ABSTRACT
The imipenem-relebactam combination is in development as a potential treatment regimen for infections caused by Enterobacteriaceae possessing complex β-lactamase backgrounds. Relebactam is a β-lactamase inhibitor that possesses the diazabicyclooctane core, as in avibactam; however, the R1 side chain of relebactam also includes a piperidine ring, whereas that of avibactam is a carboxyamide. Here, we investigated the inactivation of the Klebsiella pneumoniae carbapenemase KPC-2, the most widespread class A carbapenemase, by relebactam and performed susceptibility testing with imipenem-relebactam using KPC-producing clinical isolates of Enterobacteriaceae. MIC measurements using agar dilution methods revealed that all 101 clinical isolates of KPC-producing Enterobacteriaceae (K. pneumoniae, Klebsiella oxytoca, Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii, Citrobacter koseri, and Escherichia coli) were highly susceptible to imipenem-relebactam (MICs ≤ 2 mg/liter). Relebactam inhibited KPC-2 with a second-order onset of acylation rate constant (k2/K) value of 24,750 M−1 s−1 and demonstrated a slow off-rate constant (koff) of 0.0002 s−1. Biochemical analysis using time-based mass spectrometry to map intermediates revealed that the KPC-2–relebactam acyl-enzyme complex was stable for up to 24 h. Importantly, desulfation of relebactam was not observed using mass spectrometry. Desulfation and subsequent deacylation have been observed during the reaction of KPC-2 with avibactam. Upon molecular dynamics simulations of relebactam in the KPC-2 active site, we found that the positioning of active-site water molecules is less favorable for desulfation in the KPC-2 active site than it is in the KPC-2–avibactam complex. In the acyl complexes, the water molecules are within 2.5 to 3 Å of the avibactam sulfate; however, they are more than 5 to 6 Å from the relebactam sulfate. As a result, we propose that the KPC-2–relebactam acyl complex is more stable than the KPC-2–avibactam complex. The clinical implications of this difference are not currently known.
KEYWORDS: β-lactams, β-lactamases, relebactam, carbapenemase, β-lactamase inhibitor
INTRODUCTION
The KPC-2 β-lactamase is the most prevalent carbapenemase in carbapenem-resistant Enterobacteriaceae (CRE) in the United States and is disseminated worldwide. KPC-2's spectrum of activity includes all currently available β-lactams and clavulanic acid, sulbactam, and tazobactam (1). Fortunately, KPC-2 is found to be susceptible to inhibition by diazabicyclooctane (DBO) β-lactamase inhibitors (2, 3). Relebactam, formerly MK-7655, is a DBO that promises to contribute to the renaissance in antimicrobial chemotherapy (Fig. 1) (2). When combined with imipenem, relebactam is effective against Enterobacteriaceae with known KPC carbapenemases, AmpCs, and/or extended-spectrum β-lactamases (ESBLs) (2, 4, 5). Imipenem-relebactam also demonstrates potent activity against Enterobacteriaceae that express AmpCs or ESBLs and that have impermeability phenotypes (e.g., loss of porins) (5). Investigators showed that the imipenem-relebactam combination proved to be very effective in a hollow-fiber infection model due to a KPC-producing Klebsiella pneumoniae strain (4, 6). Furthermore, in mouse models of infection with imipenem-resistant Klebsiella strains, imipenem-relebactam performed favorably compared to imipenem alone (7). The goal of this investigation was to assess the efficacy of imipenem-relebactam against a panel of clinical isolates of KPC-producing Enterobacteriaceae and determine the inhibition parameters for purified KPC-2 with relebactam.
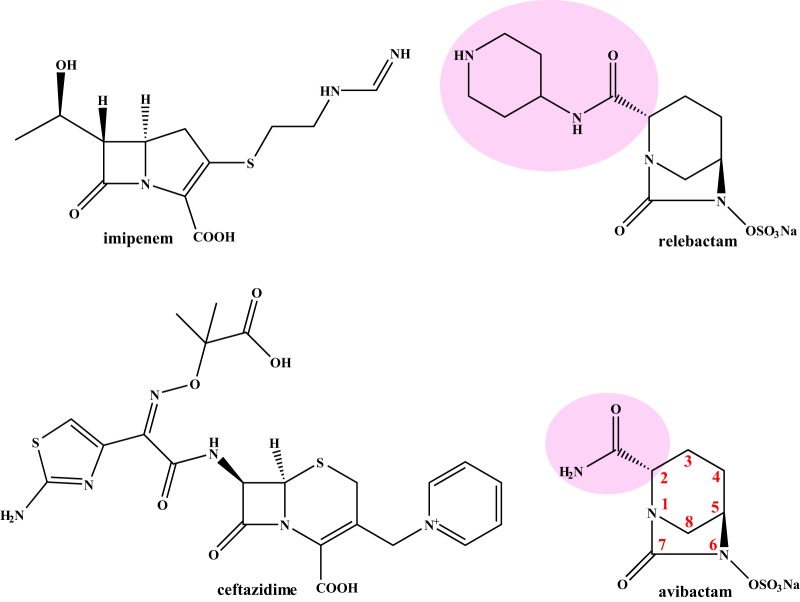
FIG 1.
Chemical structures of the compounds used in this study; the R1 side chains of the DBOs are highlighted in pink.
RESULTS AND DISCUSSION
Imipenem-relebactam demonstrates potent antimicrobial activity against KPC producers.
As the combination of imipenem-relebactam is being developed to target Enterobacteriaceae with blaKPCs, we tested the activity of imipenem and imipenem-relebactam as well as ceftazidime and ceftazidime-avibactam for comparison against a select panel of 101 isolates of the Enterobacteriaceae expressing blaKPCs (Table 1). All isolates tested were susceptible to both ceftazidime-avibactam and imipenem-relebactam (Table 2).
TABLE 1.
Modal MICs for 101 clinical isolates of Enterobacteriaceae producing KPCs and 2 control strainsa
| Strain | Modal MIC (mg/liter) |
|||
|---|---|---|---|---|
| CAZ | CAZ-AVI | IMI | IMI-REL | |
| Klebsiella pneumoniae KPC-2 | 64 | 1 | 16 | 1 |
| E. coli DH10B/pBR322-catI-blaKPC-2 | 64 | 1 | 8 | 0.5 |
| K. pneumoniae VA-398 | 64 | 1 | 2 | 0.25 |
| K. pneumoniae VA-400 | >128 | 1 | 8 | 0.25 |
| K. pneumoniae VA-184 | >128 | 2 | 8 | 0.5 |
| K. pneumoniae VA-237 | 128 | 0.25 | 8 | 0.125 |
| K. pneumoniae VA-267 | >128 | 1 | 8 | 0.125 |
| K. pneumoniae VA-357 | >128 | 2 | 16 | 0.25 |
| K. pneumoniae VA-360 | >128 | 4 | 64 | 0.25 |
| K. pneumoniae VA-362 | 64 | 1 | 16 | 0.5 |
| K. pneumoniae VA-364 | 64 | 1 | 32 | 1 |
| K. pneumoniae VA-367 | >128 | 2 | 2 | 0.25 |
| K. pneumoniae VA-368 | >128 | 1 | 4 | 0.25 |
| K. pneumoniae VA-373 | 32 | 1 | 4 | 0.5 |
| K. pneumoniae VA-401 | >128 | 4 | 64 | 0.5 |
| K. pneumoniae VA-376 | >128 | 2 | 32 | 0.5 |
| K. pneumoniae VA-378 | >128 | 1 | 16 | 0.5 |
| K. pneumoniae VA-380 | >128 | 2 | 16 | 0.5 |
| K. pneumoniae VA-383 | >128 | 1 | 8 | 0.5 |
| K. pneumoniae VA-384 | >128 | 1 | 64 | 1 |
| K. pneumoniae VA-387 | >128 | 2 | 128 | 2 |
| K. pneumoniae VA-402 | >128 | 4 | 16 | 0.25 |
| K. pneumoniae VA-389 | >128 | 2 | 8 | 0.25 |
| K. pneumoniae VA-390 | 64 | 1 | 64 | 0.25 |
| K. pneumoniae VA-391 | >128 | 4 | 8 | 0.125 |
| K. pneumoniae VA-392 | >128 | 1 | 16 | 0.125 |
| K. pneumoniae VA-394 | >128 | 2 | 8 | 0.25 |
| K. pneumoniae VA-395 | >128 | 2 | 16 | 0.25 |
| K. pneumoniae VA-396 | 128 | 1 | 32 | 0.125 |
| K. pneumoniae VA-413 | 64 | 1 | 64 | 0.25 |
| K. pneumoniae VA-414 | >256 | 4 | 64 | 0.125 |
| K. pneumoniae VA-416 | >256 | 4 | 64 | 0.5 |
| K. pneumoniae VA-417 | 256 | 2 | 16 | 0.5 |
| Enterobacter cloacae VA-407 | >256 | 8 | 16 | 0.25 |
| Enterobacter aerogenes VA-415 | >256 | 2 | 8 | 0.25 |
| E. coli S246579 | 256 | 1 | 16 | 0.5 |
| E. coli X173170 | 64 | 1 | 8 | 0.5 |
| E. cloacae M627513 | 256 | 2 | 4 | 0.5 |
| E. aerogenes M2084230 | 128 | 1 | 8 | 0.5 |
| Citrobacter freundii M5092134 | 64 | 1 | 4 | 0.5 |
| E. cloacae M2051712 | 16 | 1 | 4 | 0.25 |
| C. freundii M2295131 | 64 | 1 | 2 | 0.25 |
| Citrobacter koseri M3301980 | 128 | 2 | 8 | 0.5 |
| E. aerogenes M6315040 | 256 | 1 | 8 | 0.25 |
| E. coli M7123031 | 16 | 0.5 | 4 | 0.25 |
| E. coli M7123093 | 16 | 0.5 | 4 | 0.25 |
| E. cloacae 1225904003 | 64 | 2 | 128 | 1 |
| E. cloacae F17482 | >256 | 4 | 128 | 0.5 |
| Klebsiella oxytoca 1223805125 | 64 | 1 | 4 | 0.25 |
| E. cloacae 1220402851 | >256 | 4 | 8 | 0.25 |
| K. pneumoniae VA 403 | >512 | 1 | 32 | 0.25 |
| K. pneumoniae VA 404 | 256 | 0.5 | 8 | 0.25 |
| K. pneumoniae VA 408 | 256 | 1 | 8 | 0.125 |
| K. pneumoniae VA 409 | 256 | 2 | 16 | <0.06 |
| K. pneumoniae VA 410 | 128 | <0.06 | 4 | 0.125 |
| K. pneumoniae VA 412 | 256 | 0.5 | 8 | 0.125 |
| K. pneumoniae VA 361 | 256 | 0.5 | 8 | 0.25 |
| K. pneumoniae VA 406 | 256 | 32 | >128 | 2 |
| K. pneumoniae VA 375 | 256 | 1 | 8 | 0.25 |
| K. pneumoniae UNC 001 | 512 | 2 | 4 | 0.25 |
| K. pneumoniae UNC 002 | 256 | 1 | 4 | 0.25 |
| K. pneumoniae UNC 005 | 256 | 2 | 16 | 1 |
| K. pneumoniae UNC 008 | 512 | 1 | 16 | 0.25 |
| K. pneumoniae UNC 010 | 512 | 1 | 8 | 0.25 |
| K. pneumoniae UNC 011 | 64 | 0.25 | 0.5 | 0.25 |
| K. pneumoniae UNC 012 | 128 | 1 | 4 | 0.25 |
| K. pneumoniae UNC 015 | 128 | 1 | 16 | 0.5 |
| K. pneumoniae UNC 016 | 512 | 1 | 2 | 0.25 |
| K. pneumoniae UNC 018 | 256 | 1 | 8 | 0.5 |
| K. pneumoniae UNC 020 | 256 | 0.125 | 4 | 0.125 |
| K. pneumoniae UNC 027 | >512 | 1 | 8 | 0.25 |
| K. pneumoniae UNC 030 | 256 | 2 | 4 | 0.5 |
| K. pneumoniae UNC KPC 171 | 256 | 1 | 2 | 0.125 |
| K. pneumoniae 120/1020,2 | 256 | 0.5 | 32 | 0.25 |
| K. pneumoniae 140/1040,2 | 256 | 0.5 | 32 | 0.25 |
| K. pneumoniae 160/1080,2 | 256 | 0.5 | 4 | 0.125 |
| K. pneumoniae 300/1240,2 | >512 | 0.25 | 8 | 0.125 |
| K. pneumoniae 320/1260 | 32 | 0.5 | 8 | 0.25 |
| K. pneumoniae 361/1301,2 | 512 | 1 | 8 | 0.5 |
| K. pneumoniae 440/1360 | 256 | 1 | 16 | 0.125 |
| K. pneumoniae 540/1460 | 512 | 0.5 | 4 | 0.25 |
| K. pneumoniae 600/1500 | 512 | 2 | 16 | 0.125 |
| K. pneumoniae 620/1520 | 256 | 1 | 8 | 0.125 |
| K. pneumoniae 644/1566 | 128 | 1 | 64 | 0.5 |
| K. pneumoniae 646/1568 | 256 | 1 | 8 | 0.25 |
| K. pneumoniae 647/1569 | 512 | 2 | 16 | 2 |
| K. pneumoniae 648/1570 | 32 | 0.25 | 0.5 | 0.25 |
| K. pneumoniae 649/1571 | 32 | 0.5 | 8 | 0.25 |
| K. pneumoniae 651/1573 | 512 | 1 | 8 | 0.25 |
| K. pneumoniae 660/1586 | 512 | <0.06 | 0.5 | 0.125 |
| K. pneumoniae 665/1593 | 256 | 0.5 | 64 | 0.5 |
| K. pneumoniae 666/1594 | 256 | 0.5 | 8 | 0.125 |
| K. pneumoniae 670/1598 | 512 | 1 | 8 | 2 |
| K. pneumoniae 671/1599 | 256 | 2 | 128 | 1 |
| K. pneumoniae 672/1600 | 256 | 0.5 | 16 | 1 |
| K. pneumoniae 674/1603 | 256 | 2 | 8 | 0.25 |
| K. pneumoniae 676/1606 | >512 | 1 | >128 | 2 |
| K. pneumoniae 677/1607 | 256 | 0.125 | 8 | 0.25 |
| K. pneumoniae 679/1608 | 256 | 1 | 4 | 0.5 |
| K. pneumoniae 681/1612 | 64 | 0.5 | 16 | 0.5 |
| K. pneumoniae 682/1613 | 256 | 0.5 | 64 | 0.5 |
| K. pneumoniae 686/1617 | 128 | 0.5 | 16 | 0.5 |
| K. pneumoniae 691/1633 | 256 | <0.06 | 2 | 0.125 |
Data are from three experiments. The control strains were Klebsiella pneumoniae KPC-2 and E. coli DH10B/pBR322-catI-blaKPC-2. CLSI breakpoints for ceftazidime (for both ceftazidime and ceftazidime-avibactam) and imipenem (for both imipenem and imipenem-relebactam) were used to differentiate susceptibility versus resistance. CAZ, ceftazidime; AVI, avibactam; IMI, imipenem; REL, relebactam.
TABLE 2.
Rates of susceptibility of the isolates tested
| Antibiotic(s)a | No. of isolates with the following susceptibility/total no. of isolates tested (%): |
|||
|---|---|---|---|---|
| Nonsusceptible | Susceptible | Intermediate | Resistant | |
| CAZ | 103/103 (100) | 0/103 (0) | 0/103 (0) | 103/103 (100) |
| CAZ-AVI | 0/103 (0) | 103/103 (100) | 0/103 (0) | 0/103 (0) |
| IMI | 94/103 (91) | 9/103 (9) | 17/103 (16) | 77/103 (75) |
| IMI-REL | 0/103 (0) | 103/103 (100) | 0/103 (0) | 0/103 (0) |
CAZ, ceftazidime; AVI, avibactam; IMI, imipenem; REL, relebactam.
Relebactam is an effective inhibitor of KPC-2.
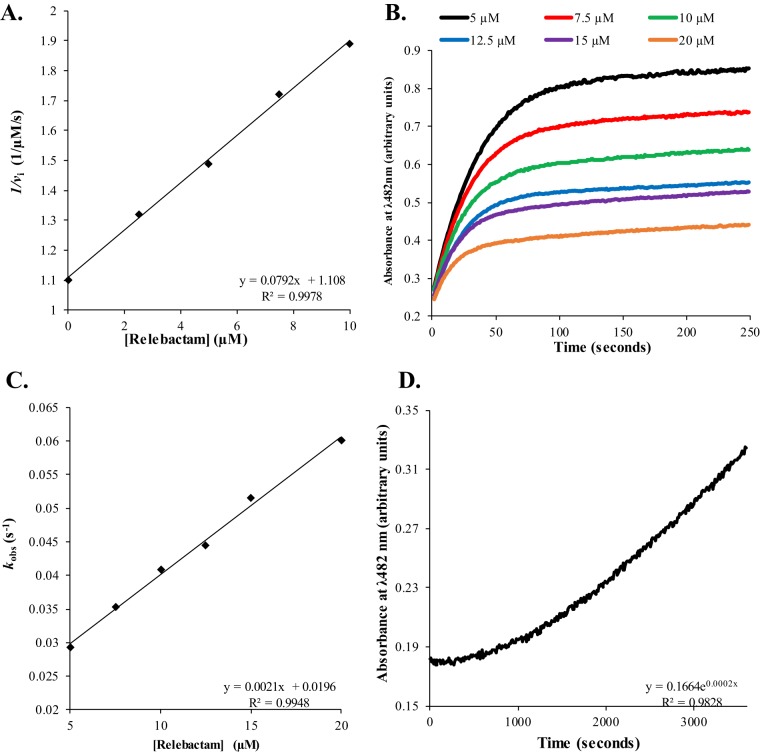
The values of the apparent Ki (Ki app), the second-order onset of the acylation rate constant (k2/K), the off-rate constant (koff), and the kcat/rate constant of enzyme inactivation (kinact) were determined for purified KPC-2 with relebactam using nitrocefin as a reporter substrate. Previously for nitrocefin, KPC-2 was found to possess a Km of 8 μM and a kcat of 130 s−1, for a kcat/Km ratio of 16 μM−1 s−1 (8). Comparatively, imipenem is a poorer substrate with a Km of 21 μM and a kcat of 21 s−1, for a kcat/Km ratio of 1.0 μM−1 s−1 (8). To determine the relative affinity of relebactam for KPC-2, a direct competition assay between 25 μM nitrocefin and increasing concentrations of relebactam measuring initial velocities for nitrocefin hydrolysis was used. The Ki app value of relebactam for KPC-2 was 2.3 ± 0.3 μM, which is similar to that of avibactam, 1.0 ± 0.1 μM (Fig. 2A and Table 3) (3, 9).
FIG 2.
(A) Determination of the Ki app value of relebactam for KPC-2 by using increasing concentrations of relebactam and inhibiting nitrocefin hydrolysis. The inverse initial velocity (vi) versus the concentration of relebactam fit to a linear equation, Ki app observed = y intercept/slope, is plotted. Ki app observed was adjusted for the use of nitrocefin to obtain the Ki app value (Table 3). (B) Progress curves showing inhibition of nitrocefin hydrolysis by KPC-2 with increasing concentrations of relebactam. The absorbance at a λ of 482 nm versus time is plotted. To obtain kobs values, progress curves were fit to y = Vf · x + (V0 − Vf) · [1 − exp(−kobs · x)]/kobs + A0, where Vf is the final velocity, V0 is the initial velocity, and A0 is initial absorbance at 482 nm. (C) Determination of the k2/K value of relebactam for KPC-2 by using increasing concentrations of relebactam and inhibiting nitrocefin hydrolysis. kobs values versus the concentration of relebactam fit to a linear equation, k2/K observed = slope, are plotted. k2/K observed was adjusted for the use of nitrocefin to obtain the k2/K value (Table 3). (D) Progress curves showing the recovery of nitrocefin hydrolysis by KPC-2 after inhibition by relebactam. The absorbance at a λ of 482 nm versus time is plotted. Progress curves were fit to a single exponential equation to obtain koff values (Table 2).
TABLE 3.
Kinetic parameters against KPC-2 using relebactam
| Kinetic parameter | Value for KPC-2 |
|---|---|
| Ki app (μM) | 2.3 ± 0.3 |
| k2/K (M−1 s−1) | 24,750 ± 2,475 |
| koff (s−1) | 0.00020 ± 0.00002 |
| Half-lifea (min) | 58 ± 6 |
| Kdb (nM) | 8 ± 1 |
| kcat/kinact | 1 |
Residence time.
Kd, dissociation constant.
To determine the k2/K or acylation rate of relebactam for KPC-2, progress curves were obtained for the hydrolysis of 50 μM nitrocefin in direct competition with increasing concentrations of relebactam (Fig. 2B). The values of the observed rate constant for inactivation (kobs) were determined and plotted versus the concentrations of inhibitor (Fig. 2C). The slope of the line corresponds to the k2/K observed value, which was corrected for the use of nitrocefin. The k2/K value for relebactam inactivating KPC-2 was similar to that obtained previously for avibactam (21,580 ± 2,200 M−1 s−1) (3, 9).
To determine the rate at which relebactam is released from KPC-2, or koff, KPC-2 (1 μM) was preincubated with relebactam (17.25 μM) at a concentration that resulted in an initial velocity value of ∼0 μM/s for 50 μM nitrocefin hydrolysis. The reaction mixture was diluted 1:10,000, and progress curves measuring the recovery of nitrocefin hydrolysis were determined. The progress curves were fit to a single exponential equation to obtain koff values (Fig. 2D). The koff value of KPC-2 for relebactam was 0.00020 ± 0.00002 s−1, which was similar to the reported koff value of KPC-2 for avibactam of 0.00014 s−1 (Table 3) (3).
The turnover number or partition ratio (kcat/kinact), which is the amount of inhibitor required to reduce the initial velocity of a β-lactamase by ≥90%, was determined for KPC-2 with relebactam. Only one molecule of relebactam was turned over by KPC-2 before the β-lactamase was inactivated (Table 3). Relebactam is as potent as avibactam when measuring kcat/kinact (9).
Mapping the intermediates of inactivation by relebactam.
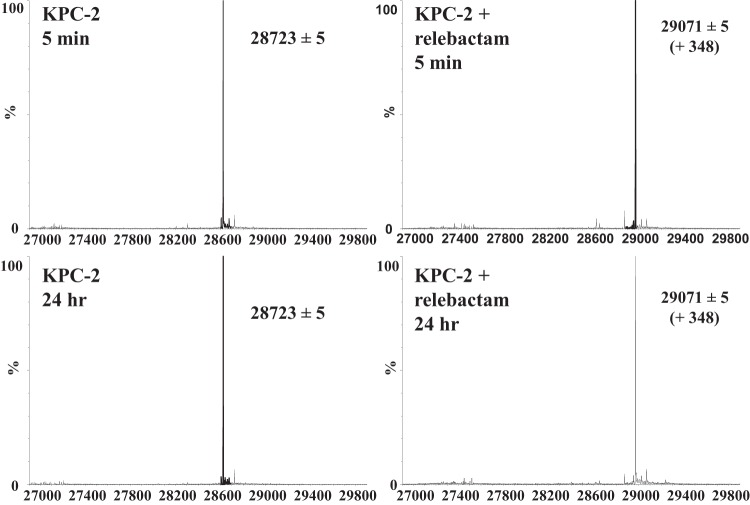
To determine if any intermediates are formed (e.g., the loss of sulfate from relebactam when it is bound to KPC-2) or if hydrolysis of relebactam occurs upon the interaction of relebactam with KPC-2, timed mass spectrometry (MS) was conducted. Five-minute and 24-h preincubations revealed that relebactam binds as the whole molecule plus 348 ± 5 Da to the KPC-2 carbapenemase and is not hydrolyzed or modified (Fig. 3). The lack of modification of relebactam upon interaction with KPC-2 is different from what was observed with avibactam. Unlike with relebactam, when KPC-2 reacts with avibactam, avibactam loses its sulfate and eventually deacylates (3).
FIG 3.
Mass spectra of KPC-2 after 5 min and 24 h of preincubation with and without relebactam. Numbers on the x axis indicate mass in atomic mass units (amu).
Modeling suggests that the positioning of active-site water molecules improves the stability of the KPC-2–relebactam complex.
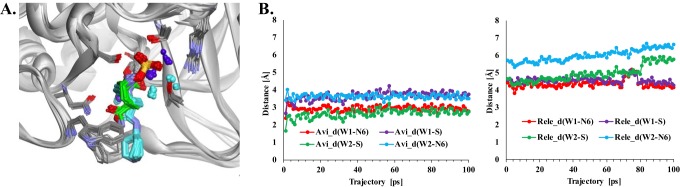
To understand why relebactam is not susceptible to desulfation during interactions with KPC-2 while avibactam is, relebactam was docked into the KPC-2 active site and the model was compared to the KPC-2–avibactam crystal structure (10). In addition, a molecular dynamics simulation (MDS) was conducted on both complexes for 0.2 ns.
Relebactam adopted two primary conformations when docked into the KPC-2 active site; the most energetically favorable complex was chosen for further analysis. Representations of the KPC-2–avibactam crystal structure (PDB accession number 4ZBE) and the KPC-2–relebactam molecular models are presented in Fig. 4A. During the course of MDS, the positions of two site water molecules (W1 and W2) were found to be closer to the DBO sulfate in the KPC-2–avibactam crystal structure than the KPC-2–relebactam model (Fig. 4A and B). Notably, the deacylation water held via hydrogen bonding interactions to E166 and N170 remained in a similar position in the KPC-2–avibactam structure and the KPC-2–relebactam model throughout the MDS (data not shown). In the KPC-2–avibactam structure, W1 maintains a close distance (≈3 Å) to the sulfate group and W2 is recruited in the proximity of the N6 sulfate group as well (Fig. 4B). However, in the KPC-2–relebactam model, the water molecules move greater than 5 to 6 Å from the N6 and/or sulfate group of relebactam (Fig. 4B). Hydrolysis is involved in both the desulfation and deacylation of avibactam from KPC-2; the mechanism of desulfation remains to be elucidated.
FIG 4.
(A) Superimposition of the conformations collected during MDS of relebactam (cyan) and avibactam (green) in the KPC-2 active site displaying water molecules (cyan, KPC-2–relebactam model; green, KPC-2–avibactam model). (B) A graphical representation of the distance (in angstroms) of active-site water molecules (W1 and W2) from the N6 and sulfate (S) of avibactam (Avi) and relebactam (Rele) during the 100-ps MDS trajectory in the KPC-2–DBO complexes reveals that W1 and W2 are closer to the DBO sulfate in avibactam (∼2 to 4 Å) than in relebactam (∼4 to 7 Å). The d in _d stands for distance.
Based on our observations, we hypothesize that the lack of critical water molecules near relebactam sulfate in the KPC-2–relebactam complex may play a role in relebactam's stability observed by mass spectrometry.
Conclusions.
The favorable inhibitory kinetic profile of relebactam against the KPC-2 β-lactamase is in agreement with the robust activity profile of relebactam against clinical isolates of KPC-producing Enterobacteriaceae, which is similar overall to the activity profiles of avibactam and the ceftazidime-avibactam combination. The one observable difference between relebactam and avibactam is that under our experimental conditions, relebactam does not desulfate or deacylate upon reacting with KPC-2. The time scale for desulfation of avibactam by KPC-2 is within a bacterial cell replication cycle; however, deacylation of avibactam requires hours, and the complete conversion to the apo-enzyme approximates nearly 24 h. Presently, the implications of desulfation and deacylation are not clear; however, with selective pressure, one can only predict that KPC-2 may evolve to desulfate/deacylate avibactam at a higher rate. In closing, it is also possible that the differential placement of W1 and W2 within the active site may have a significant impact in the future. Once imipenem-relebactam becomes commercially available, we will develop a greater understanding of whether desulfation of DBOs by β-lactamases is a clinically relevant phenotype.
MATERIALS AND METHODS
Isolates.
The strains used as controls for susceptibility testing, Klebsiella pneumoniae expressing blaKPC-2 and Escherichia coli with pBR322-catI-blaKPC-2 as well as the strain used for KPC-2 protein purification, E. coli Origami 2 DE3 with pET24(a)+blaKPC-2, were previously described (11, 12). The clinical isolates tested in this study were obtained from previous studies as well as from a collection at the University of North Carolina (13, 14).
Compounds.
Relebactam, imipenem, and avibactam were given to us as part of a Merck investigator studies program (MISP; grant number 53544); the source of avibactam was Advanced ChemBlocks. Nitrocefin was purchased from Oxoid-Remel, and ceftazidime was obtained from Sigma.
In vitro susceptibility test methods.
The MICs for the isolates were determined by the Mueller-Hinton (MH) agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines as previously described (15). Relebactam and avibactam were tested at 4 mg/liter in combination with increasing concentrations of imipenem and ceftazidime, respectively.
Purification and steady-state kinetic analysis with KPC-2.
The KPC-2 β-lactamase was purified as previously described (1). Kinetics were carried out on an Agilent 8453 diode array spectrophotometer in 10 mM phosphate-buffered saline (PBS; pH 7.4) at room temperature. Determination of the values of the kinetic constants apparent Ki (Ki app), k2/K, kcat/kinact, and koff was previously described (17).
ESI MS.
To assess the nature of any intermediates of relebactam formed during the reaction with KPC-2, 10 μM KPC-2 was incubated with 10 μM relebactam for set times (i.e., 5 min and 24 h) at room temperature in 10 mM PBS, pH 7.4. Reactions were terminated by the addition of 0.1% formic acid and 1% acetonitrile. Electrospray ionization (ESI) mass spectrometry (MS) was performed on a Waters SynaptG2-Si quadrupole-time of flight mass spectrometer equipped with a LockSpray dual electrospray ion source and an Acquity H class ultraperformance liquid chromatograph, as previously described (18).
Molecular modeling.
To obtain insight into the interactions between KPC-2 and relebactam, a molecular model of KPC-2 with relebactam was constructed using the crystal coordinates of KPC-2 (PDB accession number 2OV5) and Discovery Studio (D.S.; version 4.1; Biovia, Accelrys Inc., San Diego, CA) molecular modeling software as previously described (8, 19). Relebactam was constructed using the Fragment Builder tools, minimized, and automatically docked into the active site of KPC-2 using the LibDock module of D.S. software. The crystallographic waters were removed, the complex was solvated, the active-site crystallographic waters were added back, and the complex was minimized using the Conjugate gradient with the SHAKER algorithm. To equilibrate the complex, a 0.2-ns molecular dynamics simulation (MDS) was performed. On the basis of the calculated energies, the most energetically favorable conformation was chosen, and the complex with KPC-2 was created.
ACKNOWLEDGMENTS
These studies were supported by Merck MISP grant number 53544. The research reported in this publication was supported in part by funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program (BX002872 to K.M.P.-W. and BX001974 to R.A.B.) from the U.S. Department of Veterans Affairs Biomedical Laboratory Research and Development Service, and the Geriatric Research Education and Clinical Center (VISN 10 to R.A.B.). The National Institute of Allergy and Infectious Diseases of the National Institutes of Health provided support under award numbers R21AI114508, R01AI100560, R01AI063517, and R01AI072219 to R.A.B. and U19-AI109713-SRP to K.M.P.-W.
The contents do not represent the views of the U.S. Department of Veterans Affairs or the U.S. Government. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Papp-Wallace KM, Bethel CR, Distler AM, Kasuboski C, Taracila M, Bonomo RA. 2010. Inhibitor resistance in the KPC-2 β-lactamase, a preeminent property of this class A β-lactamase. Antimicrob Agents Chemother 54:890–897. doi: 10.1128/AAC.00693-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blizzard TA, Chen H, Kim S, Wu J, Bodner R, Gude C, Imbriglio J, Young K, Park YW, Ogawa A, Raghoobar S, Hairston N, Painter RE, Wisniewski D, Scapin G, Fitzgerald P, Sharma N, Lu J, Ha S, Hermes J, Hammond ML. 2014. Discovery of MK-7655, a β-lactamase inhibitor for combination with Primaxin(R). Bioorg Med Chem Lett 24:780–785. doi: 10.1016/j.bmcl.2013.12.101. [DOI] [PubMed] [Google Scholar]
- 3.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against class A, C, and D β-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch EB, Ledesma KR, Chang KT, Schwartz MS, Motyl MR, Tam VH. 2012. In vitro activity of MK-7655, a novel β-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56:3753–3757. doi: 10.1128/AAC.05927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Livermore DM, Warner M, Mushtaq S. 2013. Activity of MK-7655 combined with imipenem against Enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 68:2286–2290. doi: 10.1093/jac/dkt178. [DOI] [PubMed] [Google Scholar]
- 6.Bhagunde P, Chang KT, Hirsch EB, Ledesma KR, Nikolaou M, Tam VH. 2012. Novel modeling framework to guide design of optimal dosing strategies for β-lactamase inhibitors. Antimicrob Agents Chemother 56:2237–2240. doi: 10.1128/AAC.06113-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powles M, Galgoci A, Misura A, Liberator P, Hammond M.. 2010. Abstr 50th Intersci Conf Antimicrob Agents Chemother, Boston, MA, abstr F1-2140. [Google Scholar]
- 8.Papp-Wallace KM, Taracila MA, Smith KM, Xu Y, Bonomo RA. 2012. Understanding the molecular determinants of substrate and inhibitor specificities in the carbapenemase KPC-2: exploring the roles of Arg220 and Glu276. Antimicrob Agents Chemother 56:4428–4438. doi: 10.1128/AAC.05769-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of β-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan NP, Nguyen NQ, Papp-Wallace KM, Bonomo RA, van den Akker F. 2015. Inhibition of Klebsiella β-lactamases (SHV-1 and KPC-2) by avibactam: a structural study. PLoS One 10:e0136813. doi: 10.1371/journal.pone.0136813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papp-Wallace KM, Taracila M, Hornick JM, Hujer AM, Hujer KM, Distler AM, Endimiani A, Bonomo RA. 2010. Substrate selectivity and a novel role in inhibitor discrimination by residue 237 in the KPC-2 β-lactamase. Antimicrob Agents Chemother 54:2867–2877. doi: 10.1128/AAC.00197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Endimiani A, Carias LL, Hujer AM, Bethel CR, Hujer KM, Perez F, Hutton RA, Fox WR, Hall GS, Jacobs MR, Paterson DL, Rice LB, Jenkins SG, Tenover FC, Bonomo RA. 2008. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae isolates possessing blaKPC in the United States. Antimicrob Agents Chemother 52:2680–2682. doi: 10.1128/AAC.00158-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endimiani A, Hujer AM, Perez F, Bethel CR, Hujer KM, Kroeger J, Oethinger M, Paterson DL, Adams MD, Jacobs MR, Diekema DJ, Hall GS, Jenkins SG, Rice LB, Tenover FC, Bonomo RA. 2009. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the eastern USA. J Antimicrob Chemother 63:427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed CLSI document M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Reference deleted.
- 17.Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. 2014. Reclaiming the efficacy of β-lactam–β-lactamase inhibitor combinations: avibactam restores the susceptibility of ceftazidime against CMY-2-producing Escherichia coli. Antimicrob Agents Chemother 58:4290–4297. doi: 10.1128/AAC.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papp-Wallace KM, Becka SA, Taracila MA, Winkler ML, Gatta JA, Rholl DA, Schweizer HP, Bonomo RA. 2016. Exposing a β-lactamase “twist”: the mechanistic basis for the high level of ceftazidime resistance in the C69F variant of the Burkholderia pseudomallei PenI β-lactamase. Antimicrob Agents Chemother 60:777–788. doi: 10.1128/AAC.02073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levitt PS, Papp-Wallace KM, Taracila MA, Hujer AM, Winkler ML, Smith KM, Xu Y, Harris ME, Bonomo RA. 2012. Exploring the role of a conserved class A residue in the Ω-loop of KPC-2 β-lactamase: a mechanism for ceftazidime hydrolysis. J Biol Chem 287:31783–31793. doi: 10.1074/jbc.M112.348540. [DOI] [PMC free article] [PubMed] [Google Scholar]