ABSTRACT
Candida glabrata infections are increasing worldwide and exhibit greater rates of antifungal resistance than those with other species. DNA mismatch repair (MMR) gene deletions, such as msh2Δ, in C. glabrata resulting in a mutator phenotype have recently been reported to facilitate rapid acquisition of antifungal resistance. This study determined the antifungal susceptibility profiles of 210 C. glabrata isolates in 10 hospitals in India and investigated the impact of novel MSH2 polymorphisms on mutation potential. No echinocandin- or azole-resistant strains and no mutations in FKS hot spot regions were detected among the C. glabrata isolates, supporting our in vitro susceptibility testing results. CLSI antifungal susceptibility data showed that the MICs of anidulafungin (geometric mean [GM], 0.12 μg/ml) and micafungin (GM, 0.01 μg/ml) were lower and below the susceptibility breakpoint compared to that of caspofungin (CAS) (GM, 1.31 μg/ml). Interestingly, 69% of the C. glabrata strains sequenced contained six nonsynonymous mutations in MSH2, i.e., V239L and the novel mutations E459K, R847C, Q386K, T772S, and V239/D946E. Functional analysis of MSH2 mutations revealed that 49% of the tested strains (40/81) contained a partial loss-of-function MSH2 mutation. The novel MSH2 substitution Q386K produced higher frequencies of CAS-resistant colonies upon expression in the msh2Δ mutant. However, expression of two other novel MSH2 alleles, i.e., E459K or R847C, did not confer selection of resistant colonies, confirming that not all mutations in the MSH2 MMR pathway affect its function or generate a phenotype of resistance to antifungal drugs. The lack of drug resistance prevented any correlations from being drawn with respect to MSH2 genotype.
KEYWORDS: Candida glabrata, India, FKS, MSH2, echinocandins, mutator genotype, mismatch repair
INTRODUCTION
Candida species are the most common cause of fungal infections worldwide among hospitalized patients. The distribution of Candida species isolated from patients with invasive candidiasis (IC) varies geographically depending on environmental factors, the patient's age, and exposure to antimicrobial agents. The proportion of IC due to Candida glabrata has increased in Europe and North America in the last decade, and this increase had been linked to previous exposure to antifungal agents, especially azoles (1). Furthermore, in C. glabrata high rates of azole resistance coupled with a recent increase of echinocandin resistance, especially in North America, are alarming, as the latter drug is a first-line therapy for nonneutropenic patients with C. glabrata infections (2, 3). An increase in echinocandin resistance from 2% to 3% to >13% over the past 10 years has been described in one U.S. medical center in 2009 to 2010 (3). Also, a Centers for Disease Control and Prevention (CDC) survey showed echinocandin resistance rates of 3.1% to 3.6% in 4 U.S. cities in various geographic regions (2). Remarkably, echinocandin resistance was associated with cross-resistance to azole antifungals in 36% of the echinocandin-resistant strains, emphasizing the significance of multidrug-resistant (MDR) C. glabrata (2). In contrast to the case for North America, echinocandin resistance among C glabrata strains is low (<1%) in Europe (4). These geographical variations of echinocandin resistance rates may be attributed to strain variation and/or specific use of antifungal prophylaxis and therapy in particular clinical settings, which warrants further investigations. In C. glabrata, echinocandin resistance occurs due to mutations in FKS gene (FKS1 and FKS2) sequences that encode the glucan synthase enzyme, which is the target of echinocandins. Several clinical studies have shown that the presence of an FKS mutation is the most important independent risk factor in predicting echinocandin therapeutic responses among patients with IC (3, 5). Further, the presence of an FKS mutation was found to be superior to MIC values in predicting clinical responses, especially when caspofungin (CAS) was used for testing (6). The underlying molecular mechanisms promoting development of resistance in C. glabrata to multiple drug classes, including azoles and echinocandins, are multifactorial. Recently, MSH2 DNA mismatch repair (MMR) gene deletions in C. glabrata resulting in a mutator phenotype that facilitates rapid acquisition of fluconazole (FLU), echinocandins, and amphotericin B (AMB) resistance were detected in clinical isolates of C. glabrata (7). Over half of the isolates of C. glabrata collected in the United States and other countries carried mutations in MSH2 that conferred a partial hypermutable phenotype with significantly increased frequency of FKS mutations (7). MSH2 alleles were subsequently shown to be dependent upon sequence type (8). Additionally, a more recent study evaluated antifungal resistance in clinical isolates of C. glabrata in a large cohort of patients in Saint-Louis Hospital, Paris, France, and demonstrated that the mutator phenotype was not associated with FLU resistance but instead corresponded to rare and specific genotypes (9). The aim of the present study was to evaluate echinocandin resistance among 210 clinical isolates of C. glabrata collected from 10 hospitals in India during a 5-year surveillance study of candidiasis where all yeast isolates were collected from 2012 to 2016 (10). For C. glabrata strains, FKS gene profiles and mutator genotype, by sequencing of the MSH2 gene, were determined. Further, the impact of novel MSH2 polymorphisms on mutation potential in a tester strain was investigated.
RESULTS
Isolates.
Candida glabrata isolates (n = 210) from individual patients in 10 hospitals in Delhi and the adjoining National Capital Region (NCR) and one in Kochi, southern India, collected over a period of 5 years, were identified to species level by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (Bruker Daltonics, Bremen, Germany) with a score of >2. About 52.3% of the C. glabrata isolates (n = 110) were from bloodstream infections (BSIs), followed by 29% (n = 61) isolates from respiratory tract specimens (sputum, n = 35; endotracheal aspirate, n = 18; bronchoalveolar lavage fluid, n = 8). The remaining 18.5% isolates (n = 39) were obtained from urine (n = 34), vaginal swab (n = 2), pus (n = 1), transhepatic biliary drainage (n = 1), and skin swab (n = 1). Bloodstream isolates were collected primarily from oncology/hematology wards (n = 45; 41%) and medical intensive care units (ICUs) (n = 41; 37%), followed by surgical wards (n = 24; 22%), in four tertiary care private hospitals in Delhi NCR (n = 3) and in Kochi. The respiratory tract isolates and other isolates were collected from a referral chest hospital and five public hospitals in Delhi, India.
In vitro susceptibility and FKS mutation analysis.
A total of 210 C. glabrata isolates were subjected to antifungal susceptibility testing (AFST) by Clinical and Laboratory Standards Institute broth microdilution method (CLSI-BMD) M27-A3/S4 (11, 12). The in vitro susceptibility data and the MIC distribution of C. glabrata isolates are presented in Table 1. Echinocandin resistance was declared if an isolate demonstrated a resistant MIC to at least two of the three echinocandin drugs (7). The geometric mean (GM) MIC values of anidulafungin (AFG) (GM MIC, 0.12 μg/ml) and micafungin (MFG) (GM MIC, 0.01 μg/ml) were lower and below the susceptibility breakpoint compared to that of CAS (GM MIC, 1.31 μg/ml) (13). Further, 198 isolates (94%) were categorized as resistant to CAS using the 2012 CLSI M27-S4 breakpoints (>0.25 μg/ml) (12, 13). This apparent overestimate of CAS resistance relative to testing with MFG or AFG by the standard BMD-RPMI method has previously been reported, emphasizing that microdilution testing for AFG and MFG correlates well with respect to separation between wild-type and FKS mutant isolates, whereas CAS MIC distributions appear to be variable. Further, due to interlaboratory variability in the MIC ranges for CAS, susceptibility testing of other echinocandins (AFG and MFG) has been advocated as a surrogate for identifying CAS-resistant C. glabrata strains (14). Low GM MICs of azoles, i.e., isavuconazole (ISA) (0.01 μg/ml), posaconazole (POS) (0.02 μg/ml), voriconazole (VRC) (0.03 μg/ml), itraconazole (ITC) (0.05 μg/ml), and FLU (1.84 μg/ml), were noted. Also, no non-wild-type strains were observed against AMB (MIC range, 0.06 to 1 μg/ml). Of the 210 isolates, 120 that had CAS MICs of ≥0.25 μg/ml were selected for FKS gene profiling. The isolates were selected primarily from BSIs (n = 86) for which there was a record of antifungal exposure available, and the remaining isolates were respiratory tract (n = 29) and other (n = 5) isolates collected from patients who had no exposure to antifungals. Amplification of the FKS1 and FKS2 regions generated amplicons of 391 bp and 460 bp, respectively. Mutations reported for CAS-resistant C. glabrata were not observed in the FKS1 and FKS2 regions of any of the tested C. glabrata strains.
TABLE 1.
MIC distribution of C. glabrata isolates against 10 antifungal drugs tested using the CLSI-BMD method
| Druga | No. of isolates with MIC (μg/ml) of: |
MIC |
MIC50 | MIC90 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.016 | 0.032 | 0.064 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | Range | GM | |||
| Echinocandinsb | ||||||||||||||||
| CAS | 12 | 11 | 129 | 47 | 11 | 0.25–4 | 1.31 | 1 | 1 | |||||||
| MFG | 142 | 68 | ≤0.016–0.032 | 0.01 | 0.015 | 0.015 | ||||||||||
| AFG | 11 | 17 | 167 | 15 | 0.032–0.25 | 0.12 | 0.125 | 0.125 | ||||||||
| Azoles, AMB, and FCc | ||||||||||||||||
| ITC | 55 | 41 | 20 | 4 | 0.032–0.5 | 0.05 | 0.06 | 0.12 | ||||||||
| VRC | 91 | 28 | 1 | 0.032–0.5 | 0.03 | 0.03 | 0.06 | |||||||||
| ISA | 113 | 3 | 4 | ≤0.016–0.25 | 0.01 | 0.015 | 0.15 | |||||||||
| POS | 49 | 52 | 18 | 1 | ≤0.016–0.25 | 0.02 | 0.03 | 0.06 | ||||||||
| AMB | 11 | 44 | 35 | 22 | 8 | ≤0.016–1 | 0.21 | 0.25 | 0.5 | |||||||
| FLU | 2 | 2 | 30 | 71 | 9 | 2 | 3 | 1 | 0.25–32 | 1.84 | 2 | 4 | ||||
| FC | 120 | 0.125 | 0.12 | 0.12 | 0.12 | |||||||||||
CAS, caspofungin; MFG, micafungin; AFG, anidulafungin; ITC, itraconazole; VRC, voriconazole; ISA, isavuconazole; POS, posaconazole; AMB, amphotericin B; FLU, fluconazole; FC, flucytosine.
A total of 210 isolates were tested against echinocandins. The CBPs of CAS and AFG are ≤0.12 μg/ml (susceptible), 0.25 μg/ml (intermediate), and ≥0.5 μg/ml (resistant), and those of MFG are ≤0.06 μg/ml, 0.12 μg/ml, and ≥0.25 μg/ml, respectively.
A total of 120 isolates were tested against azoles, AMB, and FC. The CBPs of FLU are ≤32 μg/ml (susceptible, dose dependent) and ≥64 μg/ml (resistant).
MSH2 gene sequence analysis.
Of the 120 C. glabrata isolates that had no mutations in the FKS gene, 83 isolates were randomly selected to determine their MSH2 genotypes. These 83 C. glabrata isolates included 50 bloodstream isolates, 30 respiratory tract isolates, and single isolate each from pus, urine, and skin, and none had FKS mutations. Overall, 69% (57/83) of the C. glabrata strains contained six nonsynonymous mutations within MSH2 gene compared with the database strain, CBS138/ATCC 2001 (http://www.candidagenome.org/). Notably, 63% (n = 52) of the isolates harboring mutations had high MICs (0.5 to 4 μg/ml) of CAS. However, as expected, none of the isolates tested for MSH2 gene mutations had high MICs of AFG and MFG. Overall, 41% of the isolates had a single Msh2p substitution, i.e., V239L (n = 34), and the remaining five novel substitutions observed were E459K (n = 9), R847C (n = 6), Q386K (n = 5), T772S (n = 2), and V239L/D946E (n = 1) (Table 2). Interestingly, respiratory tract isolates exhibited the wild-type sequence in 47% of isolates (14 of 30), while 78% (39 of 50) of the bloodstream isolates tested had mutations (P < 0.05). The duration of exposure to azoles (VRC and FLU) and CAS during the hospital stay and the time of sampling were available for 120 patients whose FKS gene profiles were investigated. Out of 120 patients, only 71 (59%) were treated with antifungals, which included azoles in 61 patients and echinocandins in 10 patients (see Table S1 in the supplemental material). A total of 67 patients (94%) had samples collected between 1 and <7 days of treatment. A total of 61 patients (85%) had a history of azole exposure, and only 10 patients had exposure to CAS. About 80% (n = 57) of patients had exposure to azoles for <7 days (FLU, n = 39; VRC, n = 18), and the remaining 4 were on azoles for 2 to 4 weeks.
TABLE 2.
Distribution of MSH2 gene polymorphisms in 83 Candida glabrata isolates
| Amino acid substitution encoded in MSH2 (n) | Specimen type (n) | No. of isolates with the indicated MIC (μg/ml) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Caspofungin |
Micafungin |
Anidulafungin |
||||||||||
| 0.25 | 0.5 | 1 | 2 | 4 | 0.015 | 0.03 | 0.03 | 0.06 | 0.125 | 0.25 | ||
| Wild type (26) | Blood (11) | 6 | 5 | 6 | 5 | 1 | 8 | 2 | ||||
| Respiratory tract (14) | 1 | 8 | 5 | 11 | 3 | 1 | 11 | 2 | ||||
| Urine (1) | 1 | 1 | 1 | |||||||||
| V239L (34) | Blood (26) | 3 | 3 | 15 | 5 | 19 | 7 | 3 | 23 | |||
| Respiratory tract (7) | 1 | 2 | 2 | 2 | 6 | 1 | 1 | 1 | 5 | |||
| Pus (1) | 1 | 1 | 1 | |||||||||
| E459Ka (9) | Blood (5) | 5 | 5 | 2 | 3 | |||||||
| Respiratory tract (4) | 1 | 2 | 1 | 3 | 1 | 1 | 3 | |||||
| Q386Ka (5) | Respiratory tract (4) | 4 | 2 | 2 | 4 | |||||||
| Blood (1) | 1 | 1 | 1 | |||||||||
| R847Ca (6) | Blood (4) | 4 | 1 | 3 | 1 | 3 | ||||||
| Respiratory tract (1) | 1 | 1 | 1 | |||||||||
| Skin scraping (1) | 1 | 1 | 1 | |||||||||
| T772Sa (2) | Blood (2) | 1 | 1 | 2 | 1 | 1 | ||||||
| V239L/D946Ea (1) | Blood (1) | 1 | 1 | 1 | ||||||||
Mutation found only in Indian isolates.
MSH2 polymorphism phenotyping assay.
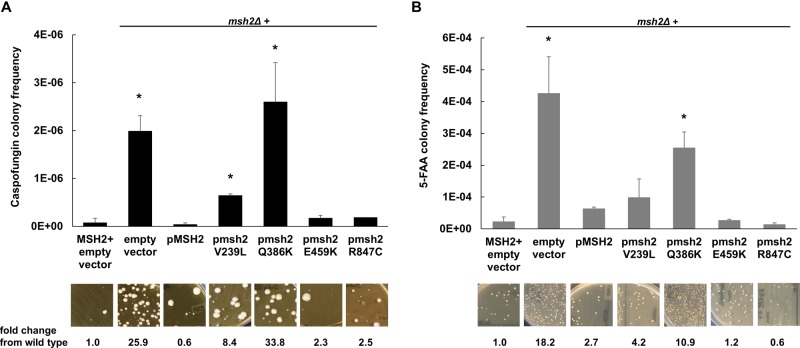
To determine the effects of DNA MMR on C. glabrata antifungal resistance, Healey et al. showed that the msh2Δ strain generated 82-, 18-, and 9-fold more CAS-, FLU-, and AMB-resistant mutants, respectively, than the wild-type strain in antifungal selections (7). Further, upon CAS selection in vitro, echinocandin-susceptible clinical isolates containing specific MSH2 mutations produced significantly higher frequencies of echinocandin-resistant colonies and FKS mutants than isolates carrying wild-type MSH2. To ascertain the significance of the novel MSH2 polymorphisms reported in this study, i.e., E459K, R847C, and Q386K, alleles carrying these polymorphisms were amplified from clinical isolates, expressed on a yeast centromere plasmid (pGRB2.0) under the control of their native promoter, and transformed into a laboratory strain carrying a deletion of MSH2 (7). Forward mutation frequencies were analyzed by measuring frequencies of colonies resistant to the echinocandin CAS (largely due to mutations in FKS1 or FKS2) or to the metabolic inhibitor 5-fluoroanthranilic acid (5-FAA) (largely due to mutations in TRP3 or TRP5). 5-FAA was used in addition to CAS as a nonantifungal alternative to distinguish frequencies of our strains. Following CAS and 5-FAA selections, we noted significantly elevated rates (33.8- and 10.9-fold, respectively) of resistant compared to wild-type colonies upon expression of pMSH2-Q386K but not with expression of pMSH2-E459K (2.3- and 1.2-fold, respectively) or pMSH2-R847C (2.5- and 0.6-fold, respectively) (Fig. 1). Similar to the results of previous studies, expression of pMSH2-V239L or an empty vector (positive control) led to elevated levels of resistant colonies, while pMSH2-wild type complemented these phenotypes (7) (Fig. 1). Overall, 49% of the strains (40/81) exhibited a partial loss-of-function MSH2 mutation. The functionality of the T772S allele, identified in only 2 strains, remains to be determined.
FIG 1.
Wild-type or msh2Δ cells expressing an empty or MSH2-containing plasmid were selected on caspofungin (A) or 5-fluoroanthranilic acid (B). Frequency data are means ± standard deviations (SD) from ≥2 independent experiments; representative images are shown. *, P < 0.05; **, P < 0.01 (Student's t test, two tailed). Scale bars, 1 cm.
DISCUSSION
This study found echinocandin- and azole-resistant strains among 210 isolates of C. glabrata collected during the course of the surveillance program from 2012 to 2016 in 10 hospitals in Delhi, NCR, and southern India. Although all C. glabrata isolates tested by the CLSI-BMD method in the present study showed CAS MICs in the intermediate and resistant categories according to published clinical breakpoints (CBPs), none of the isolates showed resistance to the other two echinocandins tested. Further, 120 isolates tested for FKS mutations in the hot spot region showed no mutations, suggesting echinocandin resistance to be unlikely in Indian isolates. Considering that the mechanism of action is shared among the echinocandins, it seems unlikely that a high percentage of isolates tested are nonsusceptible to CAS but remain susceptible to AFG and MFG (15). As previously reported, it is likely that this reflects technical issues associated with the in vitro testing of CAS rather than a true difference in antifungal activity (16). Thus, neither CLSI nor the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommends the use of CAS for antifungal susceptibility testing, while AFG and MFG are recommended as surrogate markers to predict CAS susceptibility (16, 17). It should be noted that a recent study by Pham et al. found that MFG MIC values were the strongest predictor of isolates possessing FKS mutations (2). The absence of echinocandin resistance in the present study is consistent with a report from Italy and Spain, where the resistance rate was 0% in a study analyzing 79 C. glabrata bloodstream isolates (18). Also, low echinocandin resistance rates among C. glabrata isolates obtained mainly from Lombardy, Italy (i.e., 2.1% [2 of 94]) (19), and among C. glabrata isolates obtained mainly from urine samples from France (1% [2 of 193 isolates, both with an FKS2 mutation]) (20) were reported. Similarly, 1 (2%) of the 50 C. glabrata isolates analyzed in a report from Turkey was resistant to CAS (21). In contrast, in the United States, a recent study found that 10% of C. glabrata isolates from cancer patients at the M.D. Anderson Cancer Center, Houston, TX, were resistant to CAS (22). Further, CAS resistance was associated with a higher rate of 28-day all-cause mortality, and the presences of FKS mutants was associated with therapeutic failure. A main risk factor for the development of FKS mutations in C. glabrata is pretreatment with echinocandins (23). Echinocandin resistance typically emerges after prolonged therapy, although it has been reported shortly after initiation of therapy (6). In the present study, only 10 of 120 patients (screened for FKS mutations) had <7 days of exposure to echinocandins prior to isolation of C. glabrata. The echinocandin resistance rates may be influenced by the site- or country-specific use of antifungal prophylaxis and therapy, and the frequency of resistance may vary considerably from institution to institution (24). Considering the fact that usage of echinocandins in many hospitals in India is limited due to cost constraints, only 1% of patients in this study were exposed to echinocandins, which likely accounts for the absence of echinocandin resistance in the present study. Also, usage of echinocandins in Indian hospitals is specifically restricted in private hospitals. Although they were available in early 2000 in private-sector hospitals, these antifungals have been prescribed only from 2010 in the 4 centers in the present study. A recent large, multisite, population-based candidemia surveillance program by the CDC in four metropolitan areas of the United States reported that more than half of cases with echinocandin-nonsusceptible isolates did not have known echinocandin exposure (24). Data from global surveys demonstrate that the frequency of echinocandin resistance among clinical isolates of C. glabrata ranges from 1 to 3% and is higher among isolates from North America (3%) than among those from Europe (1%), Latin America (0.0%), or the Asia-Pacific region (0.0%) (25). The data on echinocandin resistance in C. glabrata from the Indian subcontinent are limited, and this is the first study from India addressing both the susceptibility testing and molecular profiling of the FKS gene in echinocandin resistance. Importantly, in the present study, azole resistance among C. glabrata isolates was also not noted, which could be attributed to the fact that isolates were collected from patients within 2 weeks of azole therapy/prophylaxis. Previously, in a prospective multicenter study of ICU-acquired candidemia during 2011 and 2012 in 27 hospitals in India, a low prevalence of C. glabrata was reported despite prior exposure to antifungal agents in 15.7% of patients. Further, low azole resistance for FLU (1.5%), ITC (0%), and VRC (0%) was recorded in 65 C. glabrata bloodstream isolates tested for antifungal susceptibility (26). Similarly, in a prospective multicenter study of Candida bloodstream infections in three Lima-Callao hospitals during 2013 to 2015, no resistance to azoles among C. glabrata isolates was observed (27). Also, a laboratory-based candidemia survey in 20 centers in seven Latin American countries did not show an association of FLU exposure and candidemia due to C. glabrata. Furthermore, the proportion of isolates with resistance to FLU was low (6.5%) (28). Considering that C. glabrata has a complex population structure, the absence of MDR strains among Indian C. glabrata isolates suggests the possibility of genetic strain variations in the Indian subcontinent compared to North America and Europe, warranting in-depth genetic analysis of Indian strains. The defects in DNA repair may result in the emergence of drug-resistant variants in C. glabrata and, as more recently reported, in Cryptococcus neoformans (29). In the present study, 69% of the clinical strains investigated exhibited an amino acid substitution compared to the sequenced database strain CBS138/ATCC 2001, with 49% of strains carrying a partial loss-of-function mutation. The most common mutation, i.e., V239L noted in the MSH2 gene in this study has been previously reported in U.S., Switzerland, and Qatar clinical C. glabrata strains and is associated with a partial mutator phenotype (7). Similarly, the novel allele MSH2-Q386K that was noted in 5 C. glabrata strains from bloodstream and respiratory tract specimens in this study produced greater frequencies of CAS- and 5-FAA-resistant colonies upon expression in the msh2Δ mutant. However, expression of two other novel MSH2 alleles encoding the amino acid substitution E459K or R847C did not confer selection of resistant colonies, confirming that not all mutations in the MSH2 MMR pathway affect its function or generate a phenotype of resistance to antifungal drugs. Recently, comparison of mutation frequencies in strains with deletions of eight MMR pathway genes in C. neoformans showed that the loss of three of them, MSH2, MLH1, and PMS1, resulted in an increase in mutation rates (29). Also, Healey et al. showed that additional mechanisms of DNA repair, including other MMR genes, and other cellular systems may influence the mutagenic potential of C. glabrata strains (8). As all of our C. glabrata strains were fully susceptible to antifungals, the impact of the presence of MSH2 mutations on antifungal resistance could not be correlated. Similar to the 49% rate of MSH2 partial loss of function reported in our isolate population, 52% (117/225) of susceptible C. glabrata isolates demonstrated a nonsynonymous mutation within MSH2 in U.S. and non-U.S. site isolates in a previous study (7). Finally, mutator cells in the population may provide a selective advantage in vivo and enable this yeast with increased drug exposure to promote the development of antifungal resistance and/or pathogenicity (8). Despite the near absence of echinocandin or azole resistance in C. glabrata isolates in this patient population, the high prevalence of the mutator phenotype among strains, which have a heightened potential to develop resistance, is concerning. Our study calls out for continued vigilance, especially as antifungal exposure broadens at many centers.
MATERIALS AND METHODS
Fungal isolates and identification.
This study analyzed 210 C. glabrata clinical isolates collected from individual patients in 10 hospitals from Delhi and adjoining NCRs in northern India and in a single center in Kochi, Kerala, in southern India during a surveillance study of candidiasis where all yeasts isolates were collected from 2012 to 2016. All isolates were cultured on Sabouraud dextrose agar (SDA) for 24 h at 37°C for identification by MALDI-TOF MS using ethanol-formic acid extraction according to the manufacturer's protocol (10). The isolates were identified as C. glabrata with a score of >2.
AFST.
In vitro antifungal susceptibility testing (AFST) was done using the CLSI-BMD method, following the M27-A3/S4 guidelines (11, 12). The antifungals tested were AMB (Sigma, St. Louis, MO), FLU (Pfizer, Groton, CT), ITC (Lee Pharma, Hyderabad, India), VRC (Pfizer), POS (Merck, Whitehouse Station, NJ), ISA (Basilea Pharmaceutica, Basel, Switzerland), 5-flucytosine (FC) (Sigma), CAS (Merck), MFG (Astellas, Toyama, Japan), and AFG (Pfizer). Candida krusei ATCC 6258 and Candida parapsilosis ATCC 22019 were used as quality control strains. The MIC endpoints were defined as the lowest drug concentrations that caused a prominent (50%) decrease in growth vis-à-vis the controls and were read visually after 24 h for azoles and echinocandins. For AMB, the MIC was defined as the lowest concentration at which there was 100% inhibition of growth compared with the drug-free control well results. Isolates of C. glabrata were categorized as susceptible or resistant to each echinocandin using the 2012 CLSI M27-S4 breakpoints (12). The CBPs of CAS and AFG for C. glabrata are ≤0.12 μg/ml (susceptible), 0.25 μg/ml (intermediate), and ≥0.5 μg/ml (resistant), whereas those of MFG are ≤0.06 μg/ml, 0.12 μg/ml, and ≥0.25 μg/ml, respectively.
FKS gene sequencing.
The FKS1 and FKS2 genes of a total of 120 isolates were sequenced. Genomic DNA was extracted as described by Xu et al. (30). Hot spot 1 (HS1) and HS2 of FKS1 and FKS2 were amplified by PCR as previously described (31). DNA sequences were analyzed with Sequencing Analysis software version 5.3.1 (Applied Biosystems). Consensus sequences were made using BioEdit software (version 7.0.5.3) and were aligned with hot spot FKS regions of reference C. glabrata (GenBank accession no. HM366439 for FKS1 and HM366442 for FKS2).
MSH2 gene sequencing.
To assess the defects in DNA repair pathways in C. glabrata isolates, sequencing of the MMR gene MSH2 was done in 83 isolates as described previously (7). The MSH2 gene was amplified using primers CgMSH2u273F and CgMSH2d145R. The resulting amplicon (3,292 bp) was sequenced using CgMSH2u273F and CgMSH2d145R along with internal primers CgMSH2c846R, CgMSH2c1626R, and CgMSH2c2340R (7).
MSH2 cloning and drug selections.
For plasmid-based expression of MSH2, a gap repair approach was used (32). Briefly, coding regions plus promoter regions were PCR amplified from laboratory or clinical isolates containing flanking regions homologous to regions upstream and downstream of the SmaI restriction site in pGRB2.0. Strain 2001 HTU or the msh2Δ mutant was transformed with this PCR product, along with SmaI-linearized pGRB2.0, and selected on synthetically defined medium lacking uracil (SD-ura). Plasmids from transformants were sequenced for confirmation. The resulting strains were selected on 1 μg/ml CAS and 0.5 mg/ml 5-FAA (7). Briefly, strains were grown overnight in appropriate medium (yeast extract-peptone-dextrose [YPD] or SD-ura) at 37°C, and then approximately 1 × 108 CFU was plated onto echinocandin-containing drug plates and between 1 × 105 and 1 × 106 CFU was plated onto 5-FAA plates. Plates were incubated for 72 h. Dilutions were plated onto drug-free medium to determine exact CFU counts. Frequencies were calculated as the number of colonies on the drug plate divided by the total CFU plated. Frequency averages were calculated from at least two independent selections. Differences in resistance frequencies were evaluated by Student's t test. A P value of <0.05 (two tailed) was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
A.S. was supported in part by a research grant from the Council of Scientific & Industrial Research, India [F. no. 09/174(0068)/2014-EMR-I].
D.S.P. received grants from the U.S. National Institutes of Health and from Astellas. He serves on scientific advisory boards for, and receives grant support from, Astellas, Cidara, Amplyx, Scynexis, and Matinas and has been issued a U.S. patent concerning echinocandin resistance. All other authors have no conflicts to declare. The authors alone are responsible for the content and writing of the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00195-18.
REFERENCES
- 1.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pham CD, Iqbal N, Bolden CB, Kuykendall R, Harrison LH, Farley M, Schaffner W, Beldavs ZG, Chiller TM, Park BJ, Cleveland AA, Lockhart SR. 2014. Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob Agents Chemother 58:4690–4696. doi: 10.1128/AAC.03255-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander BD, Johnson MD, Pfeiffer CD, Jiménez-Ortigosa C, Catania J, Booker R, Castanheira M, Messer SA, Perlin DS, Pfaller MA. 2013. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klotz U, Schmidt D, Willinger B, Steinmann E, Buer J, Rath PM, Steinmann J. 2016. Echinocandin resistance and population structure of invasive Candida glabrata isolates from two university hospitals in Germany and Austria. Mycoses 59:312–318. doi: 10.1111/myc.12472. [DOI] [PubMed] [Google Scholar]
- 5.Shields RK, Nguyen MH, Press EG, Kwa AL, Cheng S, Du C, Clancy CJ. 2012. The presence of an FKS mutation rather than MIC is an independent risk factor for failure of echinocandin therapy among patients with invasive candidiasis due to Candida glabrata. Antimicrob Agents Chemother 256:4862–4869. doi: 10.1128/AAC.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyda ND, John J, Kilic A, Alam MJ, Lasco TM, Garey KW. 2014. FKS mutant Candida glabrata: risk factors and outcomes in patients with candidemia. Clin Infect Dis 59:819–825. doi: 10.1093/cid/ciu407. [DOI] [PubMed] [Google Scholar]
- 7.Healey KR, Zhao Y, Perez WB, Lockhart SR, Sobel JD, Farmakiotis D, Kontoyiannis DP, Sanglard D, Taj-Aldeen SJ, Alexander BD, Jimenez-Ortigosa C, Shor E, Perlin DS. 2016. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nat Commun doi: 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Healey KR, Jimenez Ortigosa C, Shor E, Perlin DS. 2016. Genetic drivers of multidrug resistance in Candida glabrata. Front Microbiol doi: 10.3389/fmicb.2016.01995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dellière S, Healey K, Gits-Muselli M, Carrara B, Barbaro A, Guigue N, Lecefel C, Touratier S, Desnos-Ollivier M, Perlin DS, Bretagne S, Alanio A. 2016. Fluconazole and echinocandin resistance of Candida glabrata correlates better with antifungal drug exposure rather than with MSH2 mutator genotype in a French cohort of patients harbouring low rates of resistance. Front Microbiol doi: 10.3389/fmicb.2016.02038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhary A, Prakash A, Sharma C, Kordalewska M, Kumar A, Sarma S, Tarai B, Singh A, Upadhyaya G, Upadhyay S, Yadav P, Singh PK, Khillan V, Sachdeva N, Perlin DS, Meis JF. 2018. A multicenter study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother 73:891–899. doi: 10.1093/jac/dkx480. [DOI] [PubMed] [Google Scholar]
- 11.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved Standard, 3rd ed CLSI document M27-A3. CLSI, Wayne, PA. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. 2012. Reference method for broth dilution antifungal susceptibility testing of yeasts: fourth informational supplement M27-S4. CLSI, Wayne, PA. [Google Scholar]
- 13.Pfaller MA, Diekema DJ, Castanheira M, Jones RN. 2011. Definitions and epidemiology of Candida species not susceptible to echinocandins. Curr Fungal Infect Rep 5:120–127. doi: 10.1007/s12281-011-0053-y. [DOI] [Google Scholar]
- 14.Shields RK, Nguyen MH, Press EG, Updike CL, Clancy CJ. 2013. Anidulafungin and micafungin MIC breakpoints are superior to that of caspofungin for identifying FKS mutant Candida glabrata strains and echinocandin resistance. Antimicrobial Agents Chemother 57:6361–6365. doi: 10.1128/AAC.01451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espinel-Ingroff A, Arendrup MC, Pfaller MA, Bonfietti LX, Bustamante B, Canton E, Chryssanthou E, Cuenca-Estrella M, Dannaoui E, Fothergill A, Fuller J, Gaustad P, Gonzalez GM, Guarro J, Lass-Flörl C, Lockhart SR, Meis JF, Moore CB, Ostrosky-Zeichner L, Pelaez T, Pukinskas SR, St-Germain G, Szeszs MW, Turnidge J. 2013. Interlaboratory variability of caspofungin MICs for Candida spp. using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother 57:5836–5842. doi: 10.1128/AAC.01519-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller MA, Messer SA, Diekema DJ, Jones RN, Castanheira M. 2014. Use of micafungin as surrogate marker to predict susceptibility and resistance to caspofungin among 3,764 clinical isolates of Candida by using CLSI methods and interpretive criteria. J Clin Microbiol 52:108–114. doi: 10.1128/JCM.02481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfaller MA, Dikeman DJ, Jones RN, Castanheira M. 2014. Use of anidulafungin as surrogate marker to predict susceptibility and resistance to caspofungin among 4,290 clinical isolates of Candida by using CLSI methods and interpretive criteria. J Clin Microbiol 52:3223–3229. doi: 10.1128/JCM.00782-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassetti M, Merelli M, Righi E, Diaz-Martin A, Rosello EM, Luzzati R, Parra A, Trecarichi EM, Sanguinetti M, Posteraro B, Garnacho-Montero J, Sartor A, Rello J, Tumbarello M. 2013. Epidemiology, species distribution, antifungal susceptibility and outcome of candidemia across five sites in Italy and Spain. J Clin Microbiol 51:4167–4172. doi: 10.1128/JCM.01998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tortorano AM, Prigitano A, Lazzarini C, Passera M, Deiana ML, Cavinato S, De Luca C, Grancini A, Lo Cascio G, Ossi C, Sala E, Montagna MT. 2013. A 1-year prospective survey of candidemia in Italy and changing epidemiology over one decade. Infection 41:655–662. doi: 10.1007/s15010-013-0455-6. [DOI] [PubMed] [Google Scholar]
- 20.Bourgeois N, Laurens C, Bertout S, Balard Y, Krasteva D, Rispail P, Lachaud L. 2014. Assessment of caspofungin susceptibility of Candida glabrata by the Etest®, CLSI and EUCAST methods, and detection of FKS1 and FKS2 mutations. Eur J Clin Microbiol Infect Dis 33:1247–1252. doi: 10.1007/s10096-014-2069-z. [DOI] [PubMed] [Google Scholar]
- 21.Kiraz N, Dag I, Oz Y, Yamac M, Kiremitci A, Kasifoglu N. 2010. Correlation between broth microdilution and disk diffusion methods for antifungal susceptibility of caspofungin, voriconazole, amphotericin B, itraconazole and fluconazole against Candida glabrata. J Microbiol Methods 82:136–140. doi: 10.1016/j.mimet.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Farmakiotis D, Tarrand JJ, Kontoyiannis DP. 2014. Drug-resistant Candida glabrata infection in cancer patients. Emerg Infect Dis 20:1833–1840. doi: 10.3201/eid2011.140685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis JS 2nd, Wiederhold NP, Wickes BL, Patterson TF, Jorgensen JH. 2013. Rapid emergence of echinocandin resistance in Candida glabrata resulting in clinical and microbiologic failure. Antimicrob Agents Chemother 57:4559–4561. doi: 10.1128/AAC.01144-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallabhaneni S, Cleveland AA, Farley MM, Harrison LH, Schaffner W, Beldavs ZG, Derado G, Pham CD, Lockhart SR, Smith RM. 2015. Epidemiology and risk factors for echinocandin nonsusceptible Candida glabrata bloodstream infections: Data from large multisite population-based Candidemia Surveillance Program, 2008–2014. Open Forum Infect Dis doi: 10.1093/ofid/ofv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. 2012. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J Clin Microbiol 50:1199–1203. doi: 10.1128/JCM.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez L, Bustamante B, Huaroto L, Agurto C, Illescas R, Ramirez R, Diaz A, Hidalgo J. 2017. A multi-centric study of Candida bloodstream infection in Lima-Callao, Peru: species distribution, antifungal resistance and clinical outcomes. PLoS One 12:e0175172. doi: 10.1371/journal.pone.0175172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nucci M, Queiroz-Telles F, Alvarado-Matute T, Tiraboschi IN, Cortes J, Zurita J, Guzman-Blanco M, Santolaya ME, Thompson L, Sifuentes-Osornio J, Echevarria JI, Colombo AL, Latin American Invasive Mycosis Network. 2013. Epidemiology of candidemia in Latin America: a laboratory-based survey. PLoS One 8:e59373. doi: 10.1371/journal.pone.0059373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boyce KJ, Wang Verma YS, Shakya VPS, Xue C, Idnurm A. 2017. Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio 8:e00595-17. doi: 10.1128/mBio.00595-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu J, Ramos AR, Vilgalys R, Mitchell TG. 2000. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J Clin Microbiol 38:1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zimbeck AJ, Iqbal N, Ahlquist AM, Farley MM, Harrison LH, Chiller T, Lockhart SR. 2010. FKS mutations and elevated echinocandin MIC values among Candida glabrata isolates from U.S. population-based surveillance. Antimicrob Agents Chemother 54:5042–5047. doi: 10.1128/AAC.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Healey KR, Katiyar SK, Raj S, Edline TD. 2012. CRS-MIS in Candida glabrata: sphinolipids modulate echinocandin-I interaction. Mol Microbiol 86:303–313. doi: 10.1111/j.1365-2958.2012.08194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.