Abstract
Purpose
Microsatellite instability (MSI) is a pattern of hypermutation that occurs at genomic microsatellites and is caused by defects in the mismatch repair system. Mismatch repair deficiency that leads to MSI has been well described in several types of human cancer, most frequently in colorectal, endometrial, and gastric adenocarcinomas. MSI is known to be both predictive and prognostic, especially in colorectal cancer; however, current clinical guidelines only recommend MSI testing for colorectal and endometrial cancers. Therefore, less is known about the prevalence and extent of MSI among other types of cancer.
Methods
Using our recently published MSI-calling software, MANTIS, we analyzed whole-exome data from 11,139 tumor-normal pairs from The Cancer Genome Atlas and Therapeutically Applicable Research to Generate Effective Treatments projects and external data sources across 39 cancer types. Within a subset of these cancer types, we assessed mutation burden, mutational signatures, and somatic variants associated with MSI.
Results
We identified MSI in 3.8% of all cancers assessed—present in 27 of tumor types—most notably adrenocortical carcinoma (ACC), cervical cancer (CESC), and mesothelioma, in which MSI has not yet been well described. In addition, MSI-high ACC and CESC tumors were observed to have a higher average mutational burden than microsatellite-stable ACC and CESC tumors.
Conclusion
We provide evidence of as-yet-unappreciated MSI in several types of cancer. These findings support an expanded role for clinical MSI testing across multiple cancer types as patients with MSI-positive tumors are predicted to benefit from novel immunotherapies in clinical trials.
INTRODUCTION
Large-scale sequencing projects of cancer genomes have opened the door to studies that have identified putative biomarkers with potential clinical and therapeutic value, among them the presence or absence of microsatellite instability (MSI). Microsatellites are defined as 10 to 60 base pair regions that contain multiple repeats of 1 to 5 base pair motifs.1 Microsatellites occur at microsatellite loci, which are widely dispersed throughout the human genome. In normal cells, repeat count of microsatellites is verified and maintained during cell division by the mismatch repair (MMR) system,2,3 one of many cellular DNA repair mechanisms. Impairment of the MMR system can render cells unable to regulate the lengths of their microsatellites during cell division, termed MSI. After multiple cycles of cell division, cells with an impaired MMR system will develop varying lengths in their microsatellite sequences.
Mismatch repair deficiency is known to occur in some tumors,2 either by somatic hypermutation of MMR genes, most commonly, MLH14,5; an inherited germline MMR pathway mutation, such as in Lynch syndrome6,7; or double somatic mutations in MMR genes. MSI has been frequently observed within several types of cancer, most commonly in colorectal, endometrial, and gastric adenocarcinomas.8,9 The clinical significance of MSI has been well described in colorectal cancer, as patients with MSI-H (MSI-high) colorectal tumors have been shown to have improved prognosis compared with those with MSS (microsatellite stable) tumors.10,11 Furthermore, MSI-H colorectal tumors have been shown to be more susceptible to immune-enhancing therapies, such as the programmed cell death 1 (PD-1) inhibitor pembrolizumab,12 which has been recently approved for any MSI-H or MMR-deficient unresectable or metastatic solid tumor.13 Thus far, MSI-H tumors have the highest response rates to PD-1 inhibitors for any cancer type and have durable responses and a statistically significant improvement in overall survival.12
MSI polymerase chain reaction (PCR) and immunohistochemistry are two molecular biology–based methods that are in routine use for clinical MSI testing. MSI-PCR analyzes the distribution of microsatellite lengths at five standardized loci (Bethesda panel),14 and immunohistochemistry detects the presence or absence of four proteins that are involved in the MMR pathway (MSH2, MSH6, MLH1, and PMS2). Recently, several computational methods have been developed that analyze next-generation sequencing (NGS) data to detect MSI. Examples of such software include mSINGS,15 MSISensor,16 and MANTIS.17 A recent study by our group17 demonstrated that MANTIS achieves high sensitivity (97%) and specificity (99%) across six cancer types—tested using samples with known MSI status by MSI-PCR—and provides stable performance with varying numbers of microsatellite loci. Because of this, MANTIS is particularly well suited for application to a wider variety of cancer types.
As clinical MSI testing is routinely performed only on colorectal and endometrial tumors,18 the prevalence of MSI in many other cancer types has been less well described. In addition, evidence exists that MSI-PCR may be less accurate in other cancer types.19 A recent study by Hause et al20 developed and applied the MSI detection tool, MOSAIC, to perform a detailed survey of MSI across 18 cancer types (n = 5,930 cases); however, many other cancer types have yet to be analyzed for MSI. The ability to detect MSI in novel cancer types would permit the investigation of immune-enhancing therapies in these cancers, with the potential to benefit previously unknown subsets of patients with cancer with MSI.
To perform a more comprehensive assessment of MSI across many additional cancer types than those analyzed by Hause et al, our study determined the prevalence of MSI in 39 distinct cancer types (n = 11,139 tumors from 11,080 patients) by using our previously published MSI-calling tool, MANTIS.
METHODS
Data Preprocessing—The Cancer Genome Atlas and Therapeutically Applicable Research to Generate Effective Treatments
For analysis, 10,701 cases of paired tumor-normal whole-exome sequencing data were obtained from The Cancer Genome Atlas (TCGA)21-44 and Therapeutically Applicable Research to Generate Effective Treatments (TARGET)45,46 projects. Data from all of these cases, with the exception of diffuse large B-cell lymphoma (DLBCL) were processed via our in-house automated pipeline, L-MAP (Landscape Microsatellite Analysis Pathway). L-MAP is implemented in Python and MySQL and was run on the Oakley supercomputer at the Ohio Supercomputing Center.47 First, the metadata for all DNA whole-exome BAM files were downloaded from the Genomic Data Commons (GDC)48 and were converted to SQL database entries. Aligned BAM files (to hg3849) were queried from GDC by L-MAP by using the slicing end point provided by the GDC REST API. Reads that covered any base within 50 base pairs of a desired microsatellite locus were downloaded. As GDC data harmonization includes duplicate marking,48 premarked duplicate reads were removed by using SAMtools (version 1.3.1).50
As a result of a GDC sample contamination issue, all 48 DLBCL paired tumor-normal cases were downloaded from the GDC Legacy Archive as whole-exome BAM files aligned to hg19 by using the GDC Data Transfer Tool. Premarked duplicate reads were removed as above.
Data Preprocessing—Other Sources
Four hundred thirty cases of paired tumor-normal whole-exome sequencing data were obtained from the Sequence Read Archive51: 338 chronic lymphocytic leukemia cases from 279 patients from Landau et al,52 32 cutaneous T-cell lymphoma cases from Choi et al,53 51 nasopharyngeal carcinoma cases from Zheng h et al,54 and 8 cholangiocarcinoma cases from Ong et al.55 Fifteen additional cholangiocarcinoma cases were obtained from the European Nucleotide Archive56 from Chan-on et al.57 All sample identifiers used are available in the Data Supplement. These cases were processed via L-MAP. Tumor and normal samples were downloaded in the FASTQ format using fastq-dump.51 Alignment to hg38 was performed by using bwa (version 0.7.12)58 with the mem algorithm. Duplicate reads were marked and removed by using Picard MarkDuplicates.59 Base quality score recalibration and indel realignment were performed by using GATK,60 and the resulting BAM files were sliced, as above, by using SAMtools.
MSI Calling
MSI analysis with MANTIS (version 1.0.3; commit #942061f) was performed as previously described17 for all cases by using an average distance threshold of 0.4 to differentiate MSI-H from MSS tumors. Coordinates for 2,539 microsatellite loci within or near the exome—originally introduced by Salipante et al15 and used by later studies17—were converted from hg19 to hg38 by using LiftOver.61 Nine unlifted loci were discarded, which left 2,530 regions that were used for analysis with MANTIS in all cohorts, with the exception of DLBCL (Data Supplement). As the DLBCL data were aligned to hg19, the original 2,539 loci were used instead. MANTIS was run with author-recommended settings for whole-exome data—minimum read quality, 20; minimum locus quality, 25; minimum locus coverage, 20; minimum repeat reads, one; all other settings left at defaults. Eight samples were observed to have fewer than 10 loci sufficiently covered and were dropped. After MSI calling, microsatellite locus performance was assessed in each type of cancer as previously described.17 Kernel density estimation functions were computed by using R (version 3.3.2) using the density() function with default settings.
Whole-Exome Analysis
For all tumor-normal pairs that were tested by MANTIS in adrenocortical carcinoma (ACC; n = 92), cervical cancer (CESC; n = 305), and mesothelioma (MESO; n = 83), we downloaded aligned reads from whole-exome sequencing. Reads were downloaded in BAM format from GDC by using the GDC Data Transfer Tool. Premarked duplicate reads were removed by using SAMtools,50 variant calling was performed using MuTect62 (see Variant Calling), and annotation was performed by using ANNOVAR (version 2016-02-01)63 and GNU Parallel.64
Variant Calling
All variant calling was performed by using MuTect (version 1.1.7).62 The target region was derived from RefSeq (release 80).65 Exon data from the refGene table of the RefSeq Genes track was downloaded in BED format on February 28, 2017, by using the University of California, Santa Cruz Table Browser66 and 100 base pair padding. Unknown contigs were excluded and overlapping regions were merged with BEDTools.67 Variant cell format output was specified for MuTect and all other options were left at default. MuTect variant cell format output was then filtered for variants marked PASS. Variant annotation was performed by using ANNOVAR (version 2016-02-01)63 and GNU Parallel.64 Somatic mutations in the repair genes MSH2, MSH6, MLH1, PMS2, EXO1, POLD1, and POLE were determined by filtering variants with a DANN68,69 pathogenicity score greater than 0.96 (included in ANNOVAR). This threshold for DANN was chosen as it was previously shown to provide optimal sensitivity and specificity.69
Mutational signature calling was performed by using the tool deconstructSigs70 with the Nature 2013 signatures set, which contains 27 signatures,71 and the exome2genome normalization method. A mutational signature is a probability vector of length 96, with each element representing a single base change, along with bases immediately flanking it. In this analysis, linear regression is used to determine the relative contribution of each signature to the observed pattern of mutations. deconstructSigs was run over every ACC, CESC, and MESO sample by using all passing variants called with MuTect, as previously described.
All other downstream analyses were performed with Perl, Python, and R (version 3.3.2). Figures were generated by using R, Excel 2010 (Microsoft, Redmond, WA), and GraphPad Prism (version 7.0a; GraphPad Software, La Jolla, CA).
RESULTS
MSI Prevalence
We analyzed paired whole-exome sequencing data from 11,139 tumor-normal samples; 10,415 from the The Cancer Genome Atlas (TCGA)72 database, 280 from the TARGET45 database, and 444 from other studies,52-55,57 representing 39 distinct cancer types. MSI was detected in 27 of these 39 types of cancer (Fig 1A; Appendix Table A1; Data Supplement). The disease-specific prevalence of MSI varied widely, from 31.4% in endometrial carcinoma to 0.25% in glioblastoma multiforme. MSI was not detected in 12 cancer types (Figs 1A and 1B). Of 27 cancer types with MSI, 12 were found to have more than a single MSI-H tumor present and MSI-H prevalence greater than 1%. The relative level of instability, as measured by MANTIS score, varied substantially among MSI-H cancer types (Fig 1B and Appendix Fig A1 In addition, we attempted to determine which specific microsatellite loci performed best across the greatest number of cancer types (Data Supplement). Of 2,530 loci, we identified 22 loci that, within at least five cohorts, had an MSI-H versus MSS difference score greater than 0.75 and were sufficiently covered by at least 50% of samples in the cohort (Appendix Table A2). Only two loci that were assessed in the Bethesda14 and Promega73 MSI-PCR panels were included in our 2,530 loci, and neither of these were within the set of 22 top-performing loci. These results indicate a striking heterogeneity of MSI patterns across various types of cancer.
Fig 1.

Prevalence of microsatellite instability (MSI) across 39 human cancer types. (A) MSI prevalence was detected across 39 tumor types. The total number of tumors and the percentage of cases called MSI-high (MSI-H) in each cohort is listed in Appendix Table A1. (B) The relative level of instability, as measured by MANTIS score, is shown across all 39 tumor types. Note that for chronic lymphocytic leukemia (CLL), the listed MSI prevalence in panel A is out of 279 patients, and all 338 tumors are shown in panel B. MANTIS threshold cutoff of 0.4 is depicted with a dashed line. ACC, adrenocortical carcinoma; AML, pediatric acute myeloid leukemia (TARGET); BLCA, bladder carcinoma; BRCA, breast carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; CTCL, cutaneous T-cell lymphoma; DLBC, diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia (TCGA); LGG, lower-grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; NBL, pediatric neuroblastoma; NPC, nasopharyngeal carcinoma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectal adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TCGT, testicular germ cell tumor; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma; WT, Wilms tumor.
All four disease types with the highest rates of MSI prevalence were Lynch syndrome–associated tumor types that have been previously known to exhibit MSI: endometrial carcinoma, colon adenocarcinoma, gastric adenocarcinoma, and rectal adenocarcinoma. Consistent with previous studies, MSI was observed to be more frequent in colon adenocarcinoma (19.7%) than rectal adenocarcinoma (5.7%).20,74 Of importance, MSI was detected in three cancer types that have not been previously well characterized, most notably ACC (4.3%), cervical squamous cell carcinoma and CESC (2.6%), and MESO (2.4%; Fig 1A). To further investigate MSI status classifications, kernel density estimation75,76 was performed on the MANTIS scores for these tumor types. This indicated clear distinctions between samples that MANTIS called MSI-H from samples called MSS (Fig 2). Kernel density estimation was also performed on all other tumor types tested (Appendix Fig A1).
Fig 2.

Kernel density plots of MANTIS scores within (A) adrenocortical carcinoma (ACC), (B) cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), and (C) mesothelioma (MESO). The dotted line denotes the average distance threshold of 0.4, used by MANTIS to differentiate microsatellite instability high from microsatellite stable tumors. ACC: n = 92, kernel bandwidth (h) = 7.6e-3; CESC: n = 305, h = 9.4e-3; MESO: n = 83, h = 3.2e-3. KD plots for the other 36 cancer types analyzed are available in Appendix Fig A1.
Comparing Mutation Burden and Signatures Between MSI-H and MSS Tumors
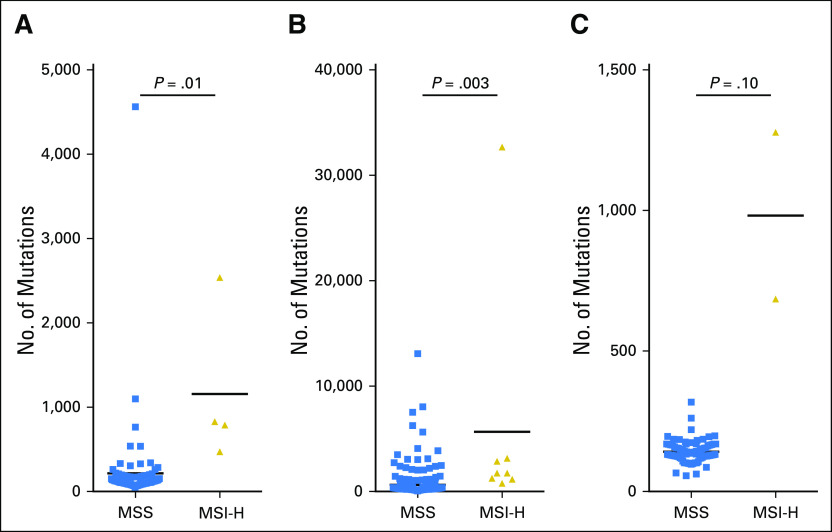
As Lynch syndrome–associated MSI-H tumors have been shown to have higher somatic mutation burden,12,77 we performed additional analyses to detect potential hypermutation in MSI-H ACC, CESC, and MESO. Somatic variant calling was performed on whole-exome samples from these four cancer types, and the mean absolute number of somatic mutations—both nonsynonymous and synonymous—was found to be increased among MSI-H versus MSS tumors within their own cohorts (Fig 3). In particular, an average of 1,157 somatic mutations were detected within MSI-H ACC samples versus 216 within MSS ACC (P = .01). An average of 5,675 somatic mutations were detected within MSI-H CESC samples versus 639 within MSS CESC (P = .003). Although statistical significance was not reached within MESO, MSI-H MESO tumors had, on average, a nearly seven-fold increase in mutational burden compared with MSS MESO tumors (982 v 142; P = .10). All P values were calculated by using Welch’s two-sample t test with log normalization. These results indicate that MSI in ACC and CESC is correlated with high mutational burden.
Fig 3.
Somatic mutational burden correlates with microsatellite instability high (MSI-H) status within adrenocortical carcinoma (ACC) and cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC). Mutational burden is listed for (A) ACC, (B) CESC, and (C) mesothelioma (MESO). P values were calculated using the Welch two-sample t test of log-normalized absolute somatic mutation counts. Variant calling was performed by using MuTect (“Variant Calling” in Methods), and all passing variants were included (nonsynonymous or synonymous).
To further investigate the observed somatic mutations in MSI-H versus MSS ACC, CESC, and MESO tumors, mutational signature analysis was performed by using a set of 27 signatures introduced by Alexandrov et al.71 A mutational signature defines a pattern of preferential somatic mutation types and may be associated with a known biologic process or type of cancer. This analysis was first performed on pooled mutations among MSI-H or MSS samples within each of these three cancer cohorts (Appendix Fig A2). No clear pattern of signature differences was evident from this pooled analysis. Next, mutational signature analysis was performed for each individual case within these cohorts without pooling (Data Supplement). Differences among signature prevalence in ACC, CESC, and MESO did not reach statistical significance. P values were calculated by using two-sided Fisher’s exact test (using signature presence or absence), with Benjamini correction for multiple hypotheses.78
MMR Pathway Alterations
MSI-H Lynch syndrome–associated tumors are known to lack the expression or function of at least one MMR protein; therefore, we analyzed somatic mutations that were predicted to be deleterious (by DANN68) in the MMR genes MSH2, MSH6, MLH1, PMS2, and EXO1, and the proofreading DNA polymerases POLD1 and POLE, among MSI-H and MSS samples within ACC, CESC, and MESO (Appendix Table A3; Data Supplement). Although POLD and POLE are not considered MMR proteins, mutations in these genes have been shown to lead to somatic hypermutation.22,79 Within these cohorts, 64% of MSI-H cases and 7% of MSS cases were found to contain at least one predicted deleterious somatic mutation in at least one of these genes; however, given that these samples were sequenced with potentially different exome captures, together with the increased mutational burden of MSI-H tumors, we could not determine the statistical significance of this finding.
DISCUSSION
In this study, we have performed, to our knowledge, the largest analysis of MSI in human cancer exomes to date, including 11,139 whole-exome tumor-normal pairs from 39 types of cancer. Compared with a study by Hause et al,20 we observed similar rates of MSI in 18 types of cancer, and we also analyzed another 5,209 whole-exome tumor-normal pairs from 21 additional types of cancer. In addition, we observed that MSI-H ACC and CESC tumors are significantly hypermutated compared with MSS ACC and CESC tumors. We identified three cohorts with significant MSI prevalence that have not been previously well described. Of particular interest, we identified MSI in 4 (4.4%) of 92 ACC cases. Previous studies of MSI in ACC have implicated Lynch syndrome as a risk factor for familial ACC80,81; however, to our knowledge, NGS-based MSI analysis has not yet been applied to ACC.
MSI-H colorectal tumors have been previously shown to be exceptionally sensitive to therapy with PD-1 immune checkpoint inhibitors.12 Identification of MSI in novel tumor types may lead to an expanded role for immunotherapy and a broader scope of clinical MSI testing.82 In addition, MSI is known to be prognostic within colorectal cancer,83 which may apply in other cancer types as well. For instance, Hause et al20 provide evidence that increasing MSI positively correlates with survival time. Clinical trials of immune checkpoint inhibitors are beginning or are underway in ACC (ClinicalTrials.gov identifier: NCT02673333), CESC (ClinicalTrials.gov identifier: NCT02635360), and MESO (ClinicalTrials.gov identifiers: NCT02784171, NCT02991482, NCT02707666, and NCT02399371), and a previous study of dendritic cell immunotherapy in ACC84 demonstrated tumor marker but not clinical response. These studies may benefit from the retrospective evaluation of MSI-H as a biomarker. Prospective expansion of clinical MSI testing to other cancer types may enlighten the prognostic and predictive value of MSI-H for noncolorectal cancers.
MMR deficiency is well recognized as the predominant cause of MSI within colorectal, endometrial, and gastric cancers. In addition, there have been anecdotal reports of ACC80,81 as a potential extracolonic manifestation of Lynch syndrome. If future studies indicate that MSI in ACC, CESC, and/or MESO is indeed a result of MMR deficiency, the findings of this study may implicate previously unappreciated cancer types as being part of Lynch syndrome. Compared with germline alterations in MMR genes, somatic events are most often a result of hypermethylation of CpG islands in the promoter region of MLH1.4 Additional investigation is needed to elucidate other molecular mechanisms that can lead to MSI, as well as the downstream effects of MSI on tumor-specific biology. In addition, of 9,569 tumors assessed in this study not within colorectal, endometrial, or gastric cancer, 77 (0.8%) were MSI-H. Only 14 of these were within ACC, CESC, or MESO, which compromised the statistical power of our mutational signature analysis. A larger cohort of MSI-H tumors would permit more comprehensive studies, including correlation with clinical data.
In summary, we have detected MSI in multiple cancer types, including ACC, CESC, and MESO, which indicates that MSI may affect non–Lynch syndrome tumor types. Within each type of cancer having MSI, we identified which loci—among 2,530—were most predictive of overall tumor MSI status. With additional analysis, these well-performing loci may form the basis of a targeted NGS panel for pancancer MSI detection. In addition, we found that MSI-H tumors in ACC and CESC have higher mutational burden than MSS tumors of these types. Given our observations of a long tail of MSI-H tumors across multiple cancer types, we propose that these and other, less common cancers undergo evaluation for MSI.
ACKNOWLEDGMENT
We thank current and past members of the Roychowdhury laboratory for their helpful insight and discussion. Data used for this analysis are available at dbGaP (accession: phs000218.v17.p6). R.B. would like to dedicate this work to his late father, Russell E. Bonneville Jr.
Appendix
Fig A1.

Kernel density plots of MANTIS scores within 36 cancer types. The dotted line denotes the average distance threshold of 0.4, used by MANTIS to differentiate microsatellite instability high from microsatellite stable tumors. Uterine corpus endometrial carcinoma (UCEC): kernel bandwidth (h) = 4.89e-02. Colon adenocarcinoma (COAD): h = 1.13e-02. Stomach adenocarcinoma (STAD): h = 7.59e-03. Rectal adenocarcinoma (READ): h = 9.16e-03. Uterine carcinosarcoma (UCS): h = 4.10e-03. Pediatric high-risk Wilms tumor (WT): h = 1.27e-02. Esophageal carcinoma (ESCA): h = 5.02e-03. Breast carcinoma (BRCA): h = 7.41e-03. Kidney renal clear cell carcinoma (KIRC): h = 6.83e-03. Ovarian serous cystadenocarcinoma (OV): h = 5.23e-03. Cholangiocarcinoma (CHOL): h = 1.17e-02. Thymoma (THYM): h = 3.08e-03. Liver hepatocellular carcinoma (LIHC): h = 4.42e-03. Head and neck squamous cell carcinoma (HNSC): h = 4.25e-03. Sarcoma (SARC): h = 7.14e-03. Skin cutaneous melanoma (SKCM): h = 5.32e-03. Lung squamous cell carcinoma (LUSC): h = 7.13e-03. Prostate adenocarcinoma (PRAD): h = 5.31e-03. Lung adenocarcinoma (LUAD:): h = 5.74e-03. Bladder carcinoma (BLCA): h = 4.40e-03. Pediatric neuroblastoma (NBL:): h = 5.47e-03. Lower-grade glioma (LGG:): h = 4.32e-03. Chronic lymphocytic leukemia (CLL): h = 2.64e-03. Glioblastoma multiforme (GBM): h = 4.38e-03. Pediatric acute myeloid leukemia (AML): h = 6.13e-03. Cutaneous T-cell lymphoma (CTCL): h = 5.86e-03. Diffuse large B-cell lymphoma (DLBC): h = 6.68e-03. Kidney chromophobe (KICH): h = 3.34e-03. Kidney renal papillary cell carcinoma (KIRP): h = 5.16e-03. Acute myeloid leukemia (LAML): h = 5.28e-03. Nasopharyngeal carcinoma (NPC): h = 6.09e-03. Pancreatic adenocarcinoma (PAAD): h = 5.36e-03. Pheochromocytoma and paraganglioma (PCPG): h = 5.04e-03. Testicular germ cell tumor (TGCT): h = 3.40e-03. Thyroid carcinoma (THCA): h = 5.09e-03. Uveal melanoma (UVM): h = 3.06e-03.
Fig A2.

Patterns of mutational signatures (S) across microsatellite instability cancers: (A) adrenocortical carcinoma (ACC), (B) cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), and (C) mesothelioma (MESO). Mutational signatures were called using deconstructSigs from pooled variants from all microsatellite instability high or microsatellite stable tumors within each cohort within ACC, CESC, and MESO. Unk., unknown.
Table A1.
Summary of MSI Landscape Analysis
Table A2.
All Microsatellite Loci With Difference Scores of > 0.75 in Five or More Cancer Types
Table A3.
Frequency of Predicted Deleterious MMR Mutations in ACC, CESC, and MESO
Footnotes
S.R. was supported by the American Cancer Society (Grant No. MRSG-12-194-01-TBG), the Prostate Cancer Foundation Young Investigator Award, the National Human Genome Research Institute (Grant No. UM1HG006508), the National Cancer Institute (Grant No. UH2CA202971), the American Lung Association, Pelotonia, and FORE Cancer Research; M.A.K. was supported by T32 Oncology Training Grant No. 5T32-CA009338; R.B. was supported by a university fellowship and M.R.W. was supported by the Helene Fuld Health Trust Nursing Scholarship. The chronic lymphocytic leukemia sequencing data (dbGaP: phs000922.v1.p1) used in this work was supported by National Human Genome Research Institute Large-Scale Sequencing Program Grant No. U54-HG003067 (to the Broad Institute).
The results published here, in whole or part, are based on data generated by The Cancer Genome Atlas managed by the National Cancer Institute (NCI) and National Human Genome Research Institute. Information about The Cancer Genome Atlas can be found at http://cancergenome.nih.gov. The results published here, in whole or part, are based on data generated by the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) initiative managed by the NCI. Information about TARGET can be found at http://ocg.cancer.gov/programs/target.
AUTHOR CONTRIBUTIONS
Conception and design: Russell Bonneville, Melanie A. Krook, Esko A. Kautto, Sameek Roychowdhury
Collection and assembly of data: Russell Bonneville, Melanie A. Krook, Sameek Roychowdhury
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or po.ascopubs.org/site/ifc.
Russell Bonneville
No relationship to disclose
Melanie A. Krook
No relationship to disclose
Esko A. Kautto
No relationship to disclose
Jharna Miya
No relationship to disclose
Michele R. Wing
No relationship to disclose
Hui-Zi Chen
No relationship to disclose
Julie W. Reeser
No relationship to disclose
Lianbo Yu
No relationship to disclose
Sameek Roychowdhury
Stock and Other Ownership Interests: Johnson & Johnson (I)
Research Funding: Takeda, Ignyta
REFERENCES
- 1.Schlötterer C: Genome evolution: Are microsatellites really simple sequences? Curr Biol 8:R132-R134, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Shia J: Evolving approach and clinical significance of detecting DNA mismatch repair deficiency in colorectal carcinoma. Semin Diagn Pathol 32:352-361, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strand M, Prolla TA, Liskay RM, et al. : Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365:274-276, 1993. [Erratum: Nature 368: 569, 1994] [DOI] [PubMed] [Google Scholar]
- 4.Armaghany T, Wilson JD, Chu Q, et al. : Genetic alterations in colorectal cancer. Gastrointest Cancer Res 5:19-27, 2012 [PMC free article] [PubMed] [Google Scholar]
- 5.Kane MF, Loda M, Gaida GM, et al. : Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res 57:808-811, 1997 [PubMed] [Google Scholar]
- 6.Aaltonen LA, Peltomäki P, Leach FS, et al. : Clues to the pathogenesis of familial colorectal cancer. Science 260:812-816, 1993 [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, Shaw MW, Magnuson CW, et al. : Hereditary factors in cancer. Study of two large midwestern kindreds. Arch Intern Med 117:206-212, 1966 [PubMed] [Google Scholar]
- 8.Imai K, Yamamoto H: Carcinogenesis and microsatellite instability: The interrelationship between genetics and epigenetics. Carcinogenesis 29:673-680, 2008 [DOI] [PubMed] [Google Scholar]
- 9.Watson P, Lynch HT: The tumor spectrum in HNPCC. Anticancer Res 14:1635-1639, 1994 [PubMed] [Google Scholar]
- 10.Buckowitz A, Knaebel HP, Benner A, et al. : Microsatellite instability in colorectal cancer is associated with local lymphocyte infiltration and low frequency of distant metastases. Br J Cancer 92:1746-1753, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benatti P, Gafà R, Barana D, et al. : Microsatellite instability and colorectal cancer prognosis. Clin Cancer Res 11:8332-8340, 2005 [DOI] [PubMed] [Google Scholar]
- 12.Le DT, Uram JN, Wang H, et al. : PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372:2509-2520, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.US Food and Drug Administration : Keytruda Biologics License Application 125514/S-14 approval letter, May 23, 2017. https://www.accessdata.fda.gov/drugsatfda_docs/appletter/2017/125514orig1s014ltr.pdf
- 14.Boland CR, Thibodeau SN, Hamilton SR, et al. : A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 58:5248-5257, 1998 [PubMed] [Google Scholar]
- 15.Salipante SJ, Scroggins SM, Hampel HL, et al. : Microsatellite instability detection by next generation sequencing. Clin Chem 60:1192-1199, 2014 [DOI] [PubMed] [Google Scholar]
- 16.Niu B, Ye K, Zhang Q, et al. : MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 30:1015-1016, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kautto EA, Bonneville R, Miya J, et al. : Performance evaluation for rapid detection of pan-cancer microsatellite instability with MANTIS. Oncotarget 8:7452-7463, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giardiello FM, Allen JI, Axilbund JE, et al. : Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology 147:502-526, 2014 [DOI] [PubMed] [Google Scholar]
- 19.Faulkner RD, Seedhouse CH, Das-Gupta EP, et al. : BAT-25 and BAT-26, two mononucleotide microsatellites, are not sensitive markers of microsatellite instability in acute myeloid leukaemia. Br J Haematol 124:160-165, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Hause RJ, Pritchard CC, Shendure J, et al. : Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 22:1342-1350, 2016 [DOI] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network : Integrated genomic analyses of ovarian carcinoma. Nature 474:609-615, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cancer Genome Atlas Network : Comprehensive molecular characterization of human colon and rectal cancer. Nature 487:330-337, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network : Comprehensive genomic characterization of squamous cell lung cancers. Nature 489:519-525, 2012. [Erratum: Nature 491: 288, 2012] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Network : Comprehensive molecular portraits of human breast tumours. Nature 490:61-70, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network : Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 368:2059-2074, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network : Integrated genomic characterization of endometrial carcinoma. Nature 497:67-73, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network : Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499:43-49, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brennan CW, Verhaak RG, McKenna A, et al. : The somatic genomic landscape of glioblastoma. Cell 155:462-477, 2013. [Erratum: Cell 157: 753, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network : Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507:315-322, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network : Comprehensive molecular profiling of lung adenocarcinoma. Nature 511:543-550, 2014. [Erratum: Nature 514: 262, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network : Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513:202-209, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoadley KA, Yau C, Wolf DM, et al. : Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158:929-944, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis CF, Ricketts CJ, Wang M, et al. : The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 26:319-330, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network : Integrated genomic characterization of papillary thyroid carcinoma. Cell 159:676-690, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Network : Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517:576-582, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network : Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481-2498, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Network : Genomic classification of cutaneous melanoma. Cell 161:1681-1696, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ciriello G, Gatza ML, Beck AH, et al. : Comprehensive molecular portraits of invasive lobular breast cancer. Cell 163:506-519, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research Network : Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 374:135-145, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network : The molecular taxonomy of primary prostate cancer. Cell 163:1011-1025, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S, Cherniack AD, Dewal N, et al. : Comprehensive pan-genomic characterization of adrenocortical carcinoma. Cancer Cell 29:723-736, 2016. [Erratum: Cancer Cell 30: 363, 2016] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network : Integrated genomic characterization of oesophageal carcinoma. Nature 541:169-175, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The Cancer Genome Atlas Research Network : Integrated genomic and molecular characterization of cervical cancer . Nature 543:378-384, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fishbein L, Leshchiner I, Walter V, et al. : Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell 31:181-193, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.National Cancer Institute : TARGET: Therapeutically Applicable Research to Generate Effective Treatments. https://ocg.cancer.gov/programs/target [Google Scholar]
- 46.Pugh TJ, Morozova O, Attiyeh EF, et al. : The genetic landscape of high-risk neuroblastoma. Nat Genet 45:279-284, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohio Supercomputer Center : Oakley. https://www.osc.edu/resources/technical_support/supercomputers/oakley
- 48.Grossman RL, Heath AP, Ferretti V, et al. : Toward a shared vision for cancer genomic data. N Engl J Med 375:1109-1112, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lander ES, Linton LM, Birren B, et al. : Initial sequencing and analysis of the human genome. Nature 409:860-921, 2001. [Erratum: Nature 411: 720, 2001] [DOI] [PubMed] [Google Scholar]
- 50.Li H, Handsaker B, Wysoker A, et al. : The sequence alignment/Map format and SAMtools. Bioinformatics 25:2078-2079, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leinonen R, Sugawara H, Shumway M: The sequence read archive. Nucleic Acids Res 39:D19-D21, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landau DA, Tausch E, Taylor-Weiner AN, et al. : Mutations driving CLL and their evolution in progression and relapse. Nature 526:525-530, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi J, Goh G, Walradt T, et al. : Genomic landscape of cutaneous T cell lymphoma. Nat Genet 47:1011-1019, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng H, Dai W, Cheung AKL, et al. : Whole-exome sequencing identifies multiple loss-of-function mutations of NF-κB pathway regulators in nasopharyngeal carcinoma. Proc Natl Acad Sci USA 113:11283-11288, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ong CK, Subimerb C, Pairojkul C, et al. : Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet 44:690-693, 2012 [DOI] [PubMed] [Google Scholar]
- 56.Leinonen R, Akhtar R, Birney E, et al. : The European Nucleotide Archive. Nucleic Acids Res 39:D28-D31, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chan-On W, Nairismägi M-L, Ong CK, et al. : Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet 45:1474-1478, 2013 [DOI] [PubMed] [Google Scholar]
- 58.Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754-1760, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Broad Institute : Picard tools. http://broadinstitute.github.io/picard
- 60.McKenna A, Hanna M, Banks E, et al. : The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297-1303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hinrichs AS, Karolchik D, Baertsch R, et al. : The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res 34:D590-D598, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cibulskis K, Lawrence MS, Carter SL, et al. : Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31:213-219, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang K, Li M, Hakonarson H: ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Usenix : GNU Parallel—The command-line power tool. https://www.usenix.org/system/files/login/articles/105438-Tange.pdf
- 65.O’Leary NA, Wright MW, Brister JR, et al. : Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733-D745, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karolchik D, Hinrichs AS, Furey TS, et al. : The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32:D493-D496, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quinlan AR, Hall IM: BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26:841-842, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quang D, Chen Y, Xie X: DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31:761-763, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jensen D: The best variant prediction method that no one is using. http://www.enlis.com/blog/2015/03/17/the-best-variant-prediction-method-that-no-one-is-using/ [Google Scholar]
- 70.Rosenthal R, McGranahan N, Herrero J, et al. : DeconstructSigs: Delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 17:31, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. : Signatures of mutational processes in human cancer. Nature 500:415-421, 2013. [Erratum: Nature 502: 502, 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weinstein JN, Collisson EA, Mills GB, et al. : The Cancer Genome Atlas pan-cancer analysis project. Nat Genet 45:1113-1120, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bacher JW, Flanagan LA, Smalley RL, et al. : Development of a fluorescent multiplex assay for detection of MSI-High tumors. Dis Markers 20:237-250, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phipps AI, Lindor NM, Jenkins MA, et al. : Colon and rectal cancer survival by tumor location and microsatellite instability: The Colon Cancer Family Registry. Dis Colon Rectum 56:937-944, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parzen E: On estimation of a probability density function and mode. Ann Math Stat 33:1065-1076, 1962 [Google Scholar]
- 76.Rosenblatt M: Remarks on some nonparametric estimates of a density function. Ann Math Stat 27:832-837, 1956 [Google Scholar]
- 77.Gatalica Z, Vranic S, Xiu J, et al. : High microsatellite instability (MSI-H) colorectal carcinoma: A brief review of predictive biomarkers in the era of personalized medicine. Fam Cancer 15:405-412, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benjamini Y, Hochberg Y: Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B (Methodological) 57:289-300, 1995 [Google Scholar]
- 79.Johnson RE, Klassen R, Prakash L, et al. : A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol Cell 59:163-175, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Challis BG, Kandasamy N, Powlson AS, et al. : Familial adrenocortical carcinoma in association with Lynch syndrome. J Clin Endocrinol Metab 101:2269-2272, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Raymond VM, Everett JN, Furtado LV, et al. : Adrenocortical carcinoma is a Lynch syndrome-associated cancer. J Clin Oncol 31:3012-3018, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dudley JC, Lin MT, Le DT, et al. : Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res 22:813-820, 2016 [DOI] [PubMed] [Google Scholar]
- 83.Kawakami H, Zaanan A, Sinicrope FA: Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol 16:30, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Papewalis C, Fassnacht M, Willenberg HS, et al. : Dendritic cells as potential adjuvant for immunotherapy in adrenocortical carcinoma. Clin Endocrinol (Oxf) 65:215-222, 2006 [DOI] [PubMed] [Google Scholar]