Abstract
Streptomycetes are multicellular filamentous microorganisms, and major producers of industrial enzymes and bioactive compounds such as antibiotics and anticancer drugs. The mycelial lifestyle plays an important role in the productivity during industrial fermentations. The hyphae of liquid-grown streptomycetes can self-aggregate into pellets, which hampers their industrial exploitation. Here we show that the Mat complex, which is required for pellet formation, catalyzes the synthesis of extracellular poly-β-1,6-N-acetylglucosamine (PNAG) in the model organisms Streptomyces coelicolor and Streptomyces lividans. Extracellular accumulation of PNAG allows Streptomyces to attach to hydrophilic surfaces, while attachment to hydrophobic surfaces requires a cellulase-degradable extracellular polymer (EPS) produced by CslA. Over-expression of matAB was sufficient to restore pellet formation to cslA null mutants of S. lividans. The two EPS systems together increase the robustness of mycelial pellets. These new insights allow better control of liquid-culture morphology of streptomycetes, which may be harnessed to improve growth and industrial exploitation of these highly versatile natural product and enzyme producers.
Keywords: Streptomyces, hyphal aggregation, adhesion, PNAG, biofilm
INTRODUCTION
Members of the genus Streptomyces are of great industrial and medical importance for their ability to produce a plethora of natural products, including antibiotics, anticancer agents and immunosuppressants 1,2, and many industrially relevant enzymes, such as polysaccharide hydrolases and proteases 3. Streptomycetes exhibit a complex multicellular life cycle 4. After germination of a single spore, a complex vegetative mycelium is formed following a process of apical growth and branching of the hyphae 5. The centrally located hyphae undergo a developmental cycle leading to the formation of reproductive aerial hyphae, which are erect and segment into long chains of spores 1. This process is accompanied by the lysis of nearby positioned mycelia 6, thereby liberating nutrients required for aerial growth and sporulation 7 and the production of antibiotics 8.
In submerged cultures, many streptomycetes form mycelial aggregates often referred to as pellets. Because of their dense architecture, which among others causes significant mass-transfer limitations, pellets are undesirable from the perspective of industrial process design. However, antibiotics are often produced more efficiently by mycelial clumps 9,10. Pellet architecture depends on genetic and environmental factors, like septum formation 11,12, shear stress 13, pH 14 and cell-wall fusion 15. Another important factor is the extracellular matrix, which embeds the mycelia 16. Omnipresent in all biofilms, the matrix facilitates cell signaling 17 and allows retaining liberated nutrients 18, while it also plays a major role in growth and morphogenesis 19,20,21. The Streptomyces extracellular matrix is a complex mixture of biopolymers containing DNA 22, amyloid proteins 23 and multiple polysaccharides 24. One polysaccharide is produced at the hyphal apex by CslA and tailored by the combined efforts of GlxA and DtpA 23,24,25,26. CslA likely produces a cellulose- or chitin-like polymer, as the polymer is specifically stained by calcofluor white, which is specific for β-1,4 glycans 24. CslA plays a role in the anchoring of fimbriae-like structures attached to the hyphae of Streptomyces coelicolor, facilitating the attachment to polystyrene surfaces by working in unison with the amphipathic chaplin proteins 23. This attachment was negated by the addition of cellulases, which releases the fimbriae from the cell surface. Interestingly, removal of cslA and glxA from wild-type S. lividans or S. coelicolor inhibits pellet formation in liquid-grown cultures 24,25. Pellets are formed by the self-aggregation of the mycelia in a submerged environment and share many characteristics with surface-grown colonies, including the lysis of internally positioned mycelia, with the co-location of antibiotic production 27. Transcriptomics analysis has also shown the activity of developmental genes in pellets of S. coelicolor, even though sporulation is absent in liquid grown cultures 28.
Recently we discovered a novel locus encompassing the matA and matB genes, which likely encode biosynthetic enzymes for an extracellular polymeric substance (EPS) and are also required for pellet formation 29. Deletion of matAB results in a submerged morphology strikingly similar to that of cslA mutants 29. Both loci are also required for spore aggregation, which contributes to the size heterogeneity of mycelia in liquid-grown cultures 30. In this study we show that the product of MatAB is a major component of the extracellular matrix and identify the exo-polysaccharide as poly-β-1,6-N-acetylglucosamine (PNAG). Attachment assays show that PNAG plays a different function in aggregation than the cellulose-like EPS produced by CslA/GlxA. The results are a major step forward in understanding how the extracellular matrix of Streptomyces facilitates adhesion and how it controls morphogenesis during submerged cultivation.
RESULTS
The Mat enzymes facilitate the formation of a granular layer on the outside of the hyphae
Previous studies have shown that the mat genes are required for mycelial aggregation and pellet formation in submerged cultures of S. coelicolor and S. lividans 29. Genome annotation suggested that matB encodes an extracellular polysaccharide synthase. To shed more light on the Mat-dependent mechanism, we investigated the cell surface of S. lividans at high resolution by cryo-scanning electron microscopy (SEM) (Fig. 1). This revealed a surface layer that decorates the entire outer surface of wild-type hyphae (Fig. 1A), which was absent in the matAB null mutant (Fig. 1B). Although the Mat proteins are expressed throughout growth 30, the Mat-dependent polymer was most apparent in young mycelia. Transmission electron microscopy (TEM) of Tungsten acid-negative stained cells, which images electron dense polymeric surface structures, further highlighted the Mat-dependent extracellular layer decorating wild-type hyphae (Fig. 1G). Between the hyphae a deposit of extracellular matrix could also be observed by SEM (Fig. 1D). Conversely, the hyphae of the matAB double mutant have instead a smooth surface, observed both with SEM (Fig. 1B) or negative staining in the TEM (Fig. 1H). We also failed to detect any extracellular material between the hyphae of matAB mutants (Fig. 1E). The cslA null mutant retained the extracellular layer on the cell surface as well as the adhesive material between the hyphae (Fig. 1C and H), again suggesting that this EPS is produced independent of CslA.
Figure 1. FIGURE 1: Electron micrographs revealing at Mat-dependent extracellular matrix.
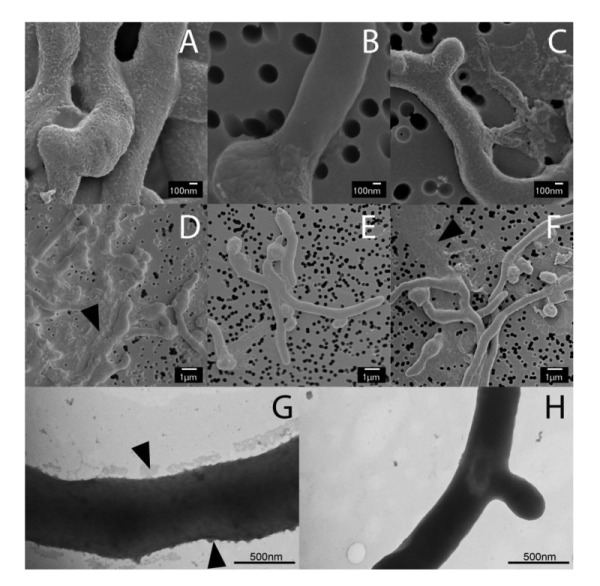
Scanning electron micrographs of young vegetative mycelium (A-F) show an abundance of extracellular material covering the hyphae of wild-type S. lividans (A) and between hyphae (D, indicated by arrow). This extracellular material was also present in the cslA null mutant (C and F, indicated by arrow), but was absent in matAB null mutants (B and E). Negatively stained hyphae with tungsten acid, specific for polymeric substances, revealed a scabrous outside coating in wild-type hyphae (G) that is absent in the mat mutant (H). All strains were grown for 8 h in TSBS media in shake flasks.
Bioinformatic analysis of the MatA and MatB enzymes
Bioinformatics analysis of MatA failed to identify known protein domains. MatB contains two functional domains, namely an intracellular glycosyltransferase type 2 (GT2) domain and an extracellular type 4 carbohydrate esterase (CE4) domain, connected by a predicted transmembrane helix. Sequences of glycosyltransferases and carbohydrate esterases were extracted from CAZy, which catalogs enzymes with characterized function, and assembled in a local database for Blast analysis. The glycosyltransferase domain of MatB returned PgaC from E. coli as the top hit (Table S1). E. coli pgaC encodes a glycosyltransferase that synthesizes PNAG 31. The next nearest homologs were enzymes with the same function in Acinetobacter baumannii, Staphylococcus epidermis, Aggregatibacter actinomycetemcomitans and Actinobacillus pleuropneumoniae, all with similar scores.
A similar blast comparison with the MatB carbohydrate esterase domain returned PgdA (BC_3618), a peptidoglycan N-acetylglucosamine deacetylase from Bacillus cereus as nearest characterized homologue (Table S1). Other top hits include a chitin deacetylase from Caldanaerobacter subterraneus and NodB proteins from Rhizobium species, all with similar scores. Interestingly, these enzymes all act on 1,4-linked oligo-chitin like substrates, in contrast to poly-β-1,6-N-acetylglucosamine glycosyltransferases. Taken together, bioinformatics analysis predicted that MatB catalyzes the formation of a poly-N-acetylglucosamine, with either a (1,4)- or (1,6)-configuration.
Figure 2. FIGURE 2: Calcofluor white staining to identify β-(1,4)-glycans.
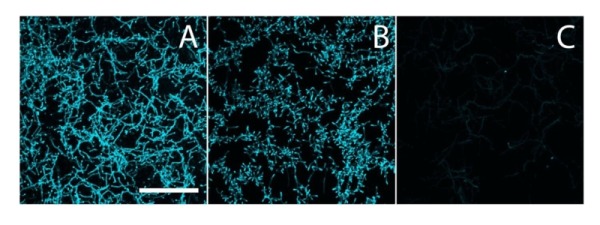
S. lividans (A), and its matAB (B) and cslA (C) mutants were stained with calcofluor white (CFW) to identify the presence of extracellular β-(1,4)-glycans. The staining patterns indicated the presence of β-(1,4)-glycans in both the parental strain and its matAB mutant, but not in the cslA mutant. Bar, 50 µM.
Biosynthesis of a PNAG-like EPS by the MatA and MatB enzymes
To analyze if the Mat proteins are involved in the biosynthesis of β-(1,4-) glycans, hyphae of S. lividans 66 and its matAB null mutant were stained with calcofluor white (CFW), which is specific for glycans in this configuration 32. Apical sites of both wild-type and matAB mutant cells were stained with equal efficiency (Fig. 2). This contrasts with the absence of staining in cslA null mutants, where the synthesis of β-(1,4)-glycans is impaired 24,25. These data suggest that only the CslA-GlxA system, and not MatA/MatB, produces β-(1,4)-glycans in S. lividans.
Figure 3. FIGURE 3: Immunofluorescence micrographs of S. lividans identifying extracellular PNAG.
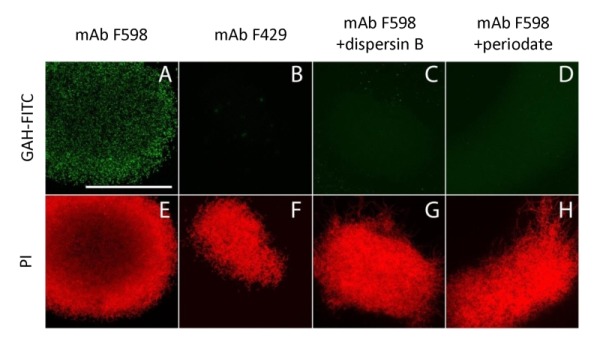
Mycelia from 18 h old cultures of S. lividans 66 were analyzed for the presence of PNAG with the monoclonal antibody mAb F598 and secondary anti-human IgG Alexa 488 conjugate (green) (A,C,D). As controls for the specificity of the primary antibody we used mAb F429, a monoclonal antibody that binds alginate (B), and samples treated with 50 µg/ml dispersin B (C) that degrades PNAG or with 0.4 M periodate (D), which degrades β-(1,6)-glycans. To visualize the hyphae, the DNA was stained with propidium iodide (red) (E-H). Bar, 100 µm.
To further characterize the product of MatA and MatB, we used monoclonal antibodies (mAb F598) that specifically recognize both intact and deacetylated PNAG 33. Mycelia obtained from 18 h liquid-grown cultures of S. lividans 66 were fixed in 4% PVA and incubated overnight with mAb F598. After washing and incubation with a fluorescently labeled secondary antibody conjugate, mounting fluid containing propidium iodide (PI) was added to stain the DNA and samples were then imaged with a confocal fluorescence microscope (Fig. 3). Wild-type cells were strongly stained with mAb F598, indicating the production of a PNAG-like polymer. Co-localization with the DNA stain PI suggests that most PNAG-like molecules are located within the pellet’s structure. Conversely staining with the control antibody mAb F429, which binds alginate 34, did not result in a fluorescent signal (Fig. 3B). Binding of mAb F598 could be negated by the PNAG-specific enzyme dispersin B 35 (Fig. 3C) or by the addition of periodate (Fig. 3D), which degrades 1,6-polysaccharides. Conversely, immunofluorescence microscopy of matA or matB null mutants or the matAB double mutant with the mAb F598 antibody only resulted in background fluorescence. The anti-PNAG signal was restored by the introduction of plasmid pMAT7, which expresses matAB from the strong gapA (SCO1947) promoter (Fig. 4). To link the Mat-dependent extracellular layer seen by SEM to the presence of PNAG, the mycelia were treated for 2 h with a suspension containing either chitinases, cellulases or dispersin B. Importantly, only dispersin B significantly affected EPS accumulation as visualized by SEM microscopy (Fig. S1). Taken together, these data strongly suggest that the MatAB-dependent extracellular matrix is indeed PNAG and that PNAG is not produced in the absence of MatA or MatB.
Figure 4. FIGURE 4: PNAG biosynthesis depends on MatA and MatB.
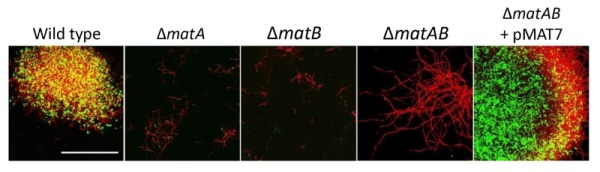
Mycelia from 18 h liquid-grown cultures were stained with mAb F598 (green), showing PNAG-specific staining for the wild-type strain S. lividans 66, while it was absent in mutants lacking matA, matB or the matAB double mutant. Genetic complementation via introduction of pMAT7 expressing matA and matB restored PNAG formation to the matAB double mutant. To visualize the hyphae, the DNA was stained with PI (red). Colocalization of PI and mAb F598 is shown in yellow. Bar: 50 µm.
MatAB expression is sufficient for pellet formation
As mentioned before, genomic disruption of either cslA or matB prevents pellet formation in shaken liquid-grown cultures, transforming the morphology in a strikingly similar fashion (Fig. 5A, E-F). Although the Mat-produced PNAG can be enzymatically dispersed by dispersin B (Fig. 3), addition of dispersin B to submerged grown mycelia failed to prevent pellet formation by S. lividans (Fig. 5B). Similarly, addition of cellulase, which antagonizes the cslA dependent glycan, failed to disrupt pellet formation (Fig. 5C). Simultaneous treatment with cellulases and dispersin B resulted in a slight reduction of the aggregation, reducing the average pellet size, but did not result in the dispersed morphology seen when either matB or cslA is disrupted (Fig. 5D). Interestingly, introduction of plasmid pMAT7 in the cslA mutant resulted in a pelleting phenotype, indicating that matAB expression by itself is sufficient for pellet formation (Fig. 5G). This MatAB-driven complementation of pellet formation in the cslA mutant could be readily antagonized by the addition of dispersin B, providing further evidence that production of PNAG is indeed the cause of the restored pellet formation (Fig. 5H). These data also indicate that native pellet formation, dependent on both cslA and matAB, results in a robust structure that cannot be easily antagonized by the addition of degrading enzymes, likely by making (parts) of the matrix inaccessible to the enzymes.
Figure 5. FIGURE 5: Effect of cellulases and dispersin B on mycelial morphology.
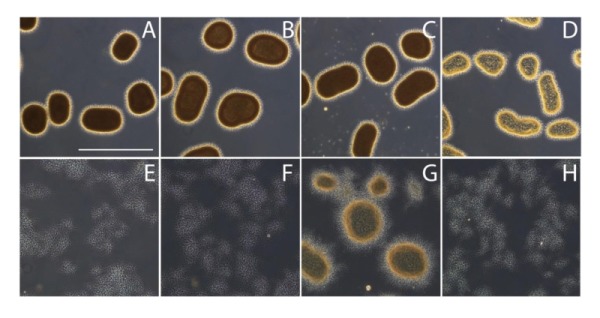
Light micrographs show S. lividans 66 (A), S. lividans 66 treated with 2 U/ml cellulase (B), 100 µg/ml dispersin B (C) or both cellulase and dispersin B (D), the matB (E) and cslA mutants (F) and the cslA mutant harboring pMAT7 without (G) or with (H) added dispersin B. All strains were grown in TSBS medium for 24 h at 30oC. The effects on morphology were visualized by wide-field microscopy. Bar, 500 µm.
Mechanistic insight into matA and matB in relation to cslA
To further assess the contribution of the two different EPSs to hyphal aggregation and pellet formation, we investigated the affinity of the hyphae for hydrophobic and hydrophilic surfaces, via adherence assays on glass and polystyrene, respectively. Attachment to glass depended on matA and matB, and was not visibly reduced in cslA mutants as compared to that of the parental strain (Fig. 6, left). Conversely, attachment to polystyrene was mostly dependent on cslA, and was less affected in matA and matB mutants (Fig. 6, right). Quantification of attachment by measuring cell-attached crystal violet spectrophotometrically, further confirmed the distinct roles for these genes (Fig. S2). This indicates that the EPS produced by CslA plays a dominant role in adherence to hydrophobic surfaces, while the PNAG produced by MatAB is particularly relevant for adherence to hydrophilic surfaces. This not only shows that the two EPSs have distinct non-redundant roles in aggregation, but also suggests that pellet formation in shaken liquid cultures depends on both hydrophilic and hydrophobic adhesive forces.
Figure 6. FIGURE 6: Adhesion assays visualizing EPS-mediated attachment to glass and polystyrene.
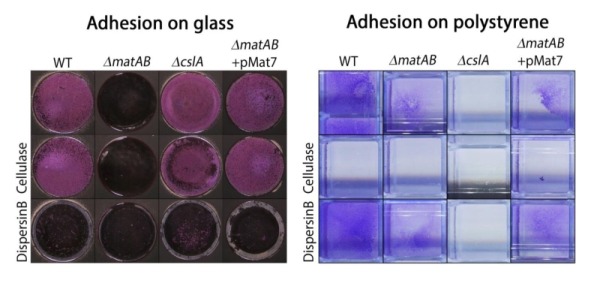
Attachment of S. lividans 66 to glass was antagonized by treatment with dispersin B (50 µg/ml) or by deletion of matAB, while addition of cellulase (0.2 U/ml) or deletion of cslA had no effect. The opposite was seen for attachment to polystyrene, which was antagonized by cellulase or by deletion of cslA and not by dispersin B or deletion of matAB. The ability to attach to glass was restored to matAB mutants by the introduction of plasmid pMAT7.
DISCUSSION
Members of the multicellular filamentous genus Streptomyces have an innate ability to self-aggregate in liquid-grown cultures. Because the multicellular mycelia are already connected, relatively little is needed for the neighboring hyphae to aggregate and start the formation of dense pellets. This propensity to self-aggregate into pellets contrasts with cells that undergo planktonic growth 4,36. Streptomycetes are the major producers of antibiotics and also produce a plethora of other medically relevant natural products as well as many industrial enzymes, and there is a strong correlation between the mycelial morphology in liquid-grown cultures and productivity 37. Enzyme production is typically favored by mycelial fragmentation 9,29, and it was shown that the Tat protein translocation machinery is located near apical sites 38. Conversely, production of erythromycin and actinorhodin requires a minimum mycelial pellet size 9,10. Therefore, it is important to understand the factors that control liquid-culture morphology.
In this study we investigated two major extracellular polysaccharides that play a crucial role in the ability of Streptomyces lividans to adhere. A polysaccharide produced by the recently discovered MatAB proteins was found to be poly-β-1,6-N-acetylglucosamine (PNAG). Extracellular polysaccharides in the matrix of bacteria with a planktonic lifestyle play a major role in biofilm formation. Although many types of exo-polysaccharides are employed by different bacteria 39,40, pathogenic bacteria often produce PNAG to adhere to their host 41,42,43,44. The soil bacterium Bacillus subtilis also produces PNAG, suggesting that this EPS is abundantly present in natural matrices 45.
The mat gene cluster resembles the PNAG biosynthetic gene cluster icaADBC from S. epidermidis, with the bifunctional MatB likely corresponding functionally to both IcaA and IcaB, forming an intracellular glycosyltransferase domain and an extracellular oligo-deacetylase domain, respectively. SCO2961, which is located directly downstream of matB, is an orthologue of icaC that might also play a role in formation of the mature polymer. The function of MatA is unclear as it lacks known functional domains, but as deletion of the matA gene reduces hyphal aggregation it may assist in efficient polymerization of the EPS, similarly to IcaD 46.
The observation that deletion of either cslA or matAB results in the disintegration of pellets, suggests a certain degree of interdependence between the two systems, and also that each system is required for pellet formation. Although this cannot be ruled out completely, our data show that pellet formation can be restored to cslA mutants by the overexpression of matAB, strongly suggesting that sufficiently high production of PNAG can compensate for the absence of the cellulase-like EPS produced by CslA. This is supported by the observation that while pellet formation by wild-type mycelia are unaffected by the addition of dispersin B or cellulases, dispersin B is sufficient to disperse pellets of cslA mutants overexpressing matAB. This suggests that although PNAG alone is sufficient, both EPSs contribute to the robustness of the pellet structure. The simultaneous addition of both dispersin B and cellulases, which should degrade both EPSs, was not sufficient to fully degrade the EPSs, as the mycelia still showed the ability to aggregate. A possible explanation may be the different roles that both systems play in maintaining the integrity of the pellet. The adhesion assays described above showed different roles of the two EPS systems. PNAG biosynthesized by MatAB mediates adhesion to a (hydrophilic) glass surface, a fast process that occurs in overnight cultures. This is supported by our earlier work showing that matAB is perhaps more important for the aggregation of germlings than the cellulose-like matrix produced by CslA 30. Adhesion mediated by the CslA-dependent EPS to the hydrophobic polystyrene is slow, requiring a week-long assay. This is likely the result of a more complex system, which likely also involves the developmentally regulated amphipathic chaplin proteins 23. Chaplins also play a role in pellet formation, which is yet poorly understood. The chaplin-based hydrophobic forces might contribute to strengthening of the pellet. Earlier work indicated that the EPS produced by CslA/GlxA plays a role in stabilization of the tip complex 24, which might explain its pleiotropic involvement in multiple systems throughout the life cycle. We hypothesize that aggregation by PNAG may also depend on proper organization of the tip complex and the presence of the cellulose-like EPS. High resolution spatial co-localization studies of CslA/GlxA, the chaplins and MatAB in native pellets, should shed more light on the involvement and interplay of these systems in the control of the mycelial architecture. This is currently being investigated in our laboratory.
Understanding how hyphal aggregation and pellet formation are controlled also brings us an important step closer to controlling the morphology of streptomycetes in liquid-grown cultures, which is highly relevant for tuning the morphology to product formation during industrial fermentations 37,47 Antibiotics such as erythromycin (produced by Saccharopolyspora erythraea) and actinorhodin (by S. coelicolor) are produced solely when a minimum pellet size is achieved, while enzyme production is typically favored by fast growing and fragmenting hyphae 9,10. Previous genetic approaches were followed to tune mycelial morphology. Over-expression of ssgA, which controls hyphal morphogenesis and activates cell division 11,12, effects fragmentation of the hyphae by enhancing cell division, resulting in increased growth and enzyme production rates 9. However, a drawback to this approach is the major effect of SsgA on the cell cycle, with enhanced sensitivity to shear stress as a result. In this respect morphological engineering targeting extracellular glue-like substances such as PNAG and the cellulose-like EPS, offers a very attractive alternative, as the effects on the internal physiology are likely minimal. Thus, besides their high relevance for our ecological understanding of how streptomycetes grow and attach to surfaces in their natural environment, the insights gained by this work may also help to develop novel technologies that improve growth and productivity of streptomycetes.
MATERIALS AND METHODS
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table S2. E. coli JM109 48 was used as a routine host for plasmid construction. The native matAB locus (SCO2963 and SCO2962) and gapA (SCO1947) promoter region were PCR-amplified from the S. coelicolor genome as described using primers SCO2963_F, SCO2962_R and PSCO1947_F, PSCO1947_R respectively (Table S1). The matAB locus was cloned as an EcoRI/BamHI fragment into the integrative vector pSET152 49 and the promoter region was placed in front of the matAB locus as an EcoRI/NdeI fragment, resulting in construct pMAT7. Conjugative plasmid transfer to Streptomyces was done using E. coli ET12567 50 harboring pUZ8002 as the host 51.
Culturing conditions
Streptomycetes were grown in shake flasks with a coiled stainless-steel spring in 30 ml tryptic soy broth (Difco) with 10% sucrose (TSBS). Cultures were inoculated with 106 cfu/ml and grown at 30°C. To assess growth in the presence of hydrolytic enzymes, strains were grown in 96-well plates where the agitation was facilitated by a Microplate Genie Digital mixer (Scientific Industries, USA) set to 1400 rpm, which was found to reproduce native morphologies at a micro scale 52. Dispersin B (50 µg/ml), cellulase (Sigma Aldrich, C1184) (2 U/ml) or a combination of both were added during growth to degrade EPS. The strains were observed after 24 h of growth by wide field microscopy or processed further for SEM analyses.
Bioinformatics
The genomes of Streptomyces coelicolor A3(2) M145 53 and S. lividans 66 54 have been published. Protein domains were annotated using the conserved domain search v3.14 55, using default settings. Homology searches were performed using the local Blast+ software v2.2.30 56. A BlastP database was built from the amino acid sequences of all characterized type 2 glycosyltransferases and type 4 carbohydrate esterases listed in the CAZy database (www.CAZy.org). The amino acid sequences were retrieved from the Uniprot database (www.uniprot.org).
Production and isolation of dispersin B
Dispersin B from Aggregatibacter actinomycetemcomitans ATCC 29522 was produced and purified as described 35. The specific activity, determined as the amount of enzyme needed to hydrolyze 1 µmol 4-nitrophenyl-β-D-N-acetylglucosaminide per minute in 50 mM sodium phosphate buffer (pH 5.5) 100 mM NaCl was 570 U /mg protein.
Calcofluor white staining
The presence of β-(1-4)-glycans was assessed by calcofluor white staining 32. Strains were grown over night in 8-well microscope chambers (LabTech II) in 300 µl TSBS medium. 30 µl calcofluor white (CFW) solution (Sigma Aldrich) was added and after 5 min incubation the samples were imaged on a Zeiss LSM5 Exciter/ Axio observer with a 405 nm laser, a 405/488 nm beam splitter and 420-480 nm bandpass filter 57.
Immunofluorescence
Immunofluorescence microscopy was performed as described 58, with small adaptations. In short, S. lividans 66 was grown in TSBS media for 18 h at 30°C. A 50 µl culture aliquot was spotted inside an eight-well microchamber with optical bottom (Lab-Tek, Thermo Scientific). After 15 min the media was removed gently and the cell layer was air dried for 10 min and fixed with 4% paraformaldehyde in PBS for 15 min. After washing samples twice with PBS, monoclonal antibodies against PNAG (mAb F598) or the control which binds alginate (mAb F429) were added to a final concentration of 10 µg/ml in PBS with 0.1% BSA-c (Aurion, the Netherlands) and samples incubated for 16 h at 4°C. The samples were then washed three times with PBS with 0.1% BSA-c and fluorescently-labeled goat-anti-human IgGs (Life Technologies) added to a final concentration of 4 µg/ml and incubated in the dark for 2 h. After washing twice with PBS with 0.1% BSA-c, some PBS with propidium iodide (PI) at a concentration 1 µg/ml was added and the samples were imaged on a Zeiss LSM5 Exciter/ Axio observer with a 488 nm and 543 nm laser, where the emission from the fluorescent antibodies was imaged between 505-545 nm and a 560 nm long-pass filter was used to monitor emission from PI. To remove PNAG, cells were submerged in Tris-buffered saline (pH 6.4) and treaded with dispersin B at a concentration of 50 µg/ml and incubated for 4 h at 37°C or with 0.4 M periodate for 2 h at 37°C. The primary antibody was subsequently added after washing.
Cryo scanning electron microscopy
Mycelia from cultures grown for 6 h and treated with either PBS (control), PBS containing 50 µg/ml dispersin B, 0.5 U/ml chitinases (Sigma) or 2U/ml cellulases (Sigma). After incubation for 4 h at 37°C the samples were fixed by 1.5% glutaraldehyde and immobilized on isopore membrane 0.8 µm filter discs (Millipore) by pushing the liquid through using a syringe and placing the filter in a filter holder. The discs were cut to size and placed on the SEM target immobilized with Tissue Tek® and quickly frozen in liquid nitrogen slush and transferred directly to the cryo-transfer attachment of the scanning electron microscope. After 10 minutes sublimation at -90°C specimens were sputter-coated with a layer of 2 nm Platinum and examined at -120°C in the JEOL JSM6700F scanning electron microscope at 3 kV as described 45.
Negative stain TEM microscopy
For negative staining, 5 µl of young mycelium was placed on a copper TEM grid and air dried for 15 min. The cellular material was stained with 3% PTA for 5 min, followed by 5 times washing with milliQ. The samples were placed in a JEOL 1010 transmission electron microscope and observed at 60 kV as described 59.
Adhesion Assays
Attachment of strains to polystyrene surfaces was tested as described 60. In short 106 CFUs/ml were inoculated into 4 ml NMMP 51 without polyethylene glycol and casamino acids, using 2% (w/v) mannitol as the sole carbon source. After 5 days at 300C the standing cultures were stained with crystal violet. After washing, the attached cells were quantified by extracting the crystal violet with 10% SDS and measuring the absorption at 570 nm 61. Attachment to glass surfaces was tested in a similar fashion, using glass bottom 96 wells plates (Greiner Bio-One, Austria) and 200 µl NMMP medium without polyethylene glycol, but with 0.5% casamino acids and 2% glucose as the carbon source. These were cultivated overnight at 300C and the attached biomass was quantified as for polystyrene. In case of enzymatic treatment, either cellulase (Sigma) at a concentration of 0.2 U/ml or Dispersin B at a concentration of 50 µg/ml was added to the medium, just prior to inoculation. All measurements are the average of five replicates.
SUPPLEMENTAL MATERIAL
All supplemental data for this article are also available online at http://microbialcell.com/researcharticles/production-of-poly-%CE%B2-16-n-acetylglucosamine-by-matab-is-required-for-hyphal-aggregation-and-hydrophilic-surface-adhesion-by-streptomyces/.
Funding Statement
We thank Marleen Janus for the supply of A.actinomycetemcomitans gDNA and Ellen de Waal for the production of dispersin B. The research was supported by VIDI grant 12957 and by VICI grant 10379 from the Netherlands Technology Foundation STW to DC and GVW, respectively.
References
- 1.Barka EA, Vatsa P, Sanchez L, Gavaut-Vaillant N, Jacquard C, Meier-Kolthoff J, Klenk HP, Clément C, Oudouch Y, van Wezel GP. Taxonomy, physiology, and natural products of the Actinobacteria. Microbiol Mol Biol Rev. 2016;80(1):1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopwood DA. How do antibiotic‐producing bacteria ensure their self‐resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol. 2007;63:937–940. doi: 10.1111/j.1365-2958.2006.05584.x. [DOI] [PubMed] [Google Scholar]
- 3.Vrancken K, Anne J. Secretory production of recombinant proteins by Streptomyces. Future Microbiol. 2009;4(2):181–188. doi: 10.2217/17460913.4.2.181. [DOI] [PubMed] [Google Scholar]
- 4.Claessen D, Rozen DE, Kuipers OP, Sogaard-Andersen L, van Wezel GP. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat Rev Microbiol. 2014;12(2):115–124. doi: 10.1038/nrmicro3178. [DOI] [PubMed] [Google Scholar]
- 5.Chater KF, Losick R. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives. 1997;In:149–182. [Google Scholar]
- 6.Wildermuth H. Development and organization of the aerial mycelium in Streptomyces coelicolor. Microbiology. 1970;60(1):43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- 7.Brana AF, Mendez C, Diaz LA, Manzanal MB, Hardisson C. Glycogen and trehalose accumulation during colony development in Streptomyces antibioticus. J Gen Microbiol. 1986;132(5):1319–1326. doi: 10.1099/00221287-132-5-1319. [DOI] [PubMed] [Google Scholar]
- 8.van Wezel GP, McKenzie NL, Nodwell JR. Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol . Methods Enzymol . 2009;458:117–141. doi: 10.1016/s0076-6879(09)04805-8. [DOI] [PubMed] [Google Scholar]
- 9.van Wezel GP, Krabben P, Traag BA, Keijser BJF, Kerste R, Vijgenboom E, Heijnen JJ, Kraal B. Unlocking Streptomyces spp. for use as sustainable industrial production platforms by morphological engineering. . Appl Environ Microbiol. 2006;72:5283–5288. doi: 10.1128/AEM.00808-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wardell JN, Stocks SM, Thomas CR, Bushell Me. Decreasing the hyphal branching rate of Saccharopolyspora erythraea NRRL 2338 leads to increased resistance to breakage and increased antibiotic production. Biotechnol Bioeng. 2002;78:141–146. doi: 10.1002/bit.10210. [DOI] [PubMed] [Google Scholar]
- 11.Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, Van Wezel GP. Loss of the controlled localization of growth stage‐specific cell‐wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol Microbiol. 2007;64:1244–1259. doi: 10.1111/j.1365-2958.2007.05732.x. [DOI] [PubMed] [Google Scholar]
- 12.Traag BA, Wezel GP. The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie van Leeuwenhoek. 2008;94:85–97. doi: 10.1007/s10482-008-9225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heydarian SM, Mirjalili N, Ison AP. Effect of shear on morphology and erythromycin production in Saccharopolyspora erythraea fermentations. Bioproc Eng. 1999;21:31–39. doi: 10.1007/PL00009062. [DOI] [Google Scholar]
- 14.Glazebrook MA, Vining LC, White RL. Growth morphology of Streptomyces akiyoshiensis in submerged culture: influence of pH, inoculum, and nutrients. Can J Microbiol. 1992;38:98–103. doi: 10.1139/m92-016. [DOI] [PubMed] [Google Scholar]
- 15.Koebsch I, Overbeck J, Piepmeyer S, Meschke H, Schrempf H. A molecular key for building hyphae aggregates: the role of the newly identified Streptomyces protein HyaS. Microb Biotechnol. 2009;2:343–360. doi: 10.1111/j.1751-7915.2009.00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrus MLC, Claessen D. Pivotal roles for Streptomyces cell surface polymers in morphological differentiation, attachment and mycelial architecture. Antonie van. 2014;Leeuwenhoek:1–13. doi: 10.1007/s10482-014-0157-9. [DOI] [PubMed] [Google Scholar]
- 17.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton Jt, Greenberg E. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280(5361):295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 18.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 19.Dragoš A, Kovács ÁT, Claessen D. The Role of Functional Amyloids in Multicellular Growth and Development of Gram-Positive Bacteria. Biomolecules. 2017;7(3):60. doi: 10.3390/biom7030060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wozniak DJ, Wyckoff TJ, Starkey M, Keyser R, Azadi P, O'Toole GA, Parsek MR. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci. 2003;100(13):7907–7912. doi: 10.1073/pnas.1231792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonas K, Tomenius H, Kader A, Normark S, Römling U, Belova LM, Melefors Ö. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 2007;7(1):70. doi: 10.1186/1471-2180-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y-M, Kim J-h. Formation and dispersion of mycelial pellets of Streptomyces coelicolor A3(2). J Microbiol (Seoul, Korea) 2004;42:64–67. [PubMed] [Google Scholar]
- 23.de Jong W, Wosten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73(6):1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Chater KF, Deng Z, Tao M. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J Bacteriol. 2008;190(14):4971–4978. doi: 10.1128/JB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaplin AK, Petrus ML, Mangiameli G, Hough MA, Svistunenko DA, Nicholls P, Claessen D, Vijgenboom E, Worrall JA. GlxA is a new structural member of the radical copper oxidase family and is required for glycan deposition at hyphal tips and morphogenesis of Streptomyces lividans. Biochem J. 2015;469(3):433–444. doi: 10.1042/BJ20150190. [DOI] [PubMed] [Google Scholar]
- 26.Petrus ML, Vijgenboom E, Chaplin AK, Worrall JA, van Wezel GP, Claessen D. The DyP-type peroxidase DtpA is a Tat-substrate required for GlxA maturation and morphogenesis in Streptomyces. Open Biol. 2016;6(1):150149. doi: 10.1098/rsob.150149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rioseras B, López-García MT, Yagüe P, Sánchez J, Manteca Á. Mycelium differentiation and development of Streptomyces coelicolor in lab-scale bioreactors: Programmed cell death, differentiation, and lysis are closely linked to undecylprodigiosin and actinorhodin production. Biores Technol. 2014;151:191–198. doi: 10.1016/j.biortech.2013.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagüe P, Rodríguez-García A, López-García MT, Rioseras B, Martín JF, Sánchez J, Manteca A. Transcriptomic analysis of liquid non-sporulating Streptomyces coelicolor cultures demonstrates the existence of a complex differentiation comparable to that occurring in solid sporulating cultures. PLoS ONE. 2014;9:e86296. doi: 10.1371/journal.pone.0086296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Dissel D, Claessen D, Roth M, van Wezel GP. A novel locus for mycelial aggregation forms a gateway to improved Streptomyces cell factories. Microb Cell Fact. 2015;14(1):44. doi: 10.1186/s12934-015-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zacchetti B, Willemse J, Recter B, van Dissel D, van Wezel GP, Wosten HA, Claessen D. Aggregation of germlings is a major contributing factor towards mycelial heterogeneity of Streptomyces. Sci Rep. 2016;6:27045. doi: 10.1038/srep27045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itoh Y, Rice JD, Goller C, Pannuri A, Taylor J, Meisner J, Beveridge TJ, Preston 3rd JF, Romeo T. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-beta-1,6-N-acetyl-D-glucosamine. J Bacteriol. 2008;190(10):3670–3680. doi: 10.1128/JB.01920-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood PJ. Specificity in the interaction of direct dyes with polysaccharides. Carb Res. 1980;85(2):271–287. doi: 10.1016/s0008-6215(00)84676-5. [DOI] [Google Scholar]
- 33.Kelly-Quintos C, Cavacini LA, Posner MR, Goldmann D, Pier GB. Characterization of the opsonic and protective activity against Staphylococcus aureus of fully human monoclonal antibodies specific for the bacterial surface polysaccharide poly-N-acetylglucosamine. Infect Immun. 2006;74(5):2742–2750. doi: 10.1128/IAI.74.5.2742-2750.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pier GB, Boyer D, Preston M, Coleman FT, Llosa N, Mueschenborn-Koglin S, Theilacker C, Goldenberg H, Uchin J, Priebe GP. Human monoclonal antibodies to Pseudomonas aeruginosa alginate that protect against infection by both mucoid and nonmucoid strains. J Immunol. 2004;173(9):5671–5678. doi: 10.4049/jimmunol.173.9.5671. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J Bacteriol. 2003;185(16):4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol. 2013;11(3):157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Dissel D, Claessen D, van Wezel GP. Morphogenesis of Streptomyces in submerged cultures. Adv Appl Microbiol. 2014;89:1–45. doi: 10.1016/B978-0-12-800259-9.00001-9. [DOI] [PubMed] [Google Scholar]
- 38.Celler K, van Wezel GP, Willemse J. Single particle tracking of dynamically localizing TatA complexes in Streptomyces coelicolor. Biochem Biophys Res Commun. 2013;438(1):38–42. doi: 10.1016/j.bbrc.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 39.Boyd A, Chakrabarty A. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J Ind Microbiol. 1995;15(3):162–168. doi: 10.1007/BF01569821. [DOI] [PubMed] [Google Scholar]
- 40.Ruas-Madiedo P, Hugenholtz J, Zoon P. An overview of the functionality of exopolysaccharides produced by lactic acid bacteria. Int Dairy J. 2002;12(2):163–171. doi: 10.1016/S0958-6946(01)00160-1. [DOI] [Google Scholar]
- 41.Wang X, Preston 3rd JF, Romeo T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J Bacteriol. 2004;186(9):2724–2734. doi: 10.1128/JB.186.9.2724-2734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178:175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(14):4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litrán T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-β-1-6-N-acetylglucosamine, which is critical for biofilm formation. J bacteriol. 2009;191(19):5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roux D, Cywes-Bentley C, Zhang Y-F, Pons S, Konkol M, Kearns DB, Little DJ, Howell PL, Skurnik D, Pier GB. Identification of Poly-N-acetylglucosamine as a Major Polysaccharide Component of the Bacillus subtilis Biofilm Matrix. J Biol Chem. 2015;290(31):19261–19272. doi: 10.1074/jbc.M115.648709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gerke C, Kraft A, Süßmuth R, Schweitzer O, Götz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273(29):18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- 47.Celler K, Picioreanu C, van Loosdrecht MC, van Wezel GP. Structured morphological modeling as a framework for rational strain design of Streptomyces species. Antonie Van Leeuwenhoek. 2012;102(3):409–423. doi: 10.1007/s10482-012-9760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook J, Russell DW. Molecular cloning: a laboratory manual 3rd edition. Coldspring-Harbour Laboratory Press, UK. 2001 [Google Scholar]
- 49.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 50.MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene. 1992;111(1):61–68. doi: 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- 51.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical streptomyces genetics. John Innes Foundation, Norwich, UK. 2000 [Google Scholar]
- 52.van Dissel D, van Wezel GP. Morphology-driven downscaling of Streptomyces lividans to micro-cultivation. Antonie van Leeuwenhoek. 2017:1–13. doi: 10.1007/s10482-017-0967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 2002;417(6885):141–147. doi: 10.1038/417141a417141a. [DOI] [PubMed] [Google Scholar]
- 54.Cruz-Morales P, Vijgenboom E, Iruegas-Bocardo F, Girard G, Yanez-Guerra LA, Ramos-Aboites HE, Pernodet JL, Anne J, van Wezel GP, Barona-Gomez F. The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic island. Genome Biol Evol. 2013;5(6):1165–1175. doi: 10.1093/gbe/evt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI. CDD: NCBI's conserved domain database. Nucleic Acids. 2014;Res:gku1221. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinf. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colson S, van Wezel GP, Craig M, Noens EE, Nothaft H, Mommaas AM, Titgemeyer F, Joris B, Rigali S. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology. 2008;154:373–382. doi: 10.1099/mic.0.2007/011940-0. [DOI] [PubMed] [Google Scholar]
- 58.Keijser BJ, Noens EE, Kraal B, Koerten HK, van Wezel GP. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol Lett. 2003;225(1):59–67. doi: 10.1016/s0378-1097(03)00481-6. [DOI] [PubMed] [Google Scholar]
- 59.Piette A, Derouaux A, Gerkens P, Noens EE, Mazzucchelli G, Vion S, Koerten HK, Titgemeyer F, De Pauw E, Leprince P, van Wezel GP, Galleni M, Rigali S. From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J Proteome Res. 2005;4(5):1699–1708. doi: 10.1021/pr050155b. [DOI] [PubMed] [Google Scholar]
- 60.van Keulen G, Jonkers HM, Claessen D, Dijkhuizen L, Wösten HA. Differentiation and anaerobiosis in standing liquid cultures of Streptomyces coelicolor. J Bacteriol. 2003;185(4):1455–1458. doi: 10.1128/JB.185.4.1455-1458.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310(91-109) doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 62.Labeda DP. Transfer of the type strain of Streptomyces erythraeus (Waksman 1923) Waksman and Henrici 1948 to the genus Saccharopolyspora Lacey and Goodfellow 1975 as Saccharopolyspora erythraea sp. nov., and designation of a neotype strain for Streptomyces erythraeus. Int J Syst Evol Microbiol . 1987;37(1):19–22. doi: 10.1099/00207713-37-1-19. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


