Abstract
The CPCA protein of the filamentous fungus Aspergillus nidulans is a member of the c-Jun-like transcriptional activator family. It acts as central transcription factor of the cross-pathway regulatory network of amino acid biosynthesis and is functionally exchangeable for the general control transcriptional activator Gcn4p of Saccharomyces cerevisiae. In contrast to GCN4, expression of cpcA is strongly regulated by two equally important mechanisms with additive effects that lead to a fivefold increased CPCA protein amount under amino acid starvation conditions. One component of cpcA regulation involves a transcriptional autoregulatory mechanism via a CPCA recognition element (CPRE) in the cpcA promoter that causes a sevenfold increased cpcA mRNA level when cells are starved for amino acids. Point mutations in the CPRE cause a constitutively low mRNA level of cpcA and a halved protein level when amino acids are limited. Moreover, two upstream open reading frames (uORFs) in the 5′ region of the cpcA mRNA are important for a translational regulatory mechanism. Destruction of both short uORFs results in a sixfold increased CPCA protein level under nonstarvation conditions and a 10-fold increase under starvation conditions. Mutations in both the CPRE and uORF regulatory elements lead to an intermediate effect, with a low cpcA mRNA level but a threefold increased CPCA protein level independent of amino acid availability. These data argue for a combined regulation of cpcA that includes a translational regulation like that of yeast GCN4 as well as a transcriptional regulation like that of the mammalian jun and fos genes.
INTRODUCTION
The availability of protein precursors as amino acids or charged tRNAs is a prerequisite for efficient translation. The system that regulates the synthesis of these precursors is known as general control of amino acid biosynthesis in the yeast Saccharomyces cerevisiae (Hinnebusch, 1984) and as cross-pathway control in the filamentous fungi Neurospora crassa and Aspergillus nidulans (Carsiotis et al., 1974; Piotrowska, 1980; Sachs, 1996; Davis, 2000). These regulatory networks, which become activated under amino acid starvation conditions, have been studied most thoroughly in yeast, in which several trans-acting factors have been identified (Hinnebusch, 1992). The transcriptional activator Gcn4p was characterized as the central control element (Hinnebusch, 1984, 1997). Gcn4p has a large activation domain in the N-terminal half of the protein, which itself is subdivided into an activation domain and a central acidic activation domain, both possessing similar activation activities (Drysdale et al., 1998). A leucine-zipper structure important for dimerization and a basic DNA binding domain are localized at the C terminus (Ellenberger et al., 1992). These sequences of the C-terminal half characterize Gcn4p as a member of the bZIP-type transcriptional activator family. Homologs of GCN4, whose products contain the typical bZIP-type structures, were isolated in A. niger (cpcA; Wanke et al., 1997), N. crassa (cpc-1; Paluh et al., 1988; Paluh and Yanofsky, 1991), and Cryphonectria parasitica (cpCPC1; Wang et al., 1998). Detailed functional analyses of domains of fungal bZIP proteins have previously been performed for Gcn4p (Kornitzer et al., 1994; Drysdale et al., 1998). bZIP proteins are also known in higher organisms. Here, proteins of the Jun and Fos family are involved in various cellular proliferation and differentiation processes (Liebermann et al., 1998; Lee et al., 1999). The characteristic bZIP domains are functionally exchangeable between Gcn4p and human c-Jun, indicating that the DNA recognition sequences for both proteins are conserved (Struhl, 1987).
The expression of Gcn4p is strongly regulated and depends on the intracellular concentration of protein precursors. When amino acids are abundant, Gcn4p is almost absent in the cell, but the amount of Gcn4p becomes strongly induced when protein precursors are limited. An increased amount of Gcn4p activates transcription of ∼50 target genes by binding to a cis-acting palindromic sequence motif 5′-ATGA(C/G)TCAT-3′ (general control recognition element) in their promoters (Hinnebusch, 1984; Thireos et al., 1984; Arndt and Fink, 1986). These target genes encode amino acid biosynthetic enzymes, aminoacyl-tRNA synthetases, and proteins involved in purine biosynthesis (Mirande and Waller, 1988; Mösch et al., 1991; Hinnebusch, 1997). Identical general control recognition elements in promoter sequences of genes involved in different biosynthetic pathways lead to an increased production of most amino acids even when only a single amino acid is deficient. The induction of Gcn4p expression in response to amino acid limitation is translational, mediated by four short open reading frames (uORFs) located 150–360 nucleotides upstream of the authentic initiation codon in the leader of the GCN4 mRNA. Eliminating translation of all four uORFs results in high-level GCN4 expression under both starvation and nonstarvation conditions without altering the GCN4 mRNA level (Mueller and Hinnebusch, 1986). The uORFs inhibit GCN4 translation in nonstarved cells when the concentration of ternary complexes is low by reduction of the reinitiation rate at the actual GCN4 ORF. The first and the fourth uORF seem to be sufficient for this regulation function. In addition to the predominant translational control of GCN4 expression, other mechanisms for regulation of the Gcn4p level and Gcn4p activity have been identified. It was shown that the Gcn4p level can be modulated by increasing its half-life under amino acid starvation conditions in auxotrophic mutants (Kornitzer et al., 1994; Prendergast et al., 1996; Meimoun et al., 2000). In addition, a negative regulator of Gcn4p protein activity, Cpc2p, has been isolated in yeast. This protein reduces Gcn4p transactivation activity under nonstarvation conditions without affecting the translational regulation of GCN4 (Hoffmann et al., 1999). The details of both mechanisms remain to be elucidated. Variations in GCN4 mRNA level are barely detectable and of minor importance (Albrecht et al., 1998).
In contrast, little is known about the regulation of the homologous transcriptional activators in filamentous fungi. Preliminary observations suggested a putative translational regulation via uORFs as well as a transcriptional regulation (Paluh et al., 1988; Luo et al., 1995; Wanke et al., 1997; Chen et al., 1998), but experiments to confirm these hypotheses have not been reported. In this study, we have isolated the homologous cross-pathway transcriptional activator gene cpcA of A. nidulans and characterized its expression by introduction of point mutations in upstream regulatory sequences. Our data suggest that regulation of CPCA expression is more complex than in yeast. Two regulatory mechanisms, one transcriptional and the other translational, are additive and dependent on each other. Only if both mechanisms are functional is A. nidulans able to provide sufficient protein precursors when availability is limited.
MATERIALS AND METHODS
Strains, Media, and Genetic Techniques
The A. nidulans strain A234 (yA2; pabaA1; veA1) was obtained from the Fungal Genetics Stock Center (University of Kansas, Lawrence, KS) and used as a wild-type control. Strain GR5 (wA3; pyrG89; pyroA4; veA1) was obtained from G. May (The University of Texas, Houston, TX). Cultivation of A. nidulans strains was performed at 30°C on minimal medium (Bennett and Lasure, 1991). Transformation was carried out as described (Punt and van den Hondel, 1992). Transformants were selected either for the presence of the ble (phleomycin-resistance) gene of Streptoalloteichus hindustanus on minimal medium containing 10 μg/ml phleomycin (Cayla, F) or on medium without uridine to select for the presence of the prototrophic marker pyrG. Expression of the alcA promoter was induced on media with 2% ethanol as sole carbon source. Amino acid starvation was induced by addition of the histidine analog 3-amino-1,2,4-triazole (3AT) at a concentration of 5 mM to solid medium and 10 mM to liquid medium. Cultures on 3AT plates were transferred at 2-day intervals to fresh 3AT plates because the amino acid analog is degraded during prolonged incubation. All yeast strains were derivatives of the S. cerevisiae strain S288C. Wild-type strain H1515 (leu2-3112 ura3-52 trp1 GAL2) was obtained from A. Hinnebusch (National Institute of Child Health and Human Development, Bethesda, MD). H1515, RH1378 (ura3Δ gcd2-1) (Mösch et al., 1990), and RH1408 (ura3-52 gcn4-103 GAL2) (Hinnebusch, 1985) were cultivated on minimal medium (Miozzari et al., 1978). Yeast cells were made competent for transformation by treatment with lithium acetate (Ito et al., 1983). Escherichia coli strain DH5α was used for plasmid propagation.
Isolation of cpcA
A full-length cDNA clone (pME1702) of cpcA was isolated by complementation of a yeast gcn4Δ deletion mutant strain (RH1408) with the use of an A. nidulans inducible cDNA expression library adapted for yeast (Krappmann et al., 1999). Of 10,000 transformants tested for growth under amino acid starvation conditions induced by 15 mM 3AT, two transformants were isolated. Both revealed the same cpcA cDNA sequence. For isolation of a full-length genomic clone, genomic DNA of A. nidulans was digested with PstI and fractionated for Southern hybridization analysis (Sambrook et al., 1989). The cDNA fragment was used as a radiolabeled probe (Feinberg and Vogelstein, 1984) and hybridized to a 2.7-kb PstI genomic DNA fragment. PstI digested DNA in the size range of 2–4 kb was subcloned into pBluescript SK+. E. coli colonies were screened and positive clones were isolated. The genomic PstI DNA fragment on plasmid pME1700 contained the whole cpcA gene and 1 kb of downstream flanking sequences.
Plasmid Construction
For construction of the cpcA deletion plasmid pME1713, a 1.2-kb region upstream of the cpcA open reading frame was amplified by polymerase chain reaction (PCR). This fragment was blunt ended with Klenow enzyme and integrated into the StuI digested plasmid pAN8–1 (Punt and van den Hondel, 1992) in front of the gpdA promoter driving the phleomycin reading frame. An ∼0.9-kb fragment downstream of the cpcA open reading frame was PCR amplified. The ends were filled with the use of Klenow enzyme and cloned into the blunt-ended XbaI site of the same plasmid downstream of the trpC terminator of the phleomycin resistance cassette.
For integration of the cpcA gene into the cpcAΔ mutant strain the cpcA gene was amplified by PCR and cloned as a 2.7-kb XbaI/BamHI DNA fragment into plasmid pRG3 bearing the pyrG gene as selectable marker (Waring et al., 1989) to give plasmid pME1707.
For in vivo analysis, cpcA gene constructs were created with single nucleotide substitutions in the CPCA recognition element (CPRE), uORFs, or both. The CPRE1 sequence 5′-TTGACTCT-3′was mutated to 5′-TTCTCTCT-3′, the CPRE2 sequence 5′-ATGACTCA-3′ to 5′-ATCTCTCA-3′. Both AUG start codons of the uORFs were converted to ACG by PCR. The PCR products were cloned as XbaI/BamHI fragments into the plasmid pRG3 (Waring et al., 1989) to give plasmids pME1708 (CPRE1−), pME1709 (CPRE2−), pME1710 (CPRE1−/2−) pME1711 (uORF1−/2−), and pME1712 (CPRE1−/2−, uORF1−/2−).
The cpcA open reading frame was amplified for overexpression analysis with the use of the cpcA cDNA as template. This was blunt ended with Klenow enzyme and cloned into the SmaI restriction site behind the inducible A. nidulans alcA promoter of plasmid pME1565. This plasmid was constructed by replacing the KpnI/BamHI green fluorescent protein fragment of plasmid pMCB32 (Fernandez-Abalos et al., 1998) with the KpnI/BamHI fragment of the multiple cloning site of pBluescipt SK+ (Stratagene, Heidelberg, Germany). The expression plasmid of cpcA was named pME1603.
Deletion of cpcA and cpcA Strain Construction
The cpcA deletion plasmid pME1713 was cut with PstI, resulting in a 5.4-kb cpcA deletion cassette and transformed into A. nidulans strain A234. Transformants were selected on minimal medium containing 10 μg/ml phleomycin (Cayla, F) and tested for homologous or ectopic integration with the use of three independent primers in PCR experiments in parallel. One primer (5′-GGAAGGCTTCGGTGAGGA-3′) was located in the cpcA open reading frame. This sequence would be deleted after homologous integration. A second primer (5′-CTCCGTAACACCCAATAC-3′) corresponded to the trpC terminator of the cpcA disruption cassette, and a third primer (5′-GTGCTATATTAAAGGGTGATGT-3′) hybridized to the A. nidulans DNA downstream of the 3′-genomic DNA fragment used for the construction of the cpcA disruption. Eighty transformants of the A. nidulans wild-type strain A234 were tested. Homologous integration of the disruption cassette resulted in a smaller PCR band than ectopic integration. Two transformants with single homologous integrations were verified by PCR and Southern hybridization. Both cpcAΔ mutant strains had identical phenotypes. Strain AGB51 (Table 1) was used for further experiments.
Table 1.
A. nidulans strains and genotypes
| Strain | Genotype | Reference |
|---|---|---|
| A234 | pabaA1; yA2; veA1 | FGSC |
| GR5 | pyroA4; pyrG89; wA3; veA1 | G. May |
| AGB51 | cpcAΔ∷phleR; pabaA1; yA2; veA1 | This work |
| AGB52 | cpcAΔ∷phleR; pyrG89, pabaA1; yA2; veA1 | This work |
| AGB54 | cpcAΔ∷phleR; cpcA∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB55 | cpcAΔ∷phleR; cpcA-CPRE1−∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB57 | cpcAΔ∷phleR; cpcA-CPRE2−∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB58 | cpcAΔ∷phleR; cpcA-CPRE1−/2−∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB60 | cpcAΔ∷phleR; cpcA-uORF1−/2−∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB61 | cpcAΔ∷phleR; cpcA-uORF1−/2− + CPRE1−/2−∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB68 | cpcAΔ∷phleR; alcA(p)-cpcA-his2B8t)∷pyrG; pyrG89; pabaA1; yA2; veA1 | This work |
| AGB121 | alcA(p)-his2B8t)∷pyrG; pyrG89; pabaA1; yA2; veA1 | Valerius et al., 2001 |
A cpcAΔ; pyrG89 mutant strain (AGB52) was isolated from a cross between AGB51 and GR5. AGB52 was then used for integration of the cpcA overexpression plasmid pME1603, resulting in strain AGB68. As a control, strain GR5 was transformed with the empty expression plasmid pME1565, yielding AGB121. Plasmids pME1707 to pME1712 containing the whole cpcA gene and the cpcA with point mutations in the CPRE1, CPRE2, CPRE1, and two, uORF1 and 2, or in all four regulatory sequences were transformed into AGB52 to yield strains AGB54, AGB55, AGB57, AGB58, AGB60, and AGB61 (Table 1). All transformants were tested in Southern hybridization experiments for single ectopic integration events without consideration of the actual integration site.
Recombinant DNA Techniques
Unless otherwise stated, standard procedures were used (Sambrook et al., 1989). DNA was sequenced with the use of the dideoxy chain-terminating method (Sanger et al., 1977), with custom oligonucleotides and the T7 sequencing kit (Amersham Pharmacia Biotech, Freiburg, Germany).
For Northern hybridization analysis, total RNA was isolated with TRIZOL reagent (Invitrogen, Carlsbad, CA). Total RNA per lane (20 μg) was separated on a formaldehyde agarose gel, electroblotted onto a nylon membrane (BiodyneB; Pall Corp., Ann Arbor, MI) and hybridized with α-[32P]dATP-labeled DNA fragments according to Feinberg and Vogelstein (1984).
Protein Methods
Protein contents were estimated according to Bradford (1976). Ornithine transcarbamylase (OTCase) activities were assayed in crude extracts as described (Davis, 1962). Appropriate liquid medium (50 ml) was inoculated with ∼2 × 108 conidia and cultivated overnight at 30°C. Mycelia was harvested by filtration and either directly used for preparation of crude extracts or used as inoculum for repressing or derepressing medium, which was then cultivated further for the desired time periods. Specific isocitrate dehydrogenase activities were measured as described (Flavell and Fincham, 1968). A. nidulans wild-type strain A234 and the cpcA mutant strain were grown overnight at 30°C on 2% glucose and 40 mM acetate, respectively. Yeast and A. nidulans crude protein extracts were prepared as described for gel retardation and Western blot analysis (Arndt et al., 1987). Detection of Gcn4p in Western blot analyses was performed with a polyclonal antibody against the 60 C-terminal amino acids of Gcn4p (generation of the antibody described in Albrecht et al., 1998).
Primer Extension
The transcriptional start site of cpcA mRNA was determined in primer extension experiments with the use of two different primers, BH80 (5′-GTCCGTCTCAACTGAGAAGAACGACGTAAC-3′) and BH81 (5′-GGTGCGCTTAGCGTCCATTTTGAGCTGGAT-3′), located 92 and 58 bp downstream of the cpcA mRNA 5′ end. For each reaction 5 μg of total RNA of A. nidulans strain A234 was used. Sequencing reactions and primer extension probes were end labeled with [γ-32P]ATP and separated on a polyacrylamide gel.
Gel Retardation Analysis
Crude protein extracts were isolated from yeast wild-type H1515 and gcd2 mutant strain RH1378 grown under nonstarvation and amino acid starvation conditions. The A. nidulans cpcA cDNA (pME1702) was expressed in the gcn4Δ mutant strain RH1408 under control of the GAL1 promoter. Protein extracts were isolated after growth for 8 h with glucose or galactose as sole carbon source, respectively. Proteins from the yeast gcn4Δ mutant strain were used as negative control. Purification of Gcn4 protein expressed in E. coli was described previously (Braus et al., 1989). Protein extracts were incubated in the presence of an end-labeled 151-bp cpcA promoter fragment spanning the nucleotides −1165 to −1015. This fragment contained either both wild-type CPREs (5′-TTGACTCT-3′ and 5′-ATGACTCA-3′), point mutations in the first CPRE (5′-TTCTCTCT-3′ and 5′-ATGACTCA-3′), point mutations in the second CPRE (5′-TTGACTCT-3′ and 5′-ATCTCTCA-3′), or mutations in both (5′-TTCTCTCT-3′ and 5′-ATCTCTCA-3′). Protein extracts (20 μg) were incubated with 10 fmol of a radiolabeled probe, separated on a native 6% polyacrylamide gel, and visualized by autoradiography.
GenBank Accession Number
The nucleotide sequence reported in this article has been submitted to the GenBank nucleotide sequence database with accession number AF302935
RESULTS
Isolation of cpcA gene of A. nidulans
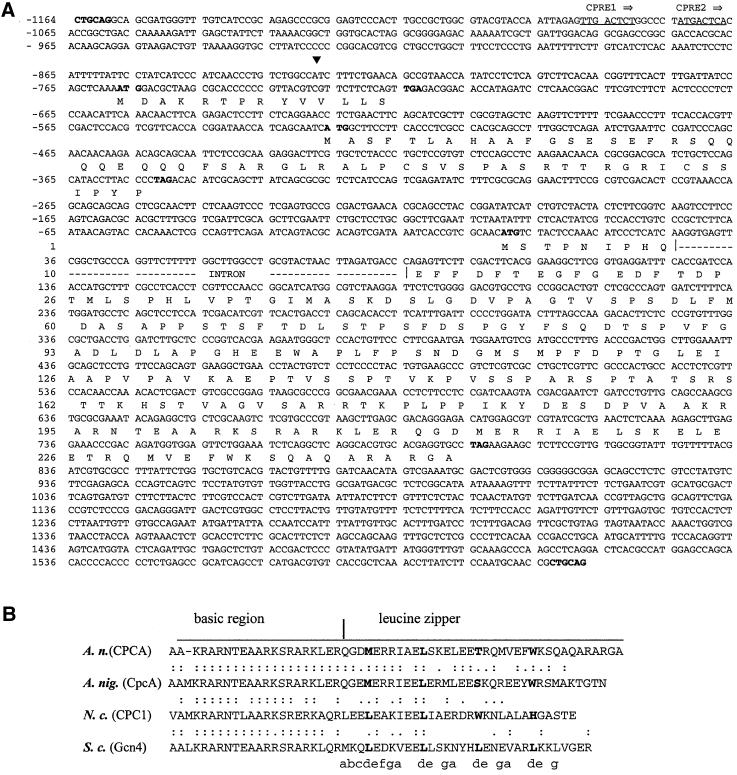
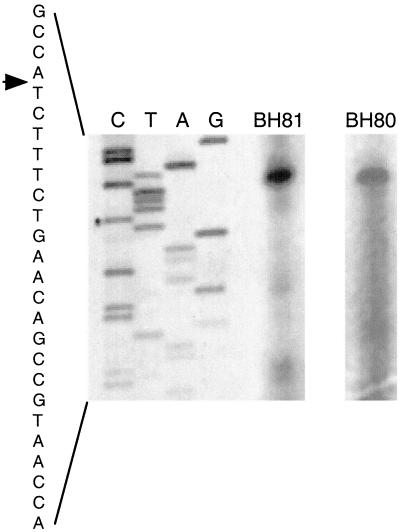
A cpcA cDNA of A. nidulans restored the ability of a yeast gcn4Δ strain to grow under amino acid starvation conditions caused by 10 mM 3AT. This cDNA was further used to isolate a genomic cpcA clone (see MATERIALS AND METHODS). Complementation of the gcn4 mutant growth phenotype with the cDNA clones resulted in a growth rate under amino acid starvation conditions identical to that seen after transformation with GCN4 itself. Comparison between the cDNA and genomic DNA sequences identified one intron in the open reading frame at position 28 relative to the AUG start codon (Figure 1A). This position in the very beginning of the cpcA ORF and the length of the intron (59 bp) are similar to the introns of the homologs in A. niger (cpcA), N. crassa (cpc-1), and C. parasitica (cpCPC1). Primer extension experiments and cDNA clones marked the transcriptional start site at position − 828 relative to the first nucleotide of the AUG start codon (Figure 2).
Figure 1.
DNA and deduced amino acid sequences of A. nidulans cpcA. (A) Nucleotide sequence of the PstI DNA fragment containing the entire cpcA gene. The transcriptional start site is indicated by a solid triangle. The CPREs are underlined. PstI restriction sites as well as the start and stop codons of uORF1, uORF2, and the cpcA open reading frame are in bold. The cpcA intron interrupting the open reading frame is indicated by a broken line. The numbers of nucleotide and amino acid sequences are shown on the left relative to the AUG start codon of cpcA. (B) Alignment of the deduced amino acid sequences of the bZIP motif from CPCA amino acid positions 189–245 at the C terminus of the deduced protein (A. nidulans), CpcA (A. niger), CPC1 (N. crassa), and Gcn4p (S. cerevisiae). The basic DNA-binding domain and the leucine-zipper region are indicated by a horizontal bar. Amino acid residues at heptad positions (d) in the leucine-zipper region are shown in bold. Amino acid residues at the e and g positions are supposed to participate in dimer stabilization and electrostatic interactions (Durr et al., 1999).
Figure 2.
Determination of the transcriptional start site of the cpcA gene. Primer-extension experiments were performed with two cpcA-specific primers, BH80 and BH81, and 5 μg of total RNA. Both primers were also used for sequencing reactions to determine the transcriptional start site by comparison. The sequencing reaction performed with primer BH81 is presented. The primer extension experiment performed with BH80 is arranged to the sequence of BH81 without changing the position concerning the nucleotide sequence. The arrow indicates the transcriptional start site in the cpcA sequence.
Known fungal transcriptional activators of amino acid regulatory networks are characterized by small uORFs that are important for translational regulation. At positions −756 and −525 there are two uORFs in the 5′-untranslated region of the cpcA gene. These uORFs encode small putative proteins of 14 and 58 amino acids, respectively. The predicted amino acid sequence of the second uORF is 45% identical to that of uORF2 of A. niger cpcA.
The main A. nidulans cpcA ORF encodes a 245 amino acid protein with strong similarities to yeast Gcn4p (40% identity), N. crassa CPC1 (40% identity), A. niger CpcAp (50% identity), and C. parasitica CPC1 (40% identity). Like Gcn4p and its other homologs, A. nidulans CPCA has a basic leucine-zipper structure at the carboxyl-terminal end. Although the basic region is highly conserved, the leucine-zipper structure is more similar to the unusual leucine-zipper of the transcriptional activator CpcAp of A. niger, which has only one leucine instead of the four to five repeated leucines in the Gcn4p and c-Jun proteins (Figure 1). The structural and functional similarities between the cpcA and its homologs suggested that it is cross-pathway control transcriptional activator gene of A. nidulans.
cpcA Deletion Results in Sensitivity to Amino Acid Analogs
We replaced the cpcA ORF in strain A234 by a phleomycin-resistance expression cassette, and afterward additionally introduced the pyrG89 marker to allow testing of the effects of various cpcA mutant alleles (see MATERIALS AND METHODS). Growth on minimal medium of the cpcAΔ mutant strains AGB51 (pyrG+) and AGB52 (pyrG89) was reduced to 80% compared with the parental wild-type strains A234 or GR5. The deletion did not result in any obvious morphological phenotype in asexual or sexual structures of the fungus.
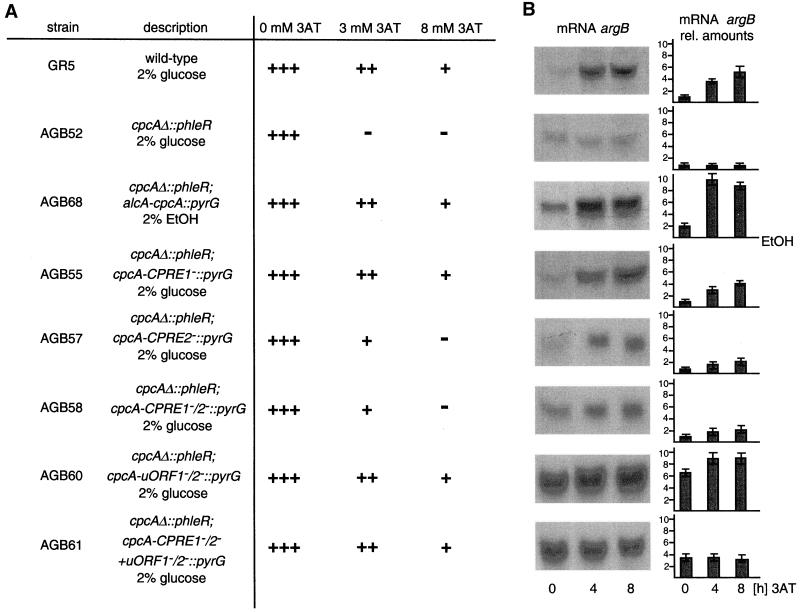
The effect of the cpcA deletion on the ability to grow under amino acid starvation conditions was analyzed (Figure 3). Starvation conditions were induced by the histidine analog 3AT, which acts as a false feedback inhibitor of the histidine biosynthetic enzyme imidazolglycerol phosphate dehydratase (EC: 4.2.1.19) and thereby induces amino acid starvation (Klopotowski and Wiater, 1965). Strain A234 was able to grow on 3AT concentrations of up to 8 mM, but in mutant strain AGB52, growth was abolished at 3 mM 3AT. In addition, amino acid starvation induced by the amino acid analog 5-methyl-tryptophan, a false feedback inhibitor of tryptophan biosynthesis, resulted in a similar phenotype. The sensitivity of the cpcA mutant strain could be reversed by the addition of histidine to the 3AT-containing growth medium. These data suggest that the growth phenotype is caused primarily by histidine starvation and not by other toxic effects of the analog.
Figure 3.
Influence of various cpcA alleles on growth and mRNA amounts of the cross-pathway regulated argB gene. (A) Conidia of A. nidulans wild-type strain A234 and the cpcA mutant AGB51 (cpcAΔ) were dropped on minimal medium containing 0, 3, or 8 mM 3AT, respectively, to induce amino acid starvation. Colony growth was scored after incubation for 5 d at 30°C and is indicated as +++ for normal growth, ++ for reduced, + for barely growing colonies, and − for no growth. In strain AGB68 (cpcAΔ; alcA-cpcA), the open reading frame of A. nidulans cpcA is expressed under the control of the alcA promoter in the cpcA mutant background. Growth of the wild-type and the cpcAΔ mutant strain were unaffected by ethanol as carbon source. Strains AGB55 (cpcAΔ; cpcA-CPRE1−), AGB57 (cpcAΔ; cpcA-CPRE2−), AGB58 (cpcAΔ; cpcA-CPRE1−/2−), AGB60 (cpcAΔ; cpcA-uORF1−/2−), and AGB61 (cpcAΔ; cpcA-CPRE1−/2− + uORF1−/2−) were analyzed similarly. (B) RNAs were isolated from the strains described in A after growth under amino acid starvation conditions for 0, 4, and 8 h, respectively. mRNA amounts were equalized in parallel with the use of the constitutively expressed gpdA gene as probe. argB mRNAs are shown as autoradiography and as fold differences setting 0 3AT level of the wild type as 1.0.
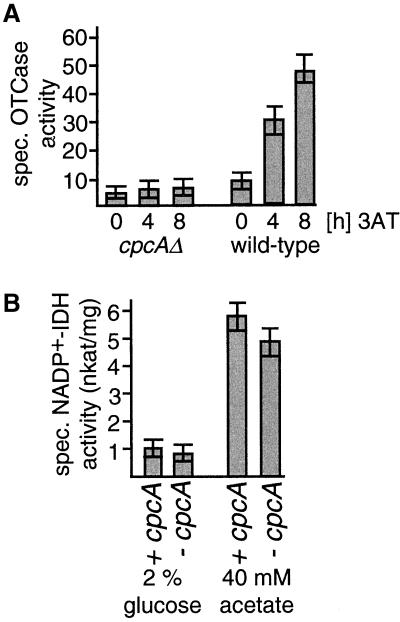
The effect of the cpcA deletion on the expression of cross-pathway regulated genes was also analyzed at the mRNA and protein levels. Strains A234 and AGB52 were cultivated in the absence or presence of 3AT. In wild-type cells, the induction of cross-pathway control by 3AT resulted in a fivefold increased mRNA level of the argB gene (required for arginine biosynthesis) (Piotrowska, 1980; Goc and Weglenski, 1988). In contrast, the argB mRNA levels of the cpcA mutant were not increased by amino acid limitation (Figure 3). In addition, the specific activity of the argB product OTCase (EC: 2.1.3.3.) was measured in mycelial extracts. Hyphae were grown either under nonstarvation conditions or in the presence of 3AT to assay whether the reduced argB transcript levels in the cpcA mutant strain correlated with the enzyme level (Figure 4). OTCase activity in strain A234 was increased by a factor of 5 within 8 h of exposure to the amino acid analog. In contrast, in strain AGB52, the basal level of specific OTCase activity was not increased by amino acid starvation. Similar results were also found for the cross-pathway regulated gene trpC.
Figure 4.
Influence of cpcA expression on enzyme activities of A. nidulans. Specific enzyme activities were determined in crude extracts (see MATERIALS AND METHODS). The values shown are the averages from three independent measurements. SDs did not exceed 15%. (A) Specific OTCase activities in the presence or absence of amino acid limitation. Wild-type strain A234 and the cpcA mutant AGB51 were grown overnight at 30°C in minimal glucose medium. Histidine starvation was induced by supplementation with 3AT to a final concentration of 10 mM for the indicated periods of time. (B) Specific isocitrate dehydrogenase (NADP+-IDH) activities under glucose and acetate growth conditions. Strains A234 and AGB51 were grown overnight at 30°C in minimal medium with 2% glucose or 40 mM acetate as carbon source.
As an additional control, the specificity of the CPCA protein as a transcriptional activator of cross-pathway control–regulated genes was tested by measuring the enzymatic activity of isocitrate dehydrogenase (EC: 1.1.1.41). This protein is involved in the tricarboxylic acid cycle and is unaffected by amino acid starvation (Barthelmess, 1982; Kelly and Hynes, 1982). As shown in Figure 4, during growth on either acetate or glucose, isocitrate dehydrogenase activity was unaffected in the cpcAΔ mutant strain compared with wild-type. Taken together, these data support the conclusion that cpcA encodes the transcriptional activator of the cross-pathway control of amino acid biosynthesis.
Overexpression of cpcA Leads to Constitutively High Transcript Levels of Target Genes
Induction of the cross-pathway control leads to increased expression of CPCA-regulated genes in A. nidulans. AlcA-regulated expression of cpcA in the cpcAΔ mutant strain AGB68 was analyzed to investigate the effect of constitutively high CPCA protein levels without induction of the cross-pathway regulatory network by amino acid limitation. alcA transcription is turned on by ethanol and is repressed by glucose (Lockington et al., 1985). Constitutively high levels of CPCA reduced the growth rate by 20% compared with wild type under nonstarvation conditions. Under starvation conditions, the growth rates were either indistinguishable or were higher for the induced strain. Expression of cpcA in the cpcA mutant strain allowed growth on medium with up to 8 mM 3AT (Figure 3). Growth of the control wild-type strain AGB121 was unaffected when the carbon source was changed from glucose to ethanol. Because argB is a target gene of CPCA, growth of the alcA-cpcA strain AGB68 under alcA induction conditions led to a 10-fold increased argB mRNA level (Figure 3).
These data show that expression of cross-pathway genes is regulated by the central transcriptional activator cpcA. In addition, a block in sexual development at the microcleistothecia stage when cpcA was overexpressed corroborated previous studies, indicating a connection between amino acid biosynthesis and sexual development in A. nidulans (Eckert et al., 1999; Hoffmann et al., 2000).
cpcA Is Transcriptionally Autoregulated by CPCA Protein
GCN4 is regulated at the translational level, and variations in its mRNA level are barely detectable and of minor importance (see INTRODUCTION). However, hints of an additional transcriptional regulation were found for the N. crassa and A. niger cpc-1 and cpcA genes. Thus, we investigated this possibility also in A. nidulans.
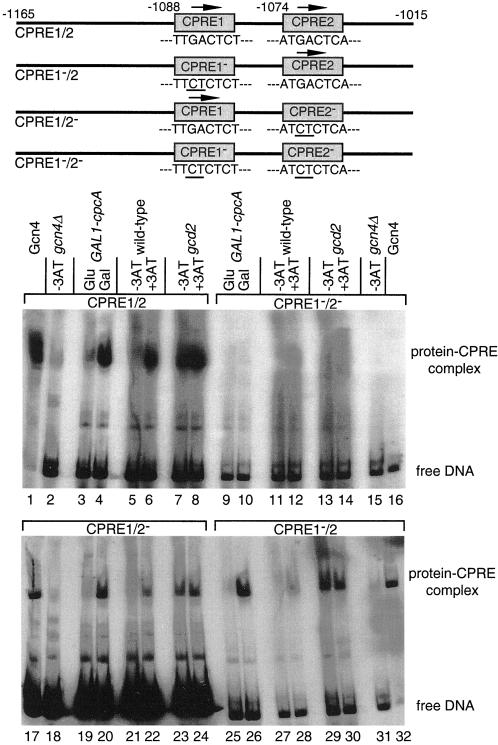
Eight hours of growth in the presence of 3AT increased the cpcA mRNA steady-state level eightfold (Figure 5A). At positions −1074 and −1088 relative to the translational start codon, cpcA has two putative binding sites for its own gene product, CPREs, similar to the consensus sequence of the binding site for Gcn4p, (A)TGACTC(AT) (Arndt and Fink, 1986; Hinnebusch, 1992). Such CPREs have also been found in the promoters of the cross-pathway–regulated genes argB, trpC, and hisHF (Hamer and Timberlake, 1987; Goc and Weglenski, 1988; Eckert et al., 1999; Valerius et al., 2001), and we further investigated in vitro those of cpcA by gel retardation experiments (Figure 6). Formation of protein–DNA complexes was investigated with CPCA and Gcn4p. Purified Gcn4p (Braus et al., 1989) and crude protein extracts of a yeast gcd2 mutant strain expressing high levels of Gcn4p served as positive controls. For a controlled expression of CPCA the gcn4Δ yeast strain (RH1408) with an extrachromosomal GAL1-cpcA cDNA-plasmid was either grown on glucose (repression) or galactose (activation). CPCA if expressed on galactose medium as well as Gcn4p of wild-type strain H1515 during growth on 3AT bound to the cpcA promoter fragment with both CPREs (Figure 6, lanes 4 and 6). Point mutations in one or the other CPRE did not prevent this complex formation (Figure 6, lanes 20, 22, 26, and 28), but mutations in both of them did (Figure 6, lanes 10 and 12). Here, no formation of specific DNA–protein complexes was observed (Figure 6, upper right set of lanes). Therefore, CPCA seems to bind to both of the CPRE elements of its own promoter in vitro.
Figure 5.
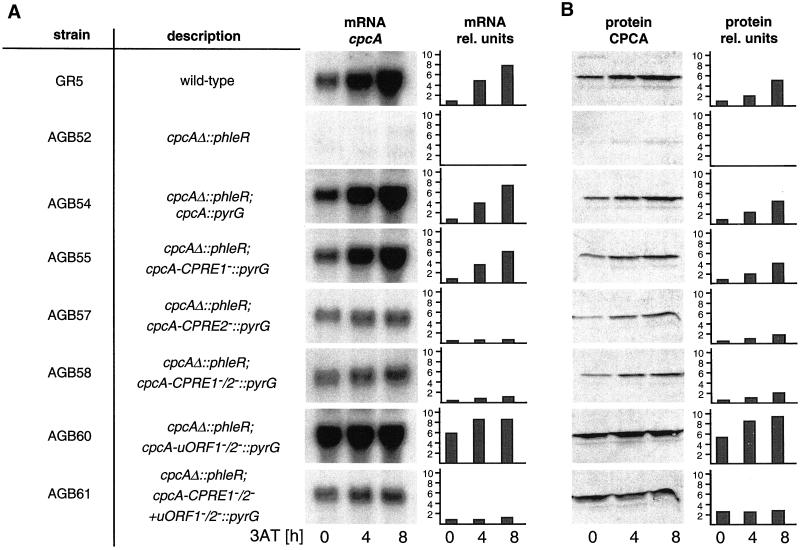
Regulation of cpcA expression in A. nidulans. (A) Analysis of cpcA mRNA levels. Wild-type strain A234 and the cpcA mutant strains AGB51, AGB54, AGB55, AGB57, AGB58, AGB60, and AGB61 (Table 1) were grown overnight at 30°C in liquid minimal medium with glucose. At time 0, the amino acid analog 3AT was added to 10 mM to induce histidine starvation. Mycelia were harvested at the indicated times. The amount of RNA per lane was equalized by phosphorimaging with the use of the constitutively expressed gpdA gene as control probe and was then probed with the use of radioactively labeled cpcA cDNA. (B) Western analysis of CPCA. Crude protein extracts of the strains described in A were isolated after growth under nonstarvation (0) and 4 and 8 h under amino acid starvation conditions. CPCA amounts were determined by cross-reaction of 20 μg of crude protein extract of each strain with the Gcn4-specific rabbit antibodies. Quantification of CPCA protein was performed by comparison with a standardized Western hybridization with the use of different amounts of purified yeast Gcn4. In addition, a nonspecific band of lower molecular weight was used as internal standard. All Northern hybridizations and Western blots were performed in parallel on the same membranes to allow a comparison between cpcA alleles. cpcA mRNA and CPCA protein amounts are shown as autoradiography and as relative units. The amounts for wild type under nonstarvation conditions were set as 1.
Figure 6.
In vitro DNA binding activity of Gcn4p and CPCA derived from various yeast extracts. DNA fragments used and the positions of integrated point mutations are shown schematically. A 151-bp DNA fragment from the cpcA promoter was used that contained both putative CPCA recognition elements (CPRE1/2, lanes 1–8). Additionally, point mutations were integrated in CPRE1 (CPRE1−/2, lanes 25–32), CPRE2 (CPRE1/2−, lanes 17–24), or both (CPRE1−/2−, lanes 9–16). All DNA fragments were radioactively labeled and incubated with 10 μg of protein extracts. Crude protein extracts of yeast wild-type H1515 and the gcd2 mutant strain RH1378 were isolated after 8 h of growth under nonstarvation (−3AT) and amino acid starvation conditions (+3AT). CPCA of A. nidulans was expressed under control of the yeast GAL1 promoter in the yeast gcn4Δ mutant strain RH1408. Protein extracts were isolated after 8 h on minimal medium containing glucose (Glu) or galactose (Gal) as sole carbon source, respectively. Purified Gcn4 protein expressed in E. coli was used as positive control; protein extracts of the gcn4Δ mutant strain RH1408 were used as negative control. Protein–DNA complexes were separated from unbound DNA by PAGE and visualized by autoradiography.
The function of the CPREs was further tested in vivo. cpcA alleles with single nucleotide substitutions in CPRE1, CPRE2, or both were ectopically integrated as single copies into the cpcAΔ mutant strain AGB52. The reintegrated wild-type cpcA allele (AGB54) as well as the cpcA allele with mutated CPRE1 (AGB55) showed cpcA transcript levels similar to wild-type strain A234 under both nonstarvation and starvation conditions (Figure 5). Growth on 3AT of strain AGB55 was similar to that of wild type (Figure 3). In contrast, strains AGB57 and AGB58, with nucleotide substitutions in CPRE2 and both CPREs, only had slightly increased cpcA mRNA levels during starvation conditions (Figure 5). As a consequence, these strains displayed an elevated sensitivity to 3AT. Also, the halved mRNA level of the CPCA target gene argB correlated with this noninducible cpcA transcription (Figure 3).
A specific anti-Gcn4p antibody (see MATERIALS AND METHODS) that also recognized CPCA on Western blots did not detect any CPCA for the cpcAΔ strain AGB52 (Figure 5). The wild-type strain A234 as well as strain AGB54 with the reintegrated cpcA wild-type allele exhibited a low level of CPCA under nonstarvation conditions, and amino acid starvation increased it fivefold (Figure 5B). Point mutations in CPRE1 (AGB55) only caused a slight reduction in the amount of CPCA under nonstarvation and starvation conditions that did not significantly affect growth under amino acid starvation conditions (Figure 3). In agreement with the cpcA transcript levels shown above, mutation of CPRE2 or both CPREs reduced CPCA levels to half of the wild type's under nonstarvation conditions. Only a twofold increase of this small amount of CPCA was detected upon starvation (Figure 5).
Taken together, these data show that cpcA is autoregulated by its own gene product via CPRE2. This autoregulation seems to be a prerequisite for the induction of the cross-pathway control and is required for growth under amino acid starvation conditions. The regulation of cpcA in A. nidulans therefore seems to be different to that of GCN4 in S. cerevisiae.
Translation of cpcA mRNA Is Inhibited in Presence of Amino Acids
The yeast transcriptional activator gene GCN4 is primarily translationally regulated (see INTRODUCTION). The possibility of a similar translational regulation of cpcA was analyzed by single nucleotide substitutions within the AUG start codons of the deduced uORFs. Strain AGB60 with substitutions from AUG to ACG in both start codons displayed an approximately fivefold increased CPCA level under nonstarvation conditions. This elevated level doubled during amino acid starvation conditions (Figure 5). The mRNA level of the CPCA target gene argB also increased approximately sixfold, and amino acid starvation even gave a further increase yielding a 10-fold higher amount of the wild-type's basal level (Figure 3). Similar results were found for the CPCA-regulated trpC gene. Probably as consequence of this high transcription of cross-pathway target genes, strain AGB60 even grew on 3AT concentrations up to 10 mM. This indicates that a translational regulation takes place controlling cpcA expression via two upstream open reading frames.
Transcriptional Autoregulation and Translational Regulation of cpcA Expression Are Dependent Additive Mechanisms
To determine whether the increased amounts of CPCA for the cpcA allele with the mutated uORFs (AGB60) were caused only by the nonfunctional translational regulation or in addition by an induced transcriptional autoregulation, cpcA mRNA levels were analyzed. Under nonstarvation conditions, cpcA mRNA amounts of the mutant strain were already sixfold higher than in wild type. There was only a slight further increase during starvation conditions (Figure 5). This indicates that translational and transcriptional regulation of cpcA are dependent on each other, and that increased CPCA levels in strain AGB60 are also a result of an induced transcriptional regulation.
To determine the effect of translational control by itself, we determined the expression of the cpcA allele with inactivated uORFs and nonfunctional CPREs (AGB61), and compared it with that of only mutated CPREs (AGB58). The cpcA mRNA levels at nonstarvation conditions of both strains were comparably low and remained almost constant during amino acid starvation. The amount of CPCA protein was low in strain AGB58 but obviously increased in strain AGB61 at nonstarvation conditions; however, because transcriptional autoregulation was impaired, less increased than for the cpcA allele with the mutated uORFs (AGB60) (Figure 5). During starvation conditions no further elevation was detectable for AGB61. Again, these observations were confirmed by a fourfold increased argB mRNA level of strain AGB61, independent of nonstarvation or amino acid starvation conditions (Figure 3).
Taken together, these data argue for an interwoven system of cpcA expression that connects a translational regulation of its mRNA with a transcriptional cpcA autoregulation. Without the transcriptional autoregulatory mechanism the ability of A. nidulans to react to starvation conditions is strongly diminished.
DISCUSSION
Amino acid starvation conditions in ascomycetes activate a complex regulatory network, the general amino acid or cross-pathway control that leads to transcriptional activation of the genes in the pathway. In yeast, the most thoroughly studied of these organisms, the pathway is controlled by GCN4, whose regulation is translational (Hinnebusch, 1997). In this article, we have characterized the homologous transcriptional activator CPCA responsible for cross-pathway control in A. nidulans. By making point mutations in regulatory sequences of cpcA, we show that its expression is not only regulated via the translational mechanism as in yeast but also by an autoregulated transcriptional mechanism. Both mechanisms are physiologically important and work together to affect the CPCA protein level under amino acid starvation conditions.
The ability of the A. nidulans CPCA to substitute for Gcn4p in yeast and to confer resistance to inhibitors of amino acid biosynthetic enzymes indicates that cpcA is a homolog of GCN4 and that the encoded transcriptional activators are functionally conserved. Both proteins share subdomains with high degrees of amino acid conservation. The highest conservation was found for the DNA binding domain. This domain is not only conserved in cross-pathway or general control transcriptional activators but also displays significant identity to other bZIP-type transcriptional activators such as the human Jun and Fos proteins. This high degree of conservation is demonstrated by the apparently identical DNA-binding specificity of Gcn4p and CPCA. In contrast, the C-terminal leucine-zipper of A. nidulans CPCA and the leucine-zipper of A. niger CpcA (Wanke et al., 1997) are the most degenerate of the bZIP-type transcriptional activators. However, mutational analysis of leucine residues in Gcn4p and N. crassa CPC-1 demonstrated that aligned heptad leucine residues are not required for dimerization and protein function (Hinnebusch, 1984; Bohmann et al., 1987; Paluh and Yanofsky, 1991).
The regulation of GCN4 expression in yeast is well studied and occurs mostly at the translational level mediated by the four uORFs present in the 5′-leader sequence (Mueller and Hinnebusch, 1986; Miller and Hinnebusch, 1989; Tzamarias et al., 1989). By demonstrating that cpc-1 transcripts become associated with larger polysomes during amino acid starvation a similar role for the two uORFs in the leader of cpc-1 has been demonstrated for N. crassa (Paluh et al., 1988; Luo et al., 1995). The point mutations in the AUG start codons of both uORFs in A. nidulans cpcA show that its regulation also takes place at the translational level. However, the nature of the mechanism behind, e.g., whether it acts by varying reinitiation rates or by controlling of scanning ribosomes, remains to be shown. Although all uORFs of yeast encode putative polypeptides of only two or three amino acids, respectively, A. nidulans cpcA uORF1 and uORF2 encode 14 and 58 amino acids. Similarly, long uORF-encoded peptides have been identified for N. crassa cpc-1 and A. niger cpcA (Paluh et al., 1988; Wanke et al., 1997). The amino acid identity of ∼50% between the second uORFs of A. niger and A. nidulans CPCA and a conserved glutamine-rich stretch found in both sequences are first hints for a translation resulting in a stable protein with a possible function in protein–protein interactions or stabilization of DNA-binding complexes (Liberati et al., 1999). However, the amino acid identity to the second uORF of N. crassa is <20%, rather low. Yet the distances between the two uORFs and between uORF2 and the cpcA/cpc-1 ORF are similar for A. nidulans and N. crassa.
The presence of a functional CPRE in the promoter of A. nidulans cpcA and the ability of CPCA and Gcn4p to bind this sequence in vitro suggest that an autoregulatory component is involved in cpcA expression. This is coincident with the strong increase of cpcA mRNA level under amino acid starvation conditions. Similar results have previously been shown for N. crassa cpc-1 mRNA (Paluh et al., 1988). The transcriptional and translational regulation of A. nidulans cpcA expression work additively and are dependent on each other. This is shown by the increased cpcA mRNA level when the translational regulation is destroyed and the reduced CPCA protein level for the allele with point mutations in uORFs and CPREs (AGB61) compared with the allele only mutated in both uORFs (AGB60). Interestingly, the CPCA protein level is further increased in strain AGB61 during amino acids starvation, although neither the transcriptional autoregulation nor the translational regulation is functional. This indicates an additional mechanism of cpcA regulation. The degradation of the yeast Gcn4 protein was shown to proceed through the ubiquitin pathway, and Gcn4p stability increases during reduced protein synthesis, e.g., caused by amino acid starvation conditions (Kornitzer et al., 1994). Beside the SCFCDC4 ubiquitination complex, the Gcn4p degradation requires the activity of the cyclin-dependent kinase Pho85p, whose activity toward the transcription factor is reduced in starved cells (Meimoun et al., 2000). A stabilization of CPCA protein stability could also be the reason for the increased protein level of strain AGB61 that has no translational and transcriptional regulation. However, this hypothesis remains to be investigated for A. nidulans.
The reasons for a more complex regulation of A. nidulans cpcA expression in comparison with yeast remain unclear. It could be caused by additional functions of cpcA as indicated for the sexual development (Hoffmann et al., 2000). The expression of several other transcription activator-encoding genes of A. nidulans, such as stuA or brlA, are supposed to be regulated by a combination of an autotranscriptional and a translational mechanism (Miller et al., 1992; Han et al., 1993). This could argue for a more general reason, e.g., increased cell size in comparison with yeast needing two regulatory mechanisms working additively to provide sufficient amounts of the transcriptional activator.
Although yeast GCN4 obviously lacks an autoregulatory transcriptional control element, the jun family homologs are strongly regulated at the transcriptional level and include autoregulation, as shown for the junD. In contrast to A. nidulans cpcA, junD transcriptional autoregulation is important for constitutive expression but not to induce the JunD protein level in reaction to changed environmental conditions (Berger and Shaul, 1994, 1998). It appears that regulation of gene expression within the class of c-Jun-like transcriptional activator-encoding genes ranges from predominantly translational control (yeast GCN4) to a combination of translational and transcriptional mechanisms (A. nidulans cpcA, N. crassa cpc-1) (Sattlegger et al., 1998) to transcriptional regulation (mammalian Jun).
ACKNOWLEDGMENTS
We thank Sabine Eckert and Oliver Draht for critical reading of the manuscript and all other members of the group for helpful discussions. This work was supported by the Deutsche Forschungsgemeinschaft, the Volkswagen-Stiftung, and the Fonds der Chemischen Industrie.
REFERENCES
- Albrecht G, Mösch HU, Hoffmann B, Reusser U, Braus GH. Monitoring the Gcn4 protein-mediated response in the yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:12696–12702. doi: 10.1074/jbc.273.21.12696. [DOI] [PubMed] [Google Scholar]
- Arndt K, Fink GR. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc Natl Acad Sci USA. 1986;83:8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt KT, Styles C, Fink GR. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Barthelmess IB. Mutants affecting amino acid cross-pathway control in Neurospora crassa. Genet Res. 1982;39:169–185. doi: 10.1017/s0016672300020863. [DOI] [PubMed] [Google Scholar]
- Bennett JW, Lasure LL. Growth media. In: Bennett JW, Lasure LL, editors. More Gene Manipulation in Fungi. San Diego: Academic Press; 1991. pp. 441–457. [Google Scholar]
- Berger I, Shaul Y. The human junD gene is positively and selectively autoregulated. DNA Cell Biol. 1994;13:249–255. doi: 10.1089/dna.1994.13.249. [DOI] [PubMed] [Google Scholar]
- Berger I, Shaul Y. c-Fos antagonizes the junD gene positive autoregulatory loop; a novel c- Fos role in promoter switching. Gene. 1998;211:375–382. doi: 10.1016/s0378-1119(98)00120-6. [DOI] [PubMed] [Google Scholar]
- Bohmann D, Bos TJ, Admon A, Nishimura T, Vogt PK, Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987;238:1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Braus G, Mösch HU, Vogel K, Hinnen A, Hütter R. Interpathway regulation of the TRP4 gene of yeast. EMBO J. 1989;8:939–945. doi: 10.1002/j.1460-2075.1989.tb03455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsiotis M, Jones RF, Wesseling AC. Cross-pathway regulation: histidine-mediated control of histidine, tryptophan, and arginine biosynthetic enzymes in Neurospora crassa. J Bacteriol. 1974;119:893–898. doi: 10.1128/jb.119.3.893-898.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Nguyen PH, Courey AJ. A role for Groucho tetramerization in transcriptional repression. Mol Cell Biol. 1998;18:7259–7268. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RH. A mutant form of ornithine transcarbamylase found in a strain of Neurospora crassa carrying a pyrimidine-proline suppressor gene. Biochim Biophys Acta. 1962;97:185–191. doi: 10.1016/0003-9861(62)90063-2. [DOI] [PubMed] [Google Scholar]
- Davis RH. Neurospora: Contributions of a Model Organism. Oxford, England: Oxford University Press; 2000. [Google Scholar]
- Drysdale CM, Jackson BM, McVeigh R, Klebanow ER, Bai Y, Kokubo T, Swanson M, Nakatani Y, Weil PA, Hinnebusch AG. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID., and the Adap-Gcn5p coactivator complex. Mol Cell Biol. 1998;18:1711–1724. doi: 10.1128/mcb.18.3.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr E, Jelesarov I, Bosshard HR. Extremely fast folding of a very stable leucine zipper with a strengthened hydrophobic core and lacking electrostatic interactions between helices. Biochemistry. 1999;38:870–880. doi: 10.1021/bi981891e. [DOI] [PubMed] [Google Scholar]
- Eckert SE, Hoffmann B, Wanke C, Braus GH. Sexual development of Aspergillus nidulans in tryptophan auxotrophic strains. Arch Microbiol. 1999;172:157–166. doi: 10.1007/s002030050755. [DOI] [PubMed] [Google Scholar]
- Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- Flavell RB, Fincham JR. Acetate-nonutilizing mutants of Neurospora rassa. II. Biochemical deficiencies and the roles of certain enzymes. J Bacteriol. 1968;95:1063–1068. doi: 10.1128/jb.95.3.1063-1068.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goc A, Weglenski P. Regulatory region of the Aspergillus nidulans argB gene. Curr Genet. 1988;14:425–429. doi: 10.1007/BF00521264. [DOI] [PubMed] [Google Scholar]
- Hamer JE, Timberlake WE. Functional organization of the Aspergillus nidulans trpC promoter. Mol Cell Biol. 1987;7:2352–2359. doi: 10.1128/mcb.7.7.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Navarro J, Greve RA, Adams TH. Translational repression of brlA expression prevents premature development in Aspergillus. EMBO J. 1993;12:2449–2457. doi: 10.1002/j.1460-2075.1993.tb05899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci USA. 1984;81:6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985;9:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Gene Expression, Vol II: The Molecular and Cellular Biology in the Yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Hinnebusch AG. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. J Biol Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Mösch HU, Sattlegger E, Barthelmess IB, Hinnebusch A, Braus GH. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino-acid starvation. Mol Microbiol. 1999;31:807–822. doi: 10.1046/j.1365-2958.1999.01219.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Wanke C, Kirchner SK, Braus GH. c-Jun and RACK1 homologs regulate a control point for sexual development in Aspergillus nidulans. Mol Microbiol. 2000;37:28–41. doi: 10.1046/j.1365-2958.2000.01954.x. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JM, Hynes MJ. The regulation of NADP-linked isocitrate dehydrogenase in Aspergillus nidulans. J Gen Microbiol. 1982;128:23–28. doi: 10.1099/00221287-128-1-23. [DOI] [PubMed] [Google Scholar]
- Klopotowski T, Wiater A. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Arch Biochem Biophys. 1965;112:562–566. doi: 10.1016/0003-9861(65)90096-2. [DOI] [PubMed] [Google Scholar]
- Kornitzer D, Raboy B, Kulka RG, Fink GR. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S, Helmstaedt K, Gerstberger T, Eckert S, Hoffmann B, Hoppert M, Schnappauf G, Braus GH. The aroC gene of Aspergillus nidulans codes for a monofunctional, allosterically regulated chorismate mutase. J Biol Chem. 1999;274:22275–22282. doi: 10.1074/jbc.274.32.22275. [DOI] [PubMed] [Google Scholar]
- Lee YH, Kim S, Kim J, Young Kim K, Kim MJ, Ryu SH, Suh P. Overexpression of phospholipase C-gamma1 U.VC-induced apoptosis through inhibition of c-fos accumulation and c-Jun N-terminal kinase activation in PC12 cells. Biochim Biophys Acta. 1999;1440:235–423. doi: 10.1016/s1388-1981(99)00128-6. [DOI] [PubMed] [Google Scholar]
- Liberati C, di Silvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J Mol Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- Liebermann DA, Gregory B, Hoffman B. AP-1 (Fos/Jun) transcription factors in hematopoietic differentiation and apoptosis. Int J Oncol. 1998;12:685–700. doi: 10.3892/ijo.12.3.685. [DOI] [PubMed] [Google Scholar]
- Lockington RA, Sealy-Lewis HM, Scazzocchio C, Davies RW. Cloning and characterization of the ethanol utilization regulon in Aspergillus nidulans. Gene. 1985;33:137–149. doi: 10.1016/0378-1119(85)90088-5. [DOI] [PubMed] [Google Scholar]
- Luo Z, Freitag M, Sachs MS. Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol Cell Biol. 1995;15:5235–5245. doi: 10.1128/mcb.15.10.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimoun A, Holtzman T, Weissmann Z, McBride HJ, Stillman DJ, Fink GR, Kornitzer D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCFCDC4 ubiquitin-ligase complex. Mol Biol Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KY, Wu J, Miller BL. StuA is required for cell pattern formation in Aspergillus. Genes Dev. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- Miller PF, Hinnebusch AG. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes Dev. 1989;3:1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- Mirande M, Waller JP. The yeast lysyl-tRNA synthetase gene. Evidence for general amino acid control of its expression and domain structure of the encoded protein. J Biol Chem. 1988;263:18443–18451. [PubMed] [Google Scholar]
- Miozzari G, Niederberger P, Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978;134:48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch HU, Graf R, Schmidheini T, Braus GH. Three GCN4 responsive elements act synergistically as upstream and as TATA-like elements in the yeast TRP4 promoter. EMBO J. 1990;9:2951–2957. doi: 10.1002/j.1460-2075.1990.tb07487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch HU, Scheier B, Lahti R, Mantsala P, Braus GH. Transcriptional activation of yeast nucleotide biosynthetic gene ADE4 by GCN4. J Biol Chem. 1991;266:20453–20456. [PubMed] [Google Scholar]
- Mueller PP, Hinnebusch AG. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Paluh JL, Orbach MJ, Legerton TL, Yanofsky C. The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc Natl Acad Sci USA. 1988;85:3728–3732. doi: 10.1073/pnas.85.11.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Yanofsky C. Characterization of Neurospora CPC1, a bZIP DNA-binding protein that does not require aligned heptad leucines for dimerization. Mol Cell Biol. 1991;11:935–944. doi: 10.1128/mcb.11.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska M. Cross-pathway regulation of ornithine carbamoyltransferase synthesis in Aspergillus nidulans. J Gen Microbiol. 1980;116:335–339. [Google Scholar]
- Prendergast JA, Ptak C, Kornitzer D, Steussy CN, Hodgins R, Goebl M, Ellison MJ. Identification of a positive regulator of the cell cycle ubiquitin- conjugating enzyme Cdc34 (Ubc3) Mol Cell Biol. 1996;16:677–684. doi: 10.1128/mcb.16.2.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt PJ, van den Hondel CAAMJ. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 1992;216:447–457. doi: 10.1016/0076-6879(92)16041-h. [DOI] [PubMed] [Google Scholar]
- Sachs MS. General and cross-pathway controls of amino acid biosynthesis. In: Brambl R, Marzluf GA, editors. The Mycota: Biochemistry and Molecular Biology. III. Heidelberg, Germany: Springer Verlag; 1996. pp. 315–345. [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattlegger E, Hinnebusch AG, Barthelmess IB. cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2 alpha kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J Biol Chem. 1998;273:20404–20416. doi: 10.1074/jbc.273.32.20404. [DOI] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987;50:841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Thireos G, Penn MD, Greer H. 5′ untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci USA. 1984;81:5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzamarias D, Roussou I, Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Valerius O, Draht O, Kübler E, Adler K, Hoffmann B, Braus GH. Regulation of hisHF transcription of Aspergillus nidulans by adenine and amino acid limitation. Fungal Genet Biol. 2001;32:21–31. doi: 10.1006/fgbi.2000.1244. [DOI] [PubMed] [Google Scholar]
- Wang P, Larson TG, Chen CH, Pawlyk DM, Clark JA, Nuss DL. Cloning and characterization of a general amino acid control transcriptional activator from the chestnut blight fungus Cryphonectria parasitica. Fungal Genet Biol. 1998;23:81–94. doi: 10.1006/fgbi.1997.1023. [DOI] [PubMed] [Google Scholar]
- Wanke C, Eckert S, Albrecht G, van Hartingsveldt W, Punt PJ, van den Hondel CA, Braus GH. The Aspergillus niger GCN4 homologue cpcA, is transcriptionally regulated and encodes an unusual leucine zipper. Mol Microbiol. 1997;23:23–33. doi: 10.1046/j.1365-2958.1997.1741549.x. [DOI] [PubMed] [Google Scholar]
- Waring RB, May GS, Morris NR. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]