Abstract
Pseudomonas syringae is one of the best studied plant pathogens and it serves as a model for understanding host-microbe interactions, bacterial virulence mechanisms, host adaptation of pathogens, as well as microbial evolution, ecology and epidemiology. Comparative genomic studies have revealed key genomic features contributing to P. syringae virulence. As an extracellular plant pathogen that lives in the intercellular space (apoplast) of aboveground tissues (phyllosphere), P. syringae has evolved two principal virulence strategies, suppression of host immunity and creation of an aqueous apoplast. In addition, P. syringae infection is profoundly influenced by external environmental conditions, such as humidity. P. syringae may serve as an excellent model to understand not only how pathogens evolve specific virulence strategies to intercept host immunity, but also how pathogenic microbes integrate external environmental conditions and endogenous plant microbiota to become ecologically robust and diverse pathogens of the plant kingdom.
Subject terms: plant immunity, pathogen effector, microbiome, stomata, plant hormone
Introduction
Pseudomonas syringae is one of the best-studied plant pathogens and serves as a model for understanding bacterial pathogenicity, molecular mechanisms of plant-microbe interactions as well as microbial ecology and epidemiology. P. syringae was originally isolated from diseased plants and was largely studied with respect to its plant pathogenic potential1, 2. So far more than 50 pathovars have been identified in the species, with each pathovar infecting a characteristic group of host plant species. Collectively, the ~50 pathovars of P. syringae infect almost all economically important crop species, making P. syringae one of most common pathogens on plants. In addition, new disease outbreaks, caused by P. syringae isolates, continue to threaten global crop production. A recent example is the devastating kiwifruit canker in New Zealand and Europe, which is caused by Pseudomonas syringae pv. actinidiae, likely originating from China3–5. Although the species was initially identified as a pathogenic bacterium, it has since been found that many isolates phylogenetically belonging to the species are non-pathogenic to plants and that they exist on plants as commensals. Understanding the genetic and phenotypic variability of P. syringae, especially by comparing with its closely-related non-pathogenic bacteria, helps elucidating what makes this organism a pathogen.
P. syringae bacteria have two interconnected phases of growth in or on plants: the epiphytic phase, when the bacteria live on the surface of plant tissues (usually the above-ground parts, such as leaves, stems and fruits, collectively known as the phyllosphere), and the endophytic phase, when bacteria enter the plant tissue and colonize the intercellular space called the apoplast (see Fig. 1, ref6). While many P. syringae strains, such as those of P. syringae pv. syringae, are strong epiphytes and had been widely used in microbial ecological studies, disease occurs only after P. syringae bacteria enter the plant and multiply in the apoplast (i.e., the endophytic phase). The initial epiphytic populations of some P. syringae strains on the plant surface can be good predictors of their later endophytic populations inside the plant tissue and disease outbreaks under favorable environmental conditions2, 7, illustrating the importance of dissecting the epiphytic phase for understanding P. syringae pathogenesis.
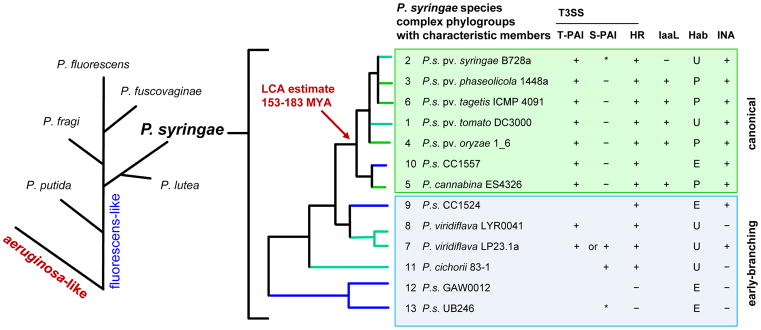
Figure 1. The phylogeny of P. syringae and common phylogroup features.
On the left, proposed phylogenetic branching order for major species groups within the P. fluorescens-like major branch of Pseudomonas14, 15. On the right, thirteen identified phylogroups (PGs) in the P. syringae species complex, based on multi-locus sequence analysis (MLSA). Phylogroups representing monophyletic species within the complex are noted. Characteristic PG members are listed along with general phylogroup-associated features when known16. S-PAI, single-part pathogenicity islands lack a canonical CEL but may carry CEL T3SS effectors within the hrp/hrc cluster. IaaL, presence of the indole acetic acid lysine synthetase gene for the inactivation of auxin29. Hab, common habitat, strains are isolated mostly from plants (P) or the environment (E), or both/ubiquitous (U). INA, reported ice nucleation capacity or the presence of the inaW ice-nucleation gene. IaaL, Hab and INA traits vary on a strain-to-strain basis. *, PG 2 clade c and PG 13 have A-typical S-PAIs (A-PAI) with distinct genomic locations12.
Genomic features that are correlated with preferably epiphytic or endophytic/pathogenic living style have been studied and discussed6,2. For example, tolerance to ultraviolet light and dry environment is generally considered important for a strong epiphytic life style. Another notable feature of P. syringae bacteria that may be important for the epiphytic phase is ice nucleation and the associated ability to cause frost injury in plants, which may lead to water and nutrient release from plants and could create openings on the plant surface to facilitate bacterial entry. The ice-nucleation ability of P. syringae depends on the ice-nucleation gene INA. INA encodes the ice-nucleating protein, which allows ice crystals to form at temperatures higher than normal freezing temperature in plants2, 8. In fact, studies of this important feature led to approaches to control frost injury in agriculture using naturally non-ice nucleating bacteria or INA- P. syringae mutant bacteria, the first recombinant microorganism allowed for release in the fields9. In addition, as one of the most effective ice nucleators in nature and ubiquitously found in precipitates and water sources, P. syringae has been proposed as an essential player in the formation of rain and snowfall, shaping the water cycle on Earth10. Readers are referred to many excellent reviews that discuss in details on the topics of microbial ecology, epidemiology, genomics and habitat interactions of both non-pathogenic and pathogenic P.syringae2, 10–13. Below, we focus on plant-pathogenic P. syringae and summarize the current understanding of virulence strategies, pathogenicity-related genomic features of P. syringae as well as effects of environmental conditions on disease outcomes.
Genomic and genetic features of P. syringae
The phylogeny of pathogenic P. syringae
P. syringae forms a monophyletic group within the P. fluorescens-like major branch of the Pseudomonas genus14, 15. Extensive efforts to collect and sequence P. syringae isolates from diverse agricultural and non-agricultural sources have driven a revolution in our understanding of P. syringae diversity and evolution. Currently, the P. syringae species complex is divided into 13 phylogroups (PGs) based on multi-locus sequence analysis (MLSA)(Fig. 1)14–16. These PGs encompass previously defined phylogenetic divisions; rarefaction curve analysis implies that the identified PGs represent the bulk of P. syringae diversity at this phylogenetic level. The 13 PGs split into two major categories, the seven late-branching canonical lineages (PGs 1–6, 10) and the six early-branching non-canonical lineages (PGs 7–9, 11–13)17. The canonical PGs are composed of strains with phenotypic characteristics traditionally associated with P. syringae (i.e., the LOPAT phenotype; see Glossary). With very few exceptions, they possess canonical tripartite pathogenicity islands (T-PAI) with the hrp/hrc-encoded type III secretion system (T3SS) gene cluster flanked by both the Conserved Effector Locus (CEL) and the Exchangeable Effector Loci (EEL)18. The CEL encodes a trio of highly conserved syntenic effector genes, hopAA1-1, hopM1 and avrE, whereas the effectors encoded by the EEL vary between pathovars and strains. The T3SS translocates a variety of bacterial effector proteins into host cells as a central mechanism of pathogenesis/symbiosis in diverse plant/animal-bacterial interactions19, 20. Other traits common among the canonical P. syringae lineages include the capacity to cause immune-associated programmed host cell death (i.e., the hypersensitive response; HR) in resistant plants, ice nucleation activity and the iaaL gene, which is involved in inactivation of the plant hormone auxin. The iaaL genes is found among the canonical PGs composed primarily of plant specialists21 (Fig. 1). The six early-branching lineages include P. syringae-like, broad-host-range plant pathogens P. viridiflava and P. cichorii, and generally have greater diversity in phenotypes as well as in the type and genomic location of PAI. Some of the early-branching lineages carry the single-part pathogenicity island (S-PAI); a genomic region that contains genes encoding the hrp/hrcT3SS but, compared with T-PAI, lack a canonical CEL and EEL.
The evolution of P. syringae into a pathogen
To answer the question of “what makes P. syringae a successful plant pathogen”, it would be important to trace a potential path of P. syringae evolution from a non-pathogenic ancestor and its relation to other plant-associated bacteria. Genetic clock estimates, calibrated with the proposed divergence rates between E. coli and Salmonella, place the last common ancestor (LCA) of the P. syringae canonical lineages between 153–183 MYA22 (Fig. 1). This time frame is roughly contemporaneous with molecular clock estimates for the emergence of angiosperms (i.e., flowering plants)23, 24.
The distribution of genetic and phenotypic traits in the P. syringae phylogeny can help us infer possible traits of the P. syringae LCA. Virulence factors common among the canonical P. syringae lineages include the T-PAI, ice nucleation, auxin synthesis, auxin inactivation (iaaL) and production of the exopolysaccharide alginate16, 17, 21, 25–29. Similar alginate synthesis and regulatory pathways are present in P. aeruginosa, P. fluorescens and P. syringae, so we can expect that the P. syringae LCA had these genes as well26. The iaaM/iaaH genes for auxin synthesis are also common among plant-associated Pseudomonas species and the phylogeny of P. syringae chromosomal iaaM/iaaH genes are largely congruent with phylogeny based on housekeeping genes, implying that they are ancestral30. The acquisition of the T-PAI and ice nucleation protein appears to have occurred prior to the divergence of the P. syringae canonical PGs and the P. viridiflava PG 7. P. viridflava PG 7 is the only early-branching PG that is composed of members with the T-PAI and ice nucleation trait that are isolated routinely from plants16, 31. Lastly, the LCA most likely did not possess plant habitat specialization or auxin inactivation, as these appear to be derived traits in the canonical P. syringae lineages16, 29. We surmise that the LCA of the canonical P. syringae lineages is likely to have been a ubiquitous strain with the capacity to synthesize alginate and auxin, possessing both ice nucleating activity and the T-PAI.
The acquisition of the T-PAI by the ancestor of canonical P. syringae appears to be a critical step towards patho-adaptation. Expansion and specialization of the virulence factor repertoire, especially T3SS effectors (T3Es), greatly shaped the host range and P. syringae diversification. More details of the T-PAI and T3E clusters are provided in Box 1. In addition to T3Es, P. syringae strains collectively produce a diverse collection of phytotoxins, such as coronatine and syringomycin, which contribute to disease by diverse mechanisms. To some degree, toxins and T3Es appear to play overlapping functional roles (see sections below). Some phytotoxin synthetic clusters have a sporadic and narrow distribution, similar to what is observed for most T3Es, while some others are much more broadly distributed. PG2 strains of P. syringae are notable for their broad host ranges, high epiphytic potential, small T3E repertoires, and their possession of a “toxin package” comprised of syringolin A as well as syringomycin and syringopeptin, both of which have membrane disruption and ion-leakage activities29. There is an overall correlation between the presence of the syringomycin synthetic cluster and a small T3E repertoire29. This extends to members of PG10, which have the smallest reported effector repertoire among P. syringae32. We propose a hypothetical and evolutionary view of a potential pathway of a Pseudomonas non-pathogen evolving into a P. syringae pathogen (Fig. 2).
Box 1. Genetic variation within the canonical P. syringae tripartite pathogenicity island (T-PAI).
The T-PAI locus of P. syringae pv. phaseolicola 1448A is shown to scale (NC_005773;1,471,435..1,510,651). The T-PAI is a virulence starter kit, and contains the hrc/hrp genes for the assembly and regulation of the T3SS, flanked by genes for both conserved and variable suits of T3Es. Both the T3SS genes as well as conserved effector functions are required for successful P. syringae infection117, 118. The P. syringae T-PAI-encoded T3SS is a member of the Hrp1 T3SS group, one of seven major groups of virulence-associated T3SS119. Presence of particular allele variants within PG member strains are noted but are not necessarily PG exclusive. The hrp/hrc T3SS gene cluster encodes all the structural genes required to assemble of the T3SS. It also encodes the upstream regulators HrpR and HrpS, which are paralogous AAA+ RpoN activator proteins, required to induce the expression of the ECF-family sigma factor HrpL, the master regulator responsible for the expression of all hrp/hrc T3SS structural genes and T3E-encoding genes120, 121. This regulatory circuitry is a defining hallmark of the Hrp1 T3SS, which is also found in the P. syringae S-PAIs, as well as in plant pathogenic Enterobacteriacea (e.g. Pantoea stewartii, Erwinia amylovora, Dickeya dadantii, etc)119, 122, 123. The HrpA pilin, which assembles to create the T3SS extracellular appendage, has undergone diversifying selection and is the only gene within the hrp/hrc cluster divided into gene family subgroups. Some PG 3 strains carry recombined hrpA3 alleles common in PG 5 P. cannabina strains124. Within the hrp/hrc gene cluster there is evidence of recombination among certain P. syringae groups in hrpR/hrpS, hrcN, hrpQ, hrcV, and hrpK1 genes125, 126. Adjacent to the hrp/hrc cluster is a conserved syntenic region, the Conserved Effector Locus (CEL), which encodes a trio of highly conserved effectors, hopAA1-1, hopM1 and avrE117. The hopAA1-1 gene is commonly pseudogenized in PG3 strains29, 127, while hopM1 and/or avrE genes have been identified within every known example of the P. syringae S-PAI and T-PAI18, 29, 123, and have been shown to play critical roles during infection (see later sections). The hrp/hrc cluster is also flanked by a second T3SS effector locus, the Exchangeable Effector Locus (EEL), and EEL effector content and loci structure vary extensively between strains and phylogroups. In strains where it has been examined, the EEL region has been extensively reworked by mutation, deletion, recombination and transposon insertion and commonly contains zero to three intact T3Es118, 128. The EEL of PG 3 strains commonly carry the effector hopX1 in a class II EEL and the effectors AvrB3 and HopZ3 are also found in other EEL classes118, 128.
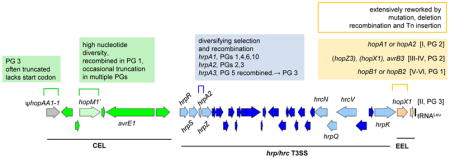
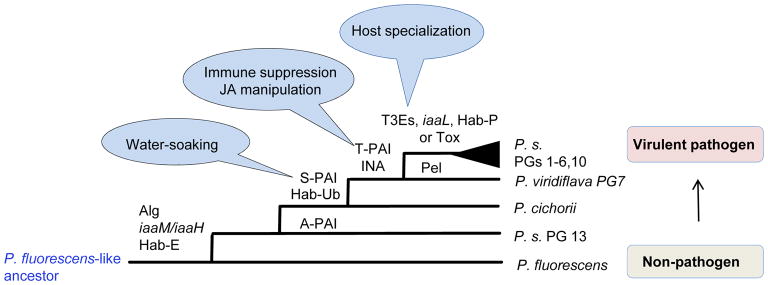
Figure 2. Potential steps to patho-adaptation in P. syringae evolution.
Hypothesized ancestry of important traits in the P. syringae species complex. The S-PAI encodes AvrE and/or HopM effectors associated with apoplast water-soaking. T-PAI effector loci genes are associated with JA manipulation and defense suppression in addition to apoplast water-soaking. In addition to trait name abbreviations in Figure 1, Alg, genes for the regulation and production of alginate. iaaM/iaaH, genes for auxin synthesis, Pel, pectate lyase. T3Es, expansion and diversification of T3E repertoires. Tox, toxin packages of broad-host-range pathogens.
Overcoming host defenses and forming a niche
As mentioned above, pathogenic P. syringae strains must make a transition from the epiphytic phase to the endophytic phase to cause disease. This involves efficient entry into the plant tissue and aggressive multiplication within the apoplast. Neither step would be easy for a microbe. In fact, most microbes (i.e., the vast number of commensal microbes) fail to do one or both of these two steps because plants have evolved ways to restrict the entry and/or multiplication of these microbes.
Overcoming stomatal closure at bacterial entry
Entering plant tissue through natural openings such as stomata represents one of the first steps of an active infection cycle. Plants have evolved defense mechanisms to reduce the entry of pathogens. Upon recognition of conserved bacterial features collectively named PAMPs (pathogen-associated molecular patterns), such as flagellin, a signaling cascade is activated in the stomatal guard cell to eventually close stomata as part of the pattern-triggered immunity (PTI) in plants33, 34,. Readers are referred to other recent reviews that summarize many secondary messengers and downstream components, including plant hormones (i.e. salicylic acid [SA] and abscisic acid [ABA]), involved in the PAMP-triggered stomatal closure pathway34, 35.
As a counter-defense strategy, P. syringae has evolved virulence factors, such as the phytotoxin coronatine and T3Es, to impair plant stomatal defense. Coronatine is a molecular mimic of the active form (jasmonoyl isoleucine; JA-Ile) of the plant hormone jasmonate (JA), directly binding to and activating the plant JA receptor36, 37. Recent studies have begun to elucidate the signaling pathways by which coronatine-mediated activation of JA signaling results in stomatal opening. Coronatine exploits the endogenous antagonistic interaction between JA signaling and SA signaling, which is downstream of PAMP signaling required for PAMP-induced stomatal closure33, 38 (Fig. 3). Key players in the coronatine-mediated stomatal opening pathway in Arabidopsis plants include canonical JA signaling components, such as the COI receptor, JAZ2 transcriptional repressor and MYC2/3/4 transcription factors, as well as ANAC19/55/72 transcription factors that regulate SA accumulation38, 39. In tomato, JA signaling components JA2L40 transcription factors are involved in coronatine-induced stomatal opening. Coronatine has also been reported to inhibit stomatal closure by suppressing guard cell NADPH oxidase-mediated ROS production41, and inhibits stomatal closure or re-opens stomata in plant leaves treated with PAMPs, ABA or darkness34, 41, 42. On the other hand, the transcription factor ANAC32, induced during P. syringae infection of Arabidopsis, has been shown to directly repress MYC2 activation, perhaps as a countermeasure of the plant to inhibit coronatine-mediated stomata opening43.
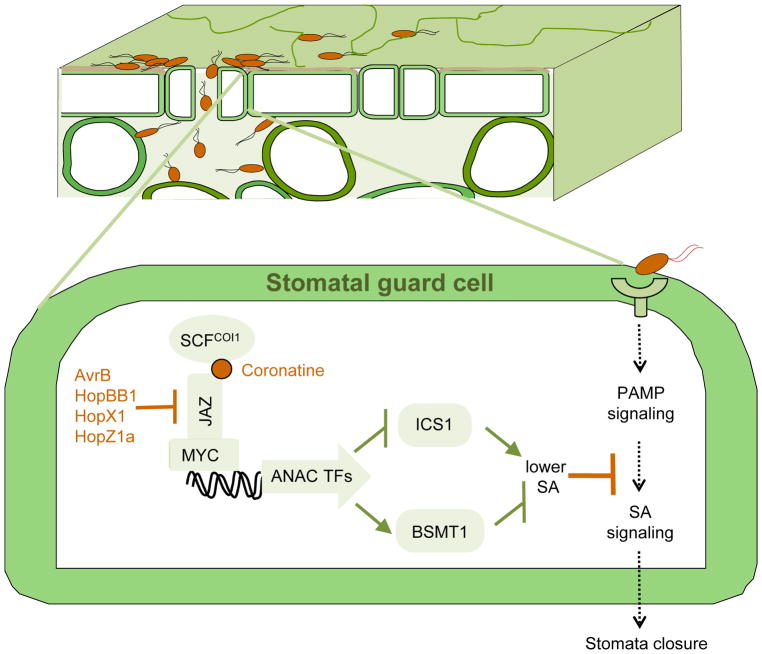
Figure 3. Battle during bacterial entry.
Upper panel, P. syringae bacteria enter a section of a plant leaf through natural opening stomata. Lower panel, perception of bacterial PAMPs stimulates PAMP immune signaling in a stomatal guard cell leading to SA signaling and eventual stomatal closure; P. syringae phytotoxin coronatine and several T3Es (i.e. AvrB, HopBB1, HopX1 and HopZ1a) target the COI1 receptor or JAZ transcriptional repressors to activate JA signaling. Activation of JA signaling leads to modulation of the expression of ANAC transcription factors and ICS1 and BSMT1, which are involved in SA biosynthesis and metabolism, respectively, resulting in lowered SA accumulation and inhibition of PAMP-triggered stomatal closure38.
In addition to coronatine, at least three P. syringae T3Es (HopX1, HopZ1a and HopBB1) have been reported to activate JA signaling by directly interacting with and/or destabilizing JAZ repressor proteins44–46. Another P. syringae T3E, AvrB, activates JA signaling by promoting JAZ protein degradation and modulates the phosphorylation of plant protein RIN4 and membrane ATPase activity, leading to stomatal opening47, 48. Finally, T3Es HopF2 and HopM1 were reported to suppress PAMP-induced oxidative burst and stomatal closure49, 50. Consistent with the observed effects of T3Es in suppression of stomatal closure, a recent in vivo imaging study showed that guard cells, which make up stomata, are target cells of type III secretion51, 52. Taken together, these studies show that P. syringae devotes a variety of virulence factors to counter stomatal closure as part of its infection strategy (Fig. 3).
Suppressing plant immunity and making a living in the apoplast
After entering the plant (e.g., leaves), P. syringae encounters the apoplast, a hostile environment and a new battlefield. In the apoplast, intricate interactions between plant immune responses and bacterial virulence strategies occur. For example, mesophyll cells inside leaves can mount (i) PTI in response to recognition of bacterial PAMPs and (ii) effector-triggered immunity (ETI) in response to recognition of T3Es delivered into the mesophyll cells. A major consequence of PTI and ETI is inhibition of bacterial multiplication53, 54. How PTI and ETI actually inhibit bacterial multiplication remains unclear. Possible mechanisms include production of anti-bacterial defense compounds, down-regulation of the T3SS, and strengthening of plant cell walls55. A recent study showed that the sugar uptake activity of the plant transporter STP13 is enhanced during PTI, which results in removal of apoplastic sugars, suggesting that restriction of nutrients in the apoplast may be one of consequences of plant immunity56.
To defeat immune responses from mesophyll cells, P. syringae again deploys T3Es and other virulence factors to intercept plant immune signaling at various steps. For example, as in the stomatal guard cell (Fig. 3), coronatine can inhibit SA-mediated defense in leaf mesophyll cells, presumably through the JA-SA antagonism38. Coronatine also induces the protein phosphatases 2C (PP2C) HAI1, which dephosphorylates and inactivates MPK3 and MPK6, two positive immune regulators57. There are other toxins, besides coronatine, produced by P. syringae. For instance, syringomycin has been shown to function as a virulence factor for P. s. pv. syringae58, 59. At least two virulence-related activities of syringomycin have been discovered: inducing pore formation on plant membranes, leading to release of plant metabolites, and acting as bio-surfactant, leading to increased wetness of plant surface and bacterial movement58.
However, coronatine and other small-molecule toxins are produced by only subsets of P. syringae pathovars29 and genetic mutations eliminating toxin production often have modest effects on virulence, especially when bacteria are inoculated directly into the apoplast33, 60. In contrast, the T3SS is conserved in all pathogenic P. syringae strains and disruption of the T3SS invariably renders P. syringae nonpathogenic even if bacteria are inoculated directly into the apoplast61. This suggests that T3Es are collectively essential for the pathogenicity of P. syringae inside the apoplast. The T3E repertoire among P. syringae strains is highly variable and relatively few effectors are conserved29, 62. An important question arises: What is the minimal repertoire of T3Es that P. syringae must possess to become a phyllosphere pathogen? This question was addressed by Cunnac and colleagues63. By an elegant combination of effector gene deletion and reconstitution experiments, a set of eight T3Es from P. s. pv. tomato (Pst) DC3000 was shown to be sufficient to rescue much of the virulence of an “effector-less” mutant strain in the plant Nicotiana benthamiana. These eight effectors include AvrPtoB, HopM1, AvrE, HopE1, HopG1, HopAM1-1, HopAA1-1, and/or HopN163. Below, we highlight the virulence functions of these eight T3Es (Fig. 4a), as they give important insights into the central question of this review: What it takes for P. syringae to become a successful pathogen? We must point out that there are many other T3Es whose intriguing virulence functions and host targets were also extensively studied. We summarized these studies and divided these T3Es in groups based on the host processes they target (Table 1). Readers are referred to other excellent reviews on this topic64–67.
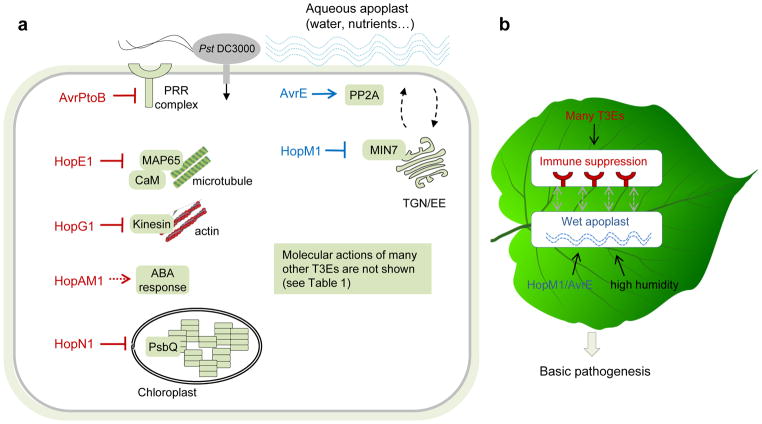
Figure 4. Battle inside the leaf apoplast after bacterial entry.
a. A diagram depicting the host targets of eight “core” T3Es in a susceptible Arabidopsis cell. AvrPtoB targets PRR complex to inhibit PTI. HopG1 and HopE1 target actin and microtubule networks through interaction with kinesin and MAP65, respectively. HopAM1 induces ABA hypersensitivity in the plant and enhances virulence on drought-condition plants, and HopN1 targets the chloroplast protein PsbQ. These five T3Es appear to be primarily involved in suppression of host immunity responses. Two conserved T3Es, HopM1 and AvrE, induce an aqueous apoplast. HopM1 targets a trans-Golgi network (TGN)/early endosome (EE)-localized ARF guanine exchange factor, MIN7, and AvrE interacts with protein phosphatase 2A (PP2A). The host target of HopAA1 (not shown) is not known. b. A conceptual model illustrating two basic aspects of host biology perturbed by P. syringae post epiphytic growth. Suppression of plant immunity and creation of an aqueous apoplast are two principal features of P. syringae infection in the leaf.
Table 1.
Host targets of P. syringae T3Es.
| T3Es | Host target(s) | Host process | References |
|---|---|---|---|
| AvrPto | FLS2, EFR, BAK1 | PTI | 70, 129, 130 |
| AvrPtoB | FLS2, CERK1, Bti9, BAK1 | PTI | 70–73, 131 |
| HopB1 | BAK1 | PTI | 132 |
| AvrPphB | BIK1/PBS1/PBLs | PTI | 133 |
| HopF2 | BAK1, MKK5 | PTI | 134–136 |
| HopAl1 | MPK3, MPK6, MPK4 | PTI | 137, 138 |
| AvrRpt2 | MPK4, MPK11 | PTI | 139 |
| AvrRps4 | WRKYs | PTI | 140, 141 |
|
| |||
| HopD1 | NTL9 | ETI | 142 |
| AvrPtoB | Fen, RHopAD1 | ETI | 69, 74 |
|
| |||
| HopX1, HopBB1, HopZ1a | JAZ | JA | 44–46, 143 |
| AvrB | JA | 48 | |
| AvrRpt2 | AUX/IAA | Auxin | 144, 145 |
| AvrPtoB | ABA | 146 | |
| HopAM1 | ABA | 79 | |
| HopQ1 | Cytokinin | 147 | |
| HopAF1 | MTN1/2 | Ethylene | 148 |
| HopI1 | SA | 149, 150 | |
|
| |||
| HopW1 | Actin | Actin | 151 |
| HopG1 | Kinesin | Actin | 75 |
| HopE1 | MAP65 | Microtubule | 77 |
| HopZ1a | Tubulin | Microtubule | 152 |
|
| |||
| HopM1 | MIN7 | Water balance | 82, 84, 85 |
| AvrE | PP2A | 82, 84 | |
|
| |||
| HopN1 | PsbQ | Chloroplast | 78 |
| HopI1 | Hsp70 | Chloroplast | 149 |
| HopK1 | Chloroplast | 153 | |
|
| |||
| AvrB | RIN4/MPK4/Hsp90/RAR1 | RIN4 complex | 47, 143 |
| AvrRpt2 | RIN4 | RIN4 complex | 154, 155 |
| AvrRpm1 | RIN4 | RIN4 complex | 156 |
| AvrPto, AvrPtoB | RIN4 | RIN4 complex | 157 |
| HopF2 | RIN4 | RIN4 complex | 158 |
|
| |||
| AvrRps4, HopA1 | EDS1 | EDS1 | 159, 160 |
|
| |||
| HopU1 | GRP7/8 | Gene transcript | 161, 162 |
|
| |||
| HopZ1a, HopZ1b | GmHID1 | Phytoalexin | 163 |
|
| |||
| HopZ4 | RPT6 | Proteasome | 164 |
| HopM1, HopG1, HopAO1, HopA1 | Proteasome | 165 | |
“Host target” denotes the plant protein that directly interacts with and/or is modified by the corresponding T3E.
Of the eight effectors in the minimal T3E repertoire of Pst DC3000, at least five have been shown to be involved in suppressing host immunity. AvrPtoB inhibits both PTI and ETI responses and is one of the first T3Es of which the host targets were identified68–70. AvrPtoB possesses an E3 ubiquitin ligase activity and targets pattern-recognition receptors (PRRs), including FLS2, CERK1 and Bti9, for protein degradation or kinase activity70–73. In tomato and N. benthamiana, certain truncated versions of AvrPtoB suppress ETI-associated plant cell death and that this activity is mediated by degradation of immune-associated kinases such as Fen in tomato69 and MAP kinase kinase 2 in N. benthamiana74. Therefore, AvrPtoB is able to inhibit both PTI and ETI.
HopG1 and HopE1 have been shown to target components of the plant cytoskeleton. HopG1 changes the actin filament architecture and interacts with a mitochondrion-localized motor protein kinesin, which is required for HopG1-mediated disease symptoms75. Transgenic expression of HopG1 inhibits PTI outputs, including immunity-associated callose deposition in the plant cell wall76. On the other hand, HopE1 targets the microtubule network through interaction with the plant calmodulin protein and microtubule-associated protein 65 (MAP65). This interaction leads to disassociation of MAP65 from microtubule and results in inhibition of multiple immune-associated responses, including callose deposition in the plant cell wall77.
HopN1 was reported to target a tomato chloroplast protein PsbQ and is able to suppress the production of reactive oxygen species (ROS) and callose deposition in Arabidopsis78. Transgenic expression of HopAM1 also suppresses callose deposition in the plant cell wall and, interestingly, enhances ABA responses in plants via an unknown mechanism79.
AvrPtoB, HopG1, HopE1, HopAM1 and HopN1 in the “minimal T3E repertoire” represent a large number of P. syringae T3Es that are capable of suppressing host immune responses under laboratory experimental conditions (Table 1). It appears that many components of the plant immune machinery are vulnerable to attacks by P. syringae T3Es. The impressive number of “immune-suppressing” T3Es in P. syringae illustrates that suppression of host immune responses is fundamentally important for P. syringae infection, a concept that echoes earlier studies80, 81.
Water soaking
Are all P. syringae T3Es in the minimal repertoire primarily involved in suppressing plant immune responses? The answer seems to be no. This is illustrated by the virulence functions of HopM1 and AvrE, which represent two of the most conserved and widely distributed T3E families within the whole P. syringae T3E repertoire29. Although studies have shown that HopM1 and AvrE are also capable of suppressing PAMP-triggered oxidative burst and/or callose deposition50, 82, 83, a recent study showed that the primary virulence function of HopM1 and AvrE appears to establish an aqueous apoplastic environment (or “water soaking” symptom, during which liquid is accumulated in the intercellular space between mesophyll cells in the leaf). The aqueous apoplast could potentially benefit bacterial multiplication in multiple ways, such as diluting anti-microbial compounds and/or making nutrients more accessible. It is possible that the previously observed effects of HopM1 and AvrE on apoplast immune responses, such as production of ROS and callose deposition in the apoplast, could be secondary effects due to a water-soaked apoplast. It was shown that transgenic expression of AvrE1 and HopM1 from Pst DC3000 is sufficient to cause severe water-soaking in Arabidopsis leaves84. AvrE1 and HopM1 share no amino acid sequence similarity, but are functionally redundant in Pst DC3000 pathogenesis 83. The Pst DC3000 avrE-hopM1− mutant, which lacks water-soaking-inducing effectors, fails to multiply aggressively in the apoplast or cause disease. However, supplementation of water to the apoplast could restore the virulence of the Pst DC3000 avrE-hopM1− mutant84, reinforcing a major role of AvrE/HopM1-mediated water soaking in bacterial pathogenesis.
HopM1 targets and degrades a plant ARF-family guanine nucleotide exchange factor, AtMIN7, which is involved in vesicle trafficking85. Correspondingly, the Arabidopsis atmin7 mutation, which partially mimics the virulence action of HopM1, promotes some spontaneous, albeit limited, water-soaked spots in certain Arabidopsis genotypes under high humidity84. These results suggest that the normal function of AtMIN7 is probably to maintain water homeostasis in the leaf apoplast and that HopM1 targets AtMIN7 as part of its mechanism to cause water-soaking. The host targets of AvrE1 include protein phosphatase 2A regulatory subunits90. It remains to be determined whether protein phosphatase 2A regulatory subunits are also involved in AvrE1-mediated establishment of an aqueous apoplast.
In summary, current studies suggest that two fundamental aspects of host biology are perturbed by the eight “minimum-repertoire” T3Es of Pst DC3000: host immunity and apoplast environment. This begs the question of whether perturbing these two host processes is sufficient to allow P. syringae pathogenesis in leaves. This critical question was investigated by Xin and colleagues84 in disease reconstitution experiments. Specifically, two high-order Arabidopsis mutants were generated, in which relevant genes involved in plant immunity (PTI) and the gene encoding AtMIN7 were mutated simultaneously. These Arabidopsis mutants allow the T3SS-defective Pst DC3000 hrcC mutant to grow significantly in the apoplast, supporting the hypothesis that perturbation of host immunity and water homeostasis in the apoplast are two principal virulence mechanisms that underlie basic P. syringae pathogenesis (Fig. 4b)84. It is likely that other virulence factors are involved in further optimizing P. syringae virulence by targeting other aspects of host biology and/or in adaptation to different environmental conditions (see below).
Suppression of host immunity and establishment of an aqueous apoplast may not be two mutually exclusive processes. A previous study showed that P. syringae strains experience different water stress levels in the leaf apoplast of susceptible and resistant Arabidopsis plants. Specifically, Pst DC3000 experiences suitable water potentials for multiplication in the leaf apoplast of the Arabidopsis Col-0 accession, whereas Pst DC3000 (avrRpm1), which activates ETI in Col-0 plants, experiences a prohibitory high water stress in the resistant plant 86. In line with this study, Pst DC3000 (avrRpt2), which also activates ETI in Col-0 plants, fails to induce water-soaking symptoms84. This finding suggests that activation of ETI in plants can block the water-soaking process, possibly as an integral part of the plant defense mechanism against bacterial pathogens. Thus, host immunity and water homeostasis in the apoplast may influence each other and future research should investigate whether the two processes may be connected at some mechanistic level.
Influence of environmental factors on P. syringae infection
In 1960, Stevens formulated the famous “disease triangle” concept in plant pathology: in addition to a virulent pathogen and a susceptible host, disease outbreaks require right environmental conditions such as optimal temperature and humidity87 (Fig. 5a). In addition, recent plant microbiome studies highlight a potential fourth vertex–the co-existing microbial communities on plants–to the “disease triangle”, as these microbial communities can potentially have a significant influence on plant immunity and/or pathogen virulence88. While host immunity and P. syringae pathogenesis mechanisms have been extensively studied in the past three decades, the molecular bases of environmental influences on diseases caused by P. syringae is under-studied.
Figure 5. Interactions between plant, P. syringae and abiotic and biotic environment.
a. A diagram illustrating the plant-pathogen-environment triangular interactions formally known as the “disease triangle”. b–d. Effects of temperature (b), humidity (c) and the microbiome (d) on P. syringae, the plant and disease outcome. Normal arrows indicate positive effects and block arrows indicate negative effects.
Humidity
High humidity has been observed to be tightly correlated with the vast majority of P. syringae disease outbreaks in crop fields. Many previous studies pointed to a role of high humidity and associated conditions (i.e., dew, fog and rain) in maintaining a high epiphytic P. syringae population on the plant surface, for which a quantitative relationship to following disease outbreaks in the field has been established2, 7, 89, 90. High humidity has also been shown to increase the formation of bacterial aggregates on leaf surfaces and bacterial swarming motility of P. s. pv. syringae91, 92, and to affect the bacterial cell length, trans-conjugation and plasmid transfer of P. s. pv. glycinea on bean leaves93. Panchal et al.94 reported that high humidity suppresses bacterium-induced stomatal closure (Fig. 5b), which could contribute to enhanced Arabidopsis susceptibility to Pst DC3000 infection.
In addition to facilitating bacterial entry, high humidity is also required for P. syringae pathogens to aggressively multiply inside the apoplast (i.e., even after bacteria are infiltrated directly into the plant leaf). As mentioned above, Pst DC3000 utilizes two conserved T3Es, AvrE and HopM1, to drive the formation of an aqueous apoplast (i.e. “water soaking”) as an essential virulence strategy. Importantly, maintenance of the aqueous apoplast requires high humidity, because, under low air humidity, apoplast water would quickly evaporate through leaf stomata. Thus, establishment of an aqueous apoplast by P. syringae not only requires specific virulence factors, such as HopM1 and AvrE1, but also high humidity, providing a critical insight into the high-humidity dependence for many P. syringae disease outbreaks84.
Temperature
Another important environmental factor is temperature (Fig. 5c). Even though ~28°C is used as an optimal growth temperature for many P. syringae strains in vitro, a generally negative effect of high temperature on the production of P. syringae virulence factors (e.g., phytotoxins, EPS or T3E secretion) have been docummented95–101. However, these studies were performed almost exclusively in vitro; whether and how high temperature might affect P. syringae virulence mechanisms in planta remains to be investigated, as studies have shown that high temperature (e.g., 28°C) leads to enhanced diseases by P. syringae102. In the context of disease, high temperature can affect plant immunity102, 103, pathogen virulence or both. Cheng and colleagues recently showed that PTI and ETI pathways respond to temperature fluctuations differently104, suggesting that different plant immunity pathways (and possibly different P. syringae virulence factors) may respond to temperature in a different manner.
Microbiome
P. syringae pathogens co-exist with numerous other microbes (i.e. microbiome) on plants. The presence of other interacting microbes could influence the virulence of a pathogen and/or the amplitude of plant immune responses, leading to different disease outcomes12, 88. There are already many studies illustrating interactions between P. syringae strains, plants and other microbes. For example, individual “bio-control” microbes could promote resistance to P. syringae through diverse mechanisms105–107, and induced systemic resistance (ISR) can be triggered by individual members of the plant root microbiome and “prime” plants for a stronger immune response against subsequent infection by P. syringae108–110 (Fig. 5d). However, current studies are largely focused on binary interactions between plants and individual strains of P. syringae and biocontrol/ISR agents. How P. syringae interacts with multiple members of the endogenous plant microbiome (i.e., in a community context) is poorly understood, as multiple microbe-microbe, microbe-pathogen and microbe-plant interactions could potentially neutralize or synergize final disease outcomes.
In summary, environmental conditions including temperature, humidity and the plant microbiome could greatly shape plant-P. syringae interactions. Yet, despite some emerging studies on these topics, we are quite far from a comprehensive and mechanistic understanding of their influences.
Conclusions and perspectives
Three decades of unprecedented mechanistic studies of P. syringae virulence factors and genomic and evolutionary insights have revealed basic features of P. syringae as a plant pathogen, putting us closer to answering the central question of “what makes P. syringae a successful plant pathogen”. Current results point to three principal strategies used by P. syringae to subvert plants: epiphytic survival and adaptation, suppression of host immunity at various stages of infection and establishment of an aqueous apoplast that promotes bacterial access to abundant water and likely nutrients. Acquisition of the T3SS and a core T3E repertoire, together with production of phytotoxins and other virulence factors, appear to be critical to the execution of these strategies and success of P. syringae as a plant pathogen.
One may wonder whether the virulence strategies employed by P. syringae are unique or common among plant pathogens that infect the phyllosphere. A clear answer to this question awaits future studies. However, there are strong indications that suppression of host immunity is a widespread virulence mechanism utilized by other pathogens. For instance, many effector proteins from other bacterial, fungal and oomycete pathogens have been shown to intercept various components of the plant immune machinery, leading to host immune suppression64–66. In addition, “water-soaking” symptom is observed in diverse diseases caused by bacteria, fungi and oomycetes111, 112. In-depth studies of Pantoea stewartii subsp. stewartii and Xanthomonas gardneri, for example, show that these bacterial pathogens can induce strong water-soaking in host plants. WtsE, an AvrE-family effector protein and an essential virulence factor in P. stewartii. subsp. stewartii, is required for water-soaking induction113, 114. On the other hand, AvrHah1, a water-soaking T3E in X. gardneri, activates the expression of plant cell wall-modifying enzymes, suggesting that plant cell wall alteration may be involved in perturbation of water homeostasis in the apoplast115. It remains to be seen whether leaf-infecting fungi and/or oomyctes also dedicate specific virulence factors to establish an aqueous apoplast as part of their infection strategy.
Because P. syringae strains typically carry dozens of T3Es62, the identification of a minimal repertoire of eight T3Es for P. syringae infection of N. benthamiana63 highlight an aspect of P. syringae biology that requires further study. Why do P. syringae strains maintain an apparently “larger-than-necessary” repertoire of T3Es and other virulence factors? Deletion of many P. syringae effectors apparently do not show a virulence loss in a given host plant, and this has been attributed to functional redundancy and the possibility that some effectors may be needed only in some other host plants116. In light of the significant influence of environmental conditions on P. syringae infection and the fact that most molecular studies of P. syringae infection have been conducted under static environmental conditions, we propose an additional possibility: many P. syringae virulence factors, including T3Es, may become necessary under natural fluctuating environmental conditions. We anticipate that understanding how environmental conditions and other biotic factors (i.e. microbiome) shape P. syringae infection will likely become an important aspect of future research. It will be particularly interesting to investigate whether, like HopM1 and AvrE1, some T3Es function to integrate different environmental conditions and microbial communities and contribute to disease development under a particular environmental and/or microbiome context. It is hoped that a complete understanding of the multi-dimensional plant-P. syringae-environment/microbiome interactions will infer innovative approaches for controlling diseases on crop plants.
Key Points.
Pseudomonas syringae is one of the most common plant pathogens that infect the phyllosphere (i.e., the aboveground plant organs). P. syringae can live on the plant surface as an epiphyte. To cause disease it enters the plant, through wounds or natural openings such as stomata, and multiplies within the intercellular space called the apoplast. In the past three decades, P. syringae has been used as an insightful model for understanding bacterial virulence mechanisms, host adaptation of pathogens, as well as microbial evolution, ecology and epidemiology.
The P. syringae species complex forms a monophyletic group in the P. fluorescens-like division of Pseudomonas. P. syringae strains are split into 13 phylogroups, which separate between early branching and canonical lineages. Members of the canonical lineages have conserved virulence-associated and phenotypic features and include several plant-specialist phylogroups. P. syringae has also been subdivided into ~50 pathovars based on host of isolation, host range and other properties.
P. syringae attacks plants using a variety of virulence factors, including “effector proteins” that are translocated into the plant cell via the type III secretion system (T3SS), small-molecule toxins, exopolysaccharides, cell wall-degrading enzymes and plant hormones (or hormone mimics). Whereas all pathogenic strains of P. syringae possess the T3SS and effectors, they may or may not produce other virulence factors.
Plants have evolved a defense mechanism (stomatal closure) to reduce bacterial entry through stomata by detection of pathogen-associated molecular patterns (PAMPs). To defeat stomatal defense, P. syringae use toxins and T3SS effector proteins to overcome PAMP-induced stomatal closure. Stomatal closure is sensitive to high atmospheric humidity, which could promote bacterial entry into the plant.
After entry into the plant, P. syringae encounters the apoplast, a potentially carbohydrate-rich but heavily defended living space for microbes. Recent advances in the identification of a minimal repertoire of T3SS effectors and host-mutation-based “disease reconstitution” experiments provide evidence that immune suppression and establishment of aqueous apoplast are two principal pathogenic processes required for P. syringae multiplication inside the apoplast.
P. syringae infection is profoundly influenced by external environmental conditions, such as air humidity, temperature and microbiota that live on healthy plants. Understanding how abiotic and biotic environmental conditions shape P. syringae infection at the mechanistic level may become an important aspect of future research. A complete understanding of the multi-dimensional plant-P. syringae-environment/microbiome interactions will infer innovative approaches for controlling diseases on crop plants.
Acknowledgments
This work was supported by grants from the Institute of Plant Physiology & Ecology, Shanghai Institute for Biological Science, Chinese Academy of Sciences (X.F.X.), National Key Laboratory of Plant Molecular Genetics, China (X.F.X.), US National Institute of Food and Agriculture (NIFA) HATCH (Project: GEO00791 to B.H.K.), the State of Georgia (B.H.K.), Gordon and Betty Moore Foundation (GBMF3037 to S.Y.H.), US National Institute of General Medical Sciences (GM109928 to S.Y.H.) and US Department of Agriculture – NIFA (2015-67017-23360 and 2017-67017-26180 to S.Y.H). The authors thank colleagues, Kyaw Aung and Christian Danve M. Castroverde, at Michigan State University for comments on this manuscript and David Baltrus at the University of Arizona for helpful discussions.
Glossary
- LOPAT
The LOPAT phenotypic scheme was developed to distinguish species of phytopathogenic fluorescent Pseudomonads. Canonical P. syringae are positive for Levan (L), negative for cytochrome C Oxidase (O), negative for Potato soft rot (P), negative for Arginine dihydrolase (A), and positive for the hypersensitive response on Tobacco (T).
- Multi-Locus Sequence Analysis (MLSA)
A technique to determine genetic relatedness and predict phylogeny based on the analysis of concatenated sequences of multiple housekeeping genes. MLSA can be used to determined phylogenetic relationships within a closely related group of organisms.
- Rarefaction curve
A tool used to estimate genetic diversity. Rarefaction curves plot total “genetic units” per analyzed idividuals. This can be set to different genetic thresholds from SNPs to species. E.g. how many total phlyogroups have been identified per individuals analyzed. As the curve flattens predictions can be made about the extent of genetic diversity yet to be identified at the particular mesured threshold.
- T3SS
Type III secretion system; a proteinaceous supramolecular complex produced by many Gram-negative bacteria infecting plants or animals. It functions as a syringe-like structure and delivers virulence proteins, called type III effectors (T3Es), into the host cell, and plays essential roles in bacterial virulence.
- hrp/hrc genes
hrp, hypersensitive response and pathogenicity. hrp genes gain their names from the phenotypes observed upon their inactivation, specifically the loss of the host hypersensitive response (HR) in resistant plants as well as the loss of pathogenic (P) potential in susceptible host plants. A subset of hrp genes were subsequently renamed to hrc (hrp conserved) genes based on conservation with Yersinia T3SS genes. Many of the hrp/hrc genes encode structural components of the T3SS.
- T3E
T3SS effectors; virulence proteins that are produced in many Gram-negative bacterial pathogens and delivered into the plant cell via the T3SS. T3Es function to manipulate various plant processes to promote infection.
- HR
Hypersensitive response, a programmed-cell death response of plants, mediated by recognition of pathogen effectors by the corresponding plant resistance proteins and activation of effector-triggered immunity (ETI).
- PTI
A branch of plant innate immunity, sometimes referred to as basal defense. PTI signaling is initiated by recognition of conserved microbial structures (e.g. flagellin) by plant membrane-localized receptors, and transduced by downstream components including the MAP kinase cascade and WRKY transcription factors, and finally leads to expression of plant immunity genes.
- ETI
Another branch of plant innate immunity, formerly called “gene-for-gene” resistance. It is triggered by recognition of specific T3E proteins by the corresponding plant resistance proteins, through direct or indirect interaction. ETI evokes strong plant immune responses, which often culminates in programed cell death (i.e., hypersensitive response).
- Stomata
Microscopic pores found in the epidermis of leaves, stems, and other plant organs, that facilitates gas exchange. The pore is bordered by a pair of specialized epidermal cells known as guard cells that are responsible for regulating the size of the stomatal opening.
- Guard cell
Specialized epidermal cells that surround the stomatal pore and enable it to open and close.
- Mesophyll cell
Cells located between the upper and lower epidermis in the plant leaf; the primary cell type for photosynthesis in the plant.
- IAA
Indole-3-acetic acid, the most common, naturally occurring, plant hormone of the auxin class.
- Coronatine
A toxin produced by Pseudomonas syringae; its chemical structure consists of two moieties, coronafacic acid (CFA) and coronamic acid (CMA).
- Syringomycins
A class of lipodepsinonapeptide molecules that are secreted by Pseudomonas syringae. Syringomycins are virulence determinants required for the manifestation of disease symptoms on a number of plants.
- EPS
Exopolysaccharide; high-molecular-weight polymers that are composed of sugar residues and are secreted by a microorganism into the surrounding environment.
- Pathovar
A bacterial strain or set of strains with the same or similar characteristics, which is differentiated at the infrasubspecific level from other strains of the same species or subspecies on the basis of distinctive pathogenicity to one or more plant hosts.
- Phylogroup
A phylogenetically related group of organisms. In the P. syringae species complex, phylogroups have been delineated based on genetic relatedness of less than 5% in conserved housekeeping genes.
- PAMP
Pathogen-associated molecular pattern, sometimes called microbe-associated molecular pattern (MAMP). These are conserved molecular structures from microbes and can elicit immune responses in the host.
- Endophyte
A microbial organism that lives within a plant for at least part of its life cycle.
- Salicylic acid
A phenolic plant defense hormone that mediates plant defense against infections by biotrophic and hemibiotrophic pathogens.
- Jasmonate
A lipid-based plant hormone that mediates plant defense against attacks by herbivory and necrotrophic pathogens as well as regulating plant growth and development.
- Abscisic acid
An isoprenoid plant stress hormone that functions in plant developmental processes such as seed dormancy and mediates plant response to water desiccation.
- Induced systemic resistance
An important mechanism by which selected plant growth–promoting bacteria and fungi in the rhizosphere prime the whole plant body for enhanced defense against a broad range of pathogens and insect herbivores.
- Oomycetes
A distinct phylogenetic lineage of fungus-like eukaryotic microorganisms. Oomycetes include some of the most notorious pathogens of plants, causing devastating diseases such as late blight of potato and sudden oak death.
Biographies
Xiu-Fang Xin is a Principal Investigator at the Institute of Plant Physiology & Ecology, Chinese Academy of Sciences (CAS), and a Group Leader at the CAS- John Innes Center (JIC) Center of Excellence for Plant and Microbial Sciences, Shanghai, China. She got her Ph.D. degree at Michigan State University, USA. Her lab is currently studying plant-pathogen-environment triangular interactions and how environmental conditions, such as temperature and humidity, influence plant-microbe interactions at the molecular level.
Brian H. Kvitko is an Assistant Professor in the Department of Plant Pathology at the University of Georgia, Athens, USA. He received his B.Sc. in Microbiology from The Ohio State University, Columbus, USA, and his Ph.D. in Microbiology from Cornell University, Ithaca, NY, USA. His research interests include the mechanisms of plant immune action on bacteria, cellular and quantitative molecular pathology of plant pathogens and the molecular host-microbe interactions of emerging bacterial plant pathogens.
Sheng Yang He is currently an Investigator at Howard Hughes Medical Institute and a University Distinguished Professor at Michigan State University, East Lansing, Michigan, USA. He received his Ph.D. degree from Cornell University, USA. His lab studies the Arabidopsis thaliana-Pseudomonas syringae interaction, focusing on molecular mechanisms that govern bacterial pathogenesis and disease susceptibility in plants. Studies from his lab have contributed to the understanding of how P. syringae deploys the type III secretion system and toxins to manipulate plant cellular processes, including innate immunity, jasmonate signaling and stomatal defense. Recent research in his lab begins to shed light on how climate conditions and phyllosphere microbiota might influence disease development. Dr. He is a fellow of the American Association for the Advancement of Science, USA, and a member of the US National Academy of Sciences.
Footnotes
Competing interest statement
The authors declare no competing interests.
References
- 1.Young JM. Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. syringae van Hall 1902. Ann Appl Biol. 1991;118:283–298. [Google Scholar]
- 2.Hirano SS, Upper CD. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol Mol Biol Rev. 2000;64:624–53. doi: 10.1128/mmbr.64.3.624-653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCann HC, et al. Genomic analysis of the Kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 2013;9:e1003503. doi: 10.1371/journal.ppat.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazzaglia A, et al. Pseudomonas syringae pv. actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS One. 2012;7:e36518. doi: 10.1371/journal.pone.0036518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler MI, et al. Pseudomonas syringae pv. actinidiae from recent outbreaks of kiwifruit bacterial canker belong to different clones that originated in China. PLoS One. 2013;8:e57464. doi: 10.1371/journal.pone.0057464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xin XF, He SY. Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol. 2013;51:473–98. doi: 10.1146/annurev-phyto-082712-102321. [DOI] [PubMed] [Google Scholar]
- 7.Rouse DI, Nordheim EV, Hirano SS, Upper CD. A model relating the probability of foliar disease incidence to the population frequencies of bacterial plant pathogens. Phytopathology. 1985;75:505–509. [Google Scholar]
- 8.Lindow SE, Arny DC, Upper CD. Bacterial ice nucleation: a factor in frost injury to plants. Plant Physiol. 1982;70:1084–9. doi: 10.1104/pp.70.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skirvina RM, et al. The use of genetically engineered bacteria to control frost on strawberries and potatoes. Whatever happened to all of that research? Sci Hortic. 2000;84:179–189. [Google Scholar]
- 10.Morris CE, Monteil CL, Berge O. The life history of Pseudomonas syringae: linking agriculture to earth system processes. Annu Rev Phytopathol. 2013;51:85–104. doi: 10.1146/annurev-phyto-082712-102402. [DOI] [PubMed] [Google Scholar]
- 11.Lindow SE, Brandl MT. Microbiology of the phyllosphere. Appl Environ Microbiol. 2003;69:1875–83. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baltrus DA, McCann HC, Guttman DS. Evolution, genomics and epidemiology of Pseudomonas syringae: Challenges in bacterial molecular plant pathology. Mol Plant Pathol. 2017;18:152–168. doi: 10.1111/mpp.12506. This is an excellent review summarizing the current knowledge on Pseudomonas syringae species from an ecological, genomic and evolutionary point of view. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vinatzer BA, Monteil CL, Clarke CR. Harnessing population genomics to understand how bacterial pathogens emerge, adapt to crop hosts, and disseminate. Annu Rev Phytopathol. 2014;52:19–43. doi: 10.1146/annurev-phyto-102313-045907. [DOI] [PubMed] [Google Scholar]
- 14.Jun SR, et al. Diversity of Pseudomonas genomes, including Populus-associated isolates, as revealed by comparative genome analysis. Appl Environ Microbiol. 2015;82:375–83. doi: 10.1128/AEM.02612-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garrido-Sanz D, et al. Genomic and genetic diversity within the Pseudomonas fluorescens Complex. PLoS One. 2016;11:e0150183. doi: 10.1371/journal.pone.0150183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berge O, et al. A user’s guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS One. 2014;9:e105547. doi: 10.1371/journal.pone.0105547. This is an important paper delineating thirteen phylogroups among the P. syringae species complex and outlining phenotypic traits common among the various phylogroups. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adamczyk L, et al. Directed flow of identified particles in Au+Au collisions at radical[SNN]=200 GeV at RHIC. Phys Rev Lett. 2012;108:202301. doi: 10.1103/PhysRevLett.108.202301. [DOI] [PubMed] [Google Scholar]
- 18.Clarke CR, Cai R, Studholme DJ, Guttman DS, Vinatzer BA. Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc Locus are common leaf colonizers equipped with an atypical type III secretion system. Mol Plant Microbe Interact. 2010;23:198–210. doi: 10.1094/MPMI-23-2-0198. [DOI] [PubMed] [Google Scholar]
- 19.Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–8. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 20.Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–64. doi: 10.1104/pp.109.139089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glickmann E, et al. Auxin production is a common feature of most pathovars of Pseudomonas syringae. Mol Plant Microbe Interact. 1998;11:156–62. doi: 10.1094/MPMI.1998.11.2.156. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien HE, et al. Extensive remodeling of the Pseudomonas syringae pv. avellanae type III secretome associated with two independent host shifts onto hazelnut. BMC Microbiol. 2012;12:141. doi: 10.1186/1471-2180-12-141. This paper describes the convergent evolution of phylogenetically distant P. syringae strains onto a common host and estimates the time of divergence for the P. syringae last common ancestor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97:1296–303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- 24.Bell CD, Soltis DE, Soltis PS. The age of the angiosperms: a molecular timescale without a clock. Evolution. 2005;59:1245–58. [PubMed] [Google Scholar]
- 25.Lindow SE, Leveau JH. Phyllosphere microbiology. Curr Opin Biotechnol. 2002;13:238–43. doi: 10.1016/s0958-1669(02)00313-0. [DOI] [PubMed] [Google Scholar]
- 26.Keith LM, Bender CL. AlgT (sigma22) controls alginate production and tolerance to environmental stress in Pseudomonas syringae. J Bacteriol. 1999;181:7176–84. doi: 10.1128/jb.181.23.7176-7184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber KJ, Desveaux D. AlgW regulates multiple Pseudomonas syringae virulence strategies. Mol Microbiol. 2011;80:364–77. doi: 10.1111/j.1365-2958.2011.07571.x. [DOI] [PubMed] [Google Scholar]
- 28.Castillo-Lizardo MG, et al. Contribution of the non-effector members of the HrpL regulon, iaaL and matE, to the virulence of Pseudomonas syringae pv. tomato DC3000 in tomato plants. BMC Microbiol. 2015;15:165. doi: 10.1186/s12866-015-0503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baltrus DA, et al. Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 2011;7:e1002132. doi: 10.1371/journal.ppat.1002132. This paper describes the diversity and distribution of T3Es and other key virulence factors in a cross section of 19 P. syringae isolates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aragon IM, Perez-Martinez I, Moreno-Perez A, Cerezo M, Ramos C. New insights into the role of indole-3-acetic acid in the virulence of Pseudomonas savastanoi pv. savastanoi. FEMS Microbiol Lett. 2014;356:184–92. doi: 10.1111/1574-6968.12413. [DOI] [PubMed] [Google Scholar]
- 31.Araki H, et al. Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:5887–92. doi: 10.1073/pnas.0601431103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hockett KL, Nishimura MT, Karlsrud E, Dougherty K, Baltrus DA. Pseudomonas syringae CC1557: a highly virulent strain with an unusually small type III effector repertoire that includes a novel effector. Mol Plant Microbe Interact. 2014;27:923–32. doi: 10.1094/MPMI-11-13-0354-R. [DOI] [PubMed] [Google Scholar]
- 33.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–80. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 34.Melotto M, Zhang L, Oblessuc PR, He SY. Stomatal Defense a Decade Later. Plant Physiol. 2017;174:561–571. doi: 10.1104/pp.16.01853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng W, Melotto M, He SY. Plant stomata: a checkpoint of host immunity and pathogen virulence. Curr Opin Biotechnol. 2010;21:599–603. doi: 10.1016/j.copbio.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melotto M, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–88. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–5. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng XY, et al. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe. 2012;11:587–96. doi: 10.1016/j.chom.2012.04.014. This paper and Du et al. (2014) show that coronatine activates JA signaling to turn on the expression of specific NAC-family transcription factors in Arabidopsis and tomato, which are required for coronatine-mediated stomatal opening and/or immune suppression in the apoplast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gimenez-Ibanez S, et al. JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 2017;213:1378–1392. doi: 10.1111/nph.14354. [DOI] [PubMed] [Google Scholar]
- 40.Du M, et al. Closely related NAC transcription factors of tomato differentially regulate stomatal closure and reopening during pathogen attack. Plant Cell. 2014;26:3167–84. doi: 10.1105/tpc.114.128272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toum L, et al. Coronatine inhibits stomatal closure through guard cell-specific inhibition of NADPH oxidase-dependent ROS production. Front Plant Sci. 2016;7:1851. doi: 10.3389/fpls.2016.01851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panchal S, et al. Coronatine facilitates Pseudomonas syringae infection of Arabidopsis Leaves at Night. Front Plant Sci. 2016;7:880. doi: 10.3389/fpls.2016.00880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allu AD, Brotman Y, Xue GP, Balazadeh S. Transcription factor ANAC032 modulates JA/SA signalling in response to Pseudomonas syringae infection. EMBO Rep. 2016;17:1578–1589. doi: 10.15252/embr.201642197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gimenez-Ibanez S, et al. The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol. 2014;12:e1001792. doi: 10.1371/journal.pbio.1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang S, et al. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013;9:e1003715. doi: 10.1371/journal.ppat.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang L, et al. Pseudomonas syringae type III effector HopBB1 promotes host rranscriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe. 2017;21:156–168. doi: 10.1016/j.chom.2017.01.003. This recent paper describes that P. syringae T3E HopBB1 directly interacts with and degrates JAZ proteins, thereby activating JA signaling and promoting disease. This represents one of an increasing number of studies (e.g., Gimenez-Ibanez et al. (2014) and Jiang et al. (2013)) showing that, like coronatine toxin, T3Es also induce JA signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee D, Bourdais G, Yu G, Robatzek S, Coaker G. Phosphorylation of the plant immune regulator RPM1-INTERACTING PROTEIN4 enhances plant plasma membrane H(+)-ATPase activity and inhibits flagellin-triggered immune responses in Arabidopsis. Plant Cell. 2015;27:2042–56. doi: 10.1105/tpc.114.132308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Z, et al. An Arabidopsis plasma membrane proton ATPase modulates JA signaling and is exploited by the Pseudomonas syringae effector protein AvrB for stomatal invasion. Plant Cell. 2015;27:2032–41. doi: 10.1105/tpc.15.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurley B, et al. The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS One. 2014;9:e114921. doi: 10.1371/journal.pone.0114921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lozano-Duran R, Bourdais G, He SY, Robatzek S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014;202:259–69. doi: 10.1111/nph.12651. [DOI] [PubMed] [Google Scholar]
- 51.Henry E, Toruno TY, Jauneau A, Deslandes L, Coaker G. Direct and indirect visualization of bacterial effector delivery into diverse plant cell types during infection. Plant Cell. 2017;29:1555–1570. doi: 10.1105/tpc.17.00027. This paper and Park et al. (2017) report an innovative GFP-based fusion approach to monitor type III translocation of P. syringae T3E in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park E, Lee HY, Woo J, Choi D, Dinesh-Kumar SP. Spatiotemporal monitoring of Pseudomonas syringae effectors via type III secretion using split fluorescent protein fragments. Plant Cell. 2017;29:1571–1584. doi: 10.1105/tpc.17.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones JD, Vance RE, Dangl JL. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354 doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- 54.Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16:537–52. doi: 10.1038/nri.2016.77. [DOI] [PubMed] [Google Scholar]
- 55.Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51:245–66. doi: 10.1146/annurev-phyto-082712-102314. [DOI] [PubMed] [Google Scholar]
- 56.Yamada K, Saijo Y, Nakagami H, Takano Y. Regulation of sugar transporter activity for antibacterial defense in Arabidopsis. Science. 2016;354:1427–1430. doi: 10.1126/science.aah5692. This paper provides intriguing evidence that activation of PTI involves direct interaction between FLS2, a PRR for bacterial flagellin, and sugar transporters STP1 and STP13, resulting in removal of sugars from the apoplast as a plant defense mechainsm. [DOI] [PubMed] [Google Scholar]
- 57.Mine A, et al. Pathogen exploitation of an abscisic acid- and jasmonate-inducible MAPK phosphatase and its interception by Arabidopsis immunity. Proc Natl Acad Sci U S A. 2017;114:7456–7461. doi: 10.1073/pnas.1702613114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hutchison ML, Tester MA, Gross DC. Role of biosurfactant and ion channel-forming activities of syringomycin in transmembrane ion flux: a model for the mechanism of action in the plant-pathogen interaction. Mol Plant Microbe Interact. 1995;8:610–20. doi: 10.1094/mpmi-8-0610. [DOI] [PubMed] [Google Scholar]
- 59.Vaughn VL, Gross DC. Characterization of salA, syrF, and syrG Genes and attendant regulatory networks involved in plant pathogenesis by Pseudomonas syringae pv. syringae B728a. PLoS One. 2016;11:e0150234. doi: 10.1371/journal.pone.0150234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bender CL, Alarcon-Chaidez F, Gross DC. Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev. 1999;63:266–92. doi: 10.1128/mmbr.63.2.266-292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roine E, et al. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci U S A. 1997;94:3459–64. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindeberg M, Cunnac S, Collmer A. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 2012;20:199–208. doi: 10.1016/j.tim.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Cunnac S, et al. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc Natl Acad Sci U S A. 2011;108:2975–80. doi: 10.1073/pnas.1013031108. This important paper describes a minimum reportoire of eight T3Es that can largely rescues the growth defect of an “effector-less” Pst DC300 strain in Nicotiana benthamiana plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toruno TY, Stergiopoulos I, Coaker G. Plant-pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol. 2016;54:419–41. doi: 10.1146/annurev-phyto-080615-100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macho AP, Zipfel C. Targeting of plant pattern recognition receptor-triggered immunity by bacterial type-III secretion system effectors. Curr Opin Microbiol. 2015;23:14–22. doi: 10.1016/j.mib.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 66.Dou D, Zhou JM. Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe. 2012;12:484–95. doi: 10.1016/j.chom.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 67.Block A, Alfano JR. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr Opin Microbiol. 2011;14:39–46. doi: 10.1016/j.mib.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.He P, et al. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 2006;125:563–75. doi: 10.1016/j.cell.2006.02.047. [DOI] [PubMed] [Google Scholar]
- 69.Rosebrock TR, et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–4. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gimenez-Ibanez S, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–9. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 72.Gohre V, et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 2008;18:1824–32. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- 73.Zeng L, Velasquez AC, Munkvold KR, Zhang J, Martin GB. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 2012;69:92–103. doi: 10.1111/j.1365-313X.2011.04773.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei HL, et al. Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell Host Microbe. 2015;17:752–62. doi: 10.1016/j.chom.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shimono M, et al. The Pseudomonas syringae type III effector HopG1 induces actin remodeling to promote symptom development and susceptibility during infection. Plant Physiol. 2016;171:2239–55. doi: 10.1104/pp.16.01593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Block A, et al. The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol. 2010;12:318–30. doi: 10.1111/j.1462-5822.2009.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo M, Kim P, Li G, Elowsky CG, Alfano JR. A bacterial effector co-opts calmodulin to rarget the plant microtubule network. Cell Host Microbe. 2016;19:67–78. doi: 10.1016/j.chom.2015.12.007. This paper describes that the T3E HopE1 dampens plant immunity by disrupting the plant microtubule network. HopE1 interacts with a microtubule-associated protein (MAP65) and causes its dissociation from microtubule. [DOI] [PubMed] [Google Scholar]
- 78.Rodriguez-Herva JJ, et al. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol. 2012;14:669–81. doi: 10.1111/j.1462-5822.2012.01749.x. [DOI] [PubMed] [Google Scholar]
- 79.Goel AK, et al. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol Plant Microbe Interact. 2008;21:361–70. doi: 10.1094/MPMI-21-3-0361. [DOI] [PubMed] [Google Scholar]
- 80.Hauck P, Thilmony R, He SY. A Pseudomonas syringae type III effector suppresses cell wall-based extracellular defense in susceptible Arabidopsis plants. Proc Natl Acad Sci U S A. 2003;100:8577–82. doi: 10.1073/pnas.1431173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jakobek JL, Smith JA, Lindgren PB. Suppression of Bean Defense Responses by Pseudomonas syringae. Plant Cell. 1993;5:57–63. doi: 10.1105/tpc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin L, et al. Direct and Indirect Targeting of PP2A by Conserved Bacterial Type-III Effector Proteins. PLoS Pathog. 2016;12:e1005609. doi: 10.1371/journal.ppat.1005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.DebRoy S, Thilmony R, Kwack YB, Nomura K, He SY. A family of conserved bacterial effectors inhibits salicylic acid-mediated basal immunity and promotes disease necrosis in plants. Proc Natl Acad Sci U S A. 2004;101:9927–32. doi: 10.1073/pnas.0401601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xin XF, et al. Bacteria establish an aqueous living space in plants crucial for virulence. Nature. 2016;539:524–529. doi: 10.1038/nature20166. This paper shows that two highly conserved T3Es, HopM1 and AvrE, induce aqueous apoplast during P. syringae infection as a critical virulence mechanism. This study also provides an important insight into the high-humidity dependence of P. syringae infection, consistent with the “disease triangle” concept. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–3. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 86.Wright CA, Beattie GA. Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:3269–74. doi: 10.1073/pnas.0400461101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stevens RB. In: Plant Pathology, an Advanced Treatise. Horsfall JG, AED, editors. Academic Press; New York: 1960. [Google Scholar]
- 88.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol. 2017;55:565–589. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
- 89.Lindemann J, Arny DC, Upper CD. Epiphytic populations of Pseudomonas syringae pv. syringae on snap bean and nonhost plants and the incidence of bacterial brown spot disease in relation to cropping patterns. Phytopathology. 1984;74:1329–1333. [Google Scholar]
- 90.Hirano SS, Upper CD. Population biology and epidemiology of Pseudomonas syringae. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- 91.Monier JM, Lindow SE. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl Environ Microbiol. 2004;70:346–55. doi: 10.1128/AEM.70.1.346-355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dechesne A, Smets BF. Pseudomonad swarming motility is restricted to a narrow range of high matric water potentials. Appl Environ Microbiol. 2012;78:2936–40. doi: 10.1128/AEM.06833-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bjorklof K, Nurmiaho-Lassila EL, Klinger N, Haahtela K, Romantschuk M. Colonization strategies and conjugal gene transfer of inoculated Pseudomonas syringae on the leaf surface. J Appl Microbiol. 2000;89:423–32. doi: 10.1046/j.1365-2672.2000.01130.x. [DOI] [PubMed] [Google Scholar]
- 94.Panchal S, et al. Regulation of stomatal defense by air relative humidity. Plant Physiol. 2016;172:2021–2032. doi: 10.1104/pp.16.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smirnova A, et al. Thermoregulated expression of virulence factors in plant-associated bacteria. Arch Microbiol. 2001;176:393–9. doi: 10.1007/s002030100344. [DOI] [PubMed] [Google Scholar]
- 96.Palmer DA, Bender CL. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol. 1993;59:1619–26. doi: 10.1128/aem.59.5.1619-1626.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weingart H, Stubner S, Schenk A, Ullrich MS. Impact of temperature on in planta expression of genes involved in synthesis of the Pseudomonas syringae phytotoxin coronatine. Mol Plant Microbe Interact. 2004;17:1095–102. doi: 10.1094/MPMI.2004.17.10.1095. [DOI] [PubMed] [Google Scholar]
- 98.Hockett KL, Burch AY, Lindow SE. Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS One. 2013;8:e59850. doi: 10.1371/journal.pone.0059850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, Schenk A, Srivastava A, Zhurina D, Ullrich MS. Thermo-responsive expression and differential secretion of the extracellular enzyme levansucrase in the plant pathogenic bacterium Pseudomonas syringae pv. glycinea. FEMS Microbiol Lett. 2006;265:178–85. doi: 10.1111/j.1574-6968.2006.00486.x. [DOI] [PubMed] [Google Scholar]
- 100.van Dijk K, et al. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol. 1999;181:4790–7. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rowley KB, Clements DE, Mandel M, Humphreys T, Patil SS. Multiple copies of a DNA sequence from Pseudomonas syringae pathovar phaseolicola abolish thermoregulation of phaseolotoxin production. Mol Microbiol. 1993;8:625–35. doi: 10.1111/j.1365-2958.1993.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 102.Wang Y, Bao Z, Zhu Y, Hua J. Analysis of temperature modulation of plant defense against biotrophic microbes. Mol Plant Microbe Interact. 2009;22:498–506. doi: 10.1094/MPMI-22-5-0498. [DOI] [PubMed] [Google Scholar]
- 103.Menna A, Nguyen D, Guttman DS, Desveaux D. Elevated temperature differentially influences effector-triggered immunity outputs in Arabidopsis. Front Plant Sci. 2015;6:995. doi: 10.3389/fpls.2015.00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cheng C, et al. Plant immune response to pathogens differs with changing temperatures. Nat Commun. 2013;4:2530. doi: 10.1038/ncomms3530. This paper shows that two major branches of plant immunity, PTI and ETI, respond to changing temperatures in distinct ways and suggests interesting interplays between plant immunity, temperature and pathogen physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rombouts S, et al. Characterization of novel bacteriophages for biocontrol of bacterial blight in leek caused by Pseudomonas syringae pv. porri. Front Microbiol. 2016;7:279. doi: 10.3389/fmicb.2016.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grosskinsky DK, et al. Cytokinin production by Pseudomonas fluorescens G20–18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci Rep. 2016;6:23310. doi: 10.1038/srep23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu K, Newman M, McInroy JA, Hu CH, Kloepper JW. Selection and assessment of plant growth-promoting Rhizobacteria for biological control of multiple plant diseases. Phytopathology. 2017;107:928–936. doi: 10.1094/PHYTO-02-17-0051-R. [DOI] [PubMed] [Google Scholar]
- 108.Pieterse CM, et al. Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol. 2014;52:347–75. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 109.Kumar AS, et al. Rhizobacteria Bacillus subtilis restricts foliar pathogen entry through stomata. Plant J. 2012;72:694–706. doi: 10.1111/j.1365-313X.2012.05116.x. [DOI] [PubMed] [Google Scholar]
- 110.Rudrappa T, Czymmek KJ, Pare PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–56. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Islam MT, et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016;14:84. doi: 10.1186/s12915-016-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Agrios GN. Plant Pathology. 5. Academic Press; 2005. [Google Scholar]
- 113.Ham JH, Majerczak DR, Arroyo-Rodriguez AS, Mackey DM, Coplin DL. WtsE, an AvrE-family effector protein from Pantoea stewartii subsp. stewartii, causes disease-associated cell death in corn and requires a chaperone protein for stability. Mol Plant Microbe Interact. 2006;19:1092–102. doi: 10.1094/MPMI-19-1092. [DOI] [PubMed] [Google Scholar]
- 114.Asselin JE, et al. Perturbation of maize phenylpropanoid metabolism by an AvrE family type III effector from Pantoea stewartii. Plant Physiol. 2015;167:1117–35. doi: 10.1104/pp.114.253120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schwartz AR, Morbitzer R, Lahaye T, Staskawicz BJ. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci U S A. 2017;114:E897–E903. doi: 10.1073/pnas.1620407114. This paper reports that a T3E from Xanthamonas gardneri induces strong water soaking in plant leaves and does so by activating plant cell wall-modifying enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lindeberg M, et al. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact. 2006;19:1151–8. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- 117.Alfano JR, et al. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci U S A. 2000;97:4856–61. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deng WL, Rehm AH, Charkowski AO, Rojas CM, Collmer A. Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J Bacteriol. 2003;185:2592–602. doi: 10.1128/JB.185.8.2592-2602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gazi AD, et al. Phylogenetic analysis of a gene cluster encoding an additional, rhizobial-like type III secretion system that is narrowly distributed among Pseudomonas syringae strains. BMC Microbiol. 2012;12:188. doi: 10.1186/1471-2180-12-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lan L, Deng X, Zhou J, Tang X. Genome-wide gene expression analysis of Pseudomonas syringae pv. tomato DC3000 reveals overlapping and distinct pathways regulated by hrpL and hrpRS. Mol Plant Microbe Interact. 2006;19:976–87. doi: 10.1094/MPMI-19-0976. [DOI] [PubMed] [Google Scholar]
- 121.Jovanovic M, et al. Regulation of the co-evolved HrpR and HrpS AAA+ proteins required for Pseudomonas syringae pathogenicity. Nat Commun. 2011;2:177. doi: 10.1038/ncomms1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brencic A, Winans SC. Detection of and response to signals involved in host-microbe interactions by plant-associated bacteria. Microbiol Mol Biol Rev. 2005;69:155–94. doi: 10.1128/MMBR.69.1.155-194.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bartoli C, et al. The Pseudomonas viridiflava phylogroups in the P. syringae species complex are characterized by genetic variability and phenotypic plasticity of pathogenicity-related traits. Environ Microbiol. 2014;16:2301–15. doi: 10.1111/1462-2920.12433. This paper explores the mututal exclusivity and variation in genetic arhitecture of the T-PAI and S-PAI among P. viridiflava phlyogroups. [DOI] [PubMed] [Google Scholar]
- 124.Guttman DS, Gropp SJ, Morgan RL, Wang PW. Diversifying selection drives the evolution of the type III secretion system pilus of Pseudomonas syringae. Mol Biol Evol. 2006;23:2342–54. doi: 10.1093/molbev/msl103. [DOI] [PubMed] [Google Scholar]
- 125.Tegli S, Gori A, Cerboneschi M, Cipriani MG, Sisto A. Type three secretion system in Pseudomonas savastanoi pathovars: does timing matter? Genes. 2011;2:957–979. doi: 10.3390/genes2040957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Monteil CL, et al. Population-genomic insights into emergence, crop adaptation and dissemination of Pseudomonas syringae pathogens. Microb Genom. 2016;2:e000089. doi: 10.1099/mgen.0.000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Munkvold KR, Russell AB, Kvitko BH, Collmer A. Pseudomonas syringae pv. tomato DC3000 type III effector HopAA1-1 functions redundantly with chlorosis-promoting factor PSPTO4723 to produce bacterial speck lesions in host tomato. Mol Plant Microbe Interact. 2009;22:1341–55. doi: 10.1094/MPMI-22-11-1341. [DOI] [PubMed] [Google Scholar]
- 128.Charity JC, Pak K, Delwiche CF, Hutcheson SW. Novel exchangeable effector loci associated with the Pseudomonas syringae hrp pathogenicity island: evidence for integron-like assembly from transposed gene cassettes. Mol Plant Microbe Interact. 2003;16:495–507. doi: 10.1094/MPMI.2003.16.6.495. [DOI] [PubMed] [Google Scholar]
- 129.Xiang T, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 130.Xiang T, et al. BAK1 is not a target of the Pseudomonas syringae effector AvrPto. Mol Plant Microbe Interact. 2011;24:100–7. doi: 10.1094/MPMI-04-10-0096. [DOI] [PubMed] [Google Scholar]
- 131.Cheng W, et al. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe. 2011;10:616–26. doi: 10.1016/j.chom.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li L, et al. Activation-dependent destruction of a co-receptor by a Pseudomonas syringae effector dampens plant immunity. Cell Host Microbe. 2016;20:504–514. doi: 10.1016/j.chom.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 133.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 134.Wu S, et al. Bacterial effector HopF2 suppresses arabidopsis innate immunity at the plasma membrane. Mol Plant Microbe Interact. 2011;24:585–93. doi: 10.1094/MPMI-07-10-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang Y, et al. A Pseudomonas syringae ADP-ribosyltransferase inhibits Arabidopsis mitogen-activated protein kinase kinases. Plant Cell. 2010;22:2033–44. doi: 10.1105/tpc.110.075697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou J, et al. The Pseudomonas syringae effector HopF2 suppresses Arabidopsis immunity by targeting BAK1. Plant J. 2013 doi: 10.1111/tpj.12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang J, et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–85. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 138.Zhang Z, et al. Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe. 2012;11:253–63. doi: 10.1016/j.chom.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 139.Eschen-Lippold L, et al. Bacterial AvrRpt2-like cysteine proteases block activation of the Arabidopsis mitogen-activated protein kinases, MPK4 and MPK11. Plant Physiol. 2016;171:2223–38. doi: 10.1104/pp.16.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sarris PF, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. This paper and Le Roux et al. (2015) show that P. syringae T3E AvrRps4 and Ralstonia solanacearum T3E Pop2 target plant immunity-associated WRKY transcription factors to dampen PTI. [DOI] [PubMed] [Google Scholar]
- 141.Le Roux C, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 142.Block A, et al. The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9. New Phytol. 2014;201:1358–70. doi: 10.1111/nph.12626. [DOI] [PubMed] [Google Scholar]
- 143.Cui H, et al. Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe. 2010;7:164–75. doi: 10.1016/j.chom.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chen Z, et al. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci U S A. 2007;104:20131–6. doi: 10.1073/pnas.0704901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cui F, et al. The Pseudomonas syringae type III effector AvrRpt2 promotes pathogen virulence via stimulating Arabidopsis auxin/indole acetic acid protein turnover. Plant Physiol. 2013;162:1018–29. doi: 10.1104/pp.113.219659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.de Torres-Zabala M, et al. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–43. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hann DR, et al. The Pseudomonas type III effector HopQ1 activates cytokinin signaling and interferes with plant innate immunity. New Phytol. 2014;201:585–98. doi: 10.1111/nph.12544. [DOI] [PubMed] [Google Scholar]
- 148.Washington EJ, et al. Pseudomonas syringae type III effector HopAF1 suppresses plant immunity by targeting methionine recycling to block ethylene induction. Proc Natl Acad Sci U S A. 2016;113:E3577–86. doi: 10.1073/pnas.1606322113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jelenska J, van Hal JA, Greenberg JT. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci U S A. 2010;107:13177–82. doi: 10.1073/pnas.0910943107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jelenska J, et al. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr Biol. 2007;17:499–508. doi: 10.1016/j.cub.2007.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kang Y, et al. HopW1 from Pseudomonas syringae disrupts the actin cytoskeleton to promote virulence in Arabidopsis. PLoS Pathog. 2014;10:e1004232. doi: 10.1371/journal.ppat.1004232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee AH, et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 2012;8:e1002523. doi: 10.1371/journal.ppat.1002523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li G, et al. Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 2014;77:310–21. doi: 10.1111/tpj.12396. [DOI] [PubMed] [Google Scholar]
- 154.Kim MG, et al. Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell. 2005;121:749–59. doi: 10.1016/j.cell.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 155.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–77. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 156.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–54. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 157.Luo Y, Caldwell KS, Wroblewski T, Wright ME, Michelmore RW. Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell. 2009;21:2458–72. doi: 10.1105/tpc.107.056044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wilton M, et al. The type III effector HopF2Pto targets Arabidopsis RIN4 protein to promote Pseudomonas syringae virulence. Proc Natl Acad Sci U S A. 2010;107:2349–54. doi: 10.1073/pnas.0904739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bhattacharjee S, Halane MK, Kim SH, Gassmann W. Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science. 2011;334:1405–8. doi: 10.1126/science.1211592. [DOI] [PubMed] [Google Scholar]
- 160.Heidrich K, et al. Arabidopsis EDS1 connects pathogen effector recognition to cell compartment-specific immune responses. Science. 2011;334:1401–4. doi: 10.1126/science.1211641. [DOI] [PubMed] [Google Scholar]
- 161.Fu ZQ, et al. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447:284–8. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 162.Nicaise V, et al. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013;32:701–12. doi: 10.1038/emboj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou H, et al. Pseudomonas syringae type III effector HopZ1 targets a host enzyme to suppress isoflavone biosynthesis and promote infection in soybean. Cell Host Microbe. 2011;9:177–86. doi: 10.1016/j.chom.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 164.Ustun S, Konig P, Guttman DS, Bornke F. HopZ4 from Pseudomonas syringae, a member of the HopZ type III effector family from the YopJ superfamily, inhibits the proteasome in plants. Mol Plant Microbe Interact. 2014;27:611–23. doi: 10.1094/MPMI-12-13-0363-R. [DOI] [PubMed] [Google Scholar]
- 165.Ustun S, et al. The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol. 2016;172:1941–1958. doi: 10.1104/pp.16.00808. [DOI] [PMC free article] [PubMed] [Google Scholar]