siderophore-beta-lactam drug conjugates. By in silico screening, we further identified a PiuA orthologue, termed PiuD, present in clinical isolates, including strain LESB58. The piuD gene in LESB58 is located at the same genetic locus as piuA in strain PAO1. PiuD has a similar crystal structure as PiuA and is involved in the transport of the siderophore-drug conjugates BAL30072, MC-1, and cefiderocol in strain LESB58. To screen for additional siderophore-drug uptake systems, we overexpressed 28 of the 34 TBDRs of strain PAO1 and identified PfuA, OptE, OptJ, and the pyochelin receptor FptA as novel TBDRs conferring increased susceptibility to siderophore-drug conjugates. The existence of a TBDR repertoire in P. aeruginosa able to transport siderophore-drug molecules potentially decreases the likelihood of resistance emergence during therapy.
KEYWORDS: Pseudomonas aeruginosa, TonB-dependent receptor, siderophore-drug conjugate
INTRODUCTION
With the shortage of novel classes of antimicrobials, alternative approaches aiming to increase antimicrobial penetration into Gram-negative bacteria have gained widespread interest. Such approaches include the inhibition of broad-spectrum efflux pumps (1), adjuvants that increase cell permeability (2), and the redirection of drug uptake through specific nutrient transport systems (3). The most prominent example of the latter approach is the hijacking of essential bacterial iron transport systems by linking antimicrobial molecules to siderophores in a Trojan horse strategy. The recent development of such compounds by all major pharmaceutical companies historically involved in antimicrobial drug development highlights the increasing interest in this appealing concept (4–7). So far, most of the efforts have focused on the design and study of beta-lactam-siderophore conjugates. Since their targets are located in the periplasmic space, the conjugates do not require further translocation across the inner membrane. Moreover, the conjugates are designed such that the siderophore moiety does not interfere with the drug target interaction and does not require prior cleavage (8). The beta-lactam scaffolds used for the design of such conjugates include penicillins (9), cephalosporins (KP736 and cefiderocol) (7, 10), and monobactams (BAL30072 and MC-1) (4, 5). The iron-binding moiety of these beta-lactam conjugates is either a catechol-type siderophore such as dihydroxypyridone or a mixed catechol/hydroxamate (11). Both the monobactam (5, 12–14) and the cephalosporin conjugates (15) showed potent activity against the Gram-negative nonfermenters Pseudomonas aeruginosa and Acinetobacter baumannii.
Two TonB-dependent receptors (TBDRs), PiuA and PirA, have been shown to be responsible for the uptake of BAL30072, MC-1, and cefiderocol in P. aeruginosa (5, 16, 17). We previously observed that some P. aeruginosa isolates did not carry the piuA gene, although they were susceptible to BAL30072 (16). Therefore, we suspected that other TonB-dependent receptors (TBDRs) might be present in these strains or that their expression differs with respect to the PAO1 reference strain. Furthermore, the expression of TBDRs is often regulated by sigma/anti-sigma factors or two-component systems (18) and is induced by the presence of the corresponding siderophore (19, 20). These receptors could potentially participate in siderophore-drug uptake, but their contribution is masked under standard noninducing conditions. Therefore, we performed an in silico screen for PiuA orthologues in the P. aeruginosa genome database, and we additionally expressed from plasmids 28 of the 34 TBDRs of PAO1. This enabled us to identify a novel TBDR, termed PiuD, sharing 60% amino acid identity with PiuA, as well as five additional TBDRs of PAO1, potentially involved in the uptake of three different siderophore-drug conjugates, including the most recent catechol-based compound, cefiderocol (21).
RESULTS
piuD and piuA encode homologous proteins and are mutually exclusive in P. aeruginosa genomes.
We and others (5) previously identified the TonB-dependent receptors (TBDR) PiuA and PirA as transporters for the uptake of siderophore-drug conjugates BAL30072 and MC-1 (Fig. 1) both in P. aeruginosa (16) and in A. baumannii (22). When performing PCR amplifications of piuA from P. aeruginosa clinical isolates, we noticed the absence of a piuA signal in 54% of genotypically nonredundant isolates collected from intensive care unit patients (data not shown). We performed a homology search for potential orthologues of PiuA in the genome of LESB58, a strain that we previously showed lacks the piuA gene (16). The BLAST algorithm identified an open reading frame (ORF) of 766 amino acids in strains LESB58 (PALES_48941) and 39016 (PA39016_000870080), showing 60% amino acid identity with PiuA of PAO1 (753 amino acids). The highest sequence identity was observed in the N terminus (99% amino acid identity in the first 84 residues, including the signal sequence) and the putative substrate binding loops (NL1 to NL3) (see Fig. S1 in the supplemental material). We termed this PiuA orthologue PiuD. The piuD gene has a GC content of 59%, which is below the average of 66% for P. aeruginosa. To determine whether piuD would be present in strains from which piuA could not be amplified, we performed a multiplex PCR with piuA and piuD primer sets on the same set of genetically distinct clinical isolates tested above for piuA. The multiplex PCR confirmed our hypothesis, showing a PCR band either for piuA or for piuD, suggesting that both genes are mutually exclusive in P. aeruginosa genomes (see Fig. S2A). The piuD gene was found to be embedded in the same genomic context as piuA (16), since the gene products of piuB (PA4513) located downstream of piuA and those of the two genes piuC (PA4515) and piuE (PA4516), transcribed in an opposite direction, shared ≥98% amino acid identities with their homologues in LESB58 (Fig. S2B).
FIG 1.
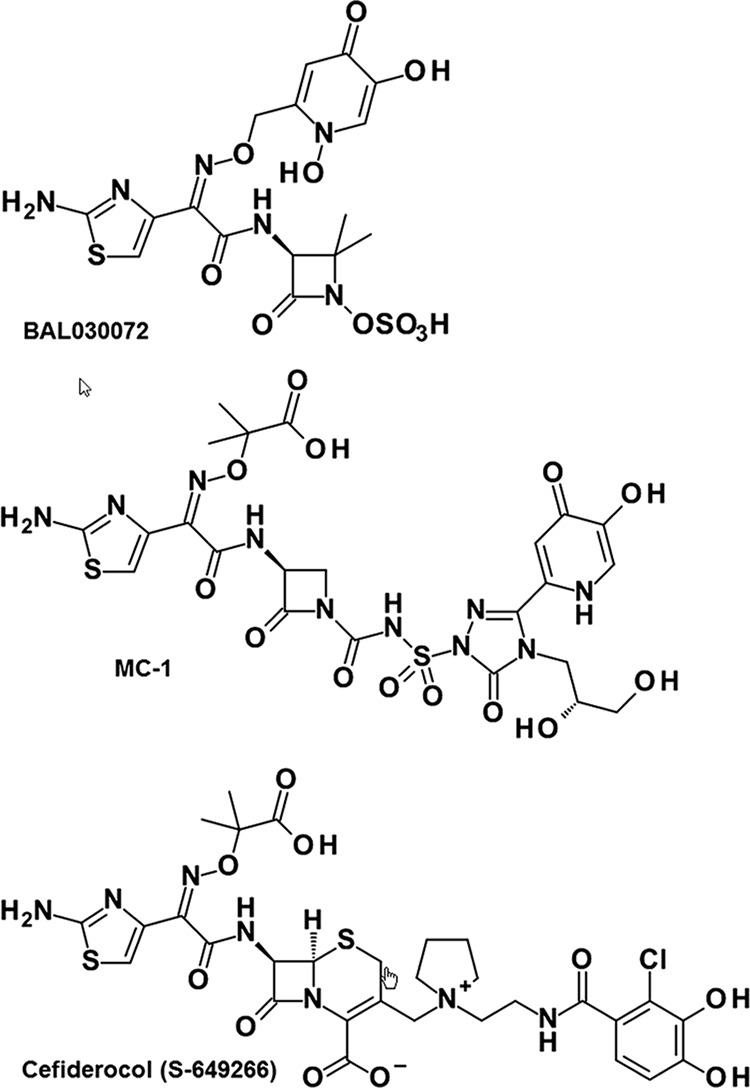
Siderophore-drug conjugates used in this study. BAL30072 and MC-1 contain a dihydroxypyridone as a siderophore, while cefiderocol contains a chlorinated catechol group.
Contribution of PiuD and PirA to the activity of siderophore-beta-lactam conjugates.
We compared the contributions of PiuA and PiuD with that of PirA, conserved in PAO1 and LESB58, to the activity of various siderophore-drug conjugates. To this end, we constructed deletion mutants in the piuD (PALES_48941) and pirA (PALES_43851) genes of LESB58. We tested the monobactam drugs BAL30072 (4) and MC-1 (5), conjugated to a hydroxypyridone siderophore, as well as the cephalosporin derivative cefiderocol, linked to a catechol siderophore (7) (Fig. 1). In the PAO1 background, both types of conjugates were strongly affected by the deletion of piuA (8- to 32-fold increase in MICs) but not by a pirA deletion. Surprisingly, the deletion of the pirA gene in the LESB58 background showed a stronger effect on the activities of the hydroxypyridone conjugates (8- to 16-fold increase in MICs) than on the catechol conjugate cefiderocol (2-fold increase in MICs). Conversely, the deletion of piuD increased cefiderocol MICs 32-fold, while MICs for MC-1 and BAL30072 increased by only 2- and 4-fold, respectively (Table 1). This could reflect the different expression levels of these receptors and/or the different affinities for the two types of siderophore-drug conjugates.
TABLE 1.
Susceptibility of piuD and pirA deletion mutants of P. aeruginosa PAO1 and LESB58
| Strain | MIC (mg/liter)a |
||||
|---|---|---|---|---|---|
| BAL | MC-1 | ATM | CFD | CAZ | |
| PAO1 | 1 | 0.5 | 4 | 0.5 | 2 |
| PAO1ΔpiuA | 8 | 8 | 4 | 8 | 2 |
| PAO1ΔpirA | 1 | 0.5 | 4 | 0.5 | 2 |
| PAO1ΔpiuAΔpirA | 16 | 16 | 4 | 16 | 2 |
| LESB58 | 1 | 1 | 16 | 0.06 | 4 |
| LESB58ΔpiuD | 4 | 2 | 16 | 2 | 4 |
| LESB58ΔpirA | 16 | 8 | 16 | 0.125 | 4 |
| LESB58ΔpiuDΔpirA | 32 | 32 | 16 | 4 | 4 |
MICs were determined in MHB-Chelex. BAL, BAL30072; ATM, aztreonam; CFD, cefiderocol; CAZ, ceftazidime.
To address this question, we extracted RNA from PAO1 and LESB58 from late exponential-phase cells grown under the same conditions as for the MIC assays and measured by reverse transcription-quantitative PCR (qRT-PCR) the expression of piuA and piuD in comparison to that of pirA. As shown in Fig. 2, pirA was expressed 3-fold less than piuA in PAO1, while the relative expression levels between pirA and piuD were comparable in strain LESB58. The low basal expression level of pirA might therefore not be sufficient to contribute to siderophore-drug uptake in PAO1, as highlighted by the identical MIC values of the pirA mutant and the wild-type strain PAO1 (Table 1). In contrast, in LESB58, PirA seemed to transport preferentially the hydroxypyridones BAL30072 and MC-1, while cefiderocol uptake occurred mainly via PiuD. Since PirA amino acid sequences from PAO1 and LESB58 (PALES_43851) are 99% identical, this difference was not due to altered substrate affinities.
FIG 2.
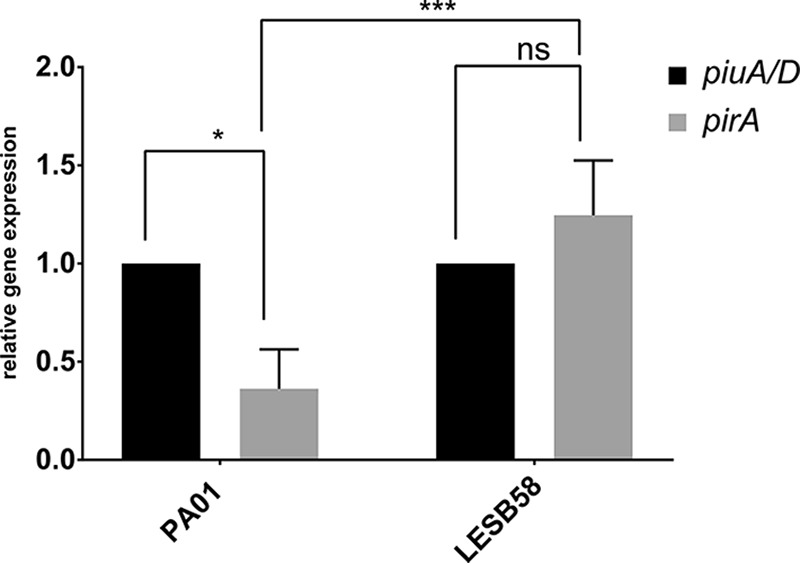
Expression analysis of pirA and piuA in PAO1 and pirA and piuD in LESB58. RNA was extracted from cells grown to late exponential phase in MHB. qPCR was performed using target-gene-specific primers. Expression of pirA is 3-fold lower than piuA in a PAO1 background, while piuD and pirA expression levels are comparable in strain LESB58. Values are the expression ratios of the target gene divided by the rpsL housekeeping gene. The expression of piuA and piuD was set to 1 (100%) in the respective strain. Values show the means from three independent experiments performed in duplicates. Error bars indicate standard deviations. *, P < 0.05 by Student t test; ***, P < 0.001 by analysis of variance (ANOVA); ns, not significant.
Crystal structure of PiuD from P. aeruginosa.
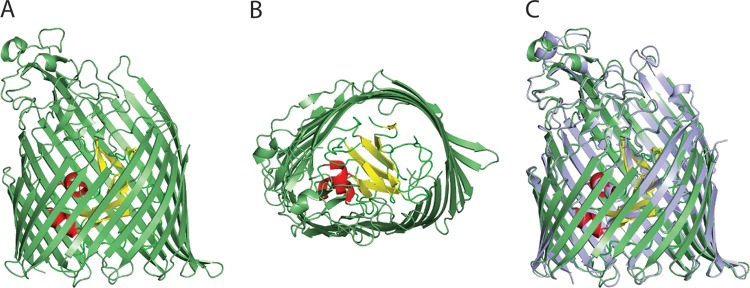
We previously determined the crystal structure of PiuA from P. aeruginosa and its orthologue from A. baumannii (22). Here, we determined the structure of PiuD from strain 39016, which shows 99.6% amino acid identity with PiuD (PALES_48941) from LESB58. The obtained PiuD structure was similar to that of PiuA from PAO1. The crystallographic asymmetric unit has two monomers (denoted A and B). PiuD comprised two domains, a 22-stranded transmembrane β-barrel and an N-terminal plug domain (residues 27 to 156) folded inside the barrel (Fig. 3). The plug domain has two β-sheets and two α-helices, which together, occluded the central pore. As often occurs in the TBDR structures, some of the extracellular loops were not experimentally located in the PiuD structure. In the B monomer, these regions, namely, NL1 (83 and 84), NL3 (113 and 114), the loop 138 to 141 of the plug domain, L7 (504 to 530), L8 (564 to 572), and L9 (609 to 624), were presumed to be disordered. The closest structural relatives were PiuA of A. baumannii (root mean square deviation [RMSD] of 1.1 Å over 701 residues) and P. aeruginosa (1.1 Å over 656 residues) and the pyochelin receptor FptA from P. aeruginosa (1.8 Å over 655 residues) (23). As a consequence of the disorder, one side of the extracellular β-barrel was absent. A “belt” of outward facing hydrophobic residues (Trp 445, 486, 541, and 594, Phe 157, 180, 217, 350, 648, and 731, and Tyr 645 and 685), sits at the periplasmic end of the barrel, a characteristic of outer membrane proteins.
FIG 3.
Crystal structure of PiuD from P. aeruginosa. Side (A) and extracellular (B) views of PiuD. The 22-stranded transmembrane β-barrel is colored in green. β-Sheets of the plug domain are colored in yellow, loops in green, and helices in red. (C) Structural comparison between PiuA (light blue) and PiuD (green).
Proteomic analysis under Fe chelation.
To identify further siderophore-drug transporters, we reasoned that under iron-limiting conditions, the expression of Fe-repressed TBDRs would be upregulated and could potentially contribute to the transport of siderophore-drug conjugates. Therefore, we performed a proteome analysis using PAO1 cells grown in Mueller-Hinton broth (MHB) and in MHB treated with Chelex, which complexes ferric iron but also divalent metal cations. The TBDRs for the endogenous siderophores pyochelin (FptA) and pyoverdin (FpvA and FpvB) showed the strongest induction in the Chelex-treated medium (40- to 130-fold increases) (Fig. 4). We also observed an induction of the heme receptors PhuR and HasR, as well as of the Zn transporter ZnuD. The expression of the known siderophore-drug transporters PiuA and PirA increased 2- and 10-fold, respectively, upon iron chelation. Among the TBDRs expected or reported to transport xenosiderophores, 11 showed a >2-fold increase in expression. CirA was below the 2-fold induction threshold, and six xenosiderophore receptors were not expressed or were expressed below the detection limit.
FIG 4.
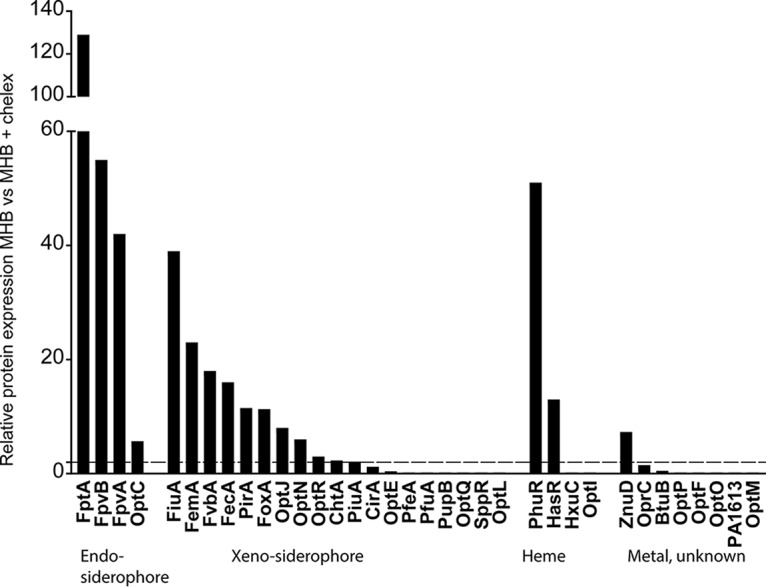
Proteomic analysis of TBDRs from P. aeruginosa PAO1. Protein expression levels were compared between cells grown for 20 h in MHB or Chelex-treated MHB. The dashed line indicates the 2-fold induction threshold level.
Constitutive expression of TonB-dependent receptors in PAO1.
To assess the possible involvement of these receptors in siderophore-drug uptake, we cloned 26 of the 34 TBDR genes from PAO1 (see Table S1), including the Chelex-induced TBDRs (FiuA, FemA, FoxA, OptJ, OptN, ChtA, and CirA) and those that were undetectable. We cloned the corresponding genes into a vector harboring a constitutively expressed promoter in P. aeruginosa and transferred the resulting plasmids into strain PAO1. We excluded the heme/hemophore transporters (PhuR, HasR, HxuC, and OptI), the cobalamin transporter BtuB (PA1271), and the citrate receptor FecA. For comparison, we included the previously identified receptor genes piuA and pirA, as well as the newly identified piuA orthologue piuD. The susceptibility data clearly showed that six TBDRs, namely, PiuD, OptJ, FemA, OptE, PfuA, and FptA, increased by at least 4-fold the susceptibility of PAO1 to the three siderophore-drug conjugates tested (shown in bold in Table 2). The strain harboring pPA0151 showed a 4-fold increase in susceptibility only for the dihydroxypyridone-containing drugs BAL30072 and MC-1, and the strain harboring ChtA only showed increased susceptibility for the catechol-based cefiderocol. Surprisingly, the overexpression of PfuA, which was undetectable by proteome analysis, produced the largest increase in susceptibility (>32-fold for BAL30072). With the exception of a 4-fold-decreased MIC for ceftazidime (pPA0151), we observed no significant changes in MICs for the nonsiderophore drugs aztreonam and ceftazidime. Since OptJ was induced to a similar level as PirA under Chelex treatment (Fig. 4), we constructed deletions in optJ in PAO1 and its piuA and pirA deletion mutants. As for a pirA deletion in PAO1, optJ deletion had no effect on siderophore-drug conjugate MICs (see Table S4). However, a consistent 2-fold increase in BAL30072 MICs in a piuA deletion background suggests a minor contribution of OptJ under uninduced conditions in a PAO1 background.
TABLE 2.
Effect of overexpression of TonB-dependent receptors on P. aeruginosa susceptibilities to three siderophore-drug conjugates
| Strain or plasmida | MIC (mg/liter)b |
||||
|---|---|---|---|---|---|
| BAL | MC-1 | ATM | CFD | CAZ | |
| PAO1 | 1 | 0.25 | 4 | 0.5 | 1 |
| pIApX2 (vector) | 1 | 0.25 | 4 | 0.5 | 1 |
| ppiuA1.1 | 0.06–0.125 | 0.06 | 4 | 0.03–0.06 | 1 |
| ppirA1.1 | 0.06–0.125 | 0.06 | 4 | 0.03–0.06 | 1 |
| ppiuD | 0.06–0.125 | 0.03 | 4 | 0.03 | 1 |
| poptJ (PA0434) | 0.06–0.125 | 0.03 | 4 | 0.03–0.06 | 1 |
| pfemA (PA1910) | 0.125 | 0.06 | 2 | 0.06 | 1 |
| poptE (PA2911) | 0.25 | 0.06 | 2 | 0.125 | 1 |
| ppfuA (PA1322) | 0.03 | 0.03 | 2 | 0.03 | 1 |
| pfptA | 0.125 | 0.06 | 2 | 0.125 | 0.5 |
| pPA0151 | 0.25 | 0.06 | 2 | 0.25 | 0.25 |
| pchtA (PA4675) | 0.5 | 0.125 | 2 | 0.125 | NDc |
| pfiuA (PA0470) | 0.5 | ND | 4 | 0.25 | 1 |
| pfoxA (PA2466) | 1 | 0.125 | 2 | 0.25 | 1 |
| ppfeA (PA2688) | 1 | 0.25 | 4 | 0.5 | ND |
| pcirA (PA1922) | 1 | 0.25 | 4 | 0.25–0.5 | 1 |
| poptN (PA1365) | 1 | 0.25 | 4 | 0.5 | ND |
| poptF (PA2590) | 1 | ND | 4 | 0.5 | 0.5 |
| poptQ (PA2289) | 1 | ND | 2 | 0.25 | 0.5 |
| poptR (PA3268) | 1 | 0.25 | 2 | 0.5 | 1 |
| pznuD (PA0781) | 1 | ND | 2 | ND | ND |
| optO (PA2335) | 0.5 | 0.25 | 2 | 0.5 | 1 |
| poptP (PA0192) | 1 | 0.25 | 2 | 0.5 | 1 |
| poptL (PA2089) | 1 | 0.25 | 4 | 0.5 | 1 |
| poptC (PA4837) | 1 | 0.125 | 2 | 0.5 | 1 |
| pPA1613 | 1 | 0.125 | 4 | 0.5 | 1 |
| poptM (PA2070) | 1 | 0.25 | 4 | 0.5 | 1 |
| psppR (PA2057) | 0.5 | 0.25 | 4 | 0.5 | 2 |
| pfvbA (PA4156) | 0.5 | 0.125 | 4 | 0.5 | 0.5 |
| pfpvA | 2 | 0.5 | 8 | 0.5 | 1 |
| pfpvB | 1 | 0.25 | 4 | 1 | 1 |
Plasmids in boldface font conferred a ≥4-fold increase in susceptibility to all siderophore-drug conjugates compared to the vector control.
aBAL, BAL30072; ATM, aztreonam; CFD, cefiderocol; CAZ, ceftazidime.
ND, not done.
To assess whether the observed changes in susceptibility could result from indirect effects on the expression level of the main siderophore-drug transporter PiuA, we introduced the relevant constructs in a PAO1ΔpiuA deletion mutant and tested the drug susceptibilities. PiuA, PirA, and PiuD expression decreased the MICs of all three siderophore-drug conjugates by 8- to ≥32-fold (Table 3). Interestingly, PirA overexpression produced only a 4- to 8-fold MIC decrease for cefiderocol compared to that of the vector control, while PiuD expression resulted in a ≥32-fold MIC decrease (Table 3). This finding is in agreement with the susceptibilities of pirA and piuD mutants in strain LESB58 (Table 1), which suggested preferential transport of cefiderocol via PiuD. The overexpression of PfuA showed MIC changes exceeding those conferred by PiuA and PirA, suggesting efficient siderophore-drug transport independent of PiuA. Similar results were obtained in a piuA-pirA double mutant (data not shown). On the other hand, FptA and OptE expression produced 2- to 8-fold decreases for MC-1 and cefiderocol, and OptJ produced a decrease only for MC-1. Finally, FemA and PA0151 expression showed no significant MIC changes in a piuA deletion mutant, suggesting an indirect effect on PiuA expression, when overexpressed in a PAO1 wild-type strain.
TABLE 3.
Effect of overexpression of TonB-dependent receptors on siderophore-drug conjugates activities in a piuA deletion mutant of P. aeruginosa
| Strain or plasmid | MIC (mg/liter)a |
||||
|---|---|---|---|---|---|
| BAL | MC-1 | ATM | CFD | CAZ | |
| PAO1ΔpiuA | 8 | 2 | 8 | 8 | 2 |
| pIApX2 (vector) | 8 | 4 | 4 | 8 | 2 |
| ppiuA | 0.25 | 0.03 | 4 | 0.06 | 2 |
| ppirA | 0.25 | 0.06 | 4 | 1–2 | 2 |
| ppiuD | 0.125 | 0.03 | 8 | 0.03–0.125 | 2 |
| ppfuA | 0.03 | 0.03 | 4 | 0.03 | 2 |
| pfptA | 4 | 0.5 | 2 | 0.25 | 2 |
| poptE | 4 | 0.5 | 4 | 2 | 2 |
| poptJ | 4 | 1 | 4 | 4–8 | 2 |
| pfemA | 8 | 2 | 4 | 8 | 2 |
| pPA0151 | 8 | 4 | 2 | 8 | 1 |
MIC changes of ≥4-fold compared to the vector control are shown in boldface font. BAL, BAL30072; ATM, aztreonam; CFD, cefiderocol; CAZ, ceftazidime.
To further evaluate if additional TBDRs would be involved in the uptake of siderophore-drug conjugates, we determined the susceptibilities under iron-limited growth conditions in MHB Chelex and in a minimal Casamino Acids medium. We observed 2-fold decreases in MICs of BAL30072 and MC-1 in PAO1 and the pirA mutant, and a 4- to 8-fold drop in the piuA mutant backgrounds. The increase in susceptibility was even more pronounced for cefiderocol (8- to 64-fold decreases) for the strains tested. MICs for the nonsiderophore drugs aztreonam and ceftazidime were not affected (see Table S5). The MICs were comparable or even lower than those obtained by the overexpression of the individual receptors from the plasmids in the piuA mutant background (Table 3), suggesting a simultaneous expression of several TBDRs besides PiuA and PirA for the uptake of siderophore-drug conjugates in P. aeruginosa under iron-limited conditions.
DISCUSSION
The Trojan horse strategy has recently gained renewed interest, as illustrated by the development of novel siderophore-beta-lactam conjugates by pharmaceutical companies (4, 12) and academic research groups (9, 24). These differ in the beta-lactam scaffolds (penicillins, monobactams, and cephems) as well as the attached siderophore moieties (mono-, tris-catechols and mixed catechol-hydroxamates). Initial investigations have identified two TBDR proteins, PiuA and PirA, in P. aeruginosa (5, 16) and their orthologues in A. baumannii (22). These are the main transporters for BAL30072 and MC-1. While the deletion of these TBDRs affected the activity of these compounds under standard MIC determination conditions, it remained unclear whether additional TBDRs expressed under iron deficiency or upon substrate-induced expression can contribute to drug susceptibility.
We have addressed these questions by screening for orthologues of PiuA in clinical strains and by overexpressing 28 of the 34 TBDRs from P. aeruginosa PAO1, thereby mimicking induction under specific physiological conditions or by natural substrates. An in silico screen identified PiuD in LESB58 and other clinical isolates as a homologue of PiuA, sharing 60% amino acid identity. The piuD gene was located in the same genetic environment as piuA, including the conserved intergenic promoter region (see Fig. S2 in the supplemental material). The lower GC content of the piuD gene (59% compared to 66% for PAO1) suggests an acquisition by horizontal gene transfer. The natural substrates of PiuA and PiuD are unknown, but the presence of conserved genes within the piu locus, including the oxidoreductase genes piuC and piuB and the ORF PA4516 (piuE), suggests that the metabolic fates of the natural substrates of these receptors are similar.
The amino acid similarity between PiuA and PiuD (Fig. S1) results in very similar crystal structures (Fig. 3). Like PiuA, PiuD also has a distinctive cluster of aromatic and positively charged residues located inside the pore at the extracellular face (see Fig. S3). This cluster is formed by Trp residues 311 and 327, Tyr 309, 710, and 714, Phe 94 (from the plug domain), His 713, and Arg 329 and 333 (Fig. S3). In PiuA, Trp 239, Tyr 307, 325, and 697, Phe 94, His 700, Lys 329, and Arg 331 form a cluster in the same position (Fig. S3). In the pyoverdin (FpvA) and pyochelin (FptA) receptors, this cluster is directly involved in the recognition of siderophores (23, 25). So far, there is no cocrystal structure available for a TBDR with its siderophore-drug conjugate, and only two cocomplexes between natural siderophores and their corresponding receptors have been solved (26–28). However, several binding and mutation studies regarding siderophore receptors and their cognate substrates have been reported (29–31), and their results are compatible with biphasic binding kinetics involving an initial binding in the loop extremities and a secondary binding at a site deeper inside the barrel, leading eventually to substrate translocation.
Our proteomic analysis revealed that divalent metal cation chelation induced the expression of 18 of the 34 TBDRs in PAO1. These include receptors for the endogenous siderophores pyoverdin (FpvA and FpvB), pyochelin (FptA), and nicotianamine (OptC) (32), as well as the heme (PhuR and HasR) and zinc (ZnuD) transporters. The other induced receptors could transport xenosiderophores that P. aeruginosa may encounter in the environment or during polymicrobial infections. A subset of these likely requires the cognate siderophore as an inducer. One example is PfeA from PAO1, sensing the presence of the exogenous siderophore enterobactin from Escherichia coli via the two-component system PfeR-PfeS (33). Similarly, the siderophore mycobactin from Mycobacterium smegmatis induces by 30-fold the expression of FemA in P. aeruginosa (19). Strikingly, the overexpression of PfuA resulted in the largest increase in susceptibility to all three siderophore-drug conjugates tested. The closest homologues of PfuA turned out to be PiuA in PAO1 and PiuD in LESB58, both sharing a 39% amino acid identity (57% similarity). The natural substrate of PfuA is unknown. A Fur binding site precedes the pfuA gene (34), suggesting iron repression; however, additional regulators and the presence of the substrate are likely required for induction of this TBDR in PAO1. Its closest orthologue in E. coli is Fiu, a TBDR also involved in the transport of BAL30072 (our unpublished data). Other receptors, undetectable by mass spectrometry (MS) analysis, may respond to other organic compounds or metal ions. Importantly, we identified the pyochelin receptor FptA as a candidate for the uptake of siderophore-drug conjugates. This receptor is the most highly induced receptor under iron limitation, as highlighted by our proteome analysis. FptA is also strongly expressed in lung and blood samples from mice and rats infected with P. aeruginosa and in human urine and respiratory samples (D. Bumann, unpublished results). The identification of the natural substrates of xenosiderophore receptors, as for instance PfuA, should provide an elegant way to induce specifically the expression of a receptor for the uptake of siderophore-drug conjugates. It also remains to be determined if siderophore-drug conjugates can act as inducers of their own transport, although this would require conjugate analogues deprived of antibiotic activity. The increased susceptibility to all three siderophore-drug conjugates under iron-limited conditions supports our findings on the plasmid-mediated expression of the individual TBDRs.
In summary, we have provided evidence for an overlapping subset of TBDRs in P. aeruginosa able to transport three different siderophore-drug conjugates, presenting two different types of iron-complexing substituents and on the basis of two classes of beta-lactams. The redundancy of TBDR recognition profiles should be an advantage during therapeutic treatments, since it should limit the risk of resistance emergence to these novel drug conjugates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table S1 in the supplemental material. E. coli and P. aeruginosa were grown in lysogeny broth (LB) at 37°C with shaking (250 rpm). E. coli DH10B was used as the cloning host and E. coli SM10 as the donor for biparental matings. Gentamicin (15 μg/ml for E. coli and 50 μg/ml for P. aeruginosa) or carbenicillin (200 μg/ml) was added for plasmid -carrying strains. MICs were determined in Mueller-Hinton broth (MHB) according to CLSI guidelines (35) and were repeated at least on three different occasions. Cation-depleted MHB was prepared by dissolving 11 g of Chelex (C7901; Sigma-Aldrich) in 100 ml MHB. After stirring for 6 h, the suspension was filtered and the filtrate autoclaved at 115°C for 15 min. The Chelex-treated MHB was supplemented with 2 mM MgSO4 and 0.2 mM CaCl2 (final concentrations). The M9 Casamino Acids medium contained 1× M9 salts, supplemented with 0.5% Casamino Acids (filter sterilized), and 2 mM MgSO4.
PCR amplifications and DNA modifications.
PCR primers are listed in Table S2. All primer sequences were based on the sequences from the pseudomonas.com website (36). For screening PCRs, bacterial cells were boiled at 95°C for 5 min and subsequently pelleted at 13,000 rpm for 1 min. Phusion DNA polymerase (Thermo Scientific) was used for high-fidelity PCRs (supplemented with 5% dimethyl sulfoxide [DMSO]). Restriction digestions were performed according to the manufacturer's instructions at the appropriate temperature. All ligation reactions were carried out at room temperature using T4 DNA ligase (Promega). DNA preparations were performed using the GeneJET PCR purification or the GeneJET gel extraction kit (Thermo Scientific).
Construction of knockout mutants.
The generation of unmarked knockout mutants was based on the protocol described by Hoang et al. (37). Briefly, DNA fragments of 500 to 700 bp were PCR-amplified using primer pairs A1/A2 and B1/B2, respectively. For the deletion of pirA in strain LESB58, the up- and downstream regions flanking the gene were PCR amplified. For the knockout of piuD in strain LESB58, the amplified fragments were located in the 5′ and 3′ regions of the genes. After amplification, the obtained A and B fragments were gel purified, and approximately 40 ng of each fragment was used in a PCR fusion amplification with primers A1 and B2, which share an 18-bp homologous region. The resulting fusion products were gel purified and further cloned into the suicide vectors pEX18Gm via HindIII/EcoRI restriction sites (pirA) and pEX18Gm via SalI/EcoRI (piuD). The cloned knockout fragments were verified by sequencing. The replacement vectors were mobilized into P. aeruginosa via biparental conjugation, and the generation of the unmarked mutants was carried out as previously described (38). The defined gene knockouts were verified by PCR amplification using the external primers and subsequent Sanger sequencing.
Construction of expression plasmids.
The coding regions, including at least 50 nucleotides (nt) upstream of the ATG initiation codon and 20 nt downstream of the STOP codon, were amplified by PCR from genomic DNA of P. aeruginosa 39016 (piuD) or PAO1. The piuD coding region was amplified with primers piuD-Xba and piuD-Hind and cloned as a 2,526-bp XbaI-HindIII DNA fragment into the expression vector pIApX2, yielding plasmid ppiuD. All other constructs were prepared in a similar way using the primers shown in Table S2. The Q5 high-fidelity DNA polymerase (NEB) was used for all amplifications. PCR conditions were as follows: denaturation at 98°C for 2 min, followed by 27 cycles of 98°C for 20 s, 57°C for 30 s, and 72°C for 2 min, and a final extension at 72°C for 4 min. The plasmids were transferred into P. aeruginosa by electroporation, and cells were spread on LB agar supplemented with carbenicillin at 200 mg/liter. All constructs were verified by Sanger sequencing.
Quantitative real-time PCR.
Overnight cultures of strains grown in LB were diluted and inoculated into fresh MHB and grown in microtiter plates (200 μl/well) until reaching late exponential phase. Three wells were combined to form one sample. RNA was extracted using the RNeasy kit (Qiagen, Germany), according to the manufacturer's protocol. Residual genomic DNA was removed by treatment with RNase-free DNase (Promega). One microgram of RNA was reverse transcribed using ImProm-II reverse transcriptase (Promega). Gene-specific primers were used for PCRs using the Rotor-Gene SYBR green PCR kit (Qiagen). qPCRs were performed in a Rotor-Gene 3000 (Corbett Research, Australia) using the following conditions: 2 min at 95°C, followed by 35 cycles of 20 s at 95°C, 30 s at 60°C, and 30 s at 72°C, followed by a final extension at 72°C for 3 min. The ribosomal rpsL gene was used as a housekeeping reference gene (39).
Cloning, overexpression, and purification of PiuD from P. aeruginosa.
The signal peptide of the proteins was predicted with Signal P4.0 (40) and excluded from cloning. The coding sequence of the mature protein was amplified from the genomic P. aeruginosa strain 39016 using KOD DNA polymerase (Novagen) and the primers piuD-39016-F and piuD-39016-R. The PCR product was digested by BspHI and XhoI restriction enzymes and cloned into the pTAMACHis6 vector using restriction sites NcoI and XhoI. The construct results in an expressed protein with an N-terminal TamA signal sequence for the outer membrane localization and a noncleavable C-terminal His6 tag. The pTAMACHis6 expression vector was obtained by replacing the PelB signal peptide of pEPELBCHIS (courtesy of Huanting Liu, University St Andrews) with the TamA signal peptide (41). PiuD was overexpressed in E. coli C43(DE3) cells. The expression and purification steps were as described for PiuA (22). The fractions were pooled and loaded on a Superdex S200 gel filtration column (GE Healthcare) equilibrated with 10 mM Tris (pH 8), 150 mM NaCl, and 0.45% (vol/vol) tetraethylene glycol monooctyl ether (C8E4). Protein fractions were pooled and concentrated to 10 mg/ml.
Crystallization and structure determination.
Crystals of PiuD appeared at 20°C after a few days by mixing 2 μl of protein solution (10 mg/ml) with 1 μl of reservoir solution containing 14% poly(ethylene glycol) methyl ether (PEG MME 5000) and 0.1 M bicine (pH 9). Crystals were frozen with the same solution containing 35% PEG MME 5000. The data were collected at ID23-1 at the ESRF. The data were processed with GrenADES (42–46). The structure of PiuD was solved by molecular replacement using P. aeruginosa PiuA coordinates (PDB code 5FOK) as a model, with the program PHASER (47). The models were adjusted with Coot (48), and the refinement was carried out using REFMAC in the CCP4 program suite with TLS parameters (49). The quality of all structures was checked with MolProbity (50). The figures were drawn using PyMOL (version 1.8; Schrödinger, LLC). The final refinement statistics are given in Table S3.
Proteomics analysis.
Sample preparation and MS analysis were performed as described previously (33). Briefly, P. aeruginosa was grown in MHB or MHB treated with Chelex (Sigma-Aldrich, Switzerland) under standard MIC determination conditions in microtiter plates without shaking at 37°C for 18 h. The cells from three wells were combined to yield sufficient material for proteome analysis. Three replicate samples were lysed, and the proteins were reduced with 5 mM Tris (2-carboxyethyl) phosphine hydrochloride and alkylated with iodoacetamide. The samples were diluted before digestion with trypsin at 37°C overnight. The peptides were desalted on a C18 reversed-phase column and dried under vacuum. One microgram of peptide was injected into a liquid chromatography-mass spectrometer (LTQ-Orbitrap Elite). The peptides were separated using an EASY nLC-1000 system (Thermo Fisher scientific) using a C18 high-performance liquid chromatography (LC) column. Tandem mass spectrometry data were exported from Progenesis LC-MS and searched against a protein decoy database of P. aeruginosa.
Statistics.
Data were analyzed and plotted using GraphPad Prism (ver 7.02).
Accession number(s).
Atomic coordinates and structure factors for PiuD have been deposited in the Protein Data Bank (accession no. 5NEC).
Supplementary Material
ACKNOWLEDGMENTS
The research leading to these results was conducted as part of the Translocation consortium (www.translocation.eu) and has received support from the Innovative Medicines Joint Undertaking under grant agreement no. 115525, resources which are composed of financial contribution from the European Union's seventh framework program (FP7/2007-2013) and EFPIA companies in kind contribution.
We thank Y. Braun and H. Weingart (Jacobs University Bremen) for help with the construction of the LESB58 mutants. We also thank E. Desarbre (Basilea Pharmaceutical Ltd.) for helpful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00097-18.
REFERENCES
- 1.Mahmood HY, Jamshidi S, Sutton JM, Rahman KM. 2016. Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr Med Chem 23:1062–1081. doi: 10.2174/0929867323666160304150522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill EE, Franco OL, Hancock RE. 2015. Antibiotic adjuvants: diverse strategies for controlling drug-resistant pathogens. Chem Biol Drug Des 85:56–78. doi: 10.1111/cbdd.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Möllmann U, Heinisch L, Bauernfeind A, Köhler T, Ankel-Fuchs D. 2009. Siderophores as drug delivery agents: application of the “Trojan horse” strategy. Biometals 22:615–624. doi: 10.1007/s10534-009-9219-2. [DOI] [PubMed] [Google Scholar]
- 4.Page MG, Dantier C, Desarbre E. 2010. In vitro properties of BAL30072, a novel siderophore sulfactam with activity against multiresistant Gram-negative bacilli. Antimicrob Agents Chemother 54:2291–2302. doi: 10.1128/AAC.01525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McPherson CJ, Aschenbrenner LM, Lacey BM, Fahnoe KC, Lemmon MM, Finegan SM, Tadakamalla B, O'Donnell JP, Mueller JP, Tomaras AP. 2012. Clinically relevant Gram-negative resistance mechanisms have no effect on the efficacy of MC-1, a novel siderophore-conjugated monocarbam. Antimicrob Agents Chemother 56:6334–6342. doi: 10.1128/AAC.01345-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim A, Kutschke A, Ehmann DE, Patey SA, Crandon JL, Gorseth E, Miller AA, McLaughlin RE, Blinn CM, Chen A, Nayar AS, Dangel B, Tsai AS, Rooney MT, Murphy-Benenato KE, Eakin AE, Nicolau DP. 2015. Pharmacodynamic profiling of a siderophore-conjugated monocarbam in Pseudomonas aeruginosa: assessing the risk for resistance and attenuated efficacy. Antimicrob Agents Chemother 59:7743–7752. doi: 10.1128/AAC.00831-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohira N, West J, Ito A, Ito-Horiyama T, Nakamura R, Sato T, Rittenhouse S, Tsuji M, Yamano Y. 2016. In vitro antimicrobial activity of siderophore cephalosporin S-649266 against Enterobacteriaceae clinical isolates including carbapenem-resistant strains. Antimicrob Agents Chemother 60:729–734. doi: 10.1128/AAC.01695-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wencewicz TA, Mollmann U, Long TE, Miller MJ. 2009. Is drug release necessary for antimicrobial activity of siderophore-drug conjugates? Syntheses and biological studies of the naturally occurring salmycin “Trojan horse” antibiotics and synthetic desferridanoxamine-antibiotic conjugates. Biometals 22:633–648. doi: 10.1007/s10534-009-9218-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji C, Miller PA, Miller MJ. 2012. Iron transport-mediated drug delivery: practical syntheses and in vitro antibacterial studies of tris-catecholate siderophore-aminopenicillin conjugates reveals selectively potent antipseudomonal activity. J Am Chem Soc 134:9898–9901. doi: 10.1021/ja303446w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maejima T, Inoue M, Mitsuhashi S. 1991. In vitro antibacterial activity of KP-736, a new cephem antibiotic. Antimicrob Agents Chemother 35:104–110. doi: 10.1128/AAC.35.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wencewicz TA, Miller MJ. 2013. Biscatecholate-monohydroxamate mixed ligand siderophore-carbacephalosporin conjugates are selective sideromycin antibiotics that target Acinetobacter baumannii. J Med Chem 56:4044–4052. doi: 10.1021/jm400265k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han S, Caspers N, Zaniewski RP, Lacey BM, Tomaras AP, Feng X, Geoghegan KF, Shanmugasundaram V. 2011. Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics. J Am Chem Soc 133:20536–20545. doi: 10.1021/ja208835z. [DOI] [PubMed] [Google Scholar]
- 13.Mima T, Kvitko BH, Rholl DA, Page MG, Desarbre E, Schweizer HP. 2011. In vitro activity of BAL30072 against Burkholderia pseudomallei. Int J Antimicrob Agents 38:157–159. doi: 10.1016/j.ijantimicag.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russo TA, Page MG, Beanan JM, Olson R, Hujer AM, Hujer KM, Jacobs M, Bajaksouzian S, Endimiani A, Bonomo RA. 2011. In vivo and in vitro activity of the siderophore monosulfactam BAL30072 against Acinetobacter baumannii. J Antimicrob Chemother 66:867–873. doi: 10.1093/jac/dkr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito A, Kohira N, Bouchillon SK, West J, Rittenhouse S, Sader HS, Rhomberg PR, Jones RN, Yoshizawa H, Nakamura R, Tsuji M, Yamano Y. 2016. In vitro antimicrobial activity of S-649266, a catechol-substituted siderophore cephalosporin, when tested against non-fermenting Gram-negative bacteria. J Antimicrob Chemother 71:670–677. doi: 10.1093/jac/dkv402. [DOI] [PubMed] [Google Scholar]
- 16.van Delden C, Page MG, Köhler T. 2013. Involvement of Fe uptake systems and AmpC beta-lactamase in susceptibility to the siderophore monosulfactam BAL30072 in Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:2095–2102. doi: 10.1128/AAC.02474-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito A, Nishikawa T, Matsumoto S, Yoshizawa H, Sato T, Nakamura R, Tsuji M, Yamano Y. 2016. Siderophore cephalosporin cefiderocol utilizes ferric iron transporter systems for antibacterial activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:7396–7401. doi: 10.1128/AAC.01405-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis P, Matthijs S, Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 19.Llamas MA, van der Sar A, Chu BC, Sparrius M, Vogel HJ, Bitter W. 2009. A novel extracytoplasmic function (ECF) sigma factor regulates virulence in Pseudomonas aeruginosa. PLoS Pathog 5:e1000572. doi: 10.1371/journal.ppat.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Llamas MA, Sparrius M, Kloet R, Jimenez CR, Vandenbroucke-Grauls C, Bitter W. 2006. The heterologous siderophores ferrioxamine B and ferrichrome activate signaling pathways in Pseudomonas aeruginosa. J Bacteriol 188:1882–1891. doi: 10.1128/JB.188.5.1882-1891.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito A, Sato T, Ota M, Takemura M, Nishikawa T, Toba S, Kohira N, Miyagawa S, Ishibashi N, Matsumoto S, Nakamura R, Tsuji M, Yamano Y. 2018. In vitro antibacterial properties of cefiderocol, a novel siderophore cephalosporin, against Gram-negative bacteria. Antimicrob Agents Chemother 62:e01454-17. doi: 10.1128/AAC.01454-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moynié L, Luscher A, Rolo D, Pletzer D, Tortajada A, Weingart H, Braun Y, Page MG, Naismith JH, Köhler T. 2017. Structure and Function of the PiuA and PirA Siderophore-Drug Receptors from Pseudomonas aeruginosa and Acinetobacter baumannii. Antimicrob Agents Chemother 61:e02531-16. doi: 10.1128/AAC.02531-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobessi D, Celia H, Pattus F. 2005. Crystal structure at high resolution of ferric-pyochelin and its membrane receptor FptA from Pseudomonas aeruginosa. J Mol Biol 352:893–904. doi: 10.1016/j.jmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Zheng T, Nolan EM. 2014. Enterobactin-mediated delivery of beta-lactam antibiotics enhances antibacterial activity against pathogenic Escherichia coli. J Am Chem Soc 136:9677–9691. doi: 10.1021/ja503911p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cobessi D, Celia H, Folschweiller N, Schalk IJ, Abdallah MA, Pattus F. 2005. The crystal structure of the pyoverdine outer membrane receptor FpvA from Pseudomonas aeruginosa at 3.6 angstroms resolution. J Mol Biol 347:121–134. doi: 10.1016/j.jmb.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 26.Ferguson AD, Hofmann E, Coulton JW, Diederichs K, Welte W. 1998. Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide. Science 282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 27.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771–778. doi: 10.1016/S0092-8674(00)81700-6. [DOI] [PubMed] [Google Scholar]
- 28.Yue WW, Grizot S, Buchanan SK. 2003. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J Mol Biol 332:353–368. doi: 10.1016/S0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 29.Payne MA, Igo JD, Cao Z, Foster SB, Newton SM, Klebba PE. 1997. Biphasic binding kinetics between FepA and its ligands. J Biol Chem 272:21950–21955. doi: 10.1074/jbc.272.35.21950. [DOI] [PubMed] [Google Scholar]
- 30.Cao Z, Qi Z, Sprencel C, Newton SM, Klebba PE. 2000. Aromatic components of two ferric enterobactin binding sites in Escherichia coli FepA. Mol Microbiol 37:1306–1317. doi: 10.1046/j.1365-2958.2000.02093.x. [DOI] [PubMed] [Google Scholar]
- 31.Annamalai R, Jin B, Cao Z, Newton SM, Klebba PE. 2004. Recognition of ferric catecholates by FepA. J Bacteriol 186:3578–3589. doi: 10.1128/JB.186.11.3578-3589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gi M, Lee KM, Kim SC, Yoon JH, Yoon SS, Choi JY. 2015. A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci Rep 5:14644. doi: 10.1038/srep14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasser V, Baco E, Cunrath O, August PS, Perraud Q, Zill N, Schleberger C, Schmidt A, Paulen A, Bumann D, Mislin GL, Schalk IJ. 2016. Catechol siderophores repress the pyochelin pathway and activate the enterobactin pathway in Pseudomonas aeruginosa: an opportunity for siderophore-antibiotic conjugates development. Environ Microbiol 18:819–832. doi: 10.1111/1462-2920.13199. [DOI] [PubMed] [Google Scholar]
- 34.Ochsner UA, Vasil ML. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci U S A 93:4409–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 8th ed Approved standard M7-A8. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 38.Pletzer D, Lafon C, Braun Y, Köhler T, Page MG, Mourez M, Weingart H. 2014. High-throughput screening of dipeptide utilization mediated by the ABC transporter DppBCDF and its substrate-binding proteins DppA1-A5 in Pseudomonas aeruginosa. PLoS One 9:e111311. doi: 10.1371/journal.pone.0111311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumas JL, van Delden C, Perron K, Köhler T. 2006. Analysis of antibiotic resistance gene expression in Pseudomonas aeruginosa by quantitative real-time-PCR. FEMS Microbiol Lett 254:217–225. doi: 10.1111/j.1574-6968.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 40.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Wolfe AJ, Eren E, Vijayaraghavan J, Indic M, van den Berg B, Movileanu L. 2012. Cation selectivity is a conserved feature in the OccD subfamily of Pseudomonas aeruginosa. Biochim Biophys Acta 1818:2908–2916. doi: 10.1016/j.bbamem.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauter NK, Grosse-Kunstleve RW, Adams PD. 2004. Robust indexing for automatic data collection. J Appl Crystallogr 37:399–409. doi: 10.1107/S0021889804005874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Sauter NK, van den Bedem H, Snell G, Deacon AM. 2006. Automated diffraction image analysis and spot searching for high-throughput crystal screening. J Appl Crystallogr 39:112–119. doi: 10.1107/S0021889805040677. [DOI] [Google Scholar]
- 44.Kabsch W. 1993. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 26:795–800. doi: 10.1107/S0021889893005588. [DOI] [Google Scholar]
- 45.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 46.Monaco S, Gordon E, Bowler MW, Delageniere S, Guijarro M, Spruce D, Svensson O, McSweeney SM, McCarthy AA, Leonard G, Nanao MH. 2013. Automatic processing of macromolecular crystallography X-ray diffraction data at the ESRF. J Appl Crystallogr 46:804–810. doi: 10.1107/S0021889813006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.