ABSTRACT
Polymyxins are increasingly used as a last-resort class of antibiotics against extensively drug-resistant (XDR) Gram-negative bacteria. However, resistance to polymyxins can emerge with monotherapy. As nephrotoxicity is the major dose-limiting factor for polymyxin monotherapy, dose escalation to suppress the emergence of polymyxin resistance is not a viable option. Therefore, novel approaches are needed to preserve this last-line class of antibiotics. This study aimed to investigate the antimicrobial synergy of polymyxin B combined with enrofloxacin against Pseudomonas aeruginosa. Static time-kill studies were conducted over 24 h with polymyxin B (1 to 4 mg/liter) and enrofloxacin (1 to 4 mg/liter) alone or in combination. Additionally, in vitro one-compartment model (IVM) and hollow-fiber infection model (HFIM) experiments were performed against P. aeruginosa 12196. Polymyxin B and enrofloxacin in monotherapy were ineffective against all of the P. aeruginosa isolates examined, whereas polymyxin B-enrofloxacin in combination was synergistic against P. aeruginosa, with ≥2 to 4 log10 kill at 24 h in the static time-kill studies. In both IVM and HFIM, the combination was synergistic, and the bacterial counting values were below the limit of quantification on day 5 in the HFIM. A population analysis profile indicated that the combination inhibited the emergence of polymyxin resistance in P. aeruginosa 12196. The mechanism-based modeling suggests that the synergistic killing is a result of the combination of mechanistic and subpopulation synergy. Overall, this is the first preclinical study to demonstrate that the polymyxin-enrofloxacin combination is of considerable utility for the treatment of XDR P. aeruginosa infections and warrants future clinical evaluations.
KEYWORDS: polymyxins, extensively drug resistant, Pseudomonas aeruginosa, enrofloxacin, antibiotic combination
INTRODUCTION
Extensively drug-resistant (XDR) Gram-negative pathogens are a major burden on the global health care system (1–5). Pseudomonas aeruginosa is at the top of the 2017 World Health Organization Priority Pathogens List, which describes the utmost needs for novel antibiotic treatments (6). Treatment options for this problematic pathogen are sparse due to the dry antibiotic discovery pipeline and the lack of new antibiotics with novel modes of action (7). Polymyxins are an old lipopeptide antibiotic class that are increasingly used as a last-line therapy for life-threatening infections caused by XDR P. aeruginosa (8, 9). Polymyxins are natural products from Paenibacillus polymyxa, and polymyxin B (PMB) and colistin (i.e., polymyxin E) are the only two clinically available polymyxins, differing by only an amino acid at position 6 (d-Phe and l-Leu, respectively) (10–13). Although polymyxin B and colistin remain effective against these Gram-negative pathogens, recent pharmacokinetic/pharmacodynamic (PK/PD) studies suggested that polymyxin monotherapy is associated with increased polymyxin resistance (12, 14–20). Worryingly, there have been increasing reports of infections caused by polymyxin-resistant XDR P. aeruginosa, which are essentially untreatable by conventional antibiotic monotherapy (21). As nephrotoxicity is the dose-limiting adverse effect of polymyxins, simply increasing the dose is not feasible (22). To preserve the efficacy of polymyxins while minimizing the emergence of resistance, novel approaches are urgently needed (23–26).
Combination therapy has been proposed as a novel strategy to maximize the antimicrobial efficacy against XDR pathogens and suppress the spread of resistance (24, 27–32). Enrofloxacin is a fluoroquinolone with broad-spectrum activity against Gram-positive and Gram-negative bacteria, and it was originally developed for veterinary use (33–36). We hypothesized that enrofloxacin displays antimicrobial synergy with polymyxin B based on the disruptive action of the latter on the Gram-negative outer membrane, thereby allowing more enrofloxacin molecules to cross the membranes and reach their intracellular DNA gyrase or topoisomerase targets. Accordingly, the primary objective of this study was to investigate the pharmacodynamics of polymyxin B and enrofloxacin alone and in combination against XDR P. aeruginosa in an in vitro static time-kill, in vitro one-compartment PK/PD model (IVM) and hollow-fiber infection PK/PD model (HFIM). This is the first preclinical study to examine the clinical potential of novel polymyxin-enrofloxacin combination for the treatment of XDR P. aeruginosa infections.
RESULTS
MICs and in vitro static time-kill.
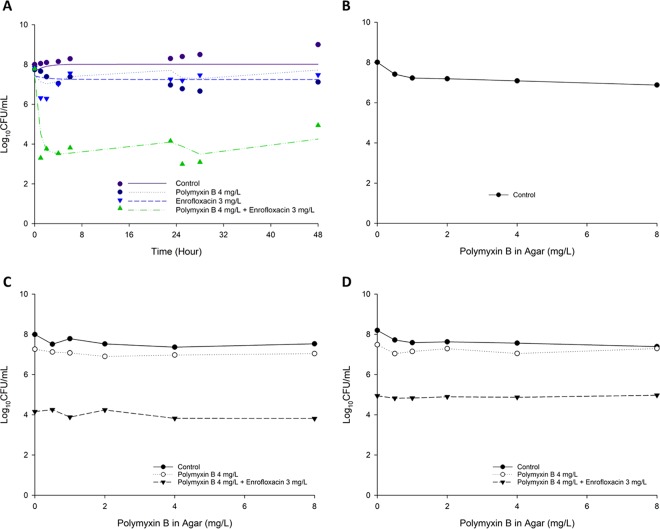
MICs of polymyxin B and enrofloxacin are summarized in Table 1. All three studied clinical isolates were resistant to both polymyxin B and enrofloxacin. The static time-kill kinetics of polymyxin B and enrofloxacin mono- and combination therapy are shown in Fig. 1, and the corresponding log10 changes in viable cell counts are summarized in Table S1 in the supplemental material. Polymyxin B monotherapy achieved moderate bactericidal activity at 1 h posttreatment, which was followed by substantial regrowth for all polymyxin B concentrations examined against P. aeruginosa 12196 (PMB MIC, 64 mg/liter) and P. aeruginosa LESB58 (PMB MIC, 8 mg/liter) (Fig. 1). Against P. aeruginosa H131300444 (PMB MIC, 128 mg/liter), no killing was evident for polymyxin B monotherapy. Similarly, enrofloxacin monotherapy resulted in a substantial killing within the first 6 h, followed by regrowth at 24 h for P. aeruginosa 12196 (ENRO MIC, 4 mg/liter) (Fig. 1). No killing was observed following enrofloxacin monotherapy for P. aeruginosa H131300444 and P. aeruginosa LESB58 (ENRO MIC, 4 mg/liter for both isolates). In contrast, all clinically achievable polymyxin B concentrations in combination with enrofloxacin resulted in extensive synergistic killing (Fig. 1 and Table S1). The lowest combination concentrations (1 mg/liter polymyxin B plus 1 mg/liter enrofloxacin) displayed synergistic killing against all strains within the first 3 h but was followed by extensive regrowth. Polymyxin B (4 mg/liter) with enrofloxacin (4 mg/liter) yielded at least a >2 log10 kill across all three isolates within 3 h (Fig. 1). Although the regrowth was observed across several concentrations of the combination, in general, a significantly reduced bacterial regrowth at 24 h was achieved with the combination compared to that for polymyxin B and enrofloxacin monotherapy (Fig. 1).
TABLE 1.
MICs of PMB and ENRO for polymyxin-resistant XDR P. aeruginosa strains examined in this studya
| Strain | PMB |
ENRO |
||
|---|---|---|---|---|
| Susceptibility | MIC (mg/liter) | Susceptibility | MIC (mg/liter) | |
| P. aeruginosa 12196 | R | 64 | R | 4 |
| P. aeruginosa H131300444 | R | 128 | R | 4 |
| P. aeruginosa LESB58 | R | 8 | R | 4 |
EUCAST breakpoints were the following. For polymyxin B (PMB), susceptibility and resistance breakpoints were defined as MICs of ≤2 mg/liter and >2 mg/liter, respectively (61). We applied the ciprofloxacin breakpoints for enrofloxacin (ENRO), i.e., ≤0.5 mg/liter for susceptibility and ≥1 mg/liter for resistance (61).
FIG 1.
Static time-kill for polymyxin B in combination with enrofloxacin against P. aeruginosa 12916 (A), P. aeruginosa H131300444 (B), and P. aeruginosa LESB58 (C). The y axis starts from the limit of detection (1.13 log10 CFU/ml). Marks represent observed viable counts, and lines represent individual fitted viable counts.
In vitro one-compartment PK/PD model.
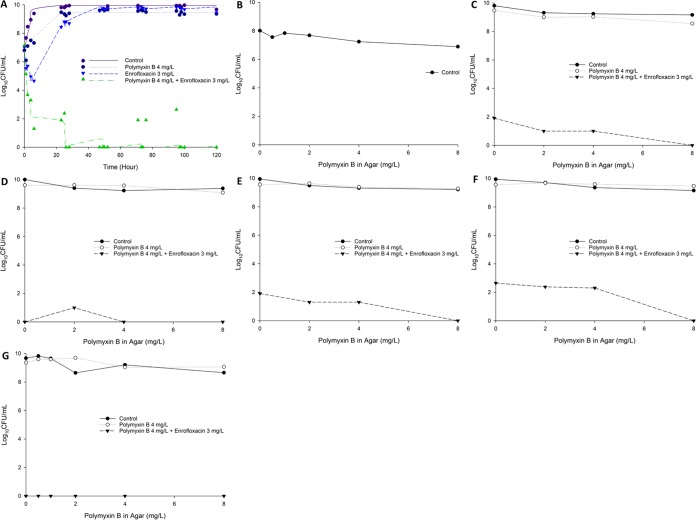
Concentrations of polymyxin B and enrofloxacin within the IVM and HFIM models were validated via liquid chromatography-mass spectrometry (LC-MS). Measured polymyxin B was 3.50 ± 1.18 mg/liter (n = 10) for the targeted concentration of 4 mg/liter. Measured enrofloxacin was 2.39 ± 0.49 mg/liter (n = 6) for the targeted concentration of 3 mg/liter and 0.67 ± 0.089 mg/liter (n = 6) for the targeted concentration of 0.6 mg/liter. The IVM time-kill curves for polymyxin B and enrofloxacin mono- and combination therapy against P. aeruginosa 12196 are shown in Fig. 2, and the corresponding log changes in viable cell counts are documented in Table S2. Polymyxin B and enrofloxacin monotherapy produced rapid initial bacterial killing during the first 6 h. However, regrowth occurred rapidly, and the killing activity of both antibiotics was diminished (Fig. 2). Even with the high inoculum (∼108 CFU/ml), the combination of polymyxin B and enrofloxacin was synergistic across 48 h and was able to result in rapid bacterial killing (>4 log10 compared to the control and >3 log10 better than any monotherapy; Fig. 2) and maintained excellent bacterial killing until 48 h (Fig. 2). Despite minor regrowth occurring after this time, the combination maintained 2 to 4 log10 greater killing than either monotherapy over 48 h (Fig. 2). Real-time population analysis profiles (PAPs) demonstrated that polymyxin B monotherapy was associated with the amplification of resistant subpopulations, and almost 100% of the remaining posttherapy population developed polymyxin resistance (Fig. 2). Excitingly, the combination therapy was able to significantly reduce the amplification of polymyxin-resistant subpopulations.
FIG 2.
(A) Killing kinetics of polymyxin B (4 mg/liter as continuous infusion) and enrofloxacin (bolus dose given every 12 h to achieve a Cmax of 3 mg/liter) alone and in combination against P. aeruginosa 12196 in the IVM with an inoculum of ∼108 CFU/ml. Population analysis profiles in the presence of 0.5, 1, 2, 4, and 8 mg/liter polymyxin B at 0 h (baseline) (B), 23 h (C), and 48 h (D) after the first dose. In panel A, lines represent individual fitted viable counts, and marks represent observed viable counts.
Hollow-fiber infection model.
Based on the very promising efficacy of the polymyxin B-enrofloxacin combination observed in the static time-kill and IVM, we further investigated the antimicrobial efficacy of the combination against XDR P. aeruginosa 12196 using an HFIM for 5 days. The HFIM results for the total and polymyxin-resistant subpopulations are shown in Fig. 3, and the corresponding log changes in viable cell counts are documented in Table S3. Polymyxin B and enrofloxacin monotherapies were associated with initial rapid bacterial killing against P. aeruginosa 12196, followed by substantial regrowth close to the control values by 50 h. From the PAPs, it is evident that most of the regrowth was due to the emergence of polymyxin-resistant subpopulations (Fig. 3). For the combination regimen, rapid initial killing occurred and the combination remained synergistic for the entire 5 days. The combination of polymyxin B and enrofloxacin was able to significantly reduce the regrowth of the polymyxin-resistant subpopulation (Fig. 3).
FIG 3.
(A) Killing kinetics of polymyxin B (4 mg/liter as continuous infusion) and enrofloxacin (bolus dose given every 12 h to achieve a Cmax of 3 mg/liter) alone and in combination against P. aeruginosa 12196 in the HFIM with an inoculum of ∼107 CFU/ml. Population analysis profiles in the presence of 0.5, 1, 2, 4, and/or 8 mg/liter polymyxin B at 0 h (baseline) (B), 23 h (C), 47 h (D), 71 h (E), 95 h (F), and 120 h (G) after the first dose. In panel A, lines represent individual fitted viable counts and marks represent observed viable counts.
MBM.
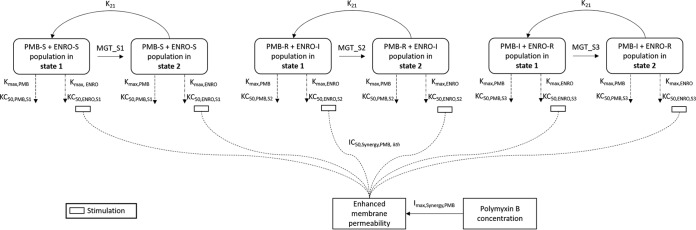
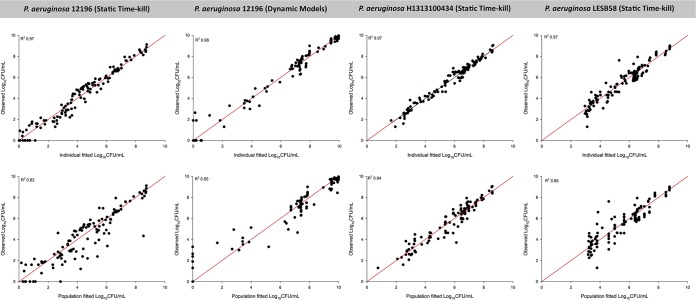
A mechanism-based model (MBM) was developed for three clinical strains to quantify and evaluate the potential mechanisms of synergy. The MBM consisted of three preexisting bacterial subpopulations with different susceptibilities (i.e., KC50, the concentration of drug causing 50% of the maximum rate of killing [Kmax]) to either polymyxin B or enrofloxacin (Fig. 4). The model-fitted parameters are summarized in Table S4, and all parameters were estimated with reasonably good precision. The synergistic combination was modeled as subpopulation and mechanistic synergy for all three isolates. Mechanistic synergy was expressed as an increase in enrofloxacin killing of the respective bacterial subpopulations with increasing polymyxin B concentrations (Table S4, manifested as IC50,Synergy). The proposed MBM simultaneously described the time course of bacterial killing and regrowth following treatment with polymyxin B and enrofloxacin mono- and combination therapies. The estimated mutation frequencies suggested that the initial inoculum consisted of a relatively larger fraction of subpopulation 2 (∼10−3 to 10−5 CFU/ml) than subpopulation 3 (∼10−7 to 10−10 CFU/ml) (Table S4). Individual and population predictions were reasonably unbiased and precise; only the population fits for the static time-kill study for P. aeruginosa 12196 and P. aeruginosa LESB58 showed slight misspecification (Fig. 5). However, this should not affect the characterization of the mechanisms of synergy. Data were distributed around the line of identity, and the coefficients of correlation for the individual fitted models were >0.95 against all three isolates (Fig. 5).
FIG 4.
Mechanism-based model of the synergistic killing of polymyxin B and in combination with enrofloxacin against P. aeruginosa. The parameters are presented in Table S4.
FIG 5.
Observed bacterial counts versus individual (upper) or population (lower) fitted bacterial counts for polymyxin B and enrofloxacin alone or in combination against P. aeruginosa isolates. The solid red lines represent the line of identity.
DISCUSSION
Polymyxin B is increasingly used as a last-line therapy for life-threatening infections caused by problematic Gram-negative pathogens such as P. aeruginosa (8, 16). However, emergence of polymyxin resistance after monotherapy has been reported both in vitro and in vivo (18, 37–39). As the incidence of XDR P. aeruginosa strains continues to increase, treatment options become very limited and there is an urgent need for rational polymyxin combination therapy with maximum killing and minimal emergence of resistance (27). Repurposing veterinary medicines for human use has gained significant interest recently, and several potential polymyxin combinations have been identified (40, 41). To date, the majority of synergistic polymyxin combinations identified involve carbapenems, rifampin, and penicillin against P. aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae, and a few studies reported synergetic combinations with fluoroquinolones against K. pneumoniae (24, 25, 28, 29, 31, 42, 43).
Enrofloxacin is a veterinary antibiotic that is active against a wide range of Gram-negative bacteria (33–36). Enrofloxacin is metabolized to ciprofloxacin via cytochrome P450 enzymes in the liver (33, 34, 44). Both enrofloxacin and its metabolite, ciprofloxacin, are pharmacologically active (45, 46). Enrofloxacin exerts its bactericidal activity by binding to topoisomerase II (DNA gyrase), thereby inhibiting the unwinding and duplication of bacterial DNA. In the present study, we evaluated the in vitro PD of polymyxin B and enrofloxacin in combination against three P. aeruginosa isolates. Enrofloxacin was selected as a potential secondary antibiotic over other fluoroquinolones, as our pilot study showed that it is synergistic against a wide range of XDR P. aeruginosa isolates. The concentrations of polymyxin B employed in this study were chosen to mimic clinically achievable unbound plasma polymyxin concentrations in critically ill patients following the currently recommended dosage regimens (47, 48). Given that enrofloxacin is partially metabolized into ciprofloxacin in vivo (33–36) and that its PK in humans is currently unknown, we employed clinically achievable ciprofloxacin concentrations in critically ill patients for enrofloxacin (49). To the best of our knowledge, this is the first preclinical study to examine polymyxin and enrofloxacin combination against XDR P. aeruginosa.
As monotherapies, neither polymyxin B nor enrofloxacin was effective against any strain of P. aeruginosa in the static time-kill studies over 24 h, while the combination substantially enhanced the antibacterial activity against all isolates, regardless of polymyxin B MICs (Fig. 1; see also Table S1 in the supplemental material). Importantly, synergy was observed even at the lowest concentrations of polymyxin B (1 mg/liter) and enrofloxacin (1 mg/liter) (Fig. 1 and Table S1). This is of clinical significance, given that nephrotoxicity is the dose-limiting adverse effect of polymyxins and dose escalation is not an option (50). Since enrofloxacin is metabolized to ciprofloxacin in vivo, the synergistic effect of polymyxin B and ciprofloxacin was also evaluated. Importantly, polymyxin B and ciprofloxacin also displayed synergy against XDR P. aeruginosa. Notably, the extent of synergy was inferior to that of enrofloxacin in most cases (Fig. S1), suggesting that enrofloxacin is a candidate for combination therapy with polymyxins. A limitation of static time-kill experiments is that the concentrations remain constant, and they do not reflect the PK of antibiotics in patients (51). Therefore, the static time-kill results (Fig. 1 and Table S1) may not truly reflect the bacterial killing of both antibiotics in humans. To investigate the therapeutic potential of the polymyxin B-enrofloxacin combination, in vitro PK/PD models IVM and HFIM were utilized to simulate the PK of both antibiotics in humans for a longer duration (2 days for IVM and 5 days for HFIM) against an XDR clinical isolate, P. aeruginosa 12196. Considering the inoculum effect of polymyxins (52), in vitro PK/PD experiments were conducted at ∼107 to 108 CFU/ml to mimic the high bacterial burden in severe bacterial pneumonia (53). In agreement with the observation in static time-kill studies with a low inoculum (∼106 CFU/ml), the polymyxin B-enrofloxacin combination was synergetic in the dynamic PK/PD models (Fig. 2 and 3 and Tables S2 and S3). The HFIM experiment was run for 5 days, as the viable counting values with the combination were below the limit of detection on day 5. A recent clinical PK/PD study revealed that the clinical efficacy and toxicity of polymyxin B are independent of the dosing regimen (e.g., continuous infusion versus intermittent dosing) (54). Therefore, it is unlikely that the continuous infusion employed in the present study has a major impact on the overall pharmacological implication of the polymyxin-enrofloxacin combination, and future PK/PD studies are warranted.
The most predictive PK/PD index for polymyxin B against P. aeruginosa after parenteral administration and inhalation is the ratio of the area under the unbound concentration-time curve over 24 h divided by the MIC (fAUC0–24/MIC) (16, 55). Applying the PK/PD index targets obtained from the thigh infection model, an fAUC0–24/MIC of >10.4 ± 2.98 would result in a 1- to 2-log10 reduction (16), and the monotherapy was in agreement with the fAUC0–24/MIC for polymyxins. In the present study, the calculated fAUC0–24/MIC for polymyxin B against P. aeruginosa 12196 was ∼1.5, smaller than the target of 10.4 ± 2.98, and not surprisingly, a 1- to 2-log10 reduction was not achieved with polymyxin B monotherapy at 24 h (Fig. 2 and 3). Combining polymyxin B with enrofloxacin produced substantially enhanced bacterial killing at 24 h, and even an fAUC0–24/MIC of ∼1.5 of polymyxin B was able to achieve a >4-log10 reduction for P. aeruginosa 12196 at 24 h and remained synergistic over 120 h in the dynamic PK/PD models. Unfortunately, the PK/PD index of enrofloxacin activity against P. aeruginosa remains unknown; hence, the antimicrobial efficacy of enrofloxacin could not be interpreted based on PK/PD index targets.
The real-time PAPs showed that even at the upper end of polymyxin B concentration achieved clinically (4 mg/liter), polymyxin B monotherapy led to rapid emergence of polymyxin resistance in P. aeruginosa 12196 in both IVM and HFIM (Fig. 2 and 3). Despite enrofloxacin monotherapy being ineffective, the addition of enrofloxacin suppressed the emergence of polymyxin resistance, unlike polymyxin B monotherapy, in IVM and HFIM (Fig. 2 and 3). The synergistic combination was able to suppress the emergence of polymyxin resistance by day 5 (Fig. 3). Translation of the in vitro PD data into the clinical setting remains challenging due to the lack of enrofloxacin PK in patients. Well-designed clinical studies are needed to validate and confirm findings.
Two models have been proposed to explain the enhanced antibacterial activity achieved with the polymyxin combination therapy (24, 28–31). Subpopulation synergy involves the killing of a subpopulation resistant to polymyxins by the second antibiotic and vice versa. Meanwhile, mechanistic synergy involves polymyxin and enrofloxacin acting on different complementary pathways to increase the rate and extent of bacterial killing. An MBM was developed for three clinical strains to quantify and evaluate the potential mechanisms of synergy. The proposed MBM well characterized the time course of bacterial growth and killing due to monotherapy and combination therapies against P. aeruginosa (Fig. 4 and 5), even though some minor misspecification existed (Fig. 1 to 3). In general, our MBM provides a satisfactory fit (overall R2 of >0.95). The MBM was developed based on the hypothesis that the disruption of the bacterial outer membrane by polymyxin B enhances the uptake and intracellular concentration of enrofloxacin. Mechanistic synergy was incorporated in the MBM as a decrease in the KC50,ENRO of respective subpopulations (Table S4). This mechanism of synergy is necessary, since bacteria were resistant to both antibiotics when polymyxin B (1 to 4 mg/liter) or enrofloxacin (1 to 4 mg/liter) was given as monotherapy. The increased permeability of the outer membrane is supported by our fractional inhibitory concentration results (Tables S5 to S7), in which the enrofloxacin MICs decreased in the presence of polymyxin B and vice versa. Additionally, the mechanistic data from polymyxin-doripenem combination against A. baumannii also showed a similar mechanism of synergy (56). The exclusion of either aspect of the synergy mechanism from the MBM resulted in a model that could not be estimated, as the simplified model provided poor curve fits (R2 of <0.50; data not shown). Interestingly, our MBM also suggested that less susceptible subpopulations (i.e., subpopulations 2 and 3) have a longer mean generation time, possibly related to the decreased biological fitness (57). Notably, our proposed MBM lacks the effect of the host immune system (58). To account for this, in vivo animal infection studies are warranted to validate and refine our MBM for the mono- and combination therapy. Once the proposed MBM is validated with future preclinical data (including those from intermittent dosing) and a population human PK model is available for enrofloxacin, our MBM can be employed in a Monte Carlo simulation to rationally optimize the dosage regimens for the combination therapy in patients (59, 60).
Despite evidence of synergistic activity at clinically relevant concentrations and the MBM supporting the proposed synergistic mechanisms, the exact mechanisms of the synergistic killing and resistance remain unknown. System biology studies are currently conducted in our laboratory to elucidate the mechanisms of synergy and potential resistance to polymyxin B and enrofloxacin against XDR P. aeruginosa 12196. In-depth knowledge of the synergistic killing and resistance mechanisms will allow optimizing the dosage regimen for clinical applications.
With the increasing incidence of infections caused by XDR Gram-negative superbugs, rationally designed polymyxin combinations with other antibiotics are of the utmost importance. To the best of our knowledge, this is the first preclinical study to demonstrate that clinically relevant dosage regimens of polymyxin B in combination with enrofloxacin significantly enhance the bacterial killing and suppressed the emergence of polymyxin resistance even in XDR isolates. Our data suggest that enrofloxacin represents a therapeutic option against XDR Gram-negative pathogens, and further safety and PK/PD investigations in murine models and humans are warranted for translation into the clinical setting.
MATERIALS AND METHODS
Chemicals and bacterial strains.
Polymyxin B (batch number BCBD1065V; Sigma-Aldrich, Castle Hill, Australia) solution was freshly prepared in sterile Milli-Q water (Millipore Australia, North Ryde, Australia) (42). Enrofloxacin was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich), and then sterile Milli-Q water was added to a final DMSO level of 10% (vol/vol) (40). Three clinical isolates of P. aeruginosa (12196 [JML Laboratories Iowa; see Table S8 in the supplemental material for the antibiogram], H131300444 [Public Health England], and LESB58 [kindly provided by Robert Hancock, from the British University of Columbia, Canada]) were employed in this study. All strains were stored in tryptone soy broth with 20% glycerol at −80°C and subcultured onto nutrient agar plates before each experiment (40, 42).
Measurements of MICs.
MICs of polymyxin B and enrofloxacin were determined for all isolates using broth microdilution in cation-adjusted Mueller-Hinton broth (CAMHB; Mg2+ at 12.2 mg/liter and Ca2+ at 23.0 mg/liter [Oxoid, Hampshire, England]) (40, 42). The susceptibility and resistance to polymyxin B were defined as MICs of ≤2 mg/liter and >2 mg/liter, respectively (42, 61). As CLSI and EUCAST do not currently provide breakpoints for enrofloxacin, we applied the ciprofloxacin breakpoints of ≤0.5 mg/liter for susceptibility and ≥1 mg/liter for resistance in this study (61).
Static time-kill experiment.
Static time-kill studies were conducted to examine the antimicrobial activity of polymyxin B and enrofloxacin monotherapy and its combination against P. aeruginosa (40, 42). All experiments were performed with an initial inoculum of ∼106 CFU/ml in 20 ml of CAMHB in 50 ml pyrogen-free and sterile polypropylene tubes. Polymyxin B and enrofloxacin mono- and combination therapies were examined at 1, 2, 3, and 4 mg/liter. Serial samples (50 μl) were collected at 0, 0.5, 1, 2, 4, 6, and 24 h for viable counting on nutrient agar plates, and the limit of detection was 20 CFU/ml (equivalent to one colony per plate). A ProtoCOL automated colony counter (Synbiosis, Cambridge, United Kingdom) was used to quantify bacteria after 24 h of incubation at 37°C.
In vitro one-compartment PK/PD model experiment.
To further evaluate the synergistic killing of the combination, an IVM (62) was employed over 48 h to assess the efficacy and the emergence of polymyxin resistance against P. aeruginosa 12196. Four reservoirs were employed: (i) a control reservoir with no antibiotic; (ii) polymyxin B monotherapy; (iii) enrofloxacin monotherapy; and (iv) polymyxin B-enrofloxacin combination therapy. Each reservoir contained 80 ml of CAMHB and was maintained at 37°C. The PK of polymyxin B in critically ill patients was simulated (47, 48). To avoid the difficulty in simulating two very different half-lives, polymyxin B was added into the diluent reservoirs (4 mg/liter) and delivered to all reservoirs as a continuous infusion to achieve a central reservoir concentration of 4 mg/liter. Enrofloxacin was added into the central reservoir every 12 h via bolus administration using an automated syringe pump to achieve a maximum concentration of drug in serum (Cmax) of 3 mg/liter. As enrofloxacin is used only in animals and no PK information in humans is available, the flow rate of sterile CAMHB was set to 0.19 ml/min to simulate the ciprofloxacin half-life of 5 h in patients (49). The same dosage regimen was simulated for the combination therapy. Serial samples were collected from the reservoirs at 0, 1, 2, 4, 6, 23, 25, 28, and 48 h. At 23 and 48 h, PAPs were carried out by plating 100 μl of the samples on polymyxin B-containing agar plates (0.5, 1, 2, 4, and 8 mg/liter) to detect the emergence of polymyxin-resistant subpopulations (42, 63). The limit of detection was 10 CFU/ml (equivalent to 1 colony per plate) for PAPs.
Hollow-fiber infection model.
An HFIM was employed to further examine the PK/PD of the synergistic combination against P. aeruginosa 12196 (64, 65). The experiment was conducted over 120 h and maintained at 35°C in a humidified incubator. A total of 4 arms were employed: (i) control, (ii) polymyxin B alone, (iii) enrofloxacin alone, and (iv) polymyxin B-enrofloxacin combination. For polymyxin B, continuous infusion was employed to achieve a central reservoir concentration of 4 mg/liter. Enrofloxacin was administered every 12 h as a bolus to achieve a Cmax of 3 mg/liter. The flow rate (0.58 ml/min) simulated the 5-h elimination half-life of ciprofloxacin in patients (49). The same dosage regimens were simulated for the combination therapy. Serial samples were taken from the cartridges for viable counting at 0, 1, 2, 4, 6, 23, 25, 26, 28, 47, 49, 50, 52, 71, 73, 74, 76, 95, 97, 98, 100, and 120 h. At 23, 47, 71, 95, and 120 h, PAPs were carried out on polymyxin B-containing plates at 0.5, 1, 2, 4, and/or 8 mg/liter to detect the emergence of polymyxin resistance (42, 63).
Pharmacokinetics validation.
Concentrations of polymyxin B and enrofloxacin in broth samples from IVM and HFIM were measured using a Waters Acquity H-class ultra high-performance liquid chromatography (UHPLC) system triple-quadrupole LC-MS/MS system (TQS). Polymyxin B concentrations were quantified using a previously established method (66). For enrofloxacin, the chromatographic separation was achieved on a Phenomenex Kinetex C18 column (internal diameter, 3 × 50 mm; 2.6-μm particle size) with a Phenomenex Krudekatcher guard cartridge. Column temperature was maintained at 40°C. Mobile phases consisted of 0.1% trifluoroacetic acid in water (solvent A) and 0.1% trifluoroacetic acid in acetonitrile (solvent B). The enrofloxacin was eluted in a step gradient (hold at 2% solvent B for 0.5 min, 2 to 40% solvent B for 0.1 min, 40 to 100% solvent B for 1.4 min) at 0.4 ml/min, followed by a wash (100% solvent B for 2 min) and an equilibration step (2% solvent B for 3 min) at 0.6 ml/min. The autosampler temperature was maintained at 4°C, the injection volume was 2 μl, and the run time was 7 min. Clinafloxacin was used as the internal standard (IS), and the analysis was performed in an electrospray ionization-positive model. The MS settings were the following: capillary voltage, 1 kV; desolvation temperature, 650°C; desolvation gas flow (nitrogen), 1,000 liters/h; cone gas flow (nitrogen), 150 liters/h; nebulizer pressure, 700 kPa; collision gas flow (argon), 0.14 ml/min. Multiple reaction monitoring (MRM) was used to detect the analytes, and the optimized settings were the following: the cone voltage was 10 V for enrofloxacin and 20 V for clinafloxacin, and the collision energies were 20 and 22, respectively. The transitions of m/z 360.29 →316.26 and m/z 366.17→305.18 were monitored for enrofloxacin and clinafloxacin, respectively. The limit of quantification was 0.10 mg/liter for enrofloxacin. The accuracy and precision of the assays were −1.28% and 7.50%, respectively, for enrofloxacin.
MBM of monotherapy and combination therapy.
An MBM was developed to quantitatively describe the time course of bacterial killing and synergy by polymyxin B and enrofloxacin mono- and combination therapy.
Life cycle model.
The proposed MBM consisted of a life cycle growth model that quantitatively describes the underlying biological processes (57, 59, 60). Bacterial replication was described by two states: state 1, representing bacterial cells that are growing and preparing for replication, and state 2, representing bacterial cells immediately before replication. In the proposed model, bacterial cells were partitioned into three preexisting subpopulations: subpopulation 1 was susceptible to both polymyxin B and enrofloxacin (CFUS1), subpopulation 2 was more resistant to polymyxin B and intermediately susceptible to enrofloxacin (CFUS2), and subpopulation 3 was intermediately susceptible to polymyxin B and more resistant to enrofloxacin (CFUS3). Intermediately susceptible and more resistant subpopulations were defined for modeling purposes as subpopulations with MICs greater than the resistance breakpoints (e.g., MIC of 2 mg/liter for polymyxin B). The number of subpopulations needed to describe the data was tested using the log-likelihood ratio (reported as −1× log likelihood in S-ADAPT), with visual inspection of the fitted function and parameter estimates, especially the mutation frequency. The subpopulations differed in antibiotic susceptibility (KC50,ii) and initial individual inoculum (CFU0). The total concentration of bacteria (CFUTotal) was represented by equation 1:
| (1) |
The differential equations for bacterial concentration in state 1 and state 2 were the following:
| (2) |
| (3) |
where CL is the clearance in the IVM, V is the volume of distribution for the IVM, and PRRP is the probability of replication.
The PRRP was assumed to be 100% successful at low bacterial load. The replication factor (PRRP) limits the total population (CFUALL) from exceeding the maximum allowable population size (CFUmax) (57, 59). The PRRP is defined as
| (4) |
where CFUmax is the maximum bacterial load and CFUALL is the total bacterial load in all populations at a given time.
Killing by polymyxin B and enrofloxacin was described by a sigmoidal Emax model and was assumed to have the same direct effect on the bacteria in both states 1 and 2 as those described in equations 2 and 3.
| (5) |
| (6) |
Kmax,PMB,ii and Kmax,ENRO,ii are the maximum polymyxin B and enrofloxacin killing rate constants for the iith subpopulation, KC50,PMB,ii and KC50,ENRO,ii are the polymyxin B and enrofloxacin concentrations causing 50% of Kmax for the iith subpopulation, and γ is the Hill coefficient.
Mechanism-based modeling of the synergy.
The mechanistic synergy was modeled as described previously (57, 58). Polymyxin B is known to permeabilize the outer membrane and thus enhance the intracellular concentrations of enrofloxacin (60). This synergy was incorporated into the proposed model as represented equation 7:
| (7) |
where SCE is synergistic combination effect.
Initial conditions.
The initial inoculum of all subpopulations was estimated. The initial inoculum of subpopulations 2 and 3 were estimated as a fraction of the total initial inoculum (CFU0). The initial condition for subpopulation 2 was calculated as MUT,S2 (mutation frequency for subpopulation 2) × CFU0, and the initial condition for subpopulation 3 was MUT,S3 (mutation frequency for subpopulation 3) × CFU0. The initial condition for subpopulation 1 was (1 – MUT,S2 – MUT,S3) × CFU0. All bacteria were assumed to initialize in state 1, and the initial condition for state 2 for subpopulations was set to zero (57, 58).
Observation.
All bacterial counts were transformed to log10 scale. Residual unexplained variability was described by an additive error model on the log10 scale, and the interexperiment variability was fixed to a very small value (coefficient of variation of 10%) (57). Viable counts below the limit of detection were plotted as zero on a log10 scale.
Estimation.
The time course of bacterial killing and regrowth was comodeled in S-ADAPT (version 1.57) (67) and facilitated by S-ADAPT TRAN (68, 69) using a Monte Carlo parametric expectation maximization algorithm (MC-PEM) (pmethod = 4). The MBM was extended for P. aeruginosa 12196 by simultaneously fitting the time course of the bacterial killing in both dynamic PK/PD models (i.e., IVM and HFIM). The final model was assessed by its precision and the biological plausibility of the fitted parameters (especially the mutation frequency, initial inoculum, and maximum population size), the goodness of fit, and the visual inspection of diagnostic plots (70–73).
Supplementary Material
ACKNOWLEDGMENTS
J.L., A.F., G.G.R., and T.V. are supported by a research grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965). J.L. is an Australian National Health Medical Research Council (NHMRC) Senior Research Fellow, and T.V. is an Australian NHMRC Industry Career Development Level 2 Research Fellow.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We thank Bob Hancock for providing P. aeruginosa LESB58.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00028-18.
REFERENCES
- 1.Gales AC, Jones RN, Sader HS. 2011. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother 66:2070–2074. doi: 10.1093/jac/dkr239. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 2014. Antimicrobial resistance: 2014 global report on surveillance. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Falagas ME, Kasiakou SK, Saravolatz LD. 2005. Colistin: the revival of polymyxins for the management of multidrug-resistant gram-negative bacterial infections. Clin Infect Dis 40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 4.Nasnas R, Saliba G, Hallak P. 2009. The revival of colistin: an old antibiotic for the 21st century. Pathol Biol 57:229–235. doi: 10.1016/j.patbio.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Cegielski JP, Dalton T, Yagui M, Wattanaamornkiet W, Volchenkov GV, Via LE, Van Der Walt M, Tupasi T, Smith SE, Odendaal R, Leimane V, Kvasnovsky C, Kuznetsova T, Kurbatova E, Kummik T, Kuksa L, Kliiman K, Kiryanova EV, Kim H, Kim C-K, Kazennyy BY, Jou R, Huang W-L, Ershova J, Erokhin VV, Diem L, Contreras C, Cho SN, Chernousova LN, Chen MP, Caoili JC, Bayona J, Akksilp S, Global Preserving Effective TB Treatement Study (PETTS) Investigators. 2014. Extensive drug resistance acquired during treatment of multidrug-resistant tuberculosis. Clin Infect Dis 59:1049–1063. doi: 10.1093/cid/ciu572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tacconelli E, Magrini N. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics 2017. World Health Organization, Geneva, Switzerland: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf. [Google Scholar]
- 7.Boucher HW, Talbot GH, Benjamin DK, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D, Infectious Disease America 2013. 10 ×′ 20 progress–development of new drugs active against Gram-negative bacilli: an update from the Infectious Diseases Society of America. Clin Infect Dis 56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Nation RL, Turnidge JD, Milne RW, Coulthard K, Rayner CR, Paterson DL. 2006. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis 6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Nation RL, Milne RW, Turnidge JD, Coulthard K. 2005. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int J Antimicrob Agents 25:11–25. doi: 10.1016/j.ijantimicag.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Kwa A, Kasiakou SK, Tam VH, Falagas ME. 2007. Polymyxin B: similarities to and differences from colistin (polymyxin E). Expert Rev Anti Infect 5:811–821. doi: 10.1586/14787210.5.5.811. [DOI] [PubMed] [Google Scholar]
- 11.Landman D, Georgescu C, Martin DA, Quale J. 2008. Polymyxins revisited. Clin Microbiol Rev 21:449–465. doi: 10.1128/CMR.00006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tran TB, Velkov T, Nation RL, Forrest A, Tsuji BT, Bergen PJ, Li J. 2016. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet? Int J Antimicrob Agents 48:592–597. doi: 10.1016/j.ijantimicag.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velkov T, Roberts KD, Thompson PE, Li J. 2016. Polymyxins: a new hope in combating Gram-negative superbugs? Future Med Chem 8:1017–1025. doi: 10.4155/fmc-2016-0091. [DOI] [PubMed] [Google Scholar]
- 14.Dudhani RV, Nation RL, Li J. 2010. Evaluating the stability of colistin and colistin methanesulphonate in human plasma under different conditions of storage. J Antimicrob Chemother 65:1412–1415. doi: 10.1093/jac/dkq134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheah S-E, Li J, Bergen PJ, Nation RL. 2016. Polymyxin pharmacokinetics and pharmacodynamics, p 221–260. In Rotschafer JC, Andes DR, Rodvold KA (ed), Antibiotic pharmacodynamics. Springer, New York, NY. doi: 10.1007/978-1-4939-3323-5_10. [DOI] [Google Scholar]
- 16.Cheah S-E, Wang J, Turnidge JD, Li J, Nation RL. 2015. New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J Antimicrob Chemother 70:3291–3297. doi: 10.1093/jac/dkv267. [DOI] [PubMed] [Google Scholar]
- 17.Bergen PJ, Bulitta JB, Forrest A, Tsuji BT, Li J, Nation RL. 2010. Pharmacokinetic/pharmacodynamic investigation of colistin against Pseudomonas aeruginosa using an in vitro model. Antimicrob Agents Chemother 54:3783–3789. doi: 10.1128/AAC.00903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergen PJ, Forrest A, Bulitta JB, Tsuji BT, Sidjabat HE, Paterson DL, Li J, Nation RL. 2011. Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob Agents Chemother 55:5134–5142. doi: 10.1128/AAC.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tam VH, Schilling AN, Vo G, Kabbara S, Kwa AL, Wiederhold NP, Lewis RE. 2005. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:3624–3630. doi: 10.1128/AAC.49.9.3624-3630.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ly NS, Yang J, Bulitta JB, Tsuji BT. 2012. Impact of two-component regulatory systems PhoP-PhoQ and PmrA-PmrB on colistin pharmacodynamics in Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:3453–3456. doi: 10.1128/AAC.06380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goli HR, Nahaei MR, Ahangarzadeh Rezaee M, Hasani A, Samadi Kafil H, Aghazadeh M. 2016. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iranian J Microbiol 8:62–69. [PMC free article] [PubMed] [Google Scholar]
- 22.Michalopoulos A, Tsiodras S, Rellos K, Mentzelopoulos S, Falagas M. 2005. Colistin treatment in patients with ICU-acquired infections caused by multiresistant Gram-negative bacteria: the renaissance of an old antibiotic. Clin Microbiol Infect 11:115–121. doi: 10.1111/j.1469-0691.2004.01043.x. [DOI] [PubMed] [Google Scholar]
- 23.Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U. 2015. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 24.Sharma R, Patel S, Abboud C, Diep J, Ly NS, Pogue JM, Kaye KS, Li J, Rao GG. 2017. Polymyxin B in combination with meropenem against carbapenemase-producing Klebsiella pneumoniae: pharmacodynamics and morphological changes. Int J Antimicrob Agents 49:224–232. doi: 10.1016/j.ijantimicag.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim T-P, Lee W, Tan T-Y, Sasikala S, Teo J, Hsu L-Y, Tan T-T, Syahidah N, Kwa AL. 2011. Effective antibiotics in combination against extreme drug-resistant Pseudomonas aeruginosa with decreased susceptibility to polymyxin B. PLoS One 6:e28177. doi: 10.1371/journal.pone.0028177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim TP, Tan TY, Lee W, Sasikala S, Tan TT, Hsu LY, Kwa AL. 2011. In-vitro activity of polymyxin B, rifampicin, tigecycline alone and in combination against carbapenem-resistant Acinetobacter baumannii in Singapore. PLoS One 6:e18485. doi: 10.1371/journal.pone.0018485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bergen PJ, Bulman ZP, Saju S, Bulitta JB, Landersdorfer C, Forrest A, Li J, Nation RL, Tsuji BT. 2015. Polymyxin combinations: pharmacokinetics and pharmacodynamics for rationale use. Pharmacotherapy 35:34–42. doi: 10.1002/phar.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D, Yu B, Diep JK, Sharma R, Dudley M, Monteiro J, Kaye KS, Pogue JM, Abboud CS, Rao GG. 2017. In vitro assessment of combined polymyxin B and minocycline therapy against Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae. Antimicrob Agents Chemother 61:e00073-17. doi: 10.1128/AAC.00073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diep J, Jacobs DM, Sharma R, Covelli J, Bowers DR, Russo TA, Rao GG. 2016. Polymyxin B in combination with rifampin and meropenem against polymyxin B-resistant KPC-producing Klebsiella pneumoniae. Antimicrob Agents Chemother doi: 10.1128/aac.02121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao GG, Ly NS, Bulitta JB, Soon RL, San Roman MD, Holden PN, Landersdorfer CB, Nation RL, Li J, Forrest A, Tsuji BT. 2016. Polymyxin B in combination with doripenem against heteroresistant Acinetobacter baumannii: pharmacodynamics of new dosing strategies. J Antimicrob Chemother 71:3148–3156. doi: 10.1093/jac/dkw293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao GG, Ly NS, Diep J, Forrest A, Bulitta JB, Holden PN, Nation RL, Li J, Tsuji BT. 2016. Combinatorial pharmacodynamics of polymyxin B and tigecycline against heteroresistant Acinetobacter baumannii. Int J Antimicrob Agents 48:331–336. doi: 10.1016/j.ijantimicag.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jasim R, Schneider EK, Han M, Azad MA, Hussein M, Nowell C, Baker MA, Wang J, Li J, Velkov T. 2017. A fresh shine on cystic fibrosis inhalation therapy: antimicrobial synergy of polymyxin B in combination with silver nanoparticles. J Biomed Nanotechnol 13:447–457. doi: 10.1166/jbn.2017.2355. [DOI] [PubMed] [Google Scholar]
- 33.Mengozzi G, Intorre L, Bertini S, Soldani G. 1996. Pharmacokinetics of enrofloxacin and its metabolite ciprofloxacin after intravenous and intramuscular administrations in sheep. Am J Vet Res 57:1040–1043. [PubMed] [Google Scholar]
- 34.Küng K, RIOND JL, Wanner M. 1993. Pharmacokinetics of enrofloxacin and its metabolite ciprofloxacin after intravenous and oral administration of enrofloxacin in dogs. J Vet Pharmacol Ther 16:462–468. doi: 10.1111/j.1365-2885.1993.tb00212.x. [DOI] [PubMed] [Google Scholar]
- 35.Kaartinen L, Salonen M, Älli L, Pyörälä S. 1995. Pharmacokinetics of enrofloxacin after single intravenous, intramuscular and subcutaneous injections in lactating cows. J Vet Pharmacol Ther 18:357–362. doi: 10.1111/j.1365-2885.1995.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 36.Giguere S, Sweeney R, Belanger M. 1996. Pharmacokinetics of enrofloxacin in adult horses and concentration of the drug in serum, body fluids, and endometrial tissues after repeated intragastrically administered doses. Am J Vet Res 57:1025–1030. [PubMed] [Google Scholar]
- 37.Hengzhuang W, Wu H, Ciofu O, Song Z, Høiby N. 2012. In vivo pharmacokinetics/pharmacodynamics of colistin and imipenem in Pseudomonas aeruginosa biofilm infection. Antimicrob Agents Chemother 56:2683–2690. doi: 10.1128/AAC.06486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Turnidge J, Milne R, Nation RL, Coulthard K. 2001. In vitro pharmacodynamic properties of colistin and colistin methanesulfonate against Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Antimicrob Agents Chemother 45:781–785. doi: 10.1128/AAC.45.3.781-785.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Jacob J, Sidjabat HE, Paterson DL, Nation RL, Li J. 2011. Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 55:5685–5695. doi: 10.1128/AAC.05298-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran TB, Cheah S-E, Yu HH, Bergen PJ, Nation RL, Creek DJ, Purcell A, Forrest A, Doi Y, Song J, Velkov T, Li J. 2016. Anthelmintic closantel enhances bacterial killing of polymyxin B against multidrug-resistant Acinetobacter baumannii. J Antibiot 69:415–421. doi: 10.1038/ja.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider EK, Reyes-Ortega F, Velkov T, Li J. 2017. Antibiotic–non-antibiotic combinations for combating extremely drug-resistant Gram-negative ‘superbugs.’ Essays Biochem 61:115–125. doi: 10.1042/EBC20160058. [DOI] [PubMed] [Google Scholar]
- 42.Abdul Rahim N, Cheah SE, Johnson MD, Yu H, Sidjabat HE, Boyce J, Butler MS, Cooper MA, Fu J, Paterson DL, Nation RL, Bergen PJ, Velkov T, Li J. 2015. Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two “old” antibiotics-polymyxin B and chloramphenicol. J Antimicrob Chemother 70:2589–2597. doi: 10.1093/jac/dkv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitsugui CS, Tognim MCB, Cardoso CL, Carrara-Marroni FE, Botelho Garcia L. 2011. In vitro activity of polymyxins in combination with β-lactams against clinical strains of Pseudomonas aeruginosa. Int J Antimicrob Agents 38:447–450. doi: 10.1016/j.ijantimicag.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Salvadori M, De Vito V, Owen H, Giorgi M. 2015. Pharmacokinetics of enrofloxacin and its metabolite ciprofloxacin after intracoelomic administration in tortoises (Testudo hermanni). Israel J Vet Med 70:45–48. [Google Scholar]
- 45.Fisher LM, Lawrence JM, Josty IC, Hopewell R, Margerrison EE, Cullen ME. 1989. Ciprofloxacin and the fluoroquinolones: new concepts on the mechanism of action and resistance. Am J Med 87:S2–S8. doi: 10.1016/0002-9343(89)90010-7. [DOI] [PubMed] [Google Scholar]
- 46.Blondeau JM. 2004. Fluoroquinolones: mechanism of action, classification, and development of resistance. Surv Ophthalmol 49:S73–S78. doi: 10.1016/j.survophthal.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Zavascki AP, Goldani LZ, Cao G, Superti SV, Lutz L, Barth AL, Ramos F, Boniatti MM, Nation RL, Li J. 2008. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis 47:1298–1304. doi: 10.1086/592577. [DOI] [PubMed] [Google Scholar]
- 48.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhão RC, Wang J, Forrest A. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez Navarro MD, Sayalero Marinero ML, Sánchez Navarro A. 2002. Pharmacokinetic/pharmacodynamic modelling of ciprofloxacin 250 mg/12 h versus 500 mg/24 h for urinary infections. J Antimicrob Chemother 50:67–72. doi: 10.1093/jac/dkf079. [DOI] [PubMed] [Google Scholar]
- 50.Zavascki AP, Nation RL. 2017. Nephrotoxicity of polymyxins: is there any difference between colistimethate and polymyxin B? Antimicrob Agents Chemother 61:e02319-16. doi: 10.1128/AAC.02319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaddady PK, Lee RE, Meibohm B. 2010. In vitro pharmacokinetic/pharmacodynamic models in anti-infective drug development: focus on TB. Future Med Chem 2:1355–1369. doi: 10.4155/fmc.10.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bulitta JB, Yang JC, Yohonn L, Ly NS, Brown SV, D'Hondt RE, Jusko WJ, Forrest A, Tsuji BT. 2010. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 54:2051–2062. doi: 10.1128/AAC.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Mareletsi T, Cutland C, Wong M, Khoosal M, Karstaedt A, Zhao P, Deatly A, Sidhu M, Jansen KU, Klugman KP. 2012. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis 54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teng CB, Koh PT, Lye DC, Ang BS. 2008. Continuous versus intermittent infusion of polymyxin B in the treatment of infections caused by multidrug-resistant Gram-negative bacteria. Int J Antimicrob Agents 31:80–82. doi: 10.1016/j.ijantimicag.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 55.Lin Y-W, Zhou Q, Onufrak NJ, Wirth V, Chen K, Wang J, Forrest A, Chan H-K, Li J. 2017. Aerosolized polymyxin B for treatment of respiratory tract infections: determination of pharmacokinetic/pharmacodynamic indices for aerosolized polymyxin B against Pseudomonas aeruginosa in a mouse lung infection model. Antimicrob Agents Chemother 61:e00211-17. doi: 10.1128/AAC.00211-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maifiah MHM, Creek DJ, Nation RL, Forrest A, Tsuji BT, Velkov T, Li J. 2017. Untargeted metabolomics analysis reveals key pathways responsible for the synergistic killing of colistin and doripenem combination against Acinetobacter baumannii. Sci Rep 7:45527. doi: 10.1038/srep45527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landersdorfer CB, Ly NS, Xu H, Tsuji BT, Bulitta JB. 2013. Quantifying subpopulation synergy for antibiotic combinations via mechanism-based modeling and a sequential dosing design. Antimicrob Agents Chemother 57:2343–2351. doi: 10.1128/AAC.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brill MJE, Kristoffersson AN, Zhao C, Nielsen EI, Friberg LE. 8 December 2017. Semi-mechanistic pharmacokinetic-pharmacodynamic modelling of antibiotic drug combinations. Clin Microbiol Infect doi: 10.1016/j.cmi.2017.11.023. [DOI] [PubMed] [Google Scholar]
- 59.Yadav R, Bulitta JB, Wang J, Nation RL, Landersdorfer CB. 2017. Evaluation of PK/PD model-based optimized combination regimens against multidrug-resistant Pseudomonas aeruginosa in a murine thigh infection model using humanized dosing schemes. Antimicrob Agents Chemother 61:e01268-17. doi: 10.1128/aac.01268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yadav R, Landersdorfer CB, Nation RL, Boyce JD, Bulitta JB. 2015. Novel approach to optimize synergistic carbapenem-aminoglycoside combinations against carbapenem-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 59:2286–2298. doi: 10.1128/AAC.04379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.EUCAST 2014. Breakpoint tables for interpretation of MICs and zone diameters version 4.0. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf.
- 62.Deris ZZ, Yu HH, Davis K, Soon RL, Jacob J, Ku CK, Poudyal A, Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Paterson DL, Velkov T, Li J, Nation RL. 2012. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 56:5103–5112. doi: 10.1128/AAC.01064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheah SE, Li J, Tsuji BT, Forrest A, Bulitta JB, Nation RL. 2016. Colistin and polymyxin B dosage regimens against Acinetobacter baumannii: differences in activity and the emergence of resistance. Antimicrob Agents Chemother 60:3921–3933. doi: 10.1128/AAC.02927-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bergen PJ, Bulitta JB, Kirkpatrick CM, Rogers KE, McGregor MJ, Wallis SC, Paterson DL, Lipman J, Roberts JA, Landersdorfer CB. 2016. Effect of different renal function on antibacterial effects of piperacillin against Pseudomonas aeruginosa evaluated via the hollow-fibre infection model and mechanism-based modelling. J Antimicrob Chemother 71:2509–2520. doi: 10.1093/jac/dkw153. [DOI] [PubMed] [Google Scholar]
- 65.Cadwell J. 2012. The hollow fiber infection model for antimicrobial pharmacodynamics and pharmacokinetics. Adv Pharmacoepidem Drug Safety 1:2167–1052. doi: 10.4172/2167-1052.S1-007. [DOI] [Google Scholar]
- 66.Cheah S-E, Bulitta JB, Li J, Nation RL. 2014. Development and validation of a liquid chromatography-mass spectrometry assay for polymyxin B in bacterial growth media. J Pharm Biomed Anal 92:177–182. doi: 10.1016/j.jpba.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9:E60–E83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bulitta JB, Landersdorfer CB. 2011. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bulitta JB, Ly NS, Yang JC, Forrest A, Jusko WJ, Tsuji BT. 2009. Development and qualification of a pharmacodynamic model for the pronounced inoculum effect of ceftazidime against Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:46–56. doi: 10.1128/AAC.00489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulitta JB, Zhao P, Arnold RD, Kessler DR, Daifuku R, Pratt J, Luciano G, Hanauske A-R, Gelderblom H, Awada A. 2009. Mechanistic population pharmacokinetics of total and unbound paclitaxel for a new nanodroplet formulation versus Taxol in cancer patients. Cancer Chemother Pharmacol 63:1049–1063. doi: 10.1007/s00280-008-0827-2. [DOI] [PubMed] [Google Scholar]
- 71.Tsuji BT, Okusanya OO, Bulitta JB, Forrest A, Bhavnani SM, Fernandez PB, Ambrose PG. 2011. Application of pharmacokinetic-pharmacodynamic modeling and the justification of a novel fusidic acid dosing regimen: raising Lazarus from the dead. Clin Infect Dis 52:S513–S519. doi: 10.1093/cid/cir166. [DOI] [PubMed] [Google Scholar]
- 72.Landersdorfer CB, Kirkpatrick CM, Kinzig-Schippers M, Bulitta JB, Holzgrabe U, Drusano GL, Sörgel F. 2007. Population pharmacokinetics at two dose levels and pharmacodynamic profiling of flucloxacillin. Antimicrob Agents Chemother 51:3290–3297. doi: 10.1128/AAC.01410-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meagher AK, Forrest A, Dalhoff A, Stass H, Schentag JJ. 2004. Novel pharmacokinetic-pharmacodynamic model for prediction of outcomes with an extended-release formulation of ciprofloxacin. Antimicrob Agents Chemother 48:2061–2068. doi: 10.1128/AAC.48.6.2061-2068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.