ABSTRACT
A carbapenem-resistant Klebsiella pneumoniae isolate was recovered from human blood. Its whole-genome sequence was obtained using Illumina and long-read MinION sequencing. The strain belongs to sequence type 273 (ST273), which was found recently and caused an outbreak in Southeast Asia. It has two carbapenemase genes, blaNDM-1 (carried by an ST7 IncN self-transmissible plasmid) and blaIMP-4 (located on a self-transmissible IncHI5 plasmid). Non-KPC-producing ST237 may represent a lineage of carbapenem-resistant K. pneumoniae, which warrants further monitoring.
KEYWORDS: β-lactamases, carbapenemases, resistance, plasmids, Klebsiella pneumoniae, Klebsiella, carbapenems
TEXT
Klebsiella pneumoniae is one of the most common pathogens of human infections, and carbapenem-resistant K. pneumoniae (CRKP) has emerged as a major challenge to clinical management and global public health (1). Production of carbapenem-hydrolyzing enzymes (carbapenemases) is the major mechanism mediating resistance to carbapenems in K. pneumoniae. There are a few types of carbapenemases, and the most common carbapenemase in K. pneumoniae is KPC (a group of serine β-lactamases), followed by NDM and IMP (both of which are metallo-β-lactamases). The global dissemination of CRKP is largely mediated by the high-risk clonal complex 258 (CC258), which comprises sequence type (ST) 11, ST258, and a number of closely related sequence types. However, other clones may also contribute to the international spread of CRKP. Recently, ST273 CRKP was found in several countries (2–4), which warrants further investigation. We identified an ST273 CRKP clinical strain carrying both blaNDM and blaIMP genes in our hospital and report its characterization here.
Strain WCHKP020034 was recovered from the blood of a 72-year-old male patient with pancreatitis at West China Hospital. The strain was identified as K. pneumoniae by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) (Bruker, Billerica, MA) and Vitek II (bioMérieux, Marcy-l'Étoile, France). MICs of amikacin, aztreonam, aztreonam-avibactam, ceftazidime, ciprofloxacin, colistin, imipenem, meropenem, piperacillin-tazobactam, tigecycline, and trimethoprim-sulfamethoxazole against the isolate were determined using the broth microdilution method of the Clinical and Laboratory Standards Institute (CLSI) (5). Because CLSI does not give breakpoints for colistin and tigecycline, we applied those defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://www.eucast.org/). The strain was resistant to aztreonam (MIC, 64 μg/ml), ceftazidime (MIC, >256 μg/ml), ciprofloxacin (MIC, 256 μg/ml), imipenem (MIC, 32 μg/ml), meropenem (MIC, 64 μg/ml), piperacillin-tazobactam (MIC, >512/4 μg/ml), and trimethoprim-sulfamethoxazole (MIC, 128/2,432 μg/ml) but susceptible to amikacin (MIC, 2 μg/ml), aztreonam-avibactam (MIC, 0.25/4 μg/ml), colistin (MIC, 1 μg/ml), and tigecycline (MIC, 1 μg/ml). Acquired carbapenemase genes blaGES, blaKPC, blaIMP, blaNDM, blaOXA-48, and blaVIM were screened as described previously (6–9), and the strain had blaNDM and blaIMP. blaNDM-1 and blaIMP-4 were identified by amplifying and sequencing the complete coding sequence of blaNDM and blaIMP.
The strain was subjected to whole-genome sequencing with 150× coverage using the HiSeq X Ten sequencer (Illumina, San Diego, CA), which generated 4,395,250 reads. Reads were trimmed using Trimmomatic (10) and were then assembled to 125 contigs (70 were ≥1,000 bp in length) with a 56.79% GC content using the SPAdes program (11). The wzi gene allele, which represents the capsular variation, of strain WCHKP020034 was 50, corresponding to several K types, i.e., K15, K17, K50, K51, and K52, with K15 being the best match predicted using Kaptive (12). None of the K types were K1, K2, or K5, which are proposed as the hypervirulent members of K. pneumoniae. With respect to virulence, strain WCHKP020034 had the mrk gene cluster (mrkA-B-C-D-F-H-I-J), which encodes type 3 fimbrial expression (13) and is seen in almost all K. pneumoniae isolates (1). Other known virulence genes, such as those encoding yersiniabactin, colibactin, allantoinase, and aerobactin, were absent from strain WCHKP020034.
Strain WCHKP020034 belonged to ST273, as determined by use of the de novo assembled genome sequence to query the MLST database of K. pneumoniae (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). There were 10 additional ST273 strains with the whole-genome sequence available in GenBank (see Table S1 in the supplemental material). Genome sequences of ST273 strains were retrieved from GenBank and aligned with that of strain WCHKP020034 using the Harvest Suite with default settings (14). Single nucleotide polymorphisms (SNPs) on recombination sites were removed by Gubbins (15). The filtered SNPs were then used as input for inferring a phylogenetic tree using RAxML (16) with the GTRGAMMA model and 1,000 bootstraps. Antimicrobial resistance genes in these genomes were identified using ABRicate (https://github.com/tseemann/abricate) to query the ResFinder database at the Center for Genomic Epidemiology (http://genomicepidemiology.org/), and the wzi gene allele was predicted using Kaptive (12). Five strains carrying blaNDM-7, a point mutant of blaNDM-1, were recovered in 2013 in the Philippines and belonged to a single cluster. No wzi allele was identified in these five strains. In contrast, strain WCHKP020034 was clustered with other ST273 strains (Fig. 1) and was closest to strain COL-Kpn113 (carrying no blaNDM, recovered in 2004 in Colombia) and strain K45-67 (carrying no blaNDM but blaVIM-1, recovered in 2007 in Norway), with 116 to 123 SNPs difference, respectively (see Table S2 in the supplemental material). Strains COL-Kpn113 and K45-67 had a wzi allele174, which was different from the allele 50 of strain WCHKP020034. The assembled genomes of ST273 strains were also typed using the cgMLST database (http://bigsdb.pasteur.fr/perl/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef_public&page=sequenceQuery), and a total of 951 genes were identified in all ST273 genomes. The 951 genes were identical in sequence among all ST273 strains other than WCHKP020034, whereas only 5 out of the 951 genes were different between strain WCHKP020034 and the other 10 ST273 strains (see Table S3 in the supplemental material). The relatively small number of SNPs and almost identical cgMLST results seen in strains from different geographic locations over such a long time frame suggest that ST273 might be highly clonal and merits further focused phylogenetic studies of this lineage. The wzi allele was different and even absent in ST273 strains, but it is not uncommon to find more than one capsular type for strains of a single ST due to homologous recombination of the capsular locus (17). Plasmids of the ST273 strains were predicted using PlasmidFinder, but there was no common plasmid replicon type present in all of the ST273 strains.
FIG 1.
Maximum-likelihood phylogenetic tree of K. pneumoniae ST273 strains with genome sequences available in GenBank. The phylogeny is inferred from the recombination-filtered SNP alignment obtained by aligning either a complete or draft genome of K. pneumoniae ST273 against the complete genome of WCHKP020034. The annotation denotes the presence of antimicrobial resistance genes as determined by ABRicate.
In addition to the two carbapenemase genes, strain WCHKP020034 had 24 intact antimicrobial resistance genes mediating resistance to aminoglycosides [aac(3)-IId, ant(3″)-Ih-aac(6′)-IId, aacA4, aadA1, aadA16, aph(3′)-Ia, strA, and strB], β-lactams (blaCTX-M-3 and blaSHV-115), fosfomycin (fosA), macrolides [mph(A)], phenicol (floR), quinolones (oqxA, oqxB, qnrB, and qnrS1), rifampin (arr3), tetracycline [tet(A)], sulfonamides (sul1 and sul2), and trimethoprim (dfrA7, dfrA14, and dfrA27) (Table 1).
TABLE 1.
Antimicrobial resistance genes and their locations in strain WCHKP020034
| WCHKP020034 chromosome/plasmid | Size (bp) | Replicon type, pMLST | Antimicrobial resistance genes |
|---|---|---|---|
| Chromosome | 5,295,791 | blaSHV-115, fosA, oqxA, oqxB | |
| pNDM1_LL34 | 58,953 | N (ST7) | blaNDM-1, dfrA14, qnrS |
| pIMP4_LL34 | 260,974 | IncHI5 | aacA4, blaCTX-M-3, blaIMP-4, sul1 |
| pQnrB_LL34 | 130,688 | FII (K2:A−:B−), Q1 | aac(3)-IId, ant(3″)-Ih-aac(6′)-IId, aadA1, aadA16, aph(3′)-Ia, arr3, dfrA27, floR, mph(A), qnrB, sul1, sul2, tet(A) |
Conjugation experiments were performed using filter- and broth-based methods at both 25°C and 37°C with the azide-resistant Escherichia coli strain J53 as the recipient. Transconjugants were screened using 1 μg/ml meropenem plus 150 μg/ml sodium azide, and the presence of blaNDM-1 or blaIMP-4 in transconjugants was screened by PCR. blaNDM-1 and blaIMP-4 were carried on two self-transmissible plasmids, designated pNDM1_LL34 and pIMP4_LL34, respectively. To obtain the complete sequence of the plasmids, strain WCHKP020034 was subjected to sequencing using the long-read MinION sequencer (Nanopore, Oxford, United Kingdom). The de novo hybrid assembly of both short Illumina reads and long MinION reads was performed using Unicycler under the conservative mode for increased accuracy (18). The complete circular contigs generated were then corrected using Plion with Illumina reads for several rounds until no change was detected (19). Plasmid replicon type and plasmid MLST were determined using the PlasmidFinder and pMLST tools (http://genomicepidemiology.org/). The hybrid assembly of Illumina and MinION reads revealed that strain WCHKP020034 has a 5,295,791-bp circular chromosome and three large plasmids, i.e., the 58,953-bp pNDM1_LL34 of IncN (ST7), a 260,974-bp pIMP4_LL34 carrying blaIMP-4 and blaCTX-M-3 with replicon types unidentified by PlasmidFinder, and a 130,688-bp plasmid carrying qnrB that contains an IncFII(K) and an IncQ1 replicon (designated pQnrB_LL34) (Table 1).
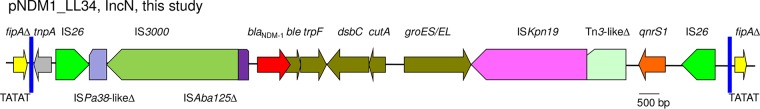
To understand the distribution of ST7 IncN plasmids, sequences of three alleles to define ST7 were concatenated and then aligned against the nucleotide database using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Plasmids of the ST7 IncN type were found in various species of Enterobacteriaceae, including Citrobacter freundii, E. coli, Enterobacter cloacae, Enterobacter hormaechei, K. pneumoniae, Klebsiella oxytoca, Morganella morganii, Raoultella ornithinolytica, and Raoultella planticola from different countries, suggesting that ST7 IncN plasmids are widely distributed. In addition, ST7 IncN plasmids have been found to mediate the dissemination of blaIMP-4 in Enterobacteriaceae in different regions of China (20). Sequences of all available ST7 IncN plasmids (n = 32) were retrieved from GenBank. Genes present on all ST7 IncN plasmids were considered backbone genes, which were identified using OrthoFinder (21). Sequences of backbone genes were concatenated and then aligned to infer a phylogenetic tree using RAxML with a 1,000-bootstrap test (16). pNDM1_LL34 is clustered with several plasmids from various species (Fig. 2), among which pNDM1_LL34 is closely related (99% coverage, 99% identity) to plasmid pNDM-BTR (GenBank accession no. KF534788), which is also an ST7 IncN plasmid carrying blaNDM-1 that was recovered from an E. coli isolate in Beijing, China, in 2013, as revealed by BLAST (blast.ncbi.nlm.nih.gov). The findings suggest interspecies spread of a common IncN plasmid. On pNDM1_LL34 and pNDM-BTR, blaNDM-1, several genes that are commonly associated with blaNDM-1, and the quinolone-resistant gene qnrS1 were bracketed by IS26 (Fig. 3). There were no 8-bp direct target repeats, which are characteristic of the insertion of IS26, flanking the two copies of IS26, suggesting that homologous recombination contributed to the formation of such a structure. Nonetheless, two copies of IS26 have the potential to form a composite transposon to mediate the mobilization of the intervening genetic components, including blaNDM-1 and qnrS1. Outside of the two IS26 copies, there was an interrupted Tn3 family transposon, in which the transposase gene tnpA and both inverted repeats remained intact, but the resolvase gene tnpR was truncated. The fipA gene that encodes a conjugal transfer inhibition protein and belongs to the plasmid backbone was interrupted into two parts by the Tn3 family transposon. The characteristic 5-bp direct target repeats flanked the Tn3 family transposon, suggesting that the transposon inserted into fipA.
FIG 2.
Phylogenetic tree of ST7 IncN plasmids. The names, host species, and accession numbers of the plasmids are shown. The tree was inferred using concatenated sequences of 26 genes belonging to the ST7 IncN backbone.
FIG 3.
The genetic context of blaNDM-1 on pNDM1_LL34. Genes between blaNDM-1 and qnrS1 are ble (mediating bleomycin resistance), trpF (encoding a phosphoribosyl anthranilate isomerase), dsbC (encoding an oxidoreductase), cutA1 (encoding an ion-tolerant protein), and groES/groEL (encoding a chaperonin). The tnpA gene (encoding a transposase) and both inverted repeats (blue bars) of a Tn3 family transposon are outside the region flanked by IS26. fipA (encoding a conjugal transfer inhibition protein) is interrupted by the insertion of the Tn3 family transposon with characteristic 5-bp direct target repeats (TATAT). Δ, truncated genes or mobile genetic elements.
blaIMP-4 was carried by a class I integron in the blaIMP-4-qacG2-aacA4 cassette array on pIMP4_LL34. Chloramphenicol resistance gene catB3 is usually seen together with blaIMP-4 in the blaIMP-4-qacG2-aacA4-catB3 cassette array but is absent from pIMP4_LL34. The integron is assigned In1498 by INTEGRALL (http://integrall.bio.ua.pt/). By BLAST, the closest match of pIMP4_LL34 was p13190-VIM (88% coverage, 99% identity) (GenBank accession no. MF344563) from K. pneumoniae in Beijing China. pIMP4_LL34 has a replicon, which has been proposed as IncHI5 but has not been included in the database of PlasmidFinder (22). By BLAST, using the 885-bp replication protein-encoding gene of the IncHI5 replicon, we identified 15 additional IncHI5 plasmids in GenBank. These plasmids were found in K. pneumoniae, K. oxytoca, K. michiganensis, R. ornithinolytica, and R. planticola, and all but one were found at various locations in China. These findings suggest that IncHI5 plasmids have been circulated in China, which warrants further investigation. blaCTX-M-3 was located downstream of ISEcp1 and upstream of a truncated orf477 gene. The ISEcp1-blaCTX-M-3-orf477Δ unit was inserted in a gene encoding a protein of the Hok/Gef family with the presence of 5-bp direct target repeats, which is characteristic of the transposition of ISEcp1. It became evident that ISEcp1 misrecognized a sequence in orf477, which has 8 out of 14 nucleotides matched with the right-hand inverted repeat (IRR), as its alternative IRR, and then realized the mobilization of blaCTX-M-3 into the Hok/Gef family protein-encoding gene.
In conclusion, we identified an ST273 CRKP carrying the carbapenemase genes blaNDM-1 and blaIMP-4. blaNDM-1 was carried by an ST7 IncN self-transmissible plasmid, and blaIMP-4 was located on an IncHI5 self-transmissible plasmid. This is yet another example of a clinical isolate containing multiple plasmids conferring resistance to carbapenems, as we described before (23). The coexistence of plasmids may generate new platforms to mediate further spread of carbapenem-resistant genes and questions our knowledge of the extent to which plasmids conferring multidrug resistance truly affect the fitness of host bacteria. The question arises as to why strains would possess multiple genes for the same resistance. The low diversity of ST273 isolates across continents and years suggests that the lineage merits further characterization.
Accession number(s).
Complete sequences of the chromosome of strain WCHKP020034, pIMP4_LL34, and pNDM1_LL34 have been deposited into GenBank under accession numbers CP025963, CP025964, and CP025965.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (project no. 81222025, 81572030, and 81772233) and a joint grant from the National Natural Science Foundation of China (project no. 81661130159) and the Newton Advanced Fellowship, Royal Society, United Kingdom (NA150363).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00160-18.
REFERENCES
- 1.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ageevets VA, Partina IV, Lisitsyna ES, Ilina EN, Lobzin YV, Shlyapnikov SA, Sidorenko SV. 2014. Emergence of carbapenemase-producing Gram-negative bacteria in Saint Petersburg, Russia. Int J Antimicrob Agents 44:152–155. doi: 10.1016/j.ijantimicag.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Bonura C, Giuffre M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, Giammanco A, MDR-GN Working Group, Palma DM, Mammina C. 2015. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One 10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou A, Roa M, Evangelista MA, Sulit AK, Lagamayo E, Torres BC, Klinzing DC, Daroy ML, Navoa-Ng J, Sucgang R, Zechiedrich L. 2016. Emergence of Klebsiella pneumoniae ST273 carrying blaNDM-7 and ST656 carrying blaNDM-1 in Manila, Philippines. Microb Drug Resist 22:585–588. doi: 10.1089/mdr.2015.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI document M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Zong Z, Zhang X. 2013. blaNDM-1-carrying Acinetobacter johnsonii detected in hospital sewage. J Antimicrob Chemother 68:1007–1010. doi: 10.1093/jac/dks505. [DOI] [PubMed] [Google Scholar]
- 7.Mendes RE, Kiyota KA, Monteiro J, Castanheira M, Andrade SS, Gales AC, Pignatari AC, Tufik S. 2007. Rapid detection and identification of metallo-β-lactamase-encoding genes by multiplex real-time PCR assay and melt curve analysis. J Clin Microbiol 45:544–547. doi: 10.1128/JCM.01728-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poirel L, Le Thomas I, Naas T, Karim A, Nordmann P. 2000. Biochemical sequence analyses of GES-1, a novel class A extended-spectrum β-lactamase, and the class 1 integron In52 from Klebsiella pneumoniae. Antimicrob Agents Chemother 44:622–632. doi: 10.1128/AAC.44.3.622-632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford PA, Bratu S, Urban C, Visalli M, Mariano N, Landman D, Rahal JJ, Brooks S, Cebular S, Quale J. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin Infect Dis 39:55–60. doi: 10.1086/421495. [DOI] [PubMed] [Google Scholar]
- 10.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlach GF, Allen BL, Clegg S. 1988. Molecular characterization of the type 3 (MR/K) fimbriae of Klebsiella pneumoniae. J Bacteriol 170:3547–3553. doi: 10.1128/jb.170.8.3547-3553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wyres KL, Holt KE. 2016. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol 24:944–956. doi: 10.1016/j.tim.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Lo WU, Lai RW, Tse CW, Lee RA, Luk WK, Cheng VC, Que TL, Chow KH, Ho PL. 2017. IncN ST7 epidemic plasmid carrying blaIMP-4 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. J Antimicrob Chemother 72:99–103. doi: 10.1093/jac/dkw353. [DOI] [PubMed] [Google Scholar]
- 21.Emms DM, Kelly S. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang Q, Yin Z, Zhao Y, Liang L, Feng J, Zhan Z, Wang H, Song Y, Tong Y, Wu W, Chen W, Wang J, Jiang L, Zhou D. 2017. Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int J Antimicrob Agents 49:709–718. doi: 10.1016/j.ijantimicag.2017.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Wu W, Feng Y, Carattoli A, Zong Z. 2015. The emergence of Enterobacter cloacae producing both KPC and NDM carbapenemases: characterization by whole genome sequencing. Antimicrob Agents Chemother 59:6625–6628. doi: 10.1128/AAC.01275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.