Abstract
Elevated blood pressure is the leading heritable risk factor for cardiovascular disease worldwide. We report genetic association of blood pressure (systolic, diastolic, pulse pressure) among UK Biobank participants of European ancestry with independent replication in other cohorts, and robust validation of 107 independent loci. We also identify new independent variants at 11 previously reported blood pressure loci. Combined with results from a range of in silico functional analyses and wet bench experiments, our findings highlight new biological pathways for blood pressure regulation enriched for genes expressed in vascular tissues and identify potential therapeutic targets for hypertension. Results from genetic risk score models raise the possibility of a precision medicine approach through early lifestyle intervention to offset the impact of blood pressure raising genetic variants on future cardiovascular disease risk.
Elevated blood pressure (BP) is a strong, heritable1–4 and modifiable driver of risk for stroke and coronary artery disease and a leading cause of global mortality and morbidity5,6. At the time of analysis, genome-wide association study (GWAS) meta-analyses, and analyses of bespoke or exome content, have identified and replicated genetic variants of mostly modest or weak effect on blood pressure at over 120 loci7–11. Here, we report association analyses between BP traits and genetic variants among ˜150,000 participants in UK Biobank, a prospective cohort study of 500,000 men and women aged 40-69 years with extensive baseline phenotypic measurements, stored biological samples12, and follow-up by electronic health record linkage13. We undertake independent replication in large international consortia and other cohorts, providing robust validation of our findings and new biological insights into BP regulation.
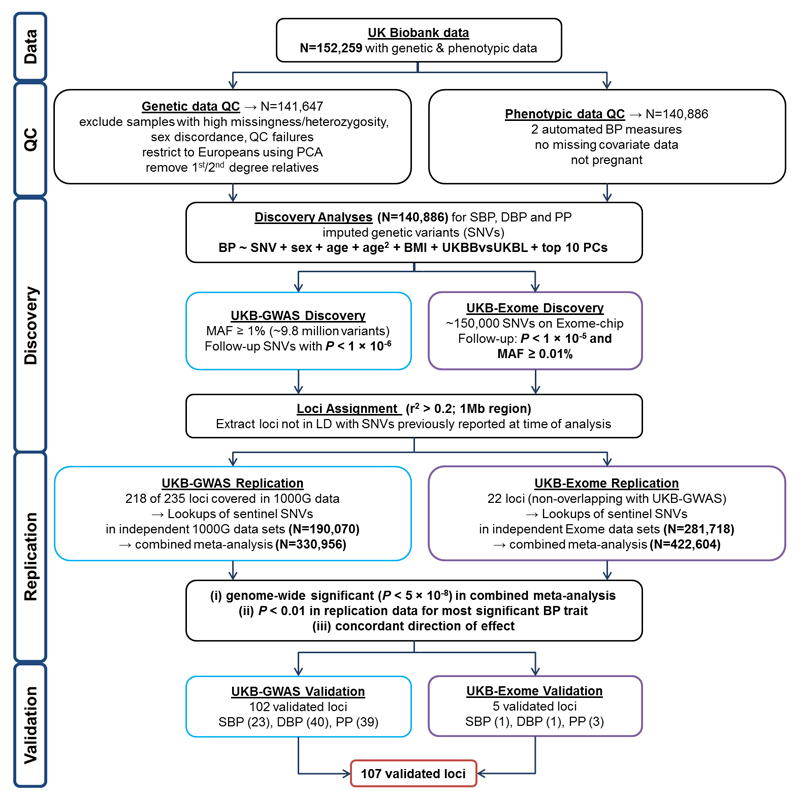
Our study design is summarized in Fig. 1. Briefly, data are available for 152,249 UK Biobank participants genotyped using a customised array (including GWAS and exome content) and with genome-wide imputation based on 1000 Genomes and UK10K sequencing data14. (Further details on the UK Biobank imputation are available at the UK Biobank website.) After quality measures and exclusions (see Online Methods), we study 140,886 unrelated individuals of European ancestry with two seated clinic BP measurements using the Omron HEM-7015IT device (Supplementary Table 1). We carry out GWAS analyses of systolic (SBP), diastolic (DBP) and pulse pressure (PP) using single-variant linear regression under an additive model, based on ˜9.8 million single nucleotide variants (SNVs) with minor allele frequency (MAF) ≥1% and imputation quality score (INFO) >0.1. For SNVs with P <1x10-6, we take forward for replication the sentinel SNV (i.e. with lowest P-value) at each locus, defined by linkage disequilibrium (LD) r2 < 0.2, within a 1Mb interval. We similarly analyze exome content for variants with MAF ≥0.01%, including rare variants, taking into replication the sentinel SNV (P < 1x10-5) from loci that are non-overlapping (r2 <0.2) with the GWAS findings. Overall we took sentinel SNVs from 240 loci into replication: 218 from GWAS and 22 from exome analysis (r2 < 0.2 and >500kb from previously reported BP SNVs at the time of analysis and not annotated to previously reported BP genes; Supplementary Table 2).
Figure 1.
Study design schematic for discovery and validation of loci. N: sample size; QC: Quality Control; PCA: Principal Component Analysis; BP: blood pressure; SBP: systolic BP; DBP: diastolic BP; PP: pulse pressure; SNVs: single nucleotide variants; BMI: body mass index; UKB: UK Biobank; UKBL: UK BiLEVE; GWAS: Genome-wide association study; MAF: Minor Allele Frequency; P: P-value; LD: Linkage Disequilibrium; 1000G: 1000 Genomes. UKBBvsUKBL: a binary indicator variable for UK Biobank vs UK BiLEVE to adjust for the different genotyping chips
The replication resources comprise individuals of European ancestry from a large BP meta-analysis consortium (ICBP cohorts listed in Supplementary Note) and further cohorts with 1000 Genomes data for GWAS (Supplementary Table 3), and two large BP exome consortia. We use P <5x10-8 to denote genome-wide significance in the combined (discovery and replication) meta-analyses, with P < 0.01 for support in the replication data alone and concordant direction of effect. Additionally, we take forward for replication potential secondary signals at 51 previously reported BP loci at the time of analysis (excluding the HLA region).
To better understand the functional consequences of our findings, we carry out a series of in silico investigations and experimental analysis of gene expression in relevant vascular tissue for selected putative functional SNVs (Supplementary Fig. 1).
Results
Genetic variants at novel and previously unvalidated loci
Of the 240 loci taken forward to replication, we validate 107 loci at P < 5x10-8, of which 102 derive from the GWAS analysis replicated and meta-analyzed in a total of 330,956 individuals (Tables 1-3; Supplementary Fig. 2a-c; Supplementary Fig. 3a), and a further five from the exome analysis in a total of 422,604 individuals (Tables 1-3 and Supplementary Fig. 3b; Supplementary Tables 4, 5 and 6). Thirty-two of these validated loci are novel findings. Since the time of analysis, the remaining 75 loci have also been reported in another study15, although at least 53 of these were previously unvalidated (Tables 1-3), hence we now validate these loci for the first time. We therefore present results here for all 107 validated loci in our study. Most SNVs also show association with hypertension in the UK Biobank data, for example 93 of the 107 validated sentinel SNVs are nominally significant (P < 0.01) (Supplementary Table 7).
Table 1. Loci validated with SBP as primary trait: combined meta-analysis results from (a) GWAS and (b) Exome for the sentinel variant.
| (a) GWAS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Chr | Pos | rsID | EA | EAF | N | Beta | SE | P | Note |
| NADK-CPSF3L | 1 | 1,685,921 | rs139385870 | D | 0.5 | 281,890 | -0.352 | 0.05 | 1.3x10-12 | GIU |
| CELA2A | 1 | 15,798,197 | rs3820068 | A | 0.81 | 310,776 | 0.425 | 0.06 | 1.1x10-12 | GIU |
| GTF2B | 1 | 89,360,158 | rs10922502 | A | 0.62 | 323,666 | -0.382 | 0.05 | 2.2x10-15 | GIU |
| FOSL2 | 2 | 28,635,740 | rs7562 | T | 0.52 | 319,942 | 0.263 | 0.05 | 1.9x10-8 | |
| PRKD3 | 2 | 37,517,566 | rs13420463 | A | 0.77 | 330,307 | 0.356 | 0.05 | 7.0x10-11 | GIU |
| METTL21A-AC079767.3 | 2 | 208,526,140 | rs55780018 | T | 0.54 | 304,567 | -0.391 | 0.05 | 5.9x10-16 | GIU |
| RYK | 3 | 134,000,025 | rs9859176 | T | 0.4 | 322,428 | 0.322 | 0.05 | 1.3x10-11 | G |
| NPNT | 4 | 106,911,742 | rs13112725 | C | 0.76 | 306,370 | 0.435 | 0.06 | 1.5x10-14 | GIU |
| TMEM161B | 5 | 87,514,515 | rs10059921 | T | 0.08 | 298,543 | -0.526 | 0.09 | 4.0x10-9 | GIU |
| FBN2 | 5 | 127,868,199 | rs6595838 | A | 0.3 | 328,401 | 0.344 | 0.05 | 7.6x10-12 | GIU |
| CASC15 | 6 | 22,130,601 | rs6911827 | T | 0.45 | 326,471 | 0.296 | 0.05 | 2.0x10-10 | GIU |
| TFAP2D | 6 | 50,683,009 | rs78648104 | T | 0.92 | 305,426 | -0.481 | 0.08 | 1.3x10-8 | |
| MKLN1 | 7 | 131,059,056 | rs13238550 | A | 0.4 | 325,647 | 0.331 | 0.05 | 1.9x10-12 | |
| HIPK2 | 7 | 139,463,264 | rs1011018 | A | 0.2 | 325,110 | -0.329 | 0.06 | 1.5x10-8 | |
| ZFAT | 8 | 135,612,745 | rs894344 | A | 0.6 | 329,834 | -0.258 | 0.05 | 3.2x10-8 | |
| PAX2 | 10 | 102,604,514 | rs112184198 | A | 0.1 | 323,791 | -0.659 | 0.08 | 3.6x10-18 | GIU |
| MCF2L | 13 | 113,636,156 | rs9549328 | T | 0.23 | 313,787 | 0.318 | 0.06 | 1.5x10-8 | GI |
| FERMT2 | 14 | 53,377,540 | rs9888615 | T | 0.29 | 326,235 | -0.318 | 0.05 | 3.5x10-10 | GIU |
| PPP2R5E | 14 | 63,928,546 | rs8016306 | A | 0.8 | 329,869 | 0.335 | 0.06 | 3.7x10-9 | |
| ABHD17C | 15 | 81,013,037 | rs35199222 | A | 0.45 | 323,407 | 0.322 | 0.05 | 5.2x10-12 | GI |
| CFDP1 | 16 | 75,331,044 | rs11643209 | T | 0.42 | 309,242 | -0.339 | 0.05 | 1.8x10-12 | GI |
| CRK | 17 | 1,333,598 | rs12941318 | T | 0.49 | 299,739 | -0.269 | 0.05 | 2.5x10-8 | GIU |
| ACOX1 | 17 | 73,949,045 | rs2467099 | T | 0.22 | 326,401 | -0.307 | 0.06 | 3.3x10-8 | GIU |
|
(b) Exome | ||||||||||
| SSPN | 12 | 26,438,189 | rs6487543 | A | 0.77 | 244,842 | 0.3 | 0.05 | 6.3x10-10 | |
Locus: named according to nearest annotated gene(s); Chr: chromosome; Pos: build 37; EA: effect allele; EAF: EA frequency in UK Biobank; Beta: effect estimate; SE: Standard Error of effect; P: P-value; N: total sample size analyzed; Note: indicates loci published since our analysis15 from GERA (G), GERA+ICBP(HapMap) (GI) or GERA+ICBP(HapMap)+UKB (GIU) analyses.
Table 3. Loci validated with PP as primary trait: combined meta-analysis results from (a) GWAS and (b) Exome for the sentinel variant.
| (a) GWAS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Chr | Pos | rsID | EA | EAF | N | Beta | SE | P | Note |
| chr1mb9 | 1 | 9,441,949 | rs9662255 | A | 0.43 | 310,618 | -0.207 | 0.03 | 1.9x10-10 | GIU |
| SF3A3 | 1 | 38,455,891 | rs4360494 | C | 0.55 | 282,851 | 0.278 | 0.03 | 3.7x10-16 | G |
| RP4-710M16.1-PPAP2B | 1 | 56,576,924 | rs112557609 | A | 0.35 | 325,952 | 0.227 | 0.03 | 6.8x10-12 | |
| FGGY | 1 | 59,653,742 | rs3889199 | A | 0.71 | 329,486 | 0.351 | 0.03 | 1.8x10-24 | G |
| C2orf43 | 2 | 20,881,840 | rs2289081 | C | 0.36 | 329,140 | -0.223 | 0.03 | 5.5x10-12 | GI |
| PRKCE | 2 | 46,363,336 | rs11690961 | A | 0.88 | 327,847 | 0.34 | 0.05 | 3.9x10-12 | GIU |
| CEP68 | 2 | 65,283,972 | rs74181299 | T | 0.62 | 324,224 | 0.23 | 0.03 | 9.6x10-13 | GIU |
| TCF7L1 | 2 | 85,491,365 | rs11689667 | T | 0.54 | 330,634 | 0.176 | 0.03 | 1.7x10-8 | GIU |
| FN1 | 2 | 216,300,482 | rs1250259 | A | 0.74 | 325,485 | -0.314 | 0.04 | 8.7x10-19 | G |
| GATA2 | 3 | 128,201,889 | rs62270945 | T | 0.03 | 279,925 | 0.607 | 0.1 | 1.8x10-9 | GIU |
| PALLD | 4 | 169,717,148 | rs1566497 | A | 0.42 | 320,948 | 0.236 | 0.03 | 1.9x10-13 | GI |
| chr4mb174 | 4 | 174,584,663 | rs17059668 | C | 0.92 | 313,277 | -0.332 | 0.06 | 2.8x10-8 | |
| LHFPL2 | 5 | 77,837,789 | rs10057188 | A | 0.46 | 325,985 | -0.205 | 0.03 | 6.7x10-11 | GIU |
| GJA1 | 6 | 121,781,390 | rs11154027 | T | 0.47 | 316,708 | 0.207 | 0.03 | 1.1x10-10 | |
| ESR1 | 6 | 152,397,912 | rs36083386 | I | 0.11 | 323,303 | 0.439 | 0.05 | 1.5x10-18 | G |
| FNDC1 | 6 | 159,699,125 | rs449789 | C | 0.14 | 325,584 | 0.359 | 0.05 | 2.4x10-15 | GIU |
| THBS2 | 6 | 169,587,103 | rs1322639 | A | 0.78 | 319,866 | 0.316 | 0.04 | 4.8x10-17 | G |
| SUGCT | 7 | 40,447,971 | rs76206723 | A | 0.1 | 328,162 | -0.346 | 0.05 | 7.4x10-12 | GIU |
| SLC20A2 | 8 | 42,324,765 | rs2978456 | T | 0.55 | 304,964 | -0.188 | 0.03 | 1.2x10-8 | GIU |
| TRAPPC9 | 8 | 141,060,027 | rs4454254 | A | 0.63 | 330,022 | -0.261 | 0.03 | 5.1x10-16 | |
| SCAI | 9 | 127,900,996 | rs72765298 | T | 0.87 | 316,271 | -0.374 | 0.05 | 2.7x10-14 | GI |
| KIAA1462 | 10 | 30,317,073 | rs9337951 | A | 0.34 | 299,646 | 0.28 | 0.04 | 2.5x10-15 | G |
| ARHGAP12 | 10 | 32,082,658 | rs10826995 | T | 0.71 | 327,373 | -0.212 | 0.03 | 1.1x10-9 | GIU |
| PRDM11 | 11 | 45,208,141 | rs11442819 | I | 0.11 | 326,483 | -0.279 | 0.05 | 7.1x10-9 | GIU |
| NOX4 | 11 | 89,224,453 | rs2289125 | A | 0.21 | 307,682 | -0.377 | 0.04 | 9.1x10-22 | G |
| CEP164 | 11 | 117,283,676 | rs8258 | T | 0.38 | 327,038 | 0.236 | 0.03 | 2.9x10-13 | G |
| CCDC41 | 12 | 94,880,742 | rs139236208 | A | 0.1 | 291,244 | -0.363 | 0.06 | 1.6x10-10 | G |
| RP11-61O1.1 | 14 | 98,587,630 | rs9323988 | T | 0.63 | 327,551 | -0.212 | 0.03 | 4.1x10-11 | GIU |
| VAC14 | 16 | 70,755,610 | rs117006983 | A | 0.01 | 250,766 | 0.986 | 0.14 | 4.1x10-12 | |
| CDH13 | 16 | 83,045,790 | rs7500448 | A | 0.75 | 321,958 | 0.329 | 0.04 | 1.1x10-19 | G |
| KIAA0753 | 17 | 6,473,828 | rs7226020 | T | 0.56 | 303,389 | -0.256 | 0.03 | 2.3x10-14 | GIU |
| TP53-SLC2A4 | 17 | 7,571,752 | rs78378222 | T | 0.99 | 294,053 | 0.904 | 0.14 | 1.8x10-10 | GIU |
| KCNH4-HSD17B1 | 17 | 40,317,241 | rs79089478 | T | 0.97 | 318,326 | 0.584 | 0.1 | 3.1x10-9 | |
| PYY | 17 | 42,060,631 | rs62080325 | A | 0.66 | 315,689 | -0.186 | 0.03 | 4.0x10-8 | |
| MRC2 | 17 | 60,767,151 | rs740698 | T | 0.56 | 311,450 | -0.228 | 0.03 | 3.1x10-12 | |
| SLC14A2 | 18 | 43,097,750 | rs7236548 | A | 0.18 | 330,075 | 0.352 | 0.04 | 2.0x10-18 | G |
| SLC24A3 | 20 | 19,465,907 | rs6081613 | A | 0.28 | 315,546 | 0.263 | 0.04 | 1.6x10-13 | GIU |
| ARVCF | 22 | 19,967,980 | rs12628032 | T | 0.3 | 310,292 | 0.24 | 0.03 | 5.5x10-12 | GIU |
| XRCC6 | 22 | 42,038,786 | rs73161324 | T | 0.05 | 267,722 | 0.496 | 0.07 | 2.8x10-11 | |
|
(b) Exome | ||||||||||
| CD34 | 1 | 208,024,820 | rs12731740 | T | 0.1 | 279,078 | -0.249 | 0.04 | 1.1x10-8 | |
| ZNF638 | 2 | 71,627,539 | rs3771371 | T | 0.57 | 280,285 | -0.16 | 0.03 | 5.8x10-9 | GIU |
| CRACR2B | 11 | 828,916 | rs7126805 | A | 0.73 | 145,162 | 0.222 | 0.04 | 3.3x10-9 | |
Locus: named according to nearest annotated gene(s); Chr: chromosome; Pos: build 37; EA: effect allele; EAF: EA frequency in UK Biobank; Beta: effect estimate; SE: Standard Error of effect; P: P-value; N: total sample size analyzed; Note: indicates loci published since our analysis15 from GERA (G), GERA+ICBP(HapMap) (GI) or GERA+ICBP(HapMap)+UKB (GIU) analyses.
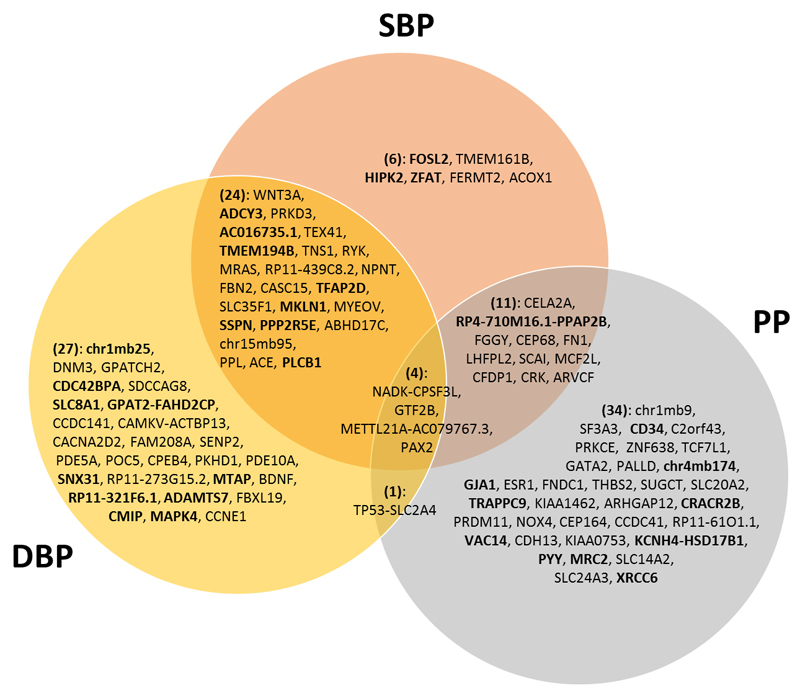
Of the 107 validated loci, 24 are reported for association with SBP as the primary trait (most significant from combined meta-analysis), 41 for DBP and 42 for PP, although many loci are associated with more than one BP trait (Supplementary Fig. 4). For example, in the combined meta-analysis, 24 validated loci are associated with both SBP and DBP, 11 with SBP and PP, one locus with DBP and PP and four loci (NADK-CPSF3L, GTF2B, METTL21A-AC079767.3 and PAX2) with all three traits at genome-wide significance (Fig. 2).
Figure 2.
Venn diagram of 107 validated loci from our study. This shows concordance of significant associations across the three blood pressure phenotypes for the 107 validated sentinel variants (Tables 1-3) from both the GWAS and exome analyses, according to genome-wide significance in the combined meta-analysis. The locus names labelled within the Venn Diagram correspond to Tables 1-3, and relate to the nearest annotated gene. The loci names in bold font highlight the 32 novel loci which are reported for the first time in our study.
After conditional analysis on the sentinel SNV we identify an independent validated secondary SNV at five of the 107 loci (Supplementary Table 8a; Supplementary Table 9). Compared with previously reported SNVs at the time of analysis, the contribution of our validated loci increases the percentage trait variance explained by ˜1%, e.g. to 3.56% for SBP.
We report signals at known hypertension drug targets, including the angiotensin converting enzyme (ACE) locus (rs4308, P = 6.8 x 10-14, ACE-inhibitors), CACNA2D2 (rs743757, P = 2.4 x 10-10, calcium channel blockers), MME (rs143112823 in the RP11-439C8.2 locus, P = 1.4 x 10-14, omapatrilat), ADRA2B (rs2579519 in the GPAT2-FAHD2CP locus, P = 4.8 x 10-12, beta blockers), SLC14A2 (rs7236548, P = 2.0 x 10-18, nifedipine), and phosphodiesterase 5A (PDE5A; rs66887589, P = 3.4 x 10-15, sildenafil).
Additionally, we evaluate our validated SNVs, where available, in cohorts of non-European ancestry9–11, while recognizing that these analyses are likely underpowered (Supplementary Table 10). We find concordance in direction of effect (P <0.05) for GWAS SNVs for all three BP traits among individuals of East Asian ancestry and for DBP for South Asian ancestry, also for exome SNVs among individuals of Hispanic ancestry, pointing to cosmopolitan effects for many of the BP associated variants.
A PhenoScanner16 search showed that 27 of our 107 validated sentinel SNVs (or proxies; r2 ≥ 0.8) exhibit genome-wide significant associations with other traits (Supplementary Fig. 5), including coronary artery disease and myocardial infarction (where BP is likely on the causal pathway17), cardiovascular risk factors (e.g. lipids, height, body mass index) and non-cardiovascular traits (e.g. lung function, cancer, Alzheimer’s).
Variants at previously reported loci at time of analysis
In conditional analyses, we identify 22 secondary SNVs (17 common, one rare, four low-frequency variants) that are conditionally independent of the BP associated SNVs at 16 previously reported loci at the time of analysis (Supplementary Table 8b; Supplementary Tables 11 and 12). One rare variant (rs138582164, MAF=0.1%) in the CDH17 locus anticipated to act as an exonic stop/gain mutation at the GEM gene is associated with a relatively large effect on PP (3.5 mm Hg per allele copy, Supplementary Table 8b). At three previously reported loci (EBF1, PDE3A, JAG1) we identify multiple independent secondary SNVs in addition to the previously reported SNVs (Supplementary Table 11).
The UK Biobank data show support (P < 0.01) for 119 of 122 previously reported BP loci at the time of analysis (159 of 163 SNVs) for one or more BP traits (Supplementary Fig. 2 a-c; Supplementary Table 13). We do not show support for one SNV (rs11066280, RPL6-ALDH1) identified from a GWAS of East Asian ancestry18, which may indicate ancestry-specific effects. We compare the MAF and effect sizes in UK Biobank with published results of previously reported variants (Supplementary Fig. 6), indicating consistency of results between the two sources of data.
We also examine findings for low-frequency and rare gene mutations previously reported to be associated with monogenic hypertension disorders19 and included on the UK Biobank gene array. Despite lack of power overall, the variant with the lowest P-value (rs387907156; KLH3; MAF=0.02%) has a seemingly large effect on BP: 8.2 mm Hg (SE=4.1, P = 0.046) per allele for SBP; 5.6 mm Hg (SE=2.6, P = 0.048) for PP (Supplementary Table 14).
Functional analyses
We annotate the 107 validated loci to 212 genes (based on LD r2 ≥0.8) and seek putative function from in silico analyses and gene expression experiments. Candidate genes with the strongest supporting evidence are indicated in the last column of Supplementary Table 4 with an indication of the supporting data source. All genome-wide significant variants in LD (r2>0.8) with the variants reported here, ranked by supporting evidence, are annotated in Supplementary Table 15. Of the 107 validated sentinel SNVs three are Indels; all other variants are single nucleotide polymorphisms (SNPs). We identify non-synonymous SNVs at 13 of the 107 validated loci (Supplementary Table 16), three of which are predicted to be damaging (ANNOVAR) in TFAP2D (rs78648104), NOX4 (rs56061986) and CCDC141 (rs17362588, reported to be associated with heart rate20) (Supplementary Fig. 5a). Beyond the coding regions we identify 29 SNVs in 3’UTRs which are predicted to significantly weaken or cause loss of miRNA regulation by altering the recognition motif in seven genes, and strengthen or create target sites for miRNA binding in 13 genes (based on miRNASNP db, Supplementary Table 16).
From our expression Quantitative Trait locus (eQTL) analysis (GTEx), 59 of the 107 validated loci contain variants with eQTLs in at least one tissue (Supplementary Table 17); arterial tissue has the largest number of loci with eQTLs (Supplementary Fig. 7), with targeted in silico analysis showing six loci with eQTLs in arterial tissue (Supplementary Table 16). For example, the GTEx tibial artery eQTL in SF3A3 (rs4360494) shows strong in silico supporting evidence, including an arterial DNase I site within which the major C allele removes a predicted AP-2 binding site (Supplementary Fig. 8). Hence we prioritized this gene for in vitro functional analysis (see below).
By considering all loci reported here (our 107 validated loci, and previously reported loci at the time of analysis), our DEPICT analysis identifies enrichment of expression across 31 tissues and cells (Supplementary Fig. 9; Supplementary Table 18), with greatest enrichment in the arteries (P = 1.9 x 10-6, false discovery rate (FDR) < 1%). We use FORGE to investigate and identify significant (FDR, P <0.05) cell type specific enrichment within DNase I hypersensitive sites in a range of tissues including dermal and lung microvascular endothelial cell types, and cardiac fibroblasts (Supplementary Fig. 10). For a set of curated candidate regulatory SNVs from our 107 validated loci (see Supplementary Note), widespread enrichment is found in microvascular endothelium, aortic smooth muscle, aortic fibroblasts, vascular epithelium, heart and skin (Supplementary Fig. 10). In addition, we identify significant enrichment of histone marks in a wide range of cell types, including strong enrichment seen for H3K4Me3 (an activating modification found near promoters) marks in umbilical vein endothelial cells (HUVEC) (Supplementary Fig. 11). To explore expression at the level of cardiovascular cell types specifically, we use Fantom5 reference transcript expression data (see Online Methods) to cluster the 212 genes annotated to our 107 validated loci according to tissue specificity (Supplementary Fig. 12), with the significantly clustered genes forming four tissue-specific clusters, including a vascular smooth muscle cell (VSMC) and fibroblast cluster, an endothelial cell cluster (including probable endothelial cells in highly vascularized tissues), and a combined vascular cell cluster.
Additionally, Ingenuity pathway analysis and upstream transcriptional analysis show enrichment of canonical pathways implicated in cardiovascular disease, including those targeted by antihypertensive drugs, such as the alpha-adrenergic, CXCR4, endothelin signalling and angiotensin receptor pathways (Supplementary Table 19). In keeping with vascular mediation of genetic influence we identify diphenyleneiodonium, an inhibitor of flavin-containing oxidases, including NAD(P)H oxidase (NOX), which is reported to reverse endothelial dysfunction (and hypertension) in a rat model21.
To identify long range target genes of non-coding variants, we use chromatin interaction (Hi-C) data from HUVEC, as enhancers and silencers often form chromatin loops with their target promoter. In most loci the strongest promoter interaction involves a gene in high LD with the SNV, but for 21 loci we find a distal potential target gene (Supplementary Table 16). Pathway analysis of the distal genes shows greatest enrichment in regulators of cardiac hypertrophy.
We evaluate pleiotropy using the Genomic Regions Enrichment of Annotations Tool (GREAT) to study enrichment of mouse phenotype and human disease ontology terms across all loci reported here. These highlight cardiovascular system abnormalities and vascular disease as the most highly enriched terms (Supplementary Fig. 5b & 5c).
Collectively evidence from eQTLs, DEPICT, DNase I sites, histone marks, Hi-C data and ontological analyses indicates predominant vascular and cardiovascular tissue involvement for genes within the BP associated loci.
We also look for association of our validated sentinel SNVs with metabolomic signatures. Three SNVs within the NOX4, KCNH4 and LHFPL2 loci show significant associations (family-wise error rate < 5%) with lipoprotein sub-fractions from 1H Nuclear Magnetic Resonance (NMR) spectroscopy analysis of 2,000 Airwave study samples (Supplementary Tables 20 and 21). The results for these variants suggest a link between BP regulation and lipid metabolism. Eleven SNVs (including at LHFPL2 locus) show association (family wise error rate < 5%) with metabolites in blood or urine from the publicly available “Metabolomics GWAS Server” resource based on mass spectrometry22,23 (Supplementary Table 21), including sugar acids, sphingolipids, fatty acids, glycerophospholipids, organic acids and benzene derivatives.
Several genes and variants with putative function are highlighted in our in silico analysis as having biological support (e.g. eQTLs or nsSNVs) and those with novelty and tractability to laboratory investigation (e.g. expression in available tissue models) are prioritized. Sentinel variants in three genes which were highly significant in the combined meta-analysis (Tables 2 and 3) are selected for experimental testing and were successfully genotyped, each for at least 100 samples. We select ADAMTS7 due to strong biological support (e.g. mouse knockout phenotype), SF3A3 due to eQTLs, and NOX4 as it contains a rare nsSNV (Supplementary Table 9) in addition to common variant associations. We use quantitative polymerase chain reaction (qPCR) to study the impact of these sentinel variants on gene expression in human VSMCs and endothelial cells (ECs) (see Online Methods). For SF3A3, the major C allele of variant rs4360494 associated with increased PP (0.278 mmHg ±0.03, P=3.7x10-16, N=307,682) is associated with SF3A3 expression in human VSMCs, although not in endothelial cells (Supplementary Fig. 13a); and the T allele of SNV rs62012628 in ADAMTS7 associated with lower DBP (0.238 mmHg ±0.03, P=5.1x10-12, N=244,143), is associated with reduced ADAMTS7 expression in human VSMCs (Supplementary Fig. 13b), while the minor A allele of SNV rs2289125 at the NOX4 locus associated with lower PP (-0.377 mmHg ±0.04, P=9.1x10-22, N=282,851) correlates with increased NOX4 expression in ECs though not VSMCs (Supplementary Fig. 13c). Our study thus finds evidence for novel cis-eQTLs in ADAMTS7 and NOX4 in addition to validating the previously reported GTEx eQTL in SF3A3, and supports the vascular expression of these genes.
Table 2. Loci validated with DBP as primary trait: combined meta-analysis results from (a) GWAS and (b) Exome for the sentinel variant.
| (a) GWAS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Chr | Pos | rsID | EA | EAF | N | Beta | SE | P | Note |
| chr1mb25 | 1 | 25,030,470 | rs6686889 | T | 0.25 | 322,575 | 0.185 | 0.03 | 3.6x10-9 | |
| DNM3 | 1 | 172,357,441 | rs12405515 | T | 0.56 | 328,543 | -0.165 | 0.03 | 1.4x10-9 | GIU |
| GPATCH2 | 1 | 217,718,789 | rs12408022 | T | 0.26 | 320,983 | 0.198 | 0.03 | 2.4x10-10 | GIU |
| CDC42BPA | 1 | 227,252,626 | rs10916082 | A | 0.73 | 327,636 | -0.177 | 0.03 | 8.4x10-9 | |
| WNT3A | 1 | 228,191,075 | rs2760061 | A | 0.47 | 312,761 | 0.23 | 0.03 | 2.1x10-16 | GIU |
| SDCCAG8 | 1 | 243,471,192 | rs953492 | A | 0.46 | 325,253 | 0.22 | 0.03 | 7.4x10-16 | G |
| ADCY3 | 2 | 25,139,596 | rs55701159 | T | 0.89 | 321,052 | 0.285 | 0.04 | 7.2x10-11 | |
| SLC8A1 | 2 | 40,567,743 | rs4952611 | T | 0.58 | 309,395 | -0.157 | 0.03 | 4.0x10-8 | |
| AC016735.1 | 2 | 43,167,878 | rs76326501 | A | 0.91 | 318,127 | 0.419 | 0.05 | 3.6x10-18 | |
| GPAT2-FAHD2CP | 2 | 96,675,166 | rs2579519 | T | 0.63 | 311,557 | -0.197 | 0.03 | 4.8x10-12 | |
| TEX41 | 2 | 145,646,072 | rs1438896 | T | 0.3 | 329,278 | 0.234 | 0.03 | 2.0x10-15 | GIU |
| CCDC141 | 2 | 179,786,068 | rs79146658 | T | 0.91 | 321,318 | -0.311 | 0.05 | 2.4x10-10 | G |
| TMEM194B | 2 | 191,439,591 | rs7592578 | T | 0.19 | 304,672 | -0.24 | 0.04 | 9.5x10-12 | |
| TNS1 | 2 | 218,668,732 | rs1063281 | T | 0.6 | 315,354 | -0.2 | 0.03 | 1.3x10-12 | GIU |
| CAMKV-ACTBP13 | 3 | 49,913,705 | rs36022378 | T | 0.8 | 319,983 | -0.202 | 0.03 | 4.7x10-9 | GIU |
| CACNA2D2 | 3 | 50,476,378 | rs743757 | C | 0.14 | 328,836 | 0.245 | 0.04 | 2.4x10-10 | GIU |
| FAM208A | 3 | 56,726,646 | rs9827472 | T | 0.37 | 323,058 | -0.177 | 0.03 | 4.3x10-10 | GIU |
| RP11-439C8.2 | 3 | 154,707,967 | rs143112823 | A | 0.09 | 297,343 | -0.403 | 0.05 | 1.4x10-14 | GIU |
| SENP2 | 3 | 185,317,674 | rs12374077 | C | 0.35 | 327,513 | 0.163 | 0.03 | 9.2x10-9 | GIU |
| PDE5A | 4 | 120,509,279 | rs66887589 | T | 0.52 | 324,397 | -0.215 | 0.03 | 3.4x10-15 | GIU |
| POC5 | 5 | 75,038,431 | rs10078021 | T | 0.63 | 314,172 | -0.164 | 0.03 | 1.3x10-8 | G |
| CPEB4 | 5 | 173,377,636 | rs72812846 | A | 0.28 | 312,601 | -0.209 | 0.03 | 2.2x10-11 | GIU |
| PKHD1 | 6 | 51,832,494 | rs13205180 | T | 0.49 | 325,419 | 0.168 | 0.03 | 7.0x10-10 | GIU |
| PDE10A | 6 | 166,178,451 | rs147212971 | T | 0.06 | 296,010 | -0.36 | 0.06 | 1.6x10-9 | GIU |
| SLC35F1 | 6 | 118,572,486 | rs9372498 | A | 0.08 | 330,625 | 0.334 | 0.05 | 1.8x10-11 | GIU |
| SNX31 | 8 | 101,676,675 | rs2978098 | A | 0.54 | 324,424 | 0.165 | 0.03 | 1.5x10-9 | |
| RP11-273G15.2 | 8 | 144,060,955 | rs62524579 | A | 0.53 | 268,645 | -0.175 | 0.03 | 3.8x10-9 | GIU |
| MTAP | 9 | 21,801,530 | rs4364717 | A | 0.55 | 327,173 | -0.175 | 0.03 | 1.3x10-10 | |
| BDNF | 11 | 27,728,102 | rs11030119 | A | 0.31 | 330,002 | -0.163 | 0.03 | 2.9x10-8 | GIU |
| MYEOV | 11 | 69,079,707 | rs67330701 | T | 0.09 | 276,760 | -0.367 | 0.05 | 2.1x10-12 | GIU |
| RP11-321F6.1 | 15 | 66,869,072 | rs7178615 | A | 0.37 | 318,076 | -0.179 | 0.03 | 2.6x10-10 | |
| ADAMTS7 | 15 | 79,070,000 | rs62012628 | T | 0.29 | 244,143 | -0.238 | 0.03 | 5.1x10-12 | |
| chr15mb95 | 15 | 95,312,071 | rs12906962 | T | 0.68 | 319,952 | -0.221 | 0.03 | 5.6x10-14 | GIU |
| PPL | 16 | 4,943,019 | rs12921187 | T | 0.43 | 326,469 | -0.174 | 0.03 | 2.5x10-10 | G |
| FBXL19 | 16 | 30,936,743 | rs72799341 | A | 0.24 | 324,502 | 0.185 | 0.03 | 5.8x10-9 | GIU |
| CMIP | 16 | 81,574,197 | rs8059962 | T | 0.42 | 319,839 | -0.17 | 0.03 | 1.3x10-9 | |
| ACE | 17 | 61,559,625 | rs4308 | A | 0.37 | 319,394 | 0.213 | 0.03 | 6.8x10-14 | GIU |
| MAPK4 | 18 | 48,142,854 | rs745821 | T | 0.76 | 330,954 | 0.189 | 0.03 | 1.4x10-9 | |
| CCNE1 | 19 | 30,294,991 | rs62104477 | T | 0.33 | 320,347 | 0.177 | 0.03 | 1.2x10-9 | GIU |
| PLCB1 | 20 | 8,626,271 | rs6108168 | A | 0.25 | 327,368 | -0.211 | 0.03 | 1.1x10-11 | |
|
(b) Exome | ||||||||||
| MRAS | 3 | 138,119,952 | rs2306374 | T | 0.84 | 281,715 | -0.184 | 0.03 | 7.4x10-9 | GIU |
Locus: named according to nearest annotated gene(s); Chr: chromosome; Pos: build 37; EA: effect allele; EAF: EA frequency in UK Biobank; Beta: effect estimate; SE: Standard Error of effect; P: P-value; N: total sample size analyzed; Note: indicates loci published since our analysis15 from GERA (G), GERA+ICBP(HapMap) (GI) or GERA+ICBP(HapMap)+UKB (GIU) analyses.
Genetic risk score analyses
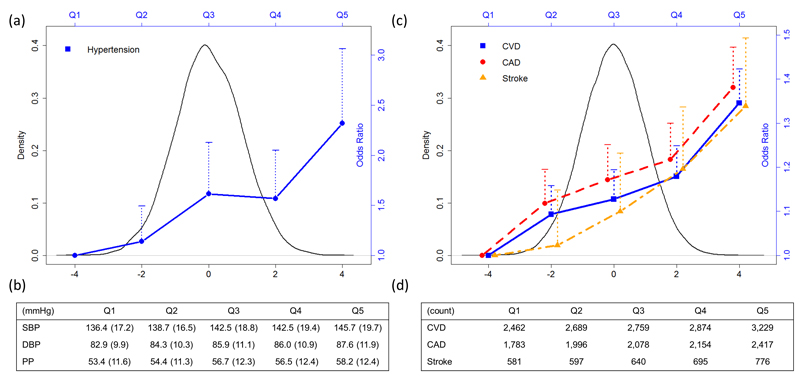
We create an unbiased genetic risk score (GRS) (Supplementary Table 22) to evaluate, in an independent cohort (Airwave, see Online Methods), the impact of the combination of all loci reported here on BP levels and risk of hypertension. When compared with the lowest quintile of the distribution of the GRS, individuals >50 years in the highest quintile have sex-adjusted mean SBP higher by 9.3 mm Hg (95% CI 6.9 to 11.7 mm Hg, P =1.0 x 10-13) and an over two-fold higher risk of hypertension (OR 2.32 95% CI 1.76 to 3.06; P=2.8 x 10-9) compared with individuals in the lowest quintile (Fig. 3; Supplementary Table 23). Similar results were obtained from GRS associations with BP and hypertension within UK Biobank (Supplementary Table 24). In UK Biobank – based on self-reported health data, record linkage to Hospital Episode Statistics and mortality follow-up data (Supplementary Table 25) – we show that the GRS is associated with increased risk of stroke, coronary heart disease and all cardiovascular outcomes; comparing the upper and lower fifths of the GRS distribution, sex-adjusted odds ratios are 1.34 (95% CI 1.20 to 1.49, P =1.5×10-7), 1.38 (95% CI 1.30 to 1.47, P= 4.3×10-23) and 1.35 (95% CI 1.27 to 1.42, P=1.3×10-25) respectively (Fig. 3; Supplementary Table 26). Results are also provided for incident-only cases (Supplementary Table 27).
Figure 3.
Distribution of Genetic Risk Score (GRS) and its relationship with blood pressure, hypertension and CVD outcomes. The GRS is based on all reported loci: both previously reported loci at the time of analysis; and all validated blood pressure variants from this study. (a) Distribution of GRS in Airwave and sex-adjusted odds ratio of hypertension in age 50+ comparing each of the upper four GRS quintiles with the lowest quintile; dotted lines represent the upper 95% confidence intervals. (b) Mean blood pressures and standard deviation in bracket in Airwave age 50+ across GRS quintiles. (c) Distribution of GRS in UKB and sex-adjusted odds ratio of CVD, CAD and stroke comparing each of the upper four GRS quintiles with the lowest quintile; dotted lines represent the upper 95% confidence intervals. (d) Count of CVD, CAD and stroke (events and deaths) across GRS quintiles in UKB participants
Discussion
A key attribute of this study is the combination of a large, single discovery sample with standardized BP measurement and dense 1000 Genomes/UK10K imputation, yielding a high quality dataset with ˜9.8 million variants14, taking advantage of major international consortia for parallel replication of common and low-frequency variants. In total we include GWAS data from 330,956 individuals and exonic SNVs from a total of 422,604 individuals. This strategy resulted in 107 robustly validated loci for BP traits, including 32 loci that have not previously been reported, and at least 53 further loci validated for the first time. Despite its size, our study is still under-powered to find low-frequency variants. Our findings are mostly common variants, with similarly modest effect sizes as variants previously reported at the time of analysis (Supplementary Fig. 14). The lack of rare variant discovery could also be due to the challenge of detecting rare variants from imputed data. There may be greater potential for identifying rare variants from the future release of genetic data for all 500,000 UK Biobank participants.
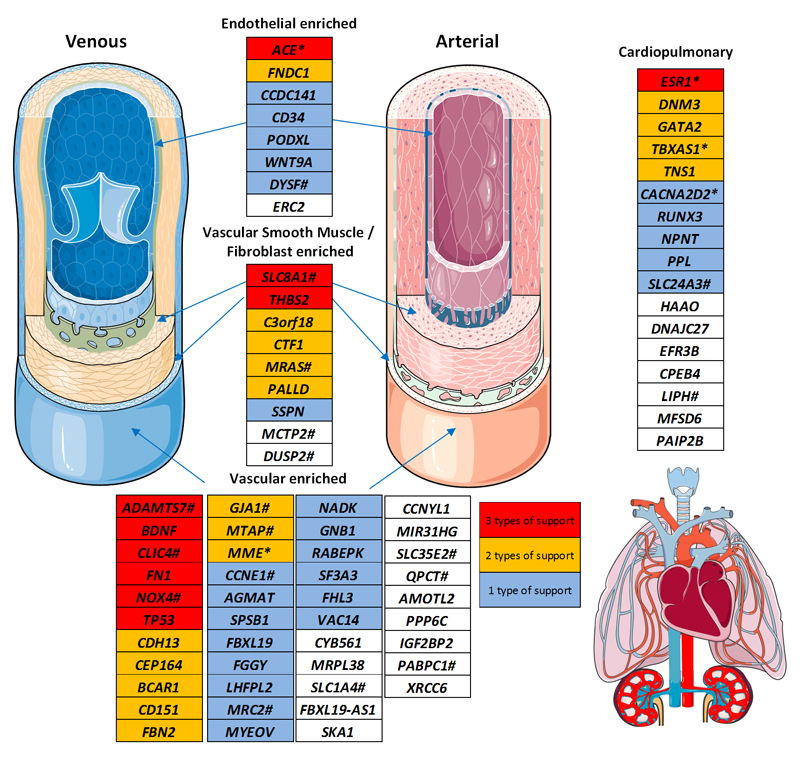
Our findings point to new biology as well as highlighting gene regions in systems that have previously been implicated in the genetics of BP. Several of our validated loci affect atherosclerosis or vascular remodelling (ADAMTS7, THBS2, CFDP1) and exhibit locus pleiotropy in prior genome-wide association studies for coronary artery disease or carotid intimal-media thickness24–26 (Supplementary Fig. 5a and Fig. 4). In previous work we have shown that expression of ADAMTS7 is upregulated and increases vascular smooth muscle cell migration in response to vascular injury in relation to a distinct coronary artery variant (rs3825807, not in LD with our sentinel SNV; r2 = 0.17)27. In endothelial cells ADAMTS7 encodes a metalloproteinase to cleave thrombospondin-1 encoded by THBS2 which leads to reduced endothelial cell migration and plays a role in neo-intimal repair in the vessel wall27. Our functional work indicates that the allele associated with lower DBP is also associated with lower ADAMTS7 expression in human VSMCs; this fits with the murine knockout that exhibits reduced atherosclerosis. SF3A3 encodes a splicing factor with no prior links to BP other than our reported association and eQTL. At the CFDP1 locus our sentinel SNV is in high LD (r2 = 0.95) with a variant previously associated with carotid intimal-medial thickness. Collectively our findings highlight a potential common mechanism among these genes in vascular remodelling that has previously been observed in small resistance arteries in essential hypertension28.
Figure 4.
Summary of gene cardiovascular expression from validated loci. Genes are shown on the basis of their tissue expression and supporting evidence summarised in Supplementary Table 16, based on Knockout (KO) phenotype, previously reported blood pressure biology or a strong functional rationale: eQTL (expression Quantitative Trait Loci), nsSNV (non-synonymous SNV), Hi-C. Multiple lines of evidence indicate the central importance of the vasculature in blood pressure regulation and we thus highlight existing drugged (*) and druggable (#) targets among these genes. Illustrations used elements with permission from Servier Medical Art. We note that some druggable genes may carry a safety liability, such as GJA1, which has known association with QT interval20
NADPH oxidase 4 (NOX4) has an established role in the endothelium where it enhances vasodilatation and reduces blood pressure in vivo29. This oxidase generates reactive oxygen species in the endothelium and may contribute to salt sensitive hypertension in the kidney and the vasculature30–32. We found that the allele of the common variant at the NOX4 locus correlates with increased tissue specific NOX4 expression in endothelial cells rather than VSMCs (Supplementary Fig. 13c). NOX4 mediates endothelial cell apoptosis and facilitates vascular collagen synthesis contributing to endothelial dysfunction and arterial stiffness, and may explain the association with PP33,34.
We identify several loci containing genes involved in vascular signalling and second messenger systems such as PDE5A and PDE10A35–37. The phosphodiesterase PDE5A hydrolyzes cyclic GMP and is inhibited by sildenafil which leads to vasodilatation38. This finding fits with our previous discoveries of a role for gene loci encoding elements of natriuretic peptide-nitric oxide pathway and guanylate cyclase signalling systems in BP regulation18,39,40. Our findings strengthen the case for evaluating the opportunity to repurpose PDE5A inhibitors for use in hypertension.
The importance of microvascular function is emphasised by the solute carrier transporters such as SLC14A2 encoding a urea transporter, which has previously been linked to autosomal dominant Streeten type orthostatic hypotensive disorder41 and BP response to nifedipine, a calcium channel blocker antihypertensive drug42. SLC8A1 encodes a sodium calcium exchanger expressed in cardiomyocytes which alters cardiac contractility and hypertrophy and shows abnormal BP in SLC8A1 transgenic mice43. Variants at SLC35F1 have previously been associated with resting heart rate and ventricular size which could contribute to BP elevation44.
We also identify loci that are involved in cardiovascular development (GATA2, KIAA1462, FBN2, FN1 and HAND2) such as fibrillin 2 (FBN2) which overlaps in action with fibrillin 1 in development of the aortic matrix45–49. In addition, fibronectin expression is increased in hypertension and in atherosclerosis but it may also play a role in the development of the heart49–51.
Our analysis validates loci containing genes with prior physiological connection to BP such as BDNF, FAM208A, and CACNA2D252–54. The neurotrophin Brain Derived Neurotrophic Factor (BDNF) modulates angiotensin 11 in the brain to elevate BP in experimental models; higher serum levels correlate with reduced risk of cardiovascular disease and mortality52. In experimental models FAM208A, which is thought to be a transcription factor, is a strong candidate for a QTL for BP54. The gene CACNA2D2 encodes a subunit of the L-type calcium channel that is most abundantly expressed in the atrium and in neurones and may be a target for negatively chronotropic and inotropic calcium channel antagonists which reduce BP55.
We examine long range genomic interactions using Hi-C, whereby the promoter region has a strong chromatin interaction with a novel SNV. One example is EPAS1, which is ˜200kb away from the SNV (rs11690961). It encodes hypoxia-inducible factor 2alpha, which affects catecholamine homeostasis, protects against heart failure and mutations in the gene are associated with pulmonary hypertension56. Another such gene is INHBA, 1.3Mb away from the SNV (rs12531683), which is elevated in pulmonary hypertension and contributes to vascular remodelling by inducing expression of endothelin-1 and plasminogen activator inhibitor-1 in pulmonary smooth muscle cells57.
Our observation of 9-10 mm Hg higher BP at age 50+ years when comparing the top vs bottom fifths of the BP GRS distribution has potential clinical and public health implications. We stratified by age due to a significant interaction of the GRS with age (P ranging between 9.96×10-11 and 1.16×10-3 for interaction with continuous BP traits, P = 0.012 for hypertension). Measuring the GRS in early life raises the possibility of adopting an early precision medicine approach to offset the genetic risk through lifestyle intervention (i.e. reduced sodium intake, increased potassium intake, maintenance of optimal weight, low adult alcohol consumption and regular exercise)58–60. Studies of non-pharmacologic approaches to BP control indicate that 10 mm Hg or more reduction in SBP is an achievable goal through lifestyle measures alone61, while recent evidence suggests that favorable lifestyle may offset the cardiovascular sequelae associated with high genetic risk62. As the above data are observational, the extent to which adherence to lifestyle recommendations amongst high genetic risk individuals might result in favorable outcomes remains uncertain; given the substantial effect of GRS on BP by middle-age, the potential for adopting early lifestyle intervention amongst individuals at high genetic risk, along with population-wide measures to lower BP, warrants further study.
Since the completion of our study, another BP GWAS using UK Biobank data has been published15, as part of a larger single-stage combined meta-analysis without replication; it reported a total of 316 loci, including 241 loci identified from the meta-analysis involving UK Biobank that were not tested for validation. Of the 107 validated loci reported in our study, 32 are discovered and validated for the first time in our analysis of UK Biobank. In addition, 75 sentinel SNVs are in LD (r2 ≥ 0.2) with the recently reported loci15 and we validate at least 53 of these for the first time in our study (indicated by “GIU” in Tables 1-3). Furthermore we note that 49 of the reported loci from the recent study15 did not validate in our large independent replication resource.
In summary we describe 107 validated loci for BP offering new biology, identifying potential new therapeutic targets and raising the possibility of a precision medicine approach to modify risk of hypertension and cardiovascular outcomes. Altogether, this represents a major advance in our understanding of the genetic architecture of BP.
Data Availability Statement
The data generated during the current study are available from the UK Biobank data repository (http://biota.osc.ox.ac.uk), which can be accessed by researchers upon application. This includes the derived GWAS analysis results summary data from our UK Biobank discovery data for all three BP traits. The genetic and phenotypic UK Biobank data are also available upon application to the UK Biobank (https://www.ukbiobank.ac.uk). All replication data generated during this study are included in the published article. For example, association results of look-up variants from our replication analyses and the subsequent combined meta-analyses are contained within all Supplementary Tables provided.
Online Methods
UK Biobank data
Our GWAS analysis is performed using data from the interim release of the first ˜150k UK Biobank (UKB) participants (Supplementary Note): ˜100k individuals from UK Biobank genotyped at ˜800,000 single nucleotide variants (SNVs) with a custom Affymetrix UK Biobank Axiom Array chip and ˜50k individuals genotyped with a custom Affymetrix UK BiLEVE Axiom Array chip from the UK BiLEVE study63, a subset of UKB. SNVs were imputed centrally by UKB using a merged UK10K sequencing + 1000G imputation reference panel. UK Biobank array design and protocols are available on the UK Biobank website.
Quality control
Following QC procedures already carried out centrally by UKB, we exclude discordant SNVs and samples with QC failures, gender discordance and high heterozygosity/missingness. We further restrict our data to a subset of individuals of European ancestry. By applying kmeans clustering to the Principal Component Analysis (PCA) data a total of N=145,315 Europeans remain (Supplementary Fig. 15). We use the kinship data to exclude 1st and 2nd degree relatives, with N=141,647 unrelated individuals remaining. Finally we restrict our data to non-pregnant individuals with two automated BP measurements available, resulting in a maximum of N=140,886 individuals for analysis (Supplementary Note).
Phenotypic data
After calculating the mean SBP and DBP values from the two BP measurements, we adjust for medication use by adding 15 and 10 mmHg to SBP and DBP, respectively, for individuals reported to be taking BP-lowering medication (21.4% of individuals)64. PP is calculated as SBP minus DBP, according to the medication-adjusted traits. Hypertension, used in secondary analyses, is defined as: (i) SBP ≥ 140 mmHg, or (ii) DBP ≥ 90 mmHg, (iii) or taking BP-lowering medication; otherwise individuals are classified as non-hypertensive. Descriptive summary statistics are provided for all individuals (Supplementary Table 1).
Statistical methods
Statistical approaches used for the discovery and replication of loci are reported in detail below. We also describe methods used for: identification of secondary signals; lookups in non-European populations and for monogenic BP genes; functional and experimental methods; construction of a genetic risk score for analysis with BP traits and cardiovascular outcomes. All P-values are from two-sided tests.
Analysis models
For the GWAS, we perform linear regression analyses of the three (untransformed) continuous, medication-adjusted BP traits (SBP, DBP, PP) for all measured and imputed genetic variants in dosage format using SNPTEST software65 under an additive genetic model. We carry out a similar analysis for the exome content. Quantile-quantile plots are shown in Supplementary Fig. 16. Each analysis includes the following covariates: sex, age, age2, body mass index, top ten PCs and a binary indicator variable for UK Biobank vs UK BiLEVE to adjust for the different genotyping chips. We also run an association analysis within UKB for validated BP SNVs and hypertension using logistic regression under an additive model with adjustments as above. There are 76,554 hypertensive cases and the 64,384 remaining participants are treated as non-hypertensive controls. This sample size is slightly larger than the N=140,866 used in the main analyses, since participants with only one BP measurement, but with reported BP-lowering medication, could be included as hypertensive.
Previously reported variants
We compile a list of all SNVs previously reported to be associated with BP at the time of analysis (Supplementary Table 13). This list includes all published SNVs which have been identified and validated from previous GWAS, CardioMetabochip and exome chip projects7–11. We augment this list to include all 34,459 SNVs in Linkage Disequilibrium (LD) with these previously reported SNVs, according to a threshold of r2 ≥ 0.2. Results for all these variants are extracted for each of the three BP traits, to check previously reported BP associations in the UKB data, according to whether the sentinel SNV or a variant at the locus in LD (r2 ≥ 0.2) with it showed evidence of support (P < 0.01) for association with at least one of the three BP traits.
Replication strategy
We use three independent external data sets for replication (Supplementary Note). First, for the GWAS analysis based on advanced 1000G imputation enhanced by UK10K data we consider SNVs with MAF ≥ 1% and perform a reciprocal replication exchange with the International Consortium of Blood Pressure (ICBP) 1000G meta-analysis (max N = 150,134). The imputation strategy for ICBP 1000G meta-analysis is based on an earlier imputation grid for the 1000G project. In addition, we recruit further cohorts with 1000G data which had not contributed to the ICBP-1000G discovery meta-analysis: ASCOT-UK (N = 3,803), ASCOT-SC (N = 2,462), BRIGHT (N = 1,791), Generation Scotland (GS) (N = 9,749), EGCUT (N = 5,468), Lifelines (N = 13,292) and PREVEND (N = 3,619). This gives a total of N = 190,318 independent replication samples for the GWAS analysis.
Second, because the UK Biobank and UK BiLEVE genotyping chips contain exome content, we sought replication from two BP exome consortia (European exome consortium and the Cohorts for Heart and Ageing research in Genome Epidemiology – CHARGE BP exome consortium), to allow validation of coding variants and variants with lower frequency. The European exome consortium (N = 161,926) and CHARGE consortium (N = 119,792) give a total of N = 281,718 independent replication samples for the exome analysis.
Note that the lookups for GWAS and exome discovery are distinct sets of SNVs. Loci are assigned sequentially, prioritising the primary GWAS discovery first, then considering any remaining loci with non-overlapping exome content for replication in the independent exome replication resources.
Statistical criteria for replication
For the GWAS discovery, there are ˜9.8 million SNVs with MAF ≥ 1% and INFO > 0.1. We consider for follow-up any SNVs with P < 1x10-6 for any of the three BP traits. For the exome discovery, there are 149,026 exome SNVs (Supplementary Note) which were polymorphic with INFO > 0.1; for follow-up we consider all SNVs with MAF ≥ 0.01% and P < 1x10-5. All such SNVs are annotated to loci according to both an LD threshold of r2 ≥ 0.2 and a 1Mb interval region (see Supplementary Note), and signals are classified either as belonging to unvalidated loci, or being potential secondary signals at previously reported loci at the time of analysis.
Selection of variants for follow-up
The sentinel (most significant) SNV from each association signal is selected for follow-up, all of which are pairwise-independent by LD (r2 < 0.2). For the GWAS discovery, we check that potential lookup SNVs are covered within the ICBP-1000G replication data (Supplementary Note; Supplementary Tables 28 and 29). Of the 235 novel loci containing previously unreported SNVs with MAF ≥ 1%, INFO > 0.1 and P<1x10-6, 218 are covered, and similarly 100 of the 123 potential secondary SNVs at 51 of the 54 previously reported BP loci are available for follow-up. For the exome discovery, by following up SNVs with MAF ≥ 0.01%, INFO > 0.1 and P < 1x10-5 across the three BP traits, we carry forward for replication sentinel SNVs at 22 unvalidated loci, and potential secondary SNVs at three previously reported loci at the time of analysis. We produce locus zoom plots for each of the lookup variants.
Replication meta-analyses
The replication and combined meta-analyses are performed within METAL software66 using fixed effects inverse variance weighted meta-analysis (Supplementary Note). The combined meta-analysis of both the UKB discovery (N = 140,886) and GWAS replication meta-analysis (max N = 190,070) include a total maximum sample size of N = 330,956. For the exome combined meta-analysis, we synthesize data from the UKB discovery exome content (max N=140,866), with the replication dataset from both exome consortia (total max N=281,718), giving a maximum sample size of N=422,604.
Validation Criteria
In our study a signal is declared validated if it satisfies ALL of the following three criteria:
-
(i)
the sentinel SNV is genome-wide significant (P < 5×10-8) in the combined meta-analysis for any of the three BP traits;
-
(ii)
the sentinel SNV shows evidence of support (P < 0.01) in the replication meta-analysis alone for association with the most significantly associated BP trait from the combined meta-analysis (NB: P < 0.01 is more stringent than a range of thresholds calculated according to False Discovery Rate (FDR), see Supplementary Methods);
-
(iii)
the sentinel SNV has concordant direction of effect between the UKB discovery and the replication meta-analysis for the most significantly associated BP trait from the combined meta-analysis.
Secondary signals
By conditional analysis within UKB data we assess all validated secondary signals from our validated and previously reported loci at the time of analysis for independence from the sentinel or previously reported SNV, respectively (Supplementary Note). We declare a secondary signal to be independent of the previously reported SNV if there is less than a 1.5 fold difference between the main association and conditional association P-values on a –log10 scale, i.e. if –log10(P) / -log10(P_cond) < 1.5. Note that the lookup criteria already ensure that the secondary variant is not in LD (r2 < 0.2) with the previously reported SNV. If more than one SNV in a region is found to be independent we undertake further rounds of iterative conditional analysis.
Lookups in non-European ancestries
As a secondary analysis, we look up 102 and 5 validated SNVs from the GWAS and exome analyses, respectively, in non-European ancestry samples. These comprise analysis of East Asian (N = 31,513) and South Asian (N = 33,115) ancestry data from the iGEN-BP consortium11 for the GWAS lookups, and South Asian (N = 25,937), African American (N = 21,488) and Hispanic (N = 4,581) ancestry data from the CHARGE BP exome consortium10 and CHD+ Exome consortium9, for the exome content lookups (Supplementary Note). We carry out a binomial (sign) test based on the number of SNVs with consistent directions of effect between UKB and each of the non-European ancestry samples.
Monogenic blood pressure gene lookups
The UKB arrays include some rare coding variants for monogenic disorders. We collate a list of all specific mutation variants within genes known to be associated with monogenic BP disorders19. Results from the UKB association analyses for all three BP traits are extracted for any of these SNVs directly covered within the UKB dataset (Supplementary Table 14). Note that a search of proxies did not augment the list of available variants, so results are reported for the specific variants only.
Functional analyses
In order to prioritize associated SNVs, we use an integrative bioinformatics approach to collate functional annotation (Supplementary Table 30) at both the variant and gene level for each SNV within the BP loci (all SNVs in LD r2 ≥ 0.8 with the BP-associated SNVs). At the variant level we use ANNOVAR67 to obtain comprehensive functional characterization of variants, including gene location, conservation and amino acid substitution impact based on a range of prediction tools including SIFT and polyphen2. All nonsynonymous variants were predicted damaging by two or more methods.
We use the University of California Santa Cruz (UCSC) genome browser to review sequence specific context of SNVs in relation to function, particularly in the Encyclopedia of DNA Elements (ENCODE) dataset68. We use the UCSC table browser to annotate SNVs in ENCODE regulatory regions. We evaluate SNVs for impact on putative micro RNA target sites in the 3’ un-translated regions (3’UTR) of transcripts by a query of the miRNASNP database69. We evaluate all SNVs in LD (r2 ≥ 0.8) with our validated sentinel SNVs for evidence of mediation of expression quantitative trait loci (eQTL) in all 44 tissues using the Genotype-Tissue Expression (GTEx) database, in order to identify validated loci which are highly expressed, and to highlight specific tissue types which show eQTLs for a large proportion of validated loci. We further seek to identify validated loci with the strongest evidence of eQTL associations in arterial tissue, in particular.
At the gene level, we use Ingenuity Pathway Analysis (IPA) software (IPA®,QIAGEN Redwood City) to review genes with prior links to BP, based on annotation with the “Blood Pressure” Medline Subject Heading (MESH) term which is annotated to 684 genes. We also use IPA to identify genes which interact with BP MESH annotated genes, and evaluate genes for evidence of small molecule druggability based on queries of Chembl and Drug Gene Interaction database.
We then perform overall enrichment testing across all loci. Firstly, we use DEPICT70 (Data-driven Expression Prioritized Integration for Complex Traits) to identify highly expressed tissues and cells within the BP loci. DEPICT uses a large number of microarrays (˜37k) to identify cells and tissues where the genes are highly expressed and uses precomputed GWAS phenotypes to adjust for co-founding sources. DEPICT provides a P-value of enrichment and false discovery rates adjusted P-values for each tissue/cells tested.
Furthermore, to investigate regulatory regions, we employ a two tiered approach to investigate cell type specific enrichment within DNase I sites using FORGE, which tests for enrichment of SNVs within DNase I sites in 123 cell types from the Epigenomics Roadmap Project and ENCODE71 (Supplementary Note). Validated sentinel SNVs from our study are analysed along with previously reported SNVs at the time of analysis and secondary signals (with P-value < 1×10-4) to evaluate the overall tissue specific enrichment of BP associated variants. In a second analysis we use FORGE (with no LD filter) to investigate directly our curated candidate regulatory SNVs for overlap with cell-specific DNase I signals.
GenomeRunner72 is used to search for enrichment of validated and previously reported sentinel SNVs with histone modification mark genomic features (Supplementary Note). Relevant cardiovascular tissue expression is investigated using Fantom5 reference transcript expression data (fantom.gsc.riken.jp/5) (Supplementary Note).
We use IPA (IPA®,QIAGEN Redwood City) to identify biological pathways and transcriptional upstream regulators enriched for genes within the BP loci. The transcriptional upstream regulator analysis aims to identify transcription factors, compounds, drugs, kinases and other molecules, for which the target is one of the BP genes under investigation.
We query SNVs against PhenoScanner16 to investigate trait pleiotropy, extracting all association results with nominal significance at P < 0.05 for full reporting (Supplementary Table 16), and then extract genome-wide significant results to highlight the validated loci with strongest evidence of association with other traits (Supplementary Fig. 5a). We also use the Genomic Regions Enrichment of Annotations Tool (GREAT) to study gene set enrichment of mouse phenotype and disease ontology terms within our validated and previously reported loci at the time of analysis, using default SNV to gene mapping settings73.
We carry out metabolomics analysis using two sets of data. First we use 1H NMR lipidomics data on plasma from a subset of 2,000 participants of the Airwave Health Monitoring Study74,75 (Supplementary Note). For each validated BP-associated SNV we ran association tests with the lipidomics data using linear regression analyses, adjusted for age and sex. We computed significance thresholds using a permutation derived family wise error rate (5%) to account for the high correlation structure of these data (ENT=35)76. We also test each validated SNV against published genome-wide vs metabolome-wide associations in plasma and urine using publicly available data from the “Metabolomics GWAS Server” to identify metabolites that have been associated with variants of interest at P < 3.0 x 10-4 (Bonferroni corrected P for validated signals)22,23.
Experimental methods
We prioritize genes for laboratory testing on the basis of evidence for SNV function (including coding variants, eQTLs and Hi-C interactions), biological support for relevance to BP (from literature review) and transgenic phenotype. We perform genotyping and Quantitative Reverse-Transcription Polymerase Chain Reaction (q RT-PCR) for the selected sentinel variants of interest using human vascular smooth muscle cells and endothelial cells and test for expression levels (Supplementary Note; Supplementary Table 31). All three SNVs were tested using an additive model.
Genetic risk scores
Genetic risk scores (GRS) are constructed using the independent Airwave study74 data to assess the combined effect of the BP-associated variants on BP and risk of hypertension (Supplementary Note), whilst avoiding bias by “winners curse”. We create weighted GRSs for all pairwise-independent, LD-filtered (r2 < 0.2) previously reported variants at the time of analysis and our validated variants (sentinel and secondary SNVs) combined, using available SNVs (Supplementary Table 22). For the previously reported variants, we weight BP increasing alleles by the beta coefficients from the UKB analysis. For our validated variants, beta coefficients of the replication meta-analysis are used as independent, unbiased weights.
For the variance explained analyses within the independent Airwave cohort, we use three trait-specific GRSs (SBP, DBP, PP). Each GRS includes all variants, but weights are trait-specific, using the beta coefficients from the analysis of each of the three different BP traits, e.g. the SBP-GRS is weighted by the beta coefficients from the SBP-GWAS. To calculate the percent of variance for each BP trait explained by its corresponding trait-specific GRS, not accounted for by known factors, we generate the residuals from the regression model of each trait against covariates of age, age2, sex and body mass index. We then fit a second linear model for the trait residuals with all the variants in the GRS plus the top 10 PCs.
For risk score analyses we calculate a single BP GRS, as the average of the SBP and DBP GRSs. We standardize the average GRS to have mean of zero and standard deviation of one. We assess the association of the continuous average GRS variable with each BP trait by simple linear regression. We also run a logistic regression to examine the association of the average GRS with risk of hypertension. We perform each analysis both with and without adjustment for sex. We test for interaction between age (< 50, and ≥50 years) and the effect of the GRS on BP. We then compare BP levels and risk of hypertension for individuals in the top and bottom 20% of the GRS distribution at ≥50 years using linear and logistic regression, respectively.
We also assess the association of the average BP GRS with cardiovascular outcomes in the UKB data. We include all pairwise-independent previously reported BP variants at the time of analysis, and our validated variants. We use logistic regression with binary outcome variables for coronary heart disease, stroke and cardiovascular disease (see Supplementary Note) and GRS as explanatory variable (with and without sex adjustment).
Supplementary Material
Acknowledgements
HRW, CPC, MR, MRB, PBM, MB and MJC were funded by the National Institutes for Health Research (NIHR) as part of the portfolio of translational research of the NIHR Biomedical Research Unit at Barts and The London School of Medicine and Dentistry.
HG was funded by the NIHR Imperial College Health Care NHS Trust and Imperial College London Biomedical Research Centre.
MR was a recipient from China Scholarship Council (No. 2011632047).
BM holds an MRC eMedLab Medical Bioinformatics Career Development Fellowship, funded from award MR/L016311/1.
JMMH was funded by the UK Medical Research Council (G0800270), British Heart Foundation (SP/09/002), UK National Institute for Health Research Cambridge Biomedical Research Centre, European Research Council (268834), European Commission Framework Programme 7 (HEALTH-F2-2012-279233).
BK holds a British Heart Foundation Personal Chair (CH/13/2/30154).
NJS holds a chair funded by the British Heart Foundation and is a NIHR Senior Investigator.
FD was funded by the MRC Unit at the University of Bristol (MC_UU_12013/1-9).
PSu was funded by the UK Medical Research Council (G0800270).
CL and AK were funded by the NHLBI intramural funding.
CNC was funded by the National Institutes of Health (HL113933, HL124262).
PVDH was funded by the ZonMw grant 90.700.441, Marie Sklodowska-Curie GF (call: H2020-MSCA-IF-2014, Project ID: 661395).
NV was supported by Marie Sklodowska-Curie GF grant (661395) and ICIN-NHI.
NP received funding from the UK National Institute for Health Research Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London and also from his Senior Investigator Award.
PS was supported by the NIHR Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London.
ST was supported by the NIHR Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London.
PFO received funding from the UK Medical Research Council (MR/N015746/1) and the Wellcome Trust (109863/Z/15/Z).
IK was supported by the EU PhenoMeNal project (Horizon 2020, 654241).
AC was funded by the National Institutes of Health (HL128782, HL086694).
MF was supported by the Wellcome Trust core award (090532/Z/09/Z) and the BHF Centre of Research Excellence, Oxford (RE/13/1/30181).
CH was funded by an MRC core grant for QTL in Health and Disease programme.
Some of this work used the ALICE and SPECTRE High Performance Computing Facilities at the University of Leicester.
MJC is a National Institute for Health Research (NIHR) senior investigator.
PE is a National Institute for Health Research (NIHR) senior investigator and acknowledges support from the NIHR Biomedical Research Centre at Imperial College Healthcare NHS Trust and Imperial College London, and the NIHR Health Protection Research Unit in Health Impact of Environmental Hazards (HPRU-2012-10141). As director of the MRC-PHE Centre for Environment and Health, PE acknowledges support from the Medical Research Council and Public Health England (MR/L01341X/1).
This work used the computing resources of the UK MEDical BIOinformatics partnership - aggregation, integration, visualisation and analysis of large, complex data (UK MED-BIO) which is supported by the Medical Research Council (MR/L01632X/1).
This research was supported by the British Heart Foundation (grant SP/13/2/30111).
Project title: Large-scale comprehensive genotyping of UK Biobank for cardiometabolic traits and diseases: UK CardioMetabolic Consortium (UKCMC).
This research has been conducted using the UK Biobank Resource under Application Number 236.
International Consortium of Blood Pressure (ICBP) 1000 Genomes Discovery Contributors
Louise V. Wain (56), Ahmad Vaez (23,58), Rick Jansen (59), Roby Joehanes (60,9), Peter J. van der Most (23), A. Mesut Erzurumluoglu (56), Paul O'Reilly (21), Claudia P. Cabrera (1,2), Helen R. Warren (1,2), Lynda M. Rose (45), Germaine C. Verwoert (61), Jouke-Jan Hottenga (62), Rona J. Strawbridge (63,64), Tonu Esko (29,65,66), Dan E. Arking (47), Shih-Jen Hwang (67,68), Xiuqing Guo (69), Zoltan Kutalik (70,71), Stella Trompet (72,73), Nick Shrine (56), Alexander Teumer (74,75), Janina S. Ried (76), Joshua C. Bis (77), Albert V. Smith (78,79), Najaf Amin (80), Ilja M. Nolte (23), Leo-Pekka Lyytikäinen (81,82), Anubha Mahajan (48), Nicholas J. Wareham (83), Edith Hofer (84,85), Peter K. Joshi (86), Kati Kristiansson (87), Michela Traglia (88), Aki S. Havulinna (87), Anuj Goel (89,48), Mike A. Nalls (90,91), Siim Sõber (92), Dragana Vuckovic (93,94), Jian'an Luan (83), Fabiola Del Greco M. (95), Kristin L. Ayers (96), Jaume Marrugat (97), Daniela Ruggiero (98), Lorna M. Lopez (99,100,101), Teemu Niiranen (87), Stefan Enroth (102), Anne U. Jackson (103), Christopher P. Nelson (54,55), Jennifer E. Huffman (19), Weihua Zhang (3,50), Jonathan Marten (19), Ilaria Gandin (94), Sarah E Harris (99,28), Tatijana Zemonik (104), Yingchang Lu (105), Evangelos Evangelou (3,4), Nabi Shah (106,107), Martin H. de Borst (36), Massimo Mangino (108,109), Bram P. Prins (110), Archie Campbell (28,111), Ruifang Li-Gao (112), Ganesh Chauhan (113,114), Christopher Oldmeadow (115), Gonçalo Abecasis (116), Maryam Abedi (117), Caterina M. Barbieri (88), Michael R. Barnes (1,2), Chiara Battini (56), BIOS Consortium (118), Tineka Blake (56), Michael Boehnke (103), Erwin P. Bottinger (105), Peter S. Braund (54,55), Morris Brown (1,2), Marco Brumat (94), Harry Campbell (86), John C. Chambers (3,50,43), Massimiliano Cocca (94), Francis Collins (119), John Connell (35), Heather J. Cordell (120), Jeffrey J. Damman (121), Gail Davies (99,122), Eco J. de Geus (62), Renée de Mutsert (112), Joris Deelen (123), Yusuf Demirkale (124), Alex S.F. Doney (106), Marcus Dörr (125,75), Martin Farrall (89,48), Teresa Ferreira (48), Mattias Frånberg (63,64,126), He Gao (3), Vilmantas Giedraitis (127), Christian Gieger (128), Franco Giulianini (45), Alan J. Gow (99,129), Anders Hamsten (63,64), Tamara B. Harris (130), Albert Hofman (61,131), Elizabeth G. Holliday (115), Marjo-Riitta Jarvelin (132,133,134,5), Åsa Johansson (102), Andrew D. Johnson (9,135), Pekka Jousilahti (87), Antti Jula (87), Mika Kähönen (136,137), Sekar Kathiresan (18,17,16), Kay-Tee Khaw (41), Ivana Kolcic (138), Seppo Koskinen (87), Claudia Langenberg (83), Marty Larson (9), Lenore J. Launer (130), Benjamin Lehne (3), David C.M. Liewald (99,122), Lifelines Cohort Study (118), Li Lin (57), Lars Lind (139), François Mach (57), Chrysovalanto Mamasoula (140), Cristina Menni (108), Borbala Mifsud (1), Yuri Milaneschi (141), Anna Morgan (94), Andrew D. Morris (142), Alanna C. Morrison (143), Peter J. Munson (124), Priyanka Nandakumar (47), Quang Tri Nguyen (124), Teresa Nutile (98), Albertine J. Oldehinkel (144), Ben A. Oostra (80), Elin Org (29), Sandosh Padmanabhan (145,111), Aarno Palotie (146), Guillaume Paré (147), Alison Pattie (122), Brenda W.J.H. Penninx (141), Neil Poulter (30), Peter P. Pramstaller (95,148,149), Olli T. Raitakari (150,151), Meixia Ren (1,152), Kenneth Rice (153), Paul M. Ridker (45,46), Harriëtte Riese (144), Samuli Ripatti (146), Antonietta Robino (154), Jerome I. Rotter (155), Igor Rudan (86), Yasaman Saba (156), Aude Saint Pierre (95,157), Cinzia F. Sala (88), Antti-Pekka Sarin (146), Reinhold Schmidt (84), Rodney Scott (115,158,159), Marc A. Seelen (36), Denis C. Shields (37), David Siscovick (160), Rossella Sorice (98,161), Alice Stanton (34), David J. Stott (33), Johan Sundström (139), Morris Swertz (162), Kent D. Taylor (163,164), Simon Thom (165), Ioanna Tzoulaki (3), Christophe Tzourio (113,114,166), André G. Uitterlinden (61,167), Understanding Society Scientific group (118), Uwe Völker (168,75), Peter Vollenweider (169), Sarah Wild (86), Gonneke Willemsen (62), Alan F. Wright (19), Jie Yao (69), Sébastien Thériault (147), David Conen (170), Attia John (115,158,159), Peter Sever (38), Stéphanie Debette (113,114,171), Dennis O. Mook-Kanamori (112,172), Eleftheria Zeggini (110), Tim D. Spector (108), Pim van der Harst (15), Colin N.A. Palmer (106), Anne-Claire Vergnaud (3), Ruth J.F. Loos (83,173,174), Ozren Polasek (138), John M. Starr (99,175), Giorgia Girotto (94,93), Caroline Hayward (19,111), Jaspal S. Kooner (38,50,43), Cecila M. Lindgren (66,48), Veronique Vitart (19), Nilesh J. Samani (54,55), Jaakko Tuomilehto (176,177,178,179), Ulf Gyllensten (102), Paul Knekt (87), Ian J. Deary (99,122), Marina Ciullo (98,161), Roberto Elosua (97), Bernard D. Keavney (52), Andrew A. Hicks (95), Robert A. Scott (83), Paolo Gasparini (93,94), Maris Laan (92,180), YongMei Liu (181), Hugh Watkins (89,48), Catharina A. Hartman (144), Veikko Salomaa (87), Daniela Toniolo (88), Markus Perola (87,146,182), James F. Wilson (86,19), Helena Schmidt (156,183), Jing Hua Zhao (83), Terho Lehtimäki (81,82), Cornelia M. van Duijn (80), Vilmundur Gudnason (78,79), Bruce M. Psaty (77,184,185,186), Annette Peters (76), Rainer Rettig (187), Alan James (188,189), J Wouter Jukema (72), David P. Strachan (190), Walter Palmas (191), Andres Metspalu (29), Erik Ingelsson (192,193), Dorret I. Boomsma (62), Oscar H. Franco (61), Murielle Bochud (70), Christopher Newton-Cheh (194,18,16,66), Patricia B. Munroe (1,2), Paul Elliott (5), Daniel I. Chasman (45,46), Aravinda Chakravarti (47), Joanne Knight (22), Andrew P. Morris (10,48), Daniel Levy (7,68), Martin D. Tobin (56), Harold Snieder (23), Mark J. Caulfield (1,2), Georg B. Ehret (47,57)
58. Research Institute for Primordial Prevention of Non-communicable Disease, Isfahan University of Medical Sciences, Isfahan, Iran
59. Department of Psychiatry, VU University Medical Center, Neuroscience Campus Amsterdam, Amsterdam, The Netherlands
60. Hebrew SeniorLife, Harvard Medical School, 1200 Centre Street Room #609, Boston, MA 02131, USA
61. Department of Epidemiology, Erasmus MC, Rotterdam, 3000CA, The Netherlands
62. Department of Biological Psychology, Vrije Universiteit, Amsterdam, EMGO+ institute, VU University medical center, Amsterdam, The Netherlands
63. Cardiovascular Medicine Unit, Department of Medicine Solna, Karolinska Institutet, Stockholm, 17176, Sweden
64. Centre for Molecular Medicine, Karolinska Universitetsjukhuset, Solna, 171 76, Sweden
65. Divisions of Endocrinology/Children's Hospital, Boston, MA 02115, USA
66. Broad Institute of Harvard and MIT, Cambridge, MA 02139 USA
67. The Population Science Branch, Division of Intramural Research, National Heart Lung and Blood Institute national Institute of Health, Bethesda, MD 20892, USA
68. The Framingham Heart Study, Framingham MA 01702, USA
69. The Institute for Translational Genomics and Population Sciences, Department of Pediatrics, LABioMed at Harbor-UCLA Medical Center, 1124 W. Carson Street, Torrance, CA 90502, USA
70. Institute of Social and Preventive Medicine, Lausanne University Hospital, Route de la Corniche 10, 1010 Lausanne, Switzerland
71. Swiss Institute of Bioinformatics, Lausanne, Switzerland
72. Department of Cardiology, Leiden University Medical Center, Leiden, 2300RC, The Netherlands
73. Department of Gerontology and Geriatrics, Leiden University Medical Center, Leiden, 2300RC, The Netherlands
74. Institute for Community Medicine, University Medicine Greifswald, Greifswald, 17475, Germany
75. DZHK (German Centre for Cardiovascular Research), partner site Greifswald, Greifswald, 17475, Germany
76. Institute of Epidemiology II, Helmholtz Zentrum München, Neuherberg 85764, Germany
77. Cardiovascular Health Research Unit, Department of Medicine, University of Washington, Seattle, WA 98101, USA
78. Icelandic Heart Assoication, Kopavogur, Iceland
79. Faculty of Medicine, University of Iceland, Reykjavik, Iceland
80. Genetic Epidemiology Unit, Department of Epidemiology, Erasmus MC, Rotterdam, 3000CA, The Netherlands
81. Department of Clinical Chemistry, Fimlab Laboratories, Tampere 33520, Finland
82. Department of Clinical Chemistry, University of Tampere School of Medicine, Tampere 33014, Finland
83. MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, Institute of Metabolic Science, Cambridge Biomedical Campus, Cambridge, CB2 0QQ, UK
84. Clinical Division of Neurogeriatrics, Department of Neurology, Medical University Graz, Auenbruggerplatz 22, 8036 Graz, Austria
85. Institute of Medical Informatics, Statistics and Documentation, Medical University Graz, Auenbruggerplatz 2, 8036 Graz, Austria
86. Centre for Global Health Research, Usher Institute of Population Health Sciences and Informatics, University of Edinburgh EH89AG, Scotland, UK
87. Department of Health, National Institute for Health and Welfare (THL), Helsinki, Finland
88. Division of Genetics and Cell Biology, San Raffaele Scientific Institute, 20132 Milano, Italy
89. Division of Cardiovascular Medicine, Radcliffe Department of Medicine, University of Oxford, Oxford, OX3 9DU, UK
90. Laboratory of Neurogenetics, National Institute on Aging, NIH, Bethesda, 20892, USA
91. Kelly Services, Rockville, MD, USA
92. Human Molecular Genetics Research Group, Institute of Molecular and Cell Biology, University of Tartu, Riia St.23, 51010 Tartu, Estonia
93. Experimental Genetics Division, Sidra Medical and Research Center, PO Box 26999, Doha, Qatar
94. Department of Medical, Surgical and Health Sciences, University of Trieste, Strada di Fiume 447, Trieste, 34100, Italy
95. Center for Biomedicine, European Academy Bozen/Bolzano (EURAC), Bolzano, Italy - Affiliated Institute of the University of Lübeck, Germany
96. Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA
97. Cardiovascular Epidemiology and Genetics, IMIM. Dr Aiguader 88, Barcelona, 08003, Spain
98. Institute of Genetics and Biophysics A. Buzzati-Traverso, CNR, via P. Castellino 111, 80131 Napoli, Italy
99. Centre for Cognitive Ageing and Cognitive Epidemiology, University of Edinburgh, 7 George Square, Edinburgh EH8 9JZ, UK
100. Department of Psychiatry, Royal College of Surgeons in Ireland, Education and Research Centre, Beaumont Hospital, Dublin, Ireland
101. University College Dublin, UCD Conway Institute, Centre for Proteome Research, UCD, Belfield, Dublin, Ireland
102. Department of Immunology, Genetics and Pathology, Uppsala Universitet, Science for Life Laboratory, Husargatan 3, Uppsala, SE-75108, Sweden
103. Department of Biostatistics and Center for Statistical Genetics, University of Michigan, Ann Arbor, MI 48109, USA
104. Department of Biology, Faculty of Medicine, University of Split, Croatia
105. The Charles Bronfman Institute for Personalized Medicine, Icachn School of Medicine at Mount Sinai, New York, NY 10029, USA
106. Medical Research Institute, University of Dundee, Ninewells Hospital and Medical School, Dundee, DD1 9SY, Scotland, UK
107. Department of Pharmacy, COMSATS Institute of Information Technology, Abbottabad, 22060, Pakistan
108. Department of Twin Research and Genetic Epidemiology, Kingâ’™s College London, Lambeth Palace Rd, London, SE1 7EH, UK
109. National Institute for Health Research Biomedical Research Centre, London SE1 9RT, UK
110. Department of Human Genetics, Wellcome Trust Sanger Institute, CB10 1HH, United Kingdom
111. Generation Scotland, Centre for Genomic and Experimental Medicine, University of Edinburgh, Edinburgh, EH4 2XU, UK
112. Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands
113. INSERM U 1219, Bordeaux Population Health center, Bordeaux, France
114. Bordeaux University, Bordeaux, France
115. Hunter Medical Research Institute, New Lambton, NSW 2305, Australia
116. Center for Statistical Genetics, Dept. of Biostatistics, SPH II, 1420 Washington Heights, Ann Arbor, MI 48109-2029, USA
117. Department of Genetics and Molecular Biology, Isfahan University of Medical Sciences, Isfahan, Iran
118. SPECIAL REFERENCE
119. Medical Genomics and Metabolic Genetics Branch, National Human Genome Research Institute, NIH, Bethesda, MD 20892, USA
120. Institute of Genetic Medicine, Newcastle University, Newcastle upon Tyne, UK
121. Department of Pathology, Amsterdam Medical Center, Meibergdreef 9, 1105 AZ, Amsterdam, The Netherlands
122. Department of Psychology, University of Edinburgh, 7 George Square, Edinburgh, EH8 9JZ, UK
123. Department of Molecular Epidemiology, Leiden University Medical Center, Leiden, 2300RC, The Netherlands
124. Center for Information Technology, NIH, USA
125. Department of Internal Medicine B, University Medicine Greifswald, Greifswald, 17475, Germany
126. Department of Numerical Analysis and Computer Science, Stockholm University, Lindstedtsvägen 3, Stockholm, 100 44, Sweden
127. Department of Public Health and Caring Sciences, Geriatrics, Uppsala 752 37, Sweden
128. Helmholtz Zentrum Muenchen, Deutsches Forschungszentrum fuer Gesundheit und Umwelt (GmbH), Ingolstaedter Landstr. 1, 85764 Neuherberg, München, Germany
129. Department of Psychology, School of Life Sciences, Heriot-Watt University, Edinburgh, EH14 4AS, UK
130. Intramural Research Program, Laboratory of Epidemiology, Demography, and Biometry, National Institute on Aging, USA
131. Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA 02115, USA
132. Center For Life-course Health Research, P.O. Box 5000, FI-90014 University of Oulu, Finland
133. Biocenter Oulu, P.O. Box 5000, Aapistie 5A, FI-90014 University of Oulu, Finland
134. Unit of Primary Care, Oulu University Hospital, Kajaanintie 50, P.O. Box 20, FI-90220 Oulu, 90029 OYS, Finland
135. National Heart, Lung and Blood Institute, Cardiovascular Epidemiology and Human Genomics Branch, Bethesda, MD 20814, USA
136. Department of Clinical Physiology, Tampere University Hospital, Tampere 33521, Finland
137. Department of Clinical Physiology, University of Tampere School of Medicine, Tampere 33014, Finland
138. Department of Public Health, Faculty of Medicine, University of Split, Croatia
139. Department of Medical Sciences, Cardiovascular Epidemiology, Uppsala University, Uppsala 751 85, Sweden
140. Institute of Health and Society, Newcastle University, Newcastle upon Tyne, UK
141. Department of Psychiatry, EMGO Institute for Health and Care Research, VU University Medical Center, A.J. Ernststraat 1187, 1081 HL Amsterdam, The Netherlands
142. School of Molecular, Genetic and Population Health Sciences, University of Edinburgh, Medical School,Teviot Place, Edinburgh, EH8 9AG, Scotland, UK
143. Department of Epidemiology, Human Genetics and Environmental Sciences, School of Public Health, University of Texas Health Science Center at Houston, 1200 Pressler St., Suite 453E, Houston, TX 77030, USA
144. Interdisciplinary Center Psychopathology and Emotion Regulation (IPCE), University of Groningen, University Medical Center Groningen, Hanzeplein 1, PO Box 30001, 9700 RB Groningen, The Netherlands
145. British Heart Foundation Glasgow Cardiovascular Research Centre, Institute of Cardiovascular and Medical Sciences, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8TA, UK
146. Institute for Molecular Medicine Finland (FIMM), University of Helsinki, Helsinki, Finland
147. Department of Pathology and Molecular Medicine, McMaster University, 1280 Main St W, Hamilton, L8S 4L8, Canada
148. Department of Neurology, General Central Hospital, Bolzano, Italy
149. Department of Neurology, University of Lübeck, Lübeck, Germany
150. Department of Clinical Physiology and Nuclear Medicine, Turku University Hospital, Turku 20521, Finland
151. Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku 20014, Finland
152. Department of Cardiology, Fujian Provincial Hospital, Fujian Medical University, Fuzhou 350001, China
153. Department of Biostatistics University of Washington, Seattle, WA 98101, USA
154. Institute for Maternal and Child Health IRCCS Burlo Garofolo, Via dell'Istria 65, Trieste, 34200, Italy
155. The Institute for Translational Genomics and Population Sciences, Departments of Pediatrics and Medicine, LABioMed at Harbor-UCLA Medical Center, 1124 W. Carson Street, Torrance, CA 90502, USA
156. Institute of Molecular Biology and Biochemistry, Centre for Molecular Medicine, Medical University of Graz, Harrachgasse 21, 8010 Graz, Austria
157. INSERM U1078, Etablissement Français du Sang, 46 rue Félix Le Dantec, CS 51819, Brest Cedex 2 29218, France
158. Faculty of Health, University of Newcastle, Callaghan NSW 2308, Australia
159. John Hunter Hospital, New Lambton NSW 2305, Australia
160. The New York Academy of Medicine. 1216 5th Ave, New York, NY 10029, USA
161. IRCCS Neuromed, Pozzilli, Isernia, Italy
162. Department of Genetics, University of Groningen, University Medical Center Groningen, PO Box 30001, 9700 RB Groningen, The Netherlands
163. Institute for Translational Genomics and Population Sciences. Los Angeles BioMedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, 90502, USA
164. Division of Genetic Outcomes, Department of Pediatrics, Harbor-UCLA Medical Center, Torrance, CA, 90502, USA
165. International Centre for Circulatory Health, Imperial College London, W2 1PG, UK
166. Department of Public Health, Bordeaux University Hospital, Bordeaux, France
167. Department of Internal Medicine, Erasmus MC, Rotterdam, 3000CA, The Netherlands
168. Interfaculty Institute for Genetics and Functional Genomics, University Medicine Greifswald, Greifswald, 17475, Germany
169. Department of Internal Medicine, Lausanne Universiyt Hospital, CHUV, 1011 Lausanne, Switzerland
170. Population Health Research Institute, McMaster University, Hamilton Ontario, Canada
171. Department of Neurology, Bordeaux University Hospital, Bordeaux, France
172. Department of Public Health and Primary Care, Leiden University Medical Center, Leiden, The Netherlands
173. The Charles Bronfman Institute for Personalized Medicine, The Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
174. Mindich Child health Development Institute, The Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
175. Alzheimer Scotland Dementia Research Centre, University of Edinburgh, 7 George Square, Edinburgh, EH8 9JZ, UK
176. Diabetes Prevention Unit, National Institute for Health and Welfare, 00271 Helsinki, Finland
177. South Ostrobothnia Central Hospital, 60220 Seinäjoki, Finland
178. Red RECAVA Grupo RD06/0014/0015, Hospital Universitario La Paz, 28046 Madrid, Spain
179. Centre for Vascular Prevention, Danube-University Krems, 3500 Krems, Austria
180. Institute of Biomedicine and Translational Medicine, University of Tartu, Ravila Str. 19, 50412 Tartu, Estonia
181. Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, 27106, USA
182. University of Tartu, Tartu, Estonia
183. Department of Neurology, Medical University Graz, Auenbruggerplatz 22, 8036 Graz, Austria
184. Department of Epidemiology University of Washington, Seattle, WA 98101, USA
185. Department of Health Services, University of Washington, Seattle, WA 98101, USA
186. Group Health Research Institute, Group Health, Seattle, WA, 98101, USA
187. Institute of Physiology, University Medicine Greifswald, Karlsburg, 17495, Germany
188. Department of Pulmonary Physiology and Sleep, Sir Charles Gairdner Hospital, Hospital Avenue, Nedlands 6009,H57, Western Australia
189. School of Medicine and Pharmacology, University of Western Australia, Australia
190. Population Health Research Institute, St George's, University of London, London SW17 0RE, UK
191. Department of Medicine, Columbia University Medical Center, 622 West 168th Street, PH 9, East, 107, New York, NY 10032, USA
192. Department of Medical Sciences, Molecular Epidemiology and Science for Life Laboratory, Uppsala University, Uppsala 752 37, Sweden
193. Department of Medicine, Division of Cardiovascular Medicine, Stanford University School of Medicine, Stanford, CA 94305, USA
194. Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA 02114, USA
Footnotes
URLs
UK Biobank: https://www.ukbiobank.ac.uk/
Genotype imputation and genetic association studies using UK Biobank data: http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=157020
UK Biobank Axion Array Content Summary: http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=146640
Exome chip design: http://genome.sph.umich.edu/wiki/Exome_Chip_Design
Genotype-Tissue Expression (GTEx) database: www.gtexportal.org
GREAT Enrichment: http://bejerano.stanford.edu/great
Ingenuity Pathway Analysis (IPA) software: www.qiagen.com/ingenuity
Chembl: www.ebi.ac.uk/chembl/
Drug Gene Interaction database: dgidb.genome.wustl.edu
FORGE (accessed 16 Aug 2016): http://browser.1000genomes.org/Homo_sapiens/UserData/Forge?db=core
Fantom5 data (accessed 16 Aug 2016): http://fantom.gsc.riken.jp/5/
ENCODE DNase I data (wgEncodeAwgDnaseMasterSites; accessed 20 Aug 2016 using Table browser)
ENCODE cell type data (accessed 20 Aug 2016), http://genome.ucsc.edu/ENCODE/cellTypes.html.
Servier Medical Art: www.servier.fr/servier-medical-art
Conflicts/Disclosures
MJC is Chief Scientist for Genomics England, a wholly owned UK government company. He leads the 100,000 Genomes Project which includes syndromic forms of blood pressure.
Author Contributions
Central analysis: HRW, CPC, HG, MRB, MPSL, MR, IT, BM, IK, EE.
Writing of the paper: HRW*, MRB, EE, CPC, HG, IT, BM, MR, MJC*, PE* (*Writing group leads).
Working group membership: MJC*, HRW, EE, IT, PBM, LVW, NJS, MT, JMMH, MDT, IN, BK, HG, MRB, CPC, JSK, PE* (*Co-Chairs).
Replication consortium contributor: [ICBP-1000G] GBE, LVW, DL, AC, MJC, MDT, POR, JK, HS; [CHD Exome+ Consortium ] PSu, RC, DSa, JMMH [ExomeBP Consortium] JPC, FD, PBM [T2D-GENES Consortium and GoT2DGenes Consortium] CML; [CHARGE] GBE, CL, AK, DL, CNC, DIC; [iGEN-BP] ML, JCC, NK, JH, EST, PE, JSK, PVDH.
Replication study contributor: [Lifelines] NV, PVDH, HS, AMS; [GS:SFHS] JM, CH, DP, SP; [EGCUT] TE, MA, RM, AM; [PREVEND] PVDH, NV, RTG, SJLB; [ASCOT] HRW, MJC, PBM, PS, NP, AS, DS, ST; [BRIGHT] HRW, MJC, PBM, MB, MF, JC; [Airwave] HG, EE, MPSL, IK, IT, PE.
All authors critically reviewed and approved the final version of the manuscript.
References
- 1.Munoz M, et al. Evaluating the contribution of genetics and familial shared environment to common disease using the UK Biobank. Nat Genet. 2016;48:980–3. doi: 10.1038/ng.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinleib M, et al. The NHLBI twin study of cardiovascular disease risk factors: methodology and summary of results. Am J Epidemiol. 1977;106:284–5. doi: 10.1093/oxfordjournals.aje.a112464. [DOI] [PubMed] [Google Scholar]
- 3.Poulter NR, Prabhakaran D, Caulfield M. Hypertension. Lancet. 2015;386:801–12. doi: 10.1016/S0140-6736(14)61468-9. [DOI] [PubMed] [Google Scholar]
- 4.Mongeau JG, Biron P, Sing CF. The influence of genetics and household environment upon the variability of normal blood pressure: the Montreal Adoption Survey. Clin Exp Hypertens A. 1986;8:653–60. doi: 10.3109/10641968609046581. [DOI] [PubMed] [Google Scholar]
- 5.Forouzanfar MH, et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;386:2287–323. doi: 10.1016/S0140-6736(15)00128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sundstrom J, et al. Blood pressure-lowering treatment based on cardiovascular risk: a meta-analysis of individual patient data. Lancet. 2014;384:591–8. doi: 10.1016/S0140-6736(14)61212-5. [DOI] [PubMed] [Google Scholar]
- 7.Cabrera CP, et al. Exploring hypertension genome-wide association studies findings and impact on pathophysiology, pathways, and pharmacogenetics. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2015;7:73–90. doi: 10.1002/wsbm.1290. [DOI] [PubMed] [Google Scholar]
- 8.Ehret GB, et al. The genetics of blood pressure regulation and its target organs from association studies in 342,415 individuals. Nat Genet. 2016;48:1171–1184. doi: 10.1038/ng.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Surendran P, et al. Trans-ancestry meta-analyses identify rare and common variants associated with blood pressure and hypertension. Nat Genet. 2016;48:1151–1161. doi: 10.1038/ng.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C, et al. Meta-analysis identifies common and rare variants influencing blood pressure and overlapping with metabolic trait loci. Nat Genet. 2016;48:1162–1170. doi: 10.1038/ng.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–93. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott P, Peakman TC. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol. 2008;37:234–44. doi: 10.1093/ije/dym276. [DOI] [PubMed] [Google Scholar]
- 13.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. doi: 10.1038/ncomms9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann TJ, et al. Genome-wide association analyses using electronic health records identify new loci influencing blood pressure variation. Nat Genet. 2016 doi: 10.1038/ng.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staley JR, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32:3207–3209. doi: 10.1093/bioinformatics/btw373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ettehad D, et al. Blood pressure lowering for prevention of cardiovascular disease and death: a systematic review and meta-analysis. Lancet. 2016;387:957–67. doi: 10.1016/S0140-6736(15)01225-8. [DOI] [PubMed] [Google Scholar]
- 18.Kato N, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–8. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munroe PB, Barnes MR, Caulfield MJ. Advances in Blood Pressure Genomics. Circulation Research. 2013;112:1365–1379. doi: 10.1161/CIRCRESAHA.112.300387. [DOI] [PubMed] [Google Scholar]
- 20.den Hoed M, et al. Identification of heart rate–associated loci and their effects on cardiac conduction and rhythm disorders. Nature genetics. 2013;45:621–631. doi: 10.1038/ng.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamilton CA, Brosnan MJ, McIntyre M, Graham D, Dominiczak AF. Superoxide excess in hypertension and aging: a common cause of endothelial dysfunction. Hypertension. 2001;37:529–34. doi: 10.1161/01.hyp.37.2.529. [DOI] [PubMed] [Google Scholar]
- 22.Shin SY, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raffler J, et al. Genome-Wide Association Study with Targeted and Non-targeted NMR Metabolomics Identifies 15 Novel Loci of Urinary Human Metabolic Individuality. PLoS Genet. 2015;11:e1005487. doi: 10.1371/journal.pgen.1005487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Setten J, et al. Genome-wide association study of coronary and aortic calcification implicates risk loci for coronary artery disease and myocardial infarction. Atherosclerosis. 2013;228:400–5. doi: 10.1016/j.atherosclerosis.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy JJ, et al. Large scale association analysis for identification of genes underlying premature coronary heart disease: cumulative perspective from analysis of 111 candidate genes. J Med Genet. 2004;41:334–41. doi: 10.1136/jmg.2003.016584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Meurs JB, et al. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am J Clin Nutr. 2013;98:668–76. doi: 10.3945/ajcn.112.044545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pu X, et al. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am J Hum Genet. 2013;92:366–74. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizzoni D, Agabiti-Rosei E. Structural abnormalities of small resistance arteries in essential hypertension. Intern Emerg Med. 2012;7:205–12. doi: 10.1007/s11739-011-0548-0. [DOI] [PubMed] [Google Scholar]
- 29.Ray R, et al. Endothelial Nox4 NADPH oxidase enhances vasodilatation and reduces blood pressure in vivo. Arterioscler Thromb Vasc Biol. 2011;31:1368–76. doi: 10.1161/ATVBAHA.110.219238. [DOI] [PubMed] [Google Scholar]
- 30.Touyz RM, Montezano AC. Vascular Nox4: a multifarious NADPH oxidase. Circ Res. 2012;110:1159–61. doi: 10.1161/CIRCRESAHA.112.269068. [DOI] [PubMed] [Google Scholar]
- 31.Steppan J, Barodka V, Berkowitz DE, Nyhan D. Vascular stiffness and increased pulse pressure in the aging cardiovascular system. Cardiol Res Pract. 2011;2011:263585. doi: 10.4061/2011/263585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan F, et al. Nox4 and redox signaling mediate TGF-beta-induced endothelial cell apoptosis and phenotypic switch. Cell Death Dis. 2014;5:e1010. doi: 10.1038/cddis.2013.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan EC, et al. Nox4 modulates collagen production stimulated by transforming growth factor beta1 in vivo and in vitro. Biochem Biophys Res Commun. 2013;430:918–25. doi: 10.1016/j.bbrc.2012.11.138. [DOI] [PubMed] [Google Scholar]
- 34.Vasa-Nicotera M, et al. miR-146a is modulated in human endothelial cell with aging. Atherosclerosis. 2011;217:326–30. doi: 10.1016/j.atherosclerosis.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 35.Tian X, et al. Phosphodiesterase 10A upregulation contributes to pulmonary vascular remodeling. PLoS One. 2011;6:e18136. doi: 10.1371/journal.pone.0018136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takimoto E, et al. Chronic inhibition of cyclic GMP phosphodiesterase 5A prevents and reverses cardiac hypertrophy. Nat Med. 2005;11:214–22. doi: 10.1038/nm1175. [DOI] [PubMed] [Google Scholar]
- 37.Perez NG, et al. Phosphodiesterase 5A inhibition induces Na+/H+ exchanger blockade and protection against myocardial infarction. Hypertension. 2007;49:1095–103. doi: 10.1161/HYPERTENSIONAHA.107.087759. [DOI] [PubMed] [Google Scholar]
- 38.Oliver JJ, Melville VP, Webb DJ. Effect of regular phosphodiesterase type 5 inhibition in hypertension. Hypertension. 2006;48:622–7. doi: 10.1161/01.HYP.0000239816.13007.c9. [DOI] [PubMed] [Google Scholar]
- 39.Levy D, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–87. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newton-Cheh C, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeStefano AL, et al. Autosomal dominant orthostatic hypotensive disorder maps to chromosome 18q. Am J Hum Genet. 1998;63:1425–30. doi: 10.1086/302096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong X, et al. Genetic polymorphisms of the urea transporter gene are associated with antihypertensive response to nifedipine GITS. Methods Find Exp Clin Pharmacol. 2007;29:3–10. doi: 10.1358/mf.2007.29.1.1063490. [DOI] [PubMed] [Google Scholar]
- 43.Takimoto E, et al. Sodium calcium exchanger plays a key role in alteration of cardiac function in response to pressure overload. FASEB J. 2002;16:373–8. doi: 10.1096/fj.01-0735com. [DOI] [PubMed] [Google Scholar]
- 44.Ronaldson PT, Davis TP. Targeting transporters: promoting blood-brain barrier repair in response to oxidative stress injury. Brain Res. 2015;1623:39–52. doi: 10.1016/j.brainres.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carta L, et al. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J Biol Chem. 2006;281:8016–23. doi: 10.1074/jbc.M511599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kazenwadel J, et al. Loss-of-function germline GATA2 mutations in patients with MDS/AML or MonoMAC syndrome and primary lymphedema reveal a key role for GATA2 in the lymphatic vasculature. Blood. 2012;119:1283–91. doi: 10.1182/blood-2011-08-374363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akashi M, Higashi T, Masuda S, Komori T, Furuse M. A coronary artery disease-associated gene product, JCAD/KIAA1462, is a novel component of endothelial cell-cell junctions. Biochem Biophys Res Commun. 2011;413:224–9. doi: 10.1016/j.bbrc.2011.08.073. [DOI] [PubMed] [Google Scholar]
- 48.Cakstina I, et al. Primary culture of avian embryonic heart forming region cells to study the regulation of vertebrate early heart morphogenesis by vitamin A. BMC Dev Biol. 2014;14:10. doi: 10.1186/1471-213X-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev Biol. 2013;382:427–35. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dietrich T, et al. ED-B fibronectin (ED-B) can be targeted using a novel single chain antibody conjugate and is associated with macrophage accumulation in atherosclerotic lesions. Basic Res Cardiol. 2007;102:298–307. doi: 10.1007/s00395-007-0652-5. [DOI] [PubMed] [Google Scholar]
- 51.Stoynev N, et al. Gene expression in peripheral blood of patients with hypertension and patients with type 2 diabetes. J Cardiovasc Med (Hagerstown) 2014;15:702–9. doi: 10.2459/JCM.0b013e32835dbcc8. [DOI] [PubMed] [Google Scholar]
- 52.Erdos B, Backes I, McCowan ML, Hayward LF, Scheuer DA. Brain-derived neurotrophic factor modulates angiotensin signaling in the hypothalamus to increase blood pressure in rats. Am J Physiol Heart Circ Physiol. 2015;308:H612–22. doi: 10.1152/ajpheart.00776.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan SH, Wu CW, Chang AY, Hsu KS, Chan JY. Transcriptional upregulation of brain-derived neurotrophic factor in rostral ventrolateral medulla by angiotensin II: significance in superoxide homeostasis and neural regulation of arterial pressure. Circ Res. 2010;107:1127–39. doi: 10.1161/CIRCRESAHA.110.225573. [DOI] [PubMed] [Google Scholar]
- 54.Crespo K, Menard A, Deng AY. Retinoblastoma-associated protein 140 as a candidate for a novel etiological gene to hypertension. Clin Exp Hypertens. 2016;38:533–40. doi: 10.3109/10641963.2016.1163373. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe Y, et al. Accumulation of common polymorphisms is associated with development of hypertension: a 12-year follow-up from the Ohasama study. Hypertens Res. 2010;33:129–34. doi: 10.1038/hr.2009.193. [DOI] [PubMed] [Google Scholar]
- 56.Gale DP, Harten SK, Reid CD, Tuddenham EG, Maxwell PH. Autosomal dominant erythrocytosis and pulmonary arterial hypertension associated with an activating HIF2 alpha mutation. Blood. 2008;112:919–21. doi: 10.1182/blood-2008-04-153718. [DOI] [PubMed] [Google Scholar]
- 57.Yndestad A, et al. Elevated levels of activin A in clinical and experimental pulmonary hypertension. J Appl Physiol (1985) 2009;106:1356–64. doi: 10.1152/japplphysiol.90719.2008. [DOI] [PubMed] [Google Scholar]
- 58.Sacks FM, et al. Effects on blood pressure of reduced dietary sodium and the Dietary Approaches to Stop Hypertension (DASH) diet. DASH-Sodium Collaborative Research Group. N Engl J Med. 2001;344:3–10. doi: 10.1056/NEJM200101043440101. [DOI] [PubMed] [Google Scholar]
- 59.Intersalt: an international study of electrolyte excretion and blood pressure. Results for 24 hour urinary sodium and potassium excretion. Intersalt Cooperative Research Group. BMJ. 1988;297:319–28. doi: 10.1136/bmj.297.6644.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whelton PK, et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. JAMA. 2002;288:1882–8. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 61.Chan Q, et al. An Update on Nutrients and Blood Pressure. J Atheroscler Thromb. 2016;23:276–89. doi: 10.5551/jat.30000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khera AV, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. New England Journal of Medicine. 2016 doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wain LV, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–81. doi: 10.1016/S2213-2600(15)00283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–35. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 65.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 66.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barnes MR. Exploring the landscape of the genome. Methods Mol Biol. 2010;628:21–38. doi: 10.1007/978-1-60327-367-1_2. [DOI] [PubMed] [Google Scholar]
- 69.Gong J, et al. Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum Mutat. 2012;33:254–63. doi: 10.1002/humu.21641. [DOI] [PubMed] [Google Scholar]
- 70.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6 doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dunham I, Kulesha E, Iotchkova V, Morganella S, Birney E. FORGE: A tool to discover cell specific enrichments of GWAS associated SNPs in regulatory regions [version 1; referees: 2 approved with reservations] F1000Research. 2015;4 [Google Scholar]
- 72.Dozmorov MG, Cara LR, Giles CB, Wren JD. GenomeRunner: automating genome exploration. Bioinformatics. 2012;28:419–20. doi: 10.1093/bioinformatics/btr666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Elliott P, et al. The Airwave Health Monitoring Study of police officers and staff in Great Britain: rationale, design and methods. Environ Res. 2014;134:280–5. doi: 10.1016/j.envres.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 75.Petersen M, et al. Quantification of lipoprotein subclasses by proton nuclear magnetic resonance-based partial least-squares regression models. Clin Chem. 2005;51:1457–61. doi: 10.1373/clinchem.2004.046748. [DOI] [PubMed] [Google Scholar]
- 76.Chadeau-Hyam M, et al. Metabolic profiling and the metabolome-wide association study: significance level for biomarker identification. J Proteome Res. 2010;9:4620–7. doi: 10.1021/pr1003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated during the current study are available from the UK Biobank data repository (http://biota.osc.ox.ac.uk), which can be accessed by researchers upon application. This includes the derived GWAS analysis results summary data from our UK Biobank discovery data for all three BP traits. The genetic and phenotypic UK Biobank data are also available upon application to the UK Biobank (https://www.ukbiobank.ac.uk). All replication data generated during this study are included in the published article. For example, association results of look-up variants from our replication analyses and the subsequent combined meta-analyses are contained within all Supplementary Tables provided.