ABSTRACT
Methicillin-resistant Staphylococcus aureus (MRSA) strains carry either a mecA- or a mecC-mediated mechanism of resistance to beta-lactam antibiotics, and the phenotypic expression of resistance shows extensive strain-to-strain variation. In recent communications, we identified the genetic determinants associated with the stringent stress response that play a major role in the antibiotic resistant phenotype of the historically earliest “archaic” clone of MRSA and in the mecC-carrying MRSA strain LGA251. Here, we sought to test whether or not the same genetic determinants also contribute to the resistant phenotype of highly and homogeneously resistant (H*R) derivatives of a major contemporary MRSA clone, USA300. We found that the resistance phenotype was linked to six genes (fruB, gmk, hpt, purB, prsA, and relA), which were most frequently targeted among the analyzed 20 H*R strains (one mutation per clone in 19 of the 20 H*R strains). Besides the strong parallels with our previous findings (five of the six genes matched), all but one of the repeatedly targeted genes were found to be linked to guanine metabolism, pointing to the key role that this pathway plays in defining the level of antibiotic resistance independent of the clonal type of MRSA.
KEYWORDS: oxacillin resistance determinants, MRSA, guanine metabolism
INTRODUCTION
In 2011, four strains belonging to the historically first methicillin-resistant Staphylococcus aureus (MRSA) clone (ST250/ST247-SCCmecI)—recovered in the United Kingdom and Denmark in the 1960s—were selected for whole-genome sequencing and detailed analysis of the antibiotic resistant phenotype (1). Each strain showed “heterogeneous resistance:” the majority of cells had oxacillin MIC values barely above that of susceptible bacteria, but cells with high-level and homogeneous resistance to oxacillin (H*R) were also present. Whole-genome sequencing performed on both the heterogeneous parental strain and its H*R derivatives identified point mutations in several genes in the H*R strains. A more recent study showed that homogeneous and high-level resistance to oxacillin can also be observed in S. aureus strain LGA251, which carries not mecA but the mecC determinant, and that homogeneous and high-level resistance requires additional genetic determinants (2).
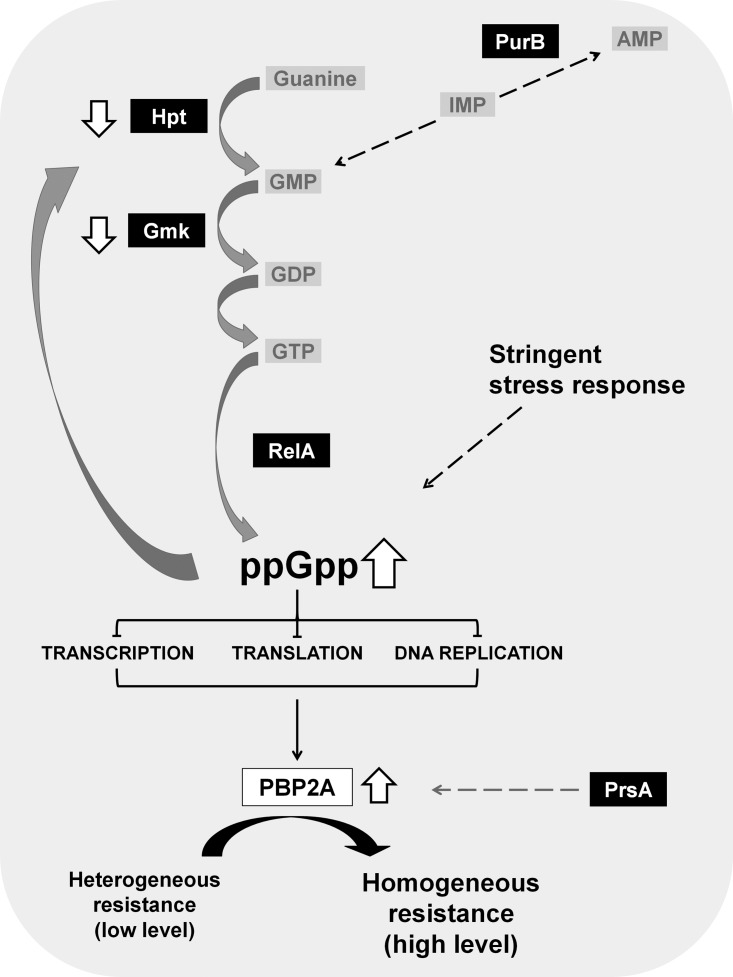
Interestingly, several of these determinants were identified in the H*R strains as part of the (p)ppGpp-dependent stringent stress-response pathway (Fig. 1) (1). Upon activation of the stringent stress response, the protein RelA is activated, leading to the synthesis of the alarmone (p)ppGpp. The accumulation of (p)ppGpp inhibits the activity of several enzymes involved in GTP biosynthesis (e.g., Hpt and Gmk), which causes a drastic reduction in the cellular GTP pool (3). Activation of the stringent stress response was shown to increase the transcription of the mecA determinant and the production of the resistance protein PBP2A, accompanied by the expression of high and homogeneous antibiotic resistance (Fig. 1) (4).
FIG 1.
Molecular pathways mediating H*R phenotype with emphasis on guanine metabolism. The activation of the stringent stress response leads to the production of the resistance protein PBP2A, leading to the expression of high and homogeneous antibiotic resistance. This phenotype may be achieved through changes affecting distinct steps of the complex guanine metabolism. Proteins targeted by mutations in H*R strains are highlighted in black boxes.
In the current work, we aimed to test if these additional genetic determinants are dependent on the clonal type of the MRSA strain. For this, we selected highly oxacillin-resistant isolates from the MRSA clone USA300, which is currently the predominant clonal type of MRSA in the United States (5–7).
A population analysis was used to characterize the resistance phenotypes, and whole-genome sequencing was used to identify mutations in the H*R derivatives of the parental MRSA strains.
RESULTS
Obtaining homogenous highly resistant subpopulations of MRSA strain USA300.
Following similar procedures as in previous studies (1), we selected as parental strains four clinical isolates obtained in 2012 from patients with skin and soft tissue infections described in a recent study in the community in New York city (6).
A population analysis was performed on the four parental strains by growing the bacteria on tryptic soy agar (TSA) supplemented with increasing concentrations of oxacillin. Five colonies were selected from plates with 100 μg/ml of oxacillin for each of the four parental isolates. The colonies were stabilized by four passages on TSA in the absence of oxacillin, and a new set of population analyses was performed to confirm their homogeneous and high-level resistance.
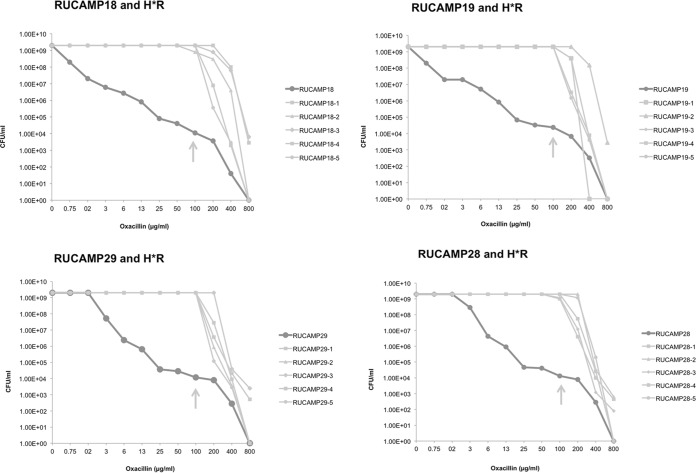
Population analysis profiles (PAPs) for the USA300 strains and their H*R derivatives demonstrate the heterogeneous phenotype of the parental strains and the homogeneous profile of the H*R strains (Fig. 2).
FIG 2.
Population analysis profiles of the USA300 parental and H*R strains. RUCAMP18 and RUCAMP19 are the wound and the nare strains recovered from one patient infected and colonized by the same USA300 strain (t008/ST8/SCCmecIVa/PVL/ACME); RUCAMP29 and RUCAMP28 are the wound and nare strains of another patient, also infected and colonized by a variant of USA300 (t052/ST8/SCCmecIVa/PVL/ACME). Five homogenously resistant colonies were selected at 100 μg/ml of oxacillin from each parental PAP plate, as indicated by the arrows. The graphs show the different profiles between parental and H*R strains.
Whole-genome sequencing of parental strains and their subpopulations.
The four parental strains, together with the 20 H*R strains, were selected for whole-genome sequencing. Each H*R strain showed a single mutation compared to the parental strain (with the exception of RUCAMP29-1 that carried two mutations). There was a great redundancy in the targeted genes, with only six genes (fruB, gmk, hpt, purB, prsA, and relA) being affected among the 20 H*R strains. All mutations were nonsynonymous or inactivating, with the exception of two mutations that targeted putative regulatory regions: the putative regulatory region of gmk (its coding DNA sequence [CDS] was also targeted by a nonsynonymous single nucleotide polymorphism [SNP] in another clone) and a gene encoding a putative DNA-binding transcriptional regulator that is contiguous with purB (also targeted by nonsynonymous or inactivating SNPs in other H*R strains). As these likely belong to the same polycistronic operon, the SNP may affect the expression of both genes. Table 1 shows the complete list of mutations observed in the H*R strains in genes that were intact in the parental strains.
TABLE 1.
Mutations found in the H*R strains
| H*R clone | Effect | Nucleotide change (5′→3′) | Amino acid change | Affected gene(s) | Locus tag(s) in referencesa | Product |
|---|---|---|---|---|---|---|
| RUCAMP18-1 | Stop gained | 383T→A | Leu128a | hpt | SAUSA300_0488/hpt; SACOL0554/hpt | Hypoxanthine-guanine phosphoribosyltransferase |
| RUCAMP18-2 | Missense variant | 500C→A | Ala167Asp | fruB/lacC_1 | SAUSA300_0684/fruB; SACOL0758/fruK | Fructose 1-phosphate kinase |
| RUCAMP18-3 | Missense variant | 317C→T | Thr106Ile | hpt | SAUSA300_0488/hpt; SACOL0554/hpt | Hypoxanthine-guanine phosphoribosyltransferase |
| RUCAMP18-4 | Stop gained | 847G→T | Glu283a | fruB/lacC_1 | SAUSA300_0684/fruB; SACOL0758/fruK | Fructose 1-phosphate kinase |
| RUCAMP18-5 | Frameshift variant | 106delG | Gly39fs | fruB/lacC_1 | SAUSA300_0684/fruB; SACOL0758/fruK | Fructose 1-phosphate kinase |
| RUCAMP19-1 | Stop gained | 730C→T | Gln244fs | purB | SAUSA300_1889/purB; SACOL1969/purB | Adenylosuccinate lyase |
| RUCAMP19-2 | Missense variant | 419C→A | Ala140Asp | hpt | SAUSA300_0488/hpt; SACOL0554/hpt | Hypoxanthine-guanine phosphoribosyltransferase |
| RUCAMP19-3 | Missense variant | 523C→T | Pro175Ser | fruB/lacC_1 | SAUSA300_0684/fruB; SACOL0758/fruK | Fructose 1-phosphate kinase |
| RUCAMP19-4 | Missense variant | 416G→A | Arg139His | purB | SAUSA300_1889/purB; SACOL1969/purB | Adenylosuccinate lyase |
| RUCAMP19-5 | Missense variant | 523C→T | Pro175Ser | fruB/lacC_1 | SAUSA300_0684/fruB; SACOL0758/fruK | Fructose 1-phosphate kinase |
| RUCAMP28-1 | Stop gained | 1860T→A | Tyr620a | relA | SAUSA300_1590; SACOL1689/relA2 | GTP pyrophosphokinase |
| RUCAMP28-2 | Missense variant | 461C→A | Ala154Glu | gmk | SAUSA300_1102/gmk; SACOL1221/gmk | Guanylate kinase |
| RUCAMP28-3 | Missense variant | 911C→T | Ala304Val | prsA | SAUSA300_0478/prs; SACOL0544/prsA | Ribose-phosphate pyrophosphokinase |
| RUCAMP28-4 | Missense variant | 394C→T | Arg132Cys | relA | SAUSA300_1590; SACOL1689/relA2 | GTP pyrophosphokinase |
| RUCAMP28-5 | Missense variant | 132T→A | Asp44Glu | prsA | SAUSA300_0478/prs; SACOL0544/prsA | Ribose-phosphate pyrophosphokinase |
| RUCAMP29-1 | SNP in IGR, putative regulatory region of gmk/SAUSA_1102 or SAUSA300_1101a | C→A (65 bp upstream gmk) | gmk | Guanylate kinase | ||
| RUCAMP29-1 | SNP in IGR, putative regulatory region of SAUSA300_1888 or the polycistronic transcript enrolling SAUSA300_1889/purBa | G→T (3 bp upstream ATG) | SAUSA300_1888 and/or purB | Conserved hypothetical protein/putative DNA-binding transcriptional regulator | ||
| RUCAMP29-2 | Missense variant | 265G→A | Ala89Thr | prsA | SAUSA300_0478/prs; SACOL0544/prsA | Ribose-phosphate pyrophosphokinase |
| RUCAMP29-3 | Missense variant | 307A→G | Thr103Ala | hpt | SAUSA300_0488/hpt; SACOL0554/hpt | Hypoxanthine-guanine phosphoribosyltransferase |
| RUCAMP29-4 | Stop gained | 2161A→T | Lys721a | relA | SAUSA300_1590; SACOL1689/relA2 | GTP pyrophosphokinase |
| RUCAMP29-5 | Missense variant | 650C→T | Ser217Leu | fruB/lacC_1 | SAUSA300_0684/fruB; SACOL0758/fruK | Fructose 1-phosphate kinase |
Guanine pathway is impacted by most of the mutations affecting resistance level.
Except for fruB, all genes mutated in the H*R strains (gmk, hpt, purB, prsA, and relA) participate in various stages of guanine metabolism (Fig. 1). Mutations in four of these genes (gmk, hpt, prsA, and relA) have already been identified in the historically earliest MRSA isolates described in the study by Dordel et al. and in S. aureus strain LGA251 (1, 2). A mutation in the adenylosuccinate lyase (purB) gene was seen for the first time in an H*R strain in this study.
Growth rate alteration associated with point mutations in fruB.
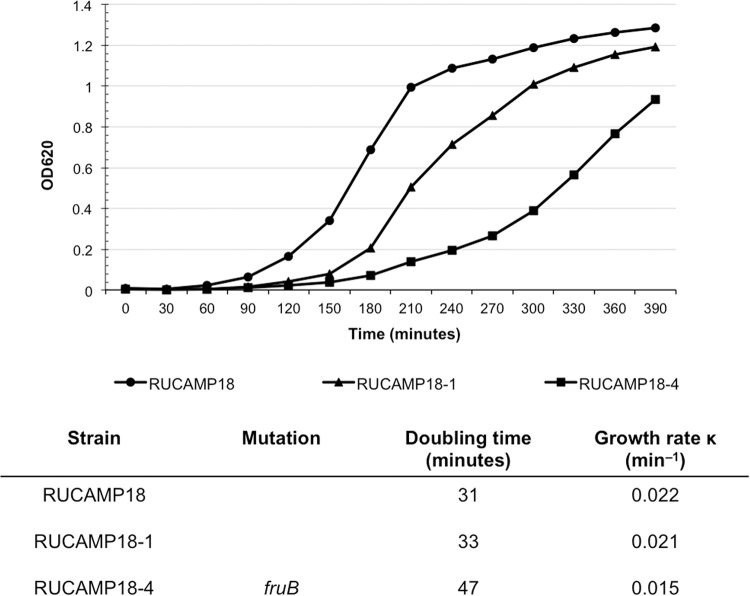
The gene fruB (lacC_1) represents one of the most frequently mutated genes in the H*R strains (in six of twenty) identified in this study. Mutations in fructose phosphate kinase are expected to affect the growth rate of the strain (8). To test this, growth curves were generated comparing one H*R strain carrying a mutated form of the gene (strain RUCAMP18-4) with its parental strain (RUCAMP18) and another H*R strain obtained from the same parental strain, but without the particular point mutation (RUCAMP18-1). The growth rate of the mutated H*R strain (RUCAMP18-4) was much lower (0.015 min−1) than the control isolates (0.021 and 0.022 min−1). For RUCAMP18-4, it took 180 min to start the exponential phase compared to 90 min for the parental strain (Fig. 3).
FIG 3.
Growth curves of mutant in fruB. Growth curves of RUCAMP18-4 (mutated in fruB), the parental strain RUCAMP18, and RUCAMP18-1, an H*R derivative of RUCAMP18 lacking the mutation in fruB. The experiment was performed twice.
DISCUSSION
In the H*R derivatives of strain USA300, the mecA gene is harbored in an SCCmecIV cassette (7). The mutated genes observed in these derivatives were virtually identical to the ones previously identified in H*R subpopulations of the “archaic clone” (ST250/ST247) which harbor a different SCCmecI cassette (1). Thus, the genetic background defining homogeneous resistance to beta-lactam antibiotics seems to be independent of the cassette harboring mecA.
In previous studies, we hypothesized that the stringent response pathway would be associated with the homogeneous high resistance to oxacillin exhibited by H*R derivatives of MRSA (1, 9, 10). In the present study, we further narrowed our suspicion to the pathway of guanine metabolism, as all but one of the repeatedly mutated genes in H*R strains (obtained from either USA300 or the “archaic clone” or even the mecC-harboring strain LGA251) were shown to affect one step or another of this particular metabolic pathway (Fig. 1).
Among the genes mutated in H*R derivatives, fruB has been associated with vancomycin resistance (11). However, none of our mutants with or without mutations in this gene showed decreased susceptibility to vancomycin (disc diffusion values in the range of 12 to 14 mm) (results not shown).
Under stress, as in the presence of high concentrations of antibiotics in the growth medium, (p)ppGpp is overexpressed, which in turn, will inhibit Gmk in S. aureus. Gmk is the enzyme responsible for the conversion of GMP to GDP during de novo synthesis of GTP (3). We hypothesize that mutations in RUCAMP28-2 or RUCAMP29-1 could lead to a constitutive inhibition of Gmk; thus, the cellular levels of GTP in these strains are constitutively low, which could help the bacteria to resist the high concentrations of antibiotic.
Mutations found in both hpt and relA genes were associated with the highly resistant phenotype of the bacteria linked to a constitutive production of (p)ppGpp (1, 12).
The chaperone PrsA was recently identified as a new auxiliary factor of oxacillin resistance in MRSA, affecting presumably the posttranscriptional maturation of penicillin-binding protein 2A (PBP2A), possibly at the stage of export and/or folding of newly synthesized PBP2A (13). More studies are needed to determine the role of mutations in psrA in high and homogeneous levels of oxacillin resistance in the mutants RUCAMP28-3, RUCAMP28-5, and RUCAMP29-2, as the mutations identified affect different domains of the protein.
Finally, in this study, purB, which is also involved in purine metabolism, has been identified as a novel gene potentially mediating the H*R phenotype. At the molecular level, this phenotype was triggered not only by inactivating nonsynonymous SNPs, but also by a SNP most likely affecting the purB mRNA.
In conclusion, of the 20 H*R strains analyzed in this study, a total of only six genes were found mutated. Five of these genes—prsA, hpt, fruB, gmk, and relA—had been identified in our previous studies associated with the homogeneous high-level resistance to oxacillin (1). The gene gmk was shown to be mutated in H*R derivatives of strain LGA251 (2). Four of the other genes (prsA, hpt, gmk, and relA), as well as purB (the additional gene mutated in USA300 H*R derivatives and identified in the present work), are involved in guanine metabolism, pointing to the key role of this metabolic pathway in defining the level of antibiotic resistance, regardless of clonal type of the MRSA isolate.
MATERIALS AND METHODS
Bacterial strains.
The parental strains were RUCAMP18, RUCAMP19, RUCAMP28, and RUCAMP29. RUCAMP18 was obtained from the active wound of one patient, and RUCAMP19 was isolated from a surveillance nasal swabbing performed on the same patient. Similarly, RUCAMP29 was obtained from the wound of a second patient, for which RUCAMP28 comes from the anterior nares. The two pairs of nasal and wound specimens from two different patients were obtained in the course of a study in which only a few patients were found to be both infected and colonized by the same strain. The goal was to study H*R strains in different isolates of the same USA300 clone but from different clinical sources recovered from different patients and to include both harmless colonization as well as active infection samples.
Population analysis profiles.
Parental strains were recovered from −80°C stocks by growing in tryptic soy broth ([TSB] Difco Laboratories, BBL, Becton Dickinson, Franklin Lakes, NJ, USA) at 37°C overnight with aeration. Plates of TSA (Difco Laboratories, BBL, Becton Dickinson, Franklin Lakes, NJ, USA) were prepared with increasing concentrations (2-fold) of oxacillin. These plates were inoculated with new dilutions of overnight cultures on TSB. CFU were counted after 48-h incubations of the plates at 37°C (14).
Selection of H*R strains: homogeneously and highly resistant subpopulations.
Five medium-size colonies capable of growing on TSA plates containing 100 μg/ml of oxacillin were picked from the PAP plates of each of the four “parental” MRSA isolates. These colonies were named “H*R” for homogeneous and high-level oxacillin resistance. The colonies were resuspended in 200 μl of TSB and used to inoculate plates of TSA and TSA with 100 μg/ml of oxacillin to verify their stability (through four passages in total). TSB cultures were also used for preparations of genomic DNA.
Whole-genome sequencing.
Genomic DNA was extracted from both the parental strains and the H*R strains using the Qiagen DNeasy blood and tissue kit (Qiagen, Ambion Inc., Austin, TX, USA). Paired-ended (2 × 250 bp) sequencing was performed at the Instituto Gulbenkian de Ciência ([IGC] Oeiras, Portugal) using the Illumina MiSeq platform. The quality control of reads, de novo assembly, contigs quality assessment, and possible contamination search were carried out using the multisoftware pipeline INNUca version 2.6 (https://github.com/B-UMMI/INNUca). Selected assemblies for parental isolates were inspected and subsequently annotated using Prokka (https://github.com/tseemann/prokka) (version 1.12). The mean depth of coverage (after quality control) ranged from 68- to 101-fold. The quality processed reads of each H*R strain were mapped against the corrected assembly of the respective parental strain using Snippy v3.1 (https://github.com/tseemann/snippy) with the following criteria: (i) a minimum mapping quality of ≥20; (ii) a minimum number of 10 quality processed reads covering the variant position; and (iii) a minimum proportion of 90% of quality processed reads at the variant position differing from the reference. All mutations were inspected and confirmed using Integrative Genomics Viewer (http://software.broadinstitute.org/software/igv/).
Growth properties of the fruB mutant.
To analyze whether a mutation in fruB caused alterations in the growth rate, strain RUCAMP18-4, its parent RUCAMP18, and RUCAMP18-1 (another H*R derivative from the same parental strain but lacking the fruB mutation) were tested for their growth rates in TSB, starting with an optical density (OD) of 0.01. The culture densities were tested every 30 min for 8 h. The growth rates were calculated by the formula k = log(Xt) − log(X0)/0.301t.
Accession number(s).
The sequences for the S. aureus isolates are in the Sequence Read Archive (SRA) under BioProject no. PRJEB24730.
ACKNOWLEDGMENTS
This work was financially supported by a US Public Health Service Award 2 R01 AI457838-15 and by project LISBOA-01-0145-FEDER-007660 (Microbiologia Molecular, Estrutural e Celular) funded by FEDER funds through COMPETE2020-Programa Operacional Competitividade e Internacionalização (POCI), by national funds through FCT-Fundação para a Ciência e a Tecnologia and project ONEIDA (LISBOA-01-0145-FEDER-016417) cofunded by FEEI-“Fundos Europeus Estruturais e de Investimento” from “Programa Operacional Regional Lisboa 2020,” and by national funds from FCT. M.P.D.L.G. was supported by PCORI grant CER-1402-10800 and by funds from the RB Roberts Bacterial Antibiotic Resistance Group (BARG). C.M. was supported by grant SFRH/BPD/111697/2015 from FCT.
The authors declare no conflict of interest.
REFERENCES
- 1.Dordel J, Kim C, Chung M, Pardos de la Gandara M, Holden MT, Parkhill J, de Lencastre H, Bentley SD, Tomasz A. 2014. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. mBio 5:e01000. doi: 10.1128/mBio.01000-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milheirico C, de Lencastre H, Tomasz A. 2017. Full-genome sequencing identifies in the genetic background several determinants that modulate the resistance phenotype in methicillin-resistant Staphylococcus aureus strains carrying the novel mecC gene. Antimicrob Agents Chemother 61:e02500-16. doi: 10.1128/AAC.02500-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corrigan RM, Bellows LE, Wood A, Grundling A. 2016. ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in Gram-positive bacteria. Proc Natl Acad Sci U S A 113:E1710–E1719. doi: 10.1073/pnas.1522179113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C, Mwangi M, Chung M, Milheirico C, de Lencastre H, Tomasz A. 2013. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One 8:e82814. doi: 10.1371/journal.pone.0082814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardos de la Gandara M, Curry M, Berger J, Burstein D, Della-Latta P, Kopetz V, Quale J, Spitzer E, Tan R, Urban C, Wang G, Whittier S, de Lencastre H, Tomasz A. 2016. MRSA causing infections in hospitals in greater metropolitan New York: major shift in the dominant clonal type between 1996 and 2014. PLoS One 11:e0156924. doi: 10.1371/journal.pone.0156924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardos de la Gandara M, Raygoza Garay JA, Mwangi M, Tobin JN, Tsang A, Khalida C, D'Orazio B, Kost RG, Leinberger-Jabari A, Coffran C, Evering TH, Coller BS, Balachandra S, Urban T, Parola C, Salvato S, Jenks N, Wu D, Burgess R, Chung M, de Lencastre H, Tomasz A. 2015. Molecular types of methicillin-resistant Staphylococcus aureus and methicillin-sensitive S. aureus strains causing skin and soft tissue infections and nasal colonization, identified in community health centers in New York City. J Clin Microbiol 53:2648–2658. doi: 10.1128/JCM.00591-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 8.Uhlemann AC, Kennedy AD, Martens C, Porcella SF, Deleo FR, Lowy FD. 2012. Toward an understanding of the evolution of Staphylococcus aureus strain USA300 during colonization in community households. Genome Biol Evol 4:1275–1285. doi: 10.1093/gbe/evs094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CK, Milheirico C, de Lencastre H, Tomasz A. 2017. Antibiotic resistance as a stress response: recovery of high-level oxacillin resistance in methicillin-resistant Staphylococcus aureus “auxiliary” (fem) mutants by induction of the stringent stress response. Antimicrob Agents Chemother 61:e00313-17. doi: 10.1128/AAC.00313-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aedo S, Tomasz A. 2016. Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 60:2311–2317. doi: 10.1128/AAC.02697-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takata T, Shirakawa T, Ito J, Okamoto A, Massi MN, Kinoshita S, Hatta M, Kawabata M. 2006. SCCmec typing and detection of VISA-related genes in methicillin-resistant Staphylococcus aureus clinical strains from Kobe University Hospital, Japan. Southeast Asian J Trop Med Public Health 37:1149–1155. [PubMed] [Google Scholar]
- 12.Mwangi MM, Kim C, Chung M, Tsai J, Vijayadamodar G, Benitez M, Jarvie TP, Du L, Tomasz A. 2013. Whole-genome sequencing reveals a link between beta-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist 19:153–159. doi: 10.1089/mdr.2013.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jousselin A, Manzano C, Biette A, Reed P, Pinho MG, Rosato AE, Kelley WL, Renzoni A. 2015. The Staphylococcus aureus chaperone PrsA is a new auxiliary factor of oxacillin resistance affecting penicillin-binding protein 2A. Antimicrob Agents Chemother 60:1656–1666. doi: 10.1128/AAC.02333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jabes D, Nachman S, Tomasz A. 1989. Penicillin-binding protein families: evidence for the clonal nature of penicillin resistance in clinical isolates of pneumococci. J Infect Dis 159:16–25. doi: 10.1093/infdis/159.1.16. [DOI] [PubMed] [Google Scholar]