Abstract
Neuroblastoma is the most common pediatric solid tumor of neural crest origin. The current treatment options for neuroblastoma produce severe side effects. Programmed death-ligand 1 (PD-L1), chronic inflammation, and non-coding RNAs are known to play a significant role in the pathogenesis of neuroblastoma. Cancer cells and the surrounding cells in the tumor microenvironment express PD-L1. Programmed death-1 (PD-1) is a co-receptor expressed predominantly by T cells. The binding of PD-1 to its ligands, PD-L1 or PD-L2, is vital for the physiologic regulation of the immune system. Chronic inflammation is involved in the recruitment of leukocytes, production of cytokines and chemokines that in turn, lead to survival, metastasis, and angiogenesis in neuroblastoma tumors. The miRNAs and long non-coding (lnc) RNAs have emerged as a novel class of non-coding RNAs that can regulate neuroblastoma associated cell-signaling pathways. The dysregulation of PD-1/PD-L1, inflammatory pathways, lncRNAs, and miRNAs have been reported in clinical and experimental samples of neuroblastoma. These signaling molecules are currently being evaluated for their potential as the biomarker and therapeutic targets in the management of neuroblastoma. A monoclonal antibody called dinutuximab (Unituxin) that attaches to a carbohydrate molecule GD2, on the surface of many neuroblastoma cells, is being used as an immunotherapy drug for neuroblastoma treatment. Atezolizumab (Tecentriq), an engineered monoclonal antibody against PD-L1, are currently in clinical trial for neuroblastoma patients. The lncRNA/miRNA-based therapeutics is being developed to deliver tumor suppressor lncRNAs/miRNAs or silencing of oncogenic lncRNAs/miRNAs. The focus of this review is to discuss the current knowledge on the immune checkpoint molecules, PD-1/PD-L1 signaling, inflammation, and non-coding RNAs in neuroblastoma.
Keywords: Neuroblastoma, PD-L1, Inflammation, non-coding RNAs, Immuno-Therapy
1. Introduction
Neuroblastoma is the most common childhood cancers that originate from neuroblast cells. During development of a fetus, neuroblasts are transformed into nerve cells. However, mutations in the small portion of immature neuroblasts can lead to neuroblastoma [1]. According to one estimate, the incidence of neuroblastoma in the United States is 1 per 100,000 children and approximately 700 children (younger than 15 years) suffer from this disease each year [2]. The majority of neuroblastoma patients are diagnosed at an advanced stage. Although surgery, radiation, and chemotherapy are common treatment options, neuroblastoma cells often develop resistance mechanisms and the disease relapses [3]. The patients with relapsed neuroblastoma are highly incurable [4]. Neuroblastoma has a poor prognosis especially when diagnosed in an advanced stage. Thus, improved detection and therapeutic methods for the diagnosis, prognosis, and therapy are required.
The immune checkpoint inhibition molecules such as programmed death protein-1 (PD-1) and its ligands (PD-L1, PD-L2), inflammatory molecules, and non-coding RNAs are known to play a significant role in the neuroblastoma pathogenesis. PD-1 is the major immune checkpoint receptor expressed on activated monocytes, B cells, T cells, dendritic cells (DCs), and natural killer T cells in humans and mice [5–8]. It plays a significant role in cell adhesion, proliferation, and cytokine signaling. It can also promote self-tolerance by suppressing T cell function [9, 10]. The inflammatory molecules (cytokines, chemokines) and transcription factors (NF-κB, STAT3) are dysregulated in many tumor types including neuroblastoma [11–13]. These inflammatory molecules play a role in modulating immunosurveillance, promoting angiogenesis and recruiting leukocytes to neuroblastoma cells [14, 15]. Non-tumor cells and factors such as tumor-associated macrophages (TAMs) and macrophage migration inhibitory factor (MIF) are also the major mediators of inflammation.
It is now clear that ~98% of the human genome accounts for non-coding sequences [16]. Furthermore, ~90% of these non-coding sequences are transcribed to produce a large number of non-coding RNAs [17–20]. There are two major classes of non-coding RNAs: microRNAs (miRNAs, 18–22 nucleotides) and long non-coding RNAs (lncRNAs, ≥200 nucleotides). Although miRNAs are well characterized, lncRNAs are relatively new. During recent years, lncRNAs have been implicated in regulating cellular functions and disease processes including neuroblastoma [21–23]. Because of specificity and ease in detection, lncRNAs can be used as a biomarker and therapeutic target [24–27]. miRNAs can also regulate different physiological and pathological processes including inflammation and cancer [28–30]. miRNAs are known to negatively regulate protein coding genes and the expression of other non-coding transcripts. miRNAs are involved in the post-transcriptional modulation of multiple genes by base-pairing to target mRNAs [31]. The binding of miRNAs to the 3’untranslated region (UTR) of mRNAs usually leads to the degradation or translational repression [32]. Accumulating evidence suggests that miRNAs play a significant role in the pathogenesis of neuroblastoma [2], and thus could be used for the diagnosis and prognosis of disease [33–37].
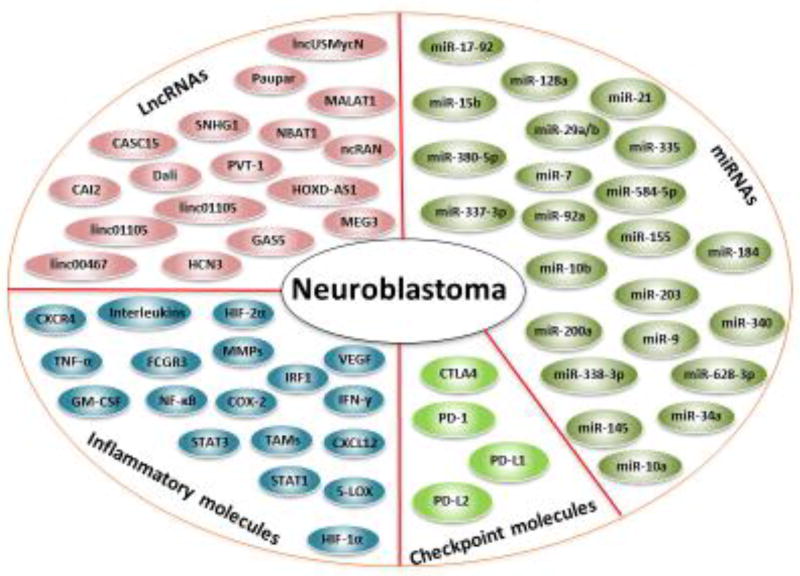
In the following sections, we discuss the role of immune checkpoint molecules, inflammatory molecules, PD-1/PD-L1 signaling, miRNAs, and lncRNAs in neuroblastoma pathogenesis. We also discuss the clinical implications of these molecules in neuroblastoma. We provide evidence that these molecules can be used as a biomarker and therapeutic target for neuroblastoma (Figure 1).
Figure 1. Potential Biomarkers and therapeutic targets of Neuroblastoma.
Abbreviations: 5-LOX: 5-lipoxygenase, CAI2: CDKN2A/ARF intron 2, CASC15: cancer susceptibility candidate15, COX-2: cyclooxygenase-2, CTLA4: cytotoxic T-lymphocyte associated protein 4, CXCL12: C-X-C motif chemokine 12, CXCR4: C-X-C chemokine receptor 4, FCGR3: Fc fragment of IgG receptor III, GAS5: growth arrest special 5, HCN3: hyperpolarization-activated cation nucleotide-gated isoform 3, HIF-1α: hypoxia-inducible factor 1-alpha, HOXD-AS1: HOXD cluster antisense RNA 1, IRF1: interferon regulatory factor 1, linc00467: long Intergenic non-protein coding RNA 467, linc01105: long intergenic non-protein coding RNA 1105, lncUSMycN: lncRNA upstream of MYCN, MALAT1: metastasis associated lung adenocarcinoma transcript 1, MEG3: maternally expressed 3, miR: microRNA, NBAT1: neuroblastoma associated transcript 1, ncRAN: non-coding RNA expressed in aggressive neuroblastoma, NF-κB: nuclear factor kappa-light-chain-enhancer of activated B cells, PD-1: programmed death-1, PD-L1: programmed death-ligand 1, PD-L2: programmed death-ligand 2, PVT-1: plasmacytoma variant translocation 1, SNHG1: small nucleolar RNA host gene 1, STAT-3: signal transducer and activator of transcription 3, TAMs: tumor-associated macrophages, TNF-α: tumor necrosis factor-alpha, VEGF: vascular endothelial growth factor.
2. Immune Checkpoint molecules
T-lymphocytes are primary effector cells of the adaptive immune response against cancer, which includes helper T cells and cytotoxic T cells. The cytotoxic T cells directly attack the tumor cells and helper T cells to propagate the anti-tumor immune response in the immune system. These cells play an important role in the recognition of tumor antigens from the major histocompatibility complex (MHC) receptors. The interaction of T cell receptor and tumor antigen MHC complex on antigen-presenting cells is called as T-cell priming/activation [38]. The priming of T-cells is firmly coordinated by immune checkpoint molecules. The most common immune checkpoint molecules are cytotoxic lymphocyte–associated protein 4 (CTLA-4) and PD-L1. CTLA-4 and PD-L1 are over expressed on most cancer types and are known to inhibit the T cell function [39].
Lymphocyte-activation gene 3 (LAG 3) are MHC class 2 ligand immunecheck point molecules, expressed in dendritic cells (DCs) and tumor-infiltrating macrophages. It augments regulatory T cells (T reg) and impede the CD 8 effector T cell to allow the cancer cell immune escape [40]. T cell membrane protein 3 (Tim-3) is an immune checkpoint receptor molecules found in natural killer T cells, macrophages and DCs [41]. It is highly expressed in many cancers including liver, lung and melanoma tumors. BTLA is a B- and T-lymphocyte attenuator protein immune checkpoint molecules, expressed in activated T-helper cells. It is associated with a ligand in herpes virus entry mediator to mediate the signal recipient cancer cells. It is expressed on the surface of melanoma cancer cells and alleviates IL-2 to impair the activation of T cells [42]. Considering the fact that immune checkpoint molecules play major role in cancer therapy, a number of inhibitors are being evaluated by clinical trials [43].
2.1. PD-1 and PD-L1 Signaling
2.1.1. Programmed Death-1 Protein (PD-1)
PD-1 also known as cluster of differentiation 279 (CD279), is a cell surface receptor that belongs to immunoglobulin gene super family [44]. PD-1 was isolated in 1992 using subtractive hybridization. It is expressed on activated monocytes, B cells, dendritic cells, NK cells, natural killer T (NKT) cells, T cells, regulatory T cells (Treg), and exhausted T cells [5, 45, 46]. PD-1 plays a crucial role in lowering the immune system through suppression of T-cell function and up-regulation of Tregs, which in turn, reduces autoimmunity and promotes self-tolerance [9, 10]. Under normal physiological conditions, PD-1 is known to interact with two ligands, PD-L1 and PD-L2; these ligands share 37% sequence homology [47–49]. The expression of PD-L1 and PDL2 is increased when cancer cells are attacked by the immune system, leading to the suppression of T cells and immune escape [50].
2.1.2. Programmed Death Ligand-1 (PD-L1)
PD-L1 is a 40-kDa type 1 transmembrane protein that suppresses the immune system during physiological and pathological events (Dong et al.,). PD-L1 is constitutively expressed on antigen presenting cells, non-lymphoid organs and non-hematopoietic cells such as heart, lung, placenta, and liver. PD-L1 is also expressed by a variety of cancer cells and by tumor-infiltrating immune cells including dendritic cells and macrophages [51]. It can be induced by various pro-inflammatory cytokines like IFN-γ, TNF-α, VEGF, granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-10. PD-L1 can also be induced by toll like receptors (TLRs), interferon regulatory factor 1 (IRF1), STAT1, STAT3, and hypoxia-inducible factor-1alpha (HIF-1α) [52]. Up-regulation of PD-L1 in tumor cells facilitates immune suppression in the tumor microenvironment [53]. Upon epidermal growth factor receptor (EGFR) stimulation, mature PD-L1 undergoes glycosylation, which leads to its stabilization [54]. In the absence of glycosylation, PD-L1 undergoes GSK-3β mediated phosphorylation which triggers its K48-ubiquitination and subsequent degradation [54]. PD-L1 can also be stabilized by COP9 signalosome (CSN5) mediated deubiquitination [55]. Furthermore, nuclear factor (NF)-κB p65 can directly regulate CSN5 expression by binding to its promoter leading to PD-L1 stabilization and immune suppression [55]. The IFN-γ-induced PD-L1 expression is dependent on the NF-κB signaling [56]. In human glioma cells, dysfunction in PTEN and activation of PI3K is associated with increased expression of PD-L1 [57]. PI3K can also increase the translation of PD-L1 mRNA [47]. These studies clearly demonstrated that PD-1 and PD-L1 contribute to cancer pathogenesis and thus could be used as biomarkers and therapeutic targets.
2.1.3. Diagnostic and Prognostic Significance of PD-1/PD-L1
Several lines of evidence have established the diagnostic and prognostic potential of PD-1/PD-L1 in neuroblastoma. For example, one study was aimed to characterize the PD-L1 expression and tumor-associated immune cells (TAICs) (lymphocytes and macrophages) in common pediatric cancers [58]. The whole slide sections and tissue microarrays were evaluated by immunohistochemistry for PD-L1 expression and the presence of TAICs. TAICs were also screened for PD-L1 expression. Thirty-nine of 451 evaluable tumors (9%) expressed PD-L1 in at least 1% of tumor cells. The common cancer types observed in these pediatric cancer patients were Burkitt lymphoma, glioblastoma multi-forms, and neuroblastoma. PD-L1 staining was associated with inferior survival in neuroblastoma patients. Furthermore, 74% of tumors contained lymphocytes and/or macrophages. The macrophages were more likely to be identified in PD-L1-positive versus PD-L1-negative tumors. The authors of this study concluded that a subset of diagnostic pediatric cancers exhibit PD-L1 expression, whereas a much larger fraction show infiltration by tumor-associated lymphocytes. Furthermore, PD-L1 expression could be used as a biomarker for poor outcome in neuroblastoma patients. However, more studies are required to define the predictive nature of PD-L1 expression in neuroblastoma both at diagnosis and after chemotherapy.
N-Myc (V-Myc Avian Myelocytomatosis Viral Oncogene Neuroblastoma-Derived Homolog) is a proto-oncogene encoded by MYCN that belongs to the Myc family of DNA binding basic helix-loop-helix leucine zipper proteins. Previous studies have demonstrated the association of MYCN amplification and N-Myc over-expression with several cancer types, most notably neuroblastoma [59–62]. MYCN is used as a biomarker to stratify and to assess neuroblastoma patients [63, 64]. MYC and MYCN are known to regulate PD-L1 in neuroblastoma [64, 65]. The functional inhibition of MYC/MYCN is known to suppress PD-L1 expression [64]. Furthermore, MYC can initiate and maintain tumorigenesis through the modulation of immune regulatory molecules [65]. Toll-like receptor 3 (TLR3) can also enhance PD-L1 and major histocompatibility complex (MHC) class I expression in neuroblastoma cells [66]. Expression of TLR3 can also serve to predict favorable prognosis in neuroblastoma [67]. Treatment of neuroblastoma cell lines with TLR3 or interferon-γ can significantly up-regulate PD-L1 and MHC class I [68].
A study was sought to evaluate the expression of PD-L1 and HLA class I on neuroblastoma cells, and PD-1 and lymphocyte activation gene 3 (LAG 3) on tumor-infiltrating lymphocytes to examine if neuroblastoma patients may benefit from therapies targeting immune checkpoint molecules [64]. The expression of PD-L1, HLA class I, PD-1 and LAG3 was assessed in 77 neuroblastoma specimens by in situ immunohistochemistry (IHC). These patients were characterized by tumor-infiltrating T-cell density that correlated with clinical outcome. A data set of 477 human primary neuroblastomas from Gene Expression Omnibus (GEO) and array expression databases was explored for PD-L1, MYC, and MYCN correlation. The combination of PD-L1 and HLA class I tumor cell density was found to be a prognostic biomarker for predicting overall survival in neuroblastoma patients. The abundance of PD-L1 transcript correlated with MYC expression in primary neuroblastoma. It was concluded that the combination of PD-L1 and HLA class I could be a novel prognostic biomarker for neuroblastoma.
Dondero and colleagues analyzed the effect of constitutive and inducible expression of PD-Ls in human neuroblastoma cell lines, ex vivo isolated neuroblasts, and lymphocytes [69]. A combination of PD-L1 and human leucocyte antigen (HLA) class I tumor cell density was identified as a prognostic biomarker for predicting overall survival in neuroblastoma patients. In another study, the infiltrating T cells were found to possess prognostic value greater than the currently used methods [70].
Relapsed/refractory neuroblastoma (rNB) after traditional chemotherapy is highly incurable. A recent clinical trial from USA (https://clinicaltrials.gov/ct2/show/study/NCT02868268) is recruiting rNB patients. This clinical trial is utilizing the multi-institutional infrastructure and Translational Genomics Research Institute GEM sequencing platform. The aim of the trial is to identify subgroups of rNB patients with potentially targetable genetic (ALK, MAPK, metabolic-related genes) and/or immunologic (tumor-associated macrophage infiltration and/or PD-L1 expression) biomarkers in rNB. It is expected that understanding the genetic and immunologic landscape of rNB will help in devising novel therapies for these patients.
2.1.4. Therapeutic Potential of PD-1/PD-L1
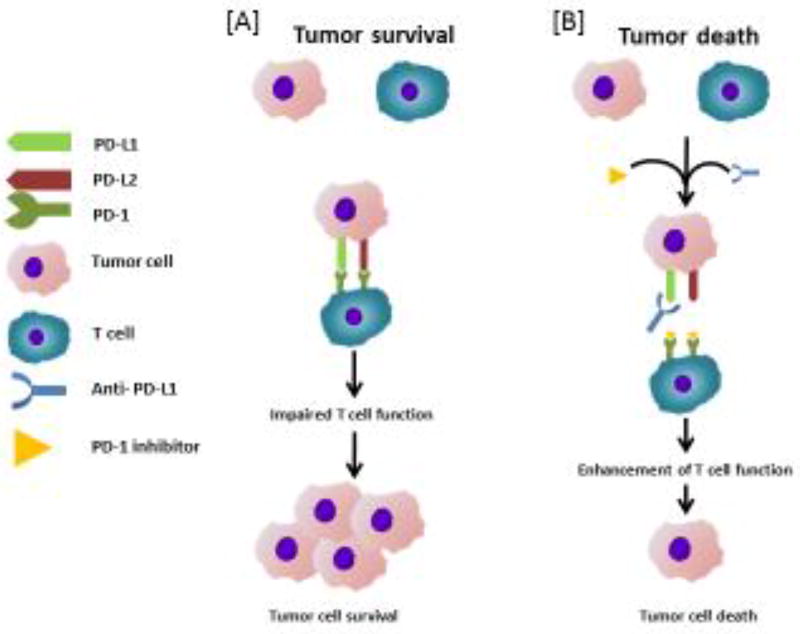
PD-1 inhibitors block the interaction of PD-L1 and PD-L2 with PD-1 on T cells and increase T cell proliferation and function [71]. The molecular basis for PD-1 and PD-L1 based therapy is presented in Figure 2. In fact, some inhibitors have been developed against PD-1/PD-Ls. For example, opdivo (nivolumab) and keytruda (pembrolizumab) are PD-1 inhibitors approved by FDA for advanced melanoma and non-small cell lung cancer (NSCLC).
Figure 2. The molecular basis for the action of PD-1and PD-L1 based therapy.
[A] PD-1 is expressed by T-cells, while PD-L1/PD-L2 is expressed by tumors. In neuroblastoma, interaction of PD-1 with PD-L1/PD-L2 suppresses T-cells function. [B] The PDL-1 antibodies act by blocking the interaction of PD-L1 with PD-1without affecting PD-L2/PD-1 interaction. This enhances the T cell function leading to anti-tumor activity.
PD-L1 antibodies act by blocking the interaction of PD-L1 with PD-1 without affecting PD-L2/PD-1 interaction. Because the PD-1/PD-L2 pathway plays a role in peripheral tolerance, the specificity of PD-L1 antibodies may help to decrease toxicity. Some monoclonal antibodies developed against PD-L1 are BMS-986559 (MDX-1105), MPDL3280A, and MEDI4736. These antibodies are currently under evaluation in clinical trials for advanced malignancy (http://www.clinicaltrial.org/). However, the efficacy of these antibodies against neuroblastoma is yet to be determined. To our knowledge, dinutuximab (unituxin), a monoclonal antibody that targets the ganglioside GD2, is the most effective immunotherapy for neuroblastoma. Dinutuximab is reported to improve the 2-year event-free survival of high-risk neuroblastoma patients from 46% to 66% [72]. Atezolizumab (Tecentriq) is an engineered monoclonal antibody designed against PD-L1. The safety, tolerability, pharmacokinetics, immunogenicity, and preliminary efficacy of this antibody is being evaluated by an early phase clinical trial (https://clinicaltrials.gov/ct2/show/study/NCT02541604). A recent report on a 6-year-old baby revealed that combination chemo-immunotherapy could be successfully used for high-risk NB relapsed after haploidentical stem cell transplantation (haplo-HSCT) [73].
In HLA class I positive neuroblastoma cell lines, PD-L1 is constitutively expressed, whereas PD-L2 is very rarely detected [69]. Furthermore, an induction in PD-L1 by IFN-γ was observed in neuroblastoma cell lines and in neuroblastoma engrafted nude mice. Importantly, PD-L1 was identified in metastatic neuroblasts isolated from bone marrow aspirates of high-risk neuroblastoma patients. PD-1 positive cells were mainly represented by αβ T cells, as well as small populations of γδ T cells and NK cells. The authors of this study concluded that PD-L1-mediated immune resistance mechanisms occur in metastatic neuroblasts and thus provide a biological rationale for blocking the PD-1/PD-Ls axis for immunotherapy.
PD-1 inhibitors have also been used in combination for neuroblastoma immunotherapy [69, 74–76]. For example, in combination with TLR ligands, the PD-L1 blockade was proposed as a promising immunotherapy for neuroblastoma [76]. Furthermore, pembrolizumab is more effective than docetaxel in PD-L1-expressing advanced NSCLC patients [77]. However, only some subsets of patients benefit from PD-1/PD-L1 immune checkpoint blockade therapies. Therefore, it is essential to identify those populations that can benefit from PD-1/PD-L1 blockade therapy. Currently, PD-L1 expression in tumor specimens by IHC is the most commonly used biomarker for selecting patients with the possibility to respond to treatments [78]. However, conflicting results have been reported on PD-L1 expression. For example, over-expression of PD-L1 in melanoma is associated with poor clinical outcome [79]. Conversely, the PD-L1 over-expression in the context of CD8-positive cells is associated with a better prognosis [80]. Although these studies suggest the efficacy of PD-1 inhibitors against melanoma and NSCLC, the potential of these inhibitors against neuroblastoma has not been determined.
4. Inflammation and Neuroblastoma
While acute inflammation is beneficial, chronic inflammation leads to various chronic diseases including cancer. Various inflammatory molecules have been identified as a molecular mediator of cancer. These include pro-inflammatory transcription factors (NF-κB, STAT3), tumor necrosis factor (TNF), interleukin (IL)-1, IL-6, chemokines, cyclooxygenase (COX)-2, 5-lipooxygenase (5-LOX), matrix metalloproteinases (MMPs), vascular endothelial growth factor (VEGF), adhesion molecules, and numerous other molecules. Apart from inflammatory molecules, non-tumor cells such as tumor-associated macrophages (TAMs) are also known mediator of inflammation. The role of TAMs, macrophage migration inhibitory factor (MIF), chemokines and transcription factors in neuroblastoma pathogenesis are discussed in following sub-sections.
4.1. Tumor Associated Macrophages (TAMs)
Tumor-associated macrophages (TAMs) are cell types that are found near or within tumor masses. M1 and M2 are two major macrophage classes that are activated in response to dynamic stimuli. These macrophages are essential modulators of the immune response. The activation of M1 macrophages is observed in response to bacterial infection or interferon-γ (IFN-γ). This leads to enhanced antigen presentation and increased production of IL-12, IL-23, and reactive oxygen species (ROS) and anti-tumor effects. M2 macrophages are induced by interleukins (IL-4, -10, -13), and glucocorticoids, and promote growth and metastasis of neuroblastoma [81]. M2 macrophages can contribute to tissue remodeling, repair, and may protect tumor cells from apoptosis [82].
TAMs are fundamental to the progression of many tumor types including neuroblastoma [83–85]. Growing evidence suggests that TAMs can facilitate progression of neuroblastoma. Metastatic neuroblastoma patients are characterized by higher infiltration of TAMs. Neuroblastoma patients with an age of ≥ 18 months have higher expression of inflammationrelated CD16, CD33, IL6R, IL10, and FCGR3 than in patients diagnosed at the age of < 18 months [86]. MYCN non-amplified stage 4 patient’s clinical reports have demonstrated that increased expression of TAM- associated genes such as CD14 and CD16 are significantly correlated with 5-year event-free survival [87]. Lymphocyte markers such as CD8A, CD4, and FOXP3 were not correlated with MYCN non-amplified stage 4 patient’s clinical reports. Neuroblastoma cells cultured with primary monocytes are associated with rapid tumor growth in a xenograft mouse model. While administration of IL-6 neutralizing antibodies reduced tumor growth, over-expression of IL-6 was associated with increased tumor growth rates. The presence of TAMs was confirmed by immunohistochemistry. Furthermore, a significant increase in IL-6 was observed in tumor-infiltrated bone marrow [87]. IL-6 derived from bone marrow stromal cells (BMSC) is enhances progression of metastatic neuroblastoma [88]. When IL-6R-positive neuroblastoma cells are cultured in the presence of BMSC or recombinant human IL-6, an increase in proliferation is observed. Furthermore, IL-6R-positive neuroblastomas are protected from etoposide-induced apoptosis in the presence of BMSC or recombinant human IL-6. However, these responses were not observed in IL-6R-negative neuroblastoma cells [88]. A major limitation of the current neuroblastoma therapy is that over time neuroblastoma cells develop resistance to therapeutic agents [89]. Previous studies have demonstrated that TAMs can promote neuroblastoma growth, metastasis and the development of drug resistance [86]. Thus TAMs represent a negative prognostic factor for neuroblastoma [90–92]. However, the molecular mechanism by which TAMs contribute to tumorigenic effects is not properly understood.
4.2. Tumor-infiltrating lymphocytes (TILs)
Tumor-infiltrating lymphocytes (TILs) are group of mononuclear T cells infiltrated from tumor tissue found in most solid tumor including breast, colon, cervical, melanoma and neuroblastoma [93]. TILs are found in stroma within tumor area and contain T cells, B cells, NK cells and macrophages. TILs are important for diagnosis and prognosis in patients with many solid tumor including neuroblastoma [70, 94, 95]. A sub population of CD4+ cells in TILs shows detrimental effect on host milieu [96]. The histopathological specimen containing TIL could provide a decisive prognostic information in a wide variety of solid tumors especially neuroblastoma. TILs are assessed by standardized methodology on hematoxylin and eosin dye stained histological section based on the international immuno-oncology biomarker working group guidelines [97]. This standardized methodology may be used to evaluate clinical validity and utility of immunotherapy in neuroblastoma patients. Genetically modified TILs are preferred over conventional therapies in treating malignancies such as neuroblastoma [98].
4.3. Tumor Associated Mesenchymal Stem Cells (TMSCs)
Mesenchymal stem cells (MSCs) are multipotent stem-like cells that can differentiate into a variety of cell types [99, 100]. MSCs are known to produce a large range of cytokines, growth factors, proteins and can regulate survival, angiogenesis, immunomodulation, and drug resistance [101]. The primary and metastatic tumors are known to attract MSCs from bone marrow and other sites in their microenvironment. Conversely, in the bone marrow, MSCs attract tumor cells and contribute to a microenvironment that promotes tumor growth.
Cancer-associated fibroblasts (CAF) originating from MSC through the mediation of STAT3 and ERK1/2 pathways [102] are known to provide a favorable environment for neuroblastoma progression [103]. MSCs have also been used as a delivery vehicle for neuroblastoma therapy [104, 105]. For example, MSCs were used as carriers of oncolytic adenovirus and improved the efficacy of virotherapy for neuroblastoma by delivering the adenovirus to tumors and recruiting T cells [105].
4.4. Macrophage Migration Inhibitory Factor (MIF)
MIF is an important pro-inflammatory mediator that links inflammation with cancer [106]. The expression of MIF in neuroblastoma is well documented [107]. MIF plays an essential role in both innate and acquired immunity. MIF is known to abrogate the functions of p53 leading to the promotion of tumor cell proliferation and angiogenesis, inhibition of apoptosis, and induction of cyclooxygenase-2 (COX-2) through enhanced endothelial cell proliferation and differentiation [106, 108].
4.5. Chemokines and Transcription Factors
Chemokines are a family of secreted proteins that play an essential role in coordinating the inflammatory and immune response by specifically controlling leukocyte trafficking. Accumulating evidence suggests that chemokines can help with the migration, proliferation, and survival of tumor cells. Neuroblastoma cells are known to express C-X-C chemokine receptor type 4 (CXCR4) and the ligand, CXCL12 [109]. Furthermore, increased expression of CXCR4 correlates with advanced clinical stage and the presence of bone marrow metastases with poorer outcome [109]. The hypoxia-inducible transcription factors (HIF-1α, HIF-2α) are correlated with metastasis and development of drug resistance [110]. These transcription factors are differentially expressed in neuroblastoma cells [111]. While HIF-1α is transiently stabilized, prolonged expression of HIF- 2α is observed in neuroblastoma cells under hypoxic conditions [111].
NF-κB is a pro-inflammatory transcription factor that is constitutively expressed in most tumor types including neuroblastoma [112, 113]. It is comprised of five subunits including NF-κB1 (p50), NF-κB2 (p52), RelA (p65), RelB, and c-Rel. Under normal conditions, NF-κB resides in the cytoplasm in association with its inhibitory subunit IκB. Upon stimulation, IκB subunit undergoes phosphorylation, ubiquitination, and degradation. This releases the p65-p50 subunit and allows it into the nucleus where it regulates the expression of NF-κB dependent target genes [114]. The phosphorylation of IκB can also lead to its dissociation from the trimeric complex without degradation. NF-κB can regulate the expression of over 500 cancer-related genes that are involved in various aspects of tumor development including transformation, survival, proliferation, invasion, angiogenesis, and metastasis [112, 115]. Therefore, NF-κB signaling pathway represents a potential target for therapeutic intervention. Agents that can inhibit protein tyrosine kinases, serine/threonine kinases, ubiquitination, proteasomes, acetylation, and DNA binding steps, have the ability to inhibit NF-κB activation. Although over 500 NF-κB inhibitors have been identified by preclinical studies, only bortezomib is used to treat multiple myeloma patients. Furthermore, the potential of bortezomib in neuroblastoma patients remains to be determined.
5. MiRNAs and Neuroblastoma
The role of miRNAs in regulating neuroblastoma pathogenesis is well established (Table 1). For example, miR-148a, miR-21, and miR-200a can modulate cell growth, migration, invasion, and apoptosis in neuroblastoma [116]. Similarly, miR-10b, miR-29a/b, miR-335, miR-7, and miR-338-3p are potentially associated with neuroblastoma progression [117]. miR-203 is known to inhibit the proliferation, migration, and invasion of neuroblastoma cells by targeting Sam 68 [118]. miR-337-3p and miR-584-5p can target matrix metalloproteinase (MMP) in neuroblastoma cells [37, 119]. miR-558 induces the transactivation of heparanase and promotes tumorigenesis in neuroblastoma [120]. The miR-17-92 polycistronic cluster is highly expressed in neuroblastoma tissues and serves as a marker for poor outcomes in patients [121]. miR-21, a well-known oncogenic miRNA, can promote proliferation and reduce chemo sensitivity in neuroblastoma cells [122]. miR-15a can also suppress the expression of reversion-inducing cysteine-rich protein with Kazal motifs (RECK) a cysteine-rich protein with Kazal motifs and promoted the migration of neuroblastoma cells [123].
Table 1.
A list of miRNAs dys-regulated in neuroblastoma and their functions
| Column1 | Column2 | Column3 | Column4 |
|---|---|---|---|
| miRNA | Expression | Function/Clinical implication |
Reference |
| miR-9 | ↓ | Inhibited invasion, metastasis, and angiogenesis of NB cells by targeting MMP-14 | [90] |
| miR-10a | Induced neural differentiation by suppressing NCOR2 and MYCN in neuroblastoma cells | [130] | |
| miR-10b | Induced neural differentiation by suppressing NCOR2 and MYCN in neuroblastoma cells | [130] | |
| miR-15a | Induced NB migration by targeting RECK and regulating MMP-9 | [123] | |
| miR-17-92 | Inhibited TGF-β signaling; induced proliferation and adhesion of NB cells | [176] | |
| miR-34a | ↓ | Inhibited NB growth when administered to the mice in conjugation with GD2 antibody | [134] |
| miR-145 | ↓ | Inhibited the growth, invasion, metastasis and angiogenesis of NB cells by targeting HIF-2α | [135] |
| miR-155 | Contributed to the development of chemoresistance by neuroblastoma cells | [116] | |
| miR-184 | ↓ | Inhibited tumor growth in an orthotopic NB murine model | [136] |
| miR-203 | ↓ | Inhibited proliferation, migration and invasion of NB cells by targeting Sam 68 | [118] |
| miR-335 | ↓ | Suppressed invasion of NB cells by targeting TGF-β signalling pathway | [177] |
| miR-337- 3p | ↓ | Suppressed nuroblastoma progression by repressing MMP-14 | [37] |
| miR-340 | ↓ | Induce differentiation and apoptosis in a context dependent manner in neuroblastoma cells | [131] |
| miR-380-5p | Repressed p53 and inhibited apoptosis in NB cells; associated with poor outcome in NBs | [128] | |
| miR-584-5b | ↓ | Exerted tumor suppressive functions in NB cells by suppressing MMP-14 | [119] |
Abbreviations: : up-regulation, ↓: down-regulation, HIF-2α: hypoxia-inducible factor 2 alpha, MMP: matrix metalloproteinase, MYCN: v-myc myelocytomatosis viral related oncogene, NB: neuroblastoma, NCOR2: nuclear receptor co-repressor 2, RECK: reversion-inducing cysteine-rich protein with kazal motifs, Sam 68: src-associated in mitosis 68, TGF-β: transforming growth factor beta
A study reported that 32 of 157 miRNAs in primary neuroblastomas are differentially expressed in favorable and unfavorable tumor subtypes [124]. miR-92a, miR-15b, miR-128a, and miR-628-3p were significantly increased in neuroblastoma cells. miRNA expression profile in low-risk and high-risk neuroblastoma patients has also been reported [125]. miR-92a, miR-15b, miR-128a and miR-628-3p are also known to be significantly altered and epigenetically silenced in metastatic neuroblastoma [126, 127]. miR-380-5p is over-expressed in neuroblastoma and is known to repress p53 and inhibit apoptosis in NB cells [128]. Furthermore, an over-expression in miR-380-5p is associated with poor outcomes in NBs [128]. Using the neuroblastoma cell line SH-SY5Y, miR-125b was found to play an important role in human neuronal differentiation [129]. Similarly, miR-10a and miR-10b are known to induce neuroblastoma cell differentiation by targeting the nuclear receptor corepressor 2 [130]. miR-340, a tumor suppressive miRNA is epigenetically silenced. Ectopic expression of this miRNA is known to induce differentiation and apoptosis in a context-dependent manner in neuroblastoma cells [131]. The inhibition of miR-18a in neuroblastoma cells led to the outgrowth of varicosity-containing neurites and the induction of sympathetic neuron differentiation markers [132]. MYCN knockdown could also induce neuroblastoma differentiation. During MYCN knockdown-mediated neuronal differentiation, 23 miRNAs were differentially expressed in neuroblastoma cells [133]. miR-21 was strongly up-regulated upon MYCN knockdown. However, miR-21 over-expression did not prevent the differentiation of neuroblastoma cells. Recently, we demonstrated that the exosomal miR-21 released from neuroblastoma cells could be transferred to human monocytes [116]. Similarly, miR-155 from human monocytes can be transferred to neuroblastoma cells. We previously shown that unique role of exosomal miR-21 and miR-155 in the cross-talk between neuroblastoma cells and human monocytes and in the development of resistance to chemotherapy[116]. Mechanistically, a novel exosomic miR-21/TLR8-NF-κB/exosomic miR-155/TERF1 signaling pathway was involved in the development of chemo resistance.
During the past decade, attempts have been made to develop miRNA-based therapeutics for neuroblastoma. One strategy is to block oncogenic miRNAs using oligonucleotides. In certain instances, nanoparticles have been used for the delivery of tumor suppressive miRNAs. For example, nanoparticles encapsulating miR-34a and conjugated to a GD2 antibody was found to facilitate tumor-specific delivery into mice [134]. Furthermore, a significant reduction in tumor growth, increased apoptosis and a reduction in vascularization were observed. miR-9 overexpression was found to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells in vivo [90]. Similarly, miR-145 can significantly inhibit the growth, invasion, metastasis, and angiogenesis of neuroblastoma cells by directly targeting HIF-2a [135]. miR-184 can significantly reduce tumor growth in an orthotopic murine model of neuroblastoma [136]. Similarly, an in vivo delivery of miR-380-5p antagonist can decrease tumor size in an orthotopic mouse model of neuroblastoma [128]. miRNAs can also be used in combination therapy for the clinical management of neuroblastoma. For example, miR-7-1 can enhance the inhibitory effects of 4-HPR [N-(4-hydroxyphenyl) retinamide] and EGCG (epigallocatechin-3-gallate) on neuroblastoma cells growth [137].
From above discussion, it is clear that miRNAs can be used as a target for neuroblastoma therapy. However, the potential of miRNAs in neuroblastoma patients remains to be explored. Extensive preclinical studies focused on safety and toxicity is necessary before a miRNA-based therapy can be considered for neuroblastoma patients.
6. LncRNAs and Neuroblastoma
Because of high specificity, dys-regulated expression pattern, and ease in the detection methods, lncRNAs can be used as a biomarker and therapeutic target for neuroblastoma. Some lncRNAs have been studied in the context of neuroblastoma (Table 2). N-Myc is a protooncogene encoded by the MYCN, which is a highly conserved and major oncogene in humans. N-Myc plays a crucial role during normal brain development [138]. However, MYCN amplification and N-Myc over-expression is associated with several cancer types, especially neuroblastoma [59–61]. The neuroblastoma patients with N-Myc amplification are prone to metastasis. N-Myc is also known to regulate the expression of lncRNAs such as T-UCRs and ncRNAs [139].
Table 2.
A list of lncRNAs dys-regulated in neuroblastoma and their functions
| Column1 | Column2 | Column3 | Column4 |
|---|---|---|---|
| lncRNA | Expression | Function/Clinical implication |
Reference |
| linc00467 | - | Reduced neuroblastoma growth through modulation of DKK1 | [140] |
| SNHG1 | Associated with event-free survival of patients | [141] | |
| GAS5 | Regulated expression of p53, BRCA1, and GADD45A in neuroblastoma cells | [142] | |
| MALAT1 | Induced migration and invasion of neuroblastoma cells | [143] | |
| MALAT1 | Modulated cell migration, invasion and vasculature formation by down-regulating FGF2 in | [144] | |
| neuroblastoma cells under hypoxic conditions | |||
| MALAT1 | Up-regulated Axl and induced invasion and migration of neuroblastoma cells | [145] | |
| MALAT1 | Induced ERK/MAPK activation and neuronal differentiation in neuroblastoma cell lines | [146] | |
| HCN3 | Regulated apoptosis and proliferation in neuroblastoma cells | [147] | |
| linc01105 | Regulated apoptosis and proliferation in neuroblastoma cells | [147] | |
| MEG3 | ↓ | Regulated apoptosis and proliferation in neuroblastoma cells | [147] |
| NBAT1 | ↓ | Regulated proliferation and invasion of neuroblastoma cells by interacting with EZH2; | [148] |
| associated with poor clinical outcome in patients | |||
| NBAT1 | ↓ | Contributed to the aggressiveness of neuroblastoma by promoting proliferation and | [21] |
| an impairment of differentiation of neuronal precursors | |||
| CASC15 | ↓ | Regulated extracellular matrix transcripts, adhesion, growth, and migration of neuroblastoma cells | [150] |
| ncRAN | Associated with poor prognosis of neuroblastoma patients | [151] | |
| Dali | - | Regulated the differentiation of neuroblastoma cells | [152] |
| PVT-1 | Regulated expression of Myc proteins | [153] | |
| CAI2 | Associated with high-risk and clinical outcome of neuroblastoma patients | [161] | |
| lncUSMycN | - | Regulated N-Myc expression and neuroblastoma oncogenesis in mice model | [162] |
| lncUSMycN | Induced NCYM expression; correlated with poor prognosis of neuroblastoma patients | [163] | |
| HOXD-AS1 | - | Regulated RA-induced differentiation and oncogenesis in mice model expression of genes associated | [164] |
| with angiogenesis and inflammation in SH-SY5Y neuroblastoma cells | |||
| Paupar | - | Induced differentiation of neuroblastoma cells | [166] |
Abbreviations: -: up-regulation, ¯: down-regulation, Axl: AXL receptor tyrosine kinase, BRCA1: breast cancer 1, DKK1: dickkopf WNT signaling pathway inhibitor 1, ERK: extracellular signal-regulated kinase, MAPK: mitogen-activated protein kinase, EZH2: enhancer of zeste homolog 2, FGF2: fibroblast growth factor 2, GADD45A: growth arrest and DNA damage-inducible protein 45 alpha, Myc: myelocytomatosis oncogene, NBAT1: neuroblastoma associated Transcript 1, RA, retinoid acid
A study was aimed to examine lncRNA expression pattern by micro array in neuroblastoma cells after transfection with control or N-Myc-specific siRNA [140]. The linc00467 was identified as the novel lncRNA target of N-Myc. N-Myc suppressed linc00467 expression by direct binding to its promoter. Furthermore, gene silencing of linc00467 up-regulated the tumor suppressor gene DKK1, suppressed the viability and increased the apoptosis in neuroblastoma cells. These effects were reversed by the use of DKK1 siRNA. Thus, linc00467 can reduce neuroblastoma growth by modulation of DKK1 expression. Another study examined the expression pattern of lncRNAs and protein-coding genes between MYCN amplified and MYCN non-amplified NB patients [141]. A total of 6 lncRNAs were differentially expressed in neuroblastoma patients. MYCN amplification was found to up-regulate the expression of the lncRNA, SNHG1. SNHG1 was co-expressed with TAF1D (coding gene), and exhibited an association with poor patient survival. Furthermore, high expression of SNHG1 was predicted as an independent prognostic marker for event-free survival of patients. A robust expression of GAS5 is also reported in MYCN-amplified and non-amplified neuroblastoma cell lines [142]. The gene silencing of GAS5 produced defects in proliferation, apoptosis, and cell cycle arrest. The loss of GAS5 also induced p53, BRCA1, and GADD45A in neuroblastoma cell lines [142].
MYCN is also known to enhance lncRNAs expression by inducing epigenetic changes. For example, JMJD1A, a histone demethylase, has the potential of demethylating the lysine 9 residue of histone H3 (H3K9) and thus can activate gene transcription. In one study, N-Myc was found to directly bind to the JMJD1A gene promoter and up-regulated the gene expression in N-Myc amplified human neuroblastoma cells [143]. Furthermore, JMJD1A up-regulated MALAT1 by inducing histone demethylation at its promoter. While JMJD1A and MALAT1 induced, use of DMOG (small molecule inhibitor of JMJD1A) was found to suppress the migration and invasion of neuroblastoma cells. Overall, these results suggest that N-Myc can modulate the neuroblastoma cell migration and invasion by modulating JMJD1A and MALAT1 expression.
Neuroblastoma cells like other cancer cells are characterized by hypoxic conditions. In one study, hypoxia was found to induce MALAT1 in neuroblastoma cell lines [144]. The gene silencing of MALAT1 was associated with a reduction in endothelial cell migration, invasion and vasculature formation, and down-regulation of the expression of fibroblast growth factor 2 (FGF2). Interestingly, an addition of recombinant FGF2 protein to the cell culture media reversed the effects of MALAT1 siRNA on vasculature formation. Overall, these data suggest that MALAT1 mediate its tumorigenic effects under hypoxic conditions by modulating the expression of FGF2 [144]. MALAT1 can also up-regulate Axl, which is a member of the receptor tyrosine kinase family and associated with neuroblastoma metastasis [145]. MALAT1 can also induce ERK/MAPK activation and neuronal differentiation in neuroblastoma cell lines [146].
The lncRNAs HCN3, linc01105, and MEG3 are known to regulate neuroblastoma pathogenesis. While a high expression of HCN3 and linc01105 was observed in neuroblastoma tissue, MEG3 expression was decreased [147]. The gene silencing of HCN3 and linc01105, and MEG3 over-expression was associated with an increase in apoptosis. Furthermore, linc01105 knockdown promoted cell proliferation, whereas MEG3 over-expression inhibited proliferation. These observations suggest that HCN3 and linc01105 act as oncogenes, while MEG3 is a tumor suppressor.
NBAT1 (neuroblastoma associated transcript 1) is a tumor suppressor lncRNA identified in neuroblastoma. It regulates cell proliferation and invasion by interacting with EZH2 (enhancer of zeste 2) [148]. Furthermore, NBAT1 can be used to predict clinical outcome of neuroblastoma patients [21, 148]. NBAT1 can also contribute to the aggressiveness of neuroblastoma by promoting proliferation and impairment in the differentiation of neuronal precursors [21].
The chromosome 6p22 has been reported as the most susceptible locus for the development of neuroblastoma [149]. This locus harbors several lncRNAs including the tumor suppressor, CASC15. The low-level expression of a short CASC15 isoform (CASC15-S) is highly associated with advanced neuroblastoma and poor patient survival [150]. Furthermore, attenuation of CASC15-S can increase extracellular matrix transcripts, cellular growth, adhesion and migration in neuroblastoma cells. The lncRNA, ncRAN is located on chromosome 17q25.1 and contains two splice variants: a long form (Nbla10727) and a short form (Nbla12061) [151]. An up-regulation in ncRAN has been reported in neuroblastoma patients that associate with poor prognosis [151]. The intergenic lncRNA loci are known to regulate the expression of adjacent protein-coding genes. Dali is an intergenic lncRNA that is transcribed downstream of the Pou3f3 transcription factor. The depletion of Dali can disrupt the differentiation of neuroblastoma cells [152]. Dali can also epigenetically regulate the expression of genes.
PVT-1 is a lncRNA that can regulate c-Myc over a long distance. One study examined the expression of PVT-1 and c-Myc in normal human tissues and transformed cells [153]. Although PVT-1 was restricted to a relatively low number of normal tissues, c-Myc mRNA was widely distributed. However, PVT-1 was highly expressed in many transformed cells including neuroblastoma that do not express c-Myc. Furthermore, PVT-1 promoter region contained two putative binding sites for Myc proteins.
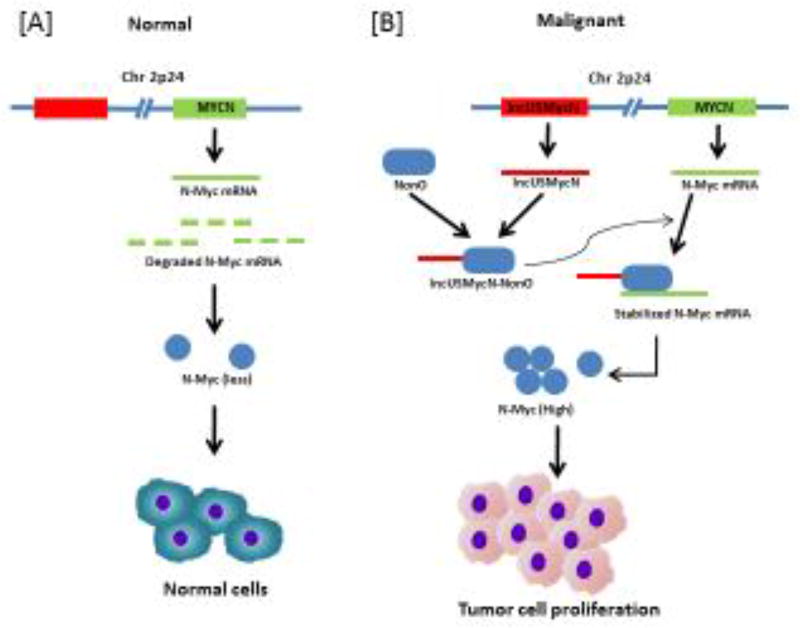
Transcribed ultra conserved regions (T-UCRs), another class of novel lncRNA [154], exhibit absolute conservation in humans, rat, and mice [155]. T-UCRs are associated with neuroblastoma pathogenesis [156]. In one study, T-UCRs were found to possess prognostic significance [157]. This novel class of lncRNAs was associated with MYCN-amplification in neuroblastoma patients. Although seven T-UCRs (uc.279, uc.347, uc.350, uc.364, uc.379, uc.446, and uc.460) were up regulated in MYCN-amplified tumors, none of the T-UCRs were down regulated. Furthermore, T-UCRs were widely associated with cancer-related pathways such as proliferation, apoptosis, and differentiation. The significance of T-UCRs in neuroblastoma pathogenesis was demonstrated by another study [158]. Some other lncRNAs reported in neuroblastoma are Gomafu [159], H19 [160], CAI2 [161], lncUSMycN [162, 163], HOXD-AS1 [164], HOTAIR [165], and Paupar [166]. The lncUSMycN inhibit degradation of N-Myc mRNA as described in Figure 3.
Figure 3. The lncUSMycN act by inhibiting degradation of N-Myc mRNA.
[A] In normal, cell, N-Myc undergoes degradation at post transcriptional and post translation level. [B] In neuroblastoma cells, MYCN oncogene harbor lncUSMycn, a lncRNA at 14 kb upstream of the gene. The lncUSMycn act as a scaffold for the RNA binding protein NonO. This facilitate the interaction between NonO and MYCN. This results in the stabilization of MYCN transcript and eventually proliferation of tumor cells
Overall, it is clear that both up-regulation and down-regulation in lncRNAs expression pattern could be of diagnostic and prognostic significance for neuroblastoma patients. Additionally, lncRNAs can also be used as therapeutic targets. However, most conclusions are based on the modulation of gene expression that could also result from non-cancer conditions. In spite of several studies, none of the lncRNAs are recommended for use in neuroblastoma patients. Future studies should be focused more towards elucidating the clinical utility of these lncRNAs in neuroblastoma patients.
8. System biology and neuroblastoma
System biology is an interdisciplinary field to study the complex interaction within the biological system using computational and mathematical modeling. In 1952, British neurophysiologists Alan Lloyd Hodgkin and Andrew Fielding Huxley were created a mathematical model to explain the neuronal cell axon’s action potential propagation [167]. This model described the interaction of cellular functions of sodium and potassium channels, and this was a landmark discovery to beginning computational system biology field [168]. It analyzed complex data sets such as genomics, proteomics, transcriptomics, metabolomics, glycomics, lipidomics from various experimental data sets to use computational tools. Cancer system biology deal with specific data sets such as patients samples, high-throughput patient’s genome, cancer cell lines, xenograft models, next-generation sequencing, siRNA based screenings, somatic mutations and genome instability [169].
The recent development of genome and proteasome level annotation of protein-protein interaction (PPI) networks facilitate functional cross-talk between genes in neuroblastoma [170]. Computational modeling was performed in neuroblastoma PPI network to elucidate the mutated genes effects on altered pathways in neuroblastoma [171]. The researchers have an opportunity to overlay patients and experimental data on to the network scaffold database such as BioGRID, STRING, and KEGG to analyze protein and gene interactions [172]. NeAT, an open source network analysis tool analyze and visualize the epigenetic, transcriptomic and metabolomics data may be an option to use neuroblastoma research [173]. Reverse-engineer networks approaches presume the interaction between data through Bayesian probabilistic models and pearson correlation coefficients[174]. This method has the capable the power to operate the human disease model like neuroblastoma and suitable for study the novel vital regulators play an essential role in neuroblastoma etiology and progression using large-scale multi-omics data [175].
It is challenging effect to apply system biology tools in human disease, but the ultimate goal was achieved through the collaborative efforts made with clinicians from all over the world. System biology promise of shedding new light to superior diagnosis to classify the virtual patient data and predict the outcome of suggested treatment on the basis of personalized neuroblastoma medicine. The system biology is more relevant to heterogeneous nature of neuroblastoma because it benefitted from the holistic and integrated approach to risk stratification. It provides realistic multi-scale in silico models of neuroblastoma and helps the clinical management and therapeutic design for the neuroblastoma patients.
9. Conclusions
The checkpoint molecules (PD-1, PD-L1), inflammatory molecules, lncRNAs, and miRNAs play a crucial role in neuroblastoma pathogenesis. These signaling molecules regulate various aspects of tumor development including transformation, survival, proliferation, invasion, angiogenesis, and metastasis of tumor cells. Opdivo (nivolumab) and keytruda (pembrolizumab) are PD-1 inhibitors approved by FDA for advanced melanoma and non-small cell lung cancer. However, the efficacy of these inhibitors for neuroblastoma patients remains to be examined. Dinutuximab (unituxin), a monoclonal antibody that targets the ganglioside GD2, is the most effective immunotherapy for neuroblastoma. Atezolizumab (Tecentriq), an engineered monoclonal antibody against PD-L1, is currently being evaluated for its potential in neuroblastoma patients.
Although bortezomib, a potent NF-κB inhibitor is approved for multiple myeloma patients, its potential for neuroblastoma patients remains to be determined. An inhibition of oncogenic miRNAs/lncRNAs or delivery of tumor suppressive miRNAs/lncRNAs is a potential strategy for neuroblastoma therapy. However, delivery, stability and off-target effects are some of the limitations associated with miRNA/lncRNA-based therapeutics. Future studies should be focused towards improving the delivery options of miRNAs and lncRNAs for prolonged therapeutic efficiency and safety. It is also imperative to examine if the crosstalk between miRNAs/lncRNAs and PD-1/PD-L signaling pathways exist. The system biology approach will probably add significantly in this direction.
In conclusion, immune-oncology has provided new hope to neuroblastoma patients. As discussed in this review, miRNA/lncRNA and immune checkpoint molecules could be used as biomarker and therapeutic target. The non-coding based biomarker has been developed for some cancer type. However, non-coding RNA based biomarker has yet to be approved for neuroblastoma. As for other cancer types, clinical trials in neuroblastoma are associated with high failure rates due to the expression of PD-L1 and other immune checkpoints in cancer cells as well as other cells of the tumor microenvironment. The use of reliable preclinical animal models will probably help to rapidly progress neuroblastoma field. New areas such as “Drug repurposing” should be explored to develop neuroblastoma therapy. We hope that the ongoing research across the scientific community will possibly help to place PD-1/PD-L1 and miRNA/lncRNA-based therapeutics as a potentially new facet for neuroblastoma therapy.
Table 3.
A list of immunotherapy based clinical trials for neuroblastoma
| Columnl | Column2 | Column3 | Column4 |
|---|---|---|---|
| Trials identifier No. | Trial status |
Drug/agent used | NB patient feature |
| NCT02169609 | Active, not recruiting | Dinutuximab (Ch 14.18) GM-CSF and IL-2 | high-risk neuroblastoma |
| NCT02573896 | Yet to recruit | ch14.18/Lenalidomide | relapsed refractory neuroblastoma |
| NCT01183897 | Active, not recruiting | Hu3F8/GM-CSF and 13-Cis-Retinoic Acid | primary refractory neuroblastoma in bone marrow |
| NCT03033303 | Recruiting | Hu3F8/GM-CSF Isotretinoin | high-risk neuroblastoma with first remission |
| NCT01183884 | Active, not recruiting | 3F8/GM-CSF 13-Cis-Retinoic Acid | high-risk neuroblastoma with second or greater remission |
| NCT01183429 | Active, not recruiting | 3F8/GM-CSF 13-Cis-Retinoic Acid | non-myeloablative therapy with high-risk neuroblastoma in first remission |
| NCT02765243 | Recruiting | 4SCAR-GD2 T cells | refractory or recurrent neuroblastoma |
| NCT02311621 | Recruiting | genetically modified T cells to express CAR | recurrent or refractory neuroblastoma |
| NCT01183416 | Active, not recruiting | High-dose 3F8/GM-CSF and 13-cis-retinoic acid | autologous stem-cell transplantation after myeloablative therapy first remission |
| NCT03242603 | Recruiting | Anti-GD2/ NK Cells | high-risk neuroblastoma |
| NCT02130869 | Recruiting | CD133+ selected autologous stem cell infusion/hu 14.18K322A | high-risk neuroblastoma |
| NCT02919046 | Recruiting | GD2-targeted CAR-T cells | Relapsed or refractory neuroblastoma |
| NCT02173093 | Recruiting | GD2 bispecific antibody | Children with neuroblastoma and osteosarcoma |
Abbreviations: GM-CSF: granulocyte-macrophage colony stimulating factor, NK: natural killer cells, GD2: disialoganglioside, Hu3F8: humanized 3F8 monoclonal antibody; CAR: chimeric antigen receptor
Acknowledgments
Dr. Challagundla’s laboratory is supported in whole or part from the National Institutes of Health (NIH) grant 1K22CA197074; the Nebraska State Department of Health & Human Services (LB506); an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the NIH under grant number P30 GM106397; the State of Nebraska Pediatric Cancer Research Grant (LB905) and the Biochemistry and Molecular Biology Department and the Fred and Pamela Buffet Cancer Center at University of Nebraska Medical Center. Dr. Gupta’s laboratory is supported by Science and Engineering Research Board (ECR/2016/000034), University Grants Commission [No.F.30-112/2015 (BSR)], and Design and Innovation Center (DIC-BHU/Project Approval/2015-16/608). We acknowledge support of NIAID R01AI129745 to SNB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contribution
All authors contributed equally to the design and writing of this manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Maris JM. Recent advances in neuroblastoma. N Engl J Med. 2010;362(23):2202–11. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3(3):203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 3.Maris JM, Matthay KK. Molecular biology of neuroblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1999;17(7):2264–79. doi: 10.1200/JCO.1999.17.7.2264. [DOI] [PubMed] [Google Scholar]
- 4.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 5.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vibhakar R, Juan G, Traganos F, Darzynkiewicz Z, Finger LR. Activation-induced expression of human programmed death-1 gene in T-lymphocytes. Exp Cell Res. 1997;232(1):25–8. doi: 10.1006/excr.1997.3493. [DOI] [PubMed] [Google Scholar]
- 7.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439(7077):682–7. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura H, Okazaki T, Tanaka Y, Nakatani K, Hara M, Matsumori A, Sasayama S, Mizoguchi A, Hiai H, Minato N, Honjo T. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291(5502):319–22. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 9.Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27(1):111–22. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francisco LM, Sage PT, Sharpe AH. The PD-1 pathway in tolerance and autoimmunity. Immunol Rev. 2010;236:219–42. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karin M. The IkappaB kinase - a bridge between inflammation and cancer. Cell Res. 2008;18(3):334–42. doi: 10.1038/cr.2008.30. [DOI] [PubMed] [Google Scholar]
- 12.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 13.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–45. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 14.Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. The Lancet. Oncology. 2014;15(11):e493–503. doi: 10.1016/S1470-2045(14)70263-3. [DOI] [PubMed] [Google Scholar]
- 15.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–81. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 16.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews. Genetics. 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 17.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS genetics. 2013;9(6):e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costa FF. Non-coding RNAs: lost in translation? Gene. 2007;386(1–2):1–10. doi: 10.1016/j.gene.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Costa FF. Non-coding RNAs and new opportunities for the private sector. Drug discovery today. 2009;14(9–10):446–52. doi: 10.1016/j.drudis.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Costa FF. Non-coding RNAs: Meet thy masters. BioEssays : news and reviews in molecular, cellular and developmental biology. 2010;32(7):599–608. doi: 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 21.Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S, Ostensson M, Akyurek LM, Abrahamsson J, Pfeifer S, Larsson E, Shi L, Peng Z, Fischer M, Martinsson T, Hedborg F, Kogner P, Kanduri C. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell. 2014;26(5):722–37. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 22.Pandey GK, Kanduri C. Long noncoding RNAs and neuroblastoma. Oncotarget. 2015;6(21):18265–75. doi: 10.18632/oncotarget.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu X, Ravindranath L, Tran N, Petrovics G, Srivastava S. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA and cell biology. 2006;25(3):135–41. doi: 10.1089/dna.2006.25.135. [DOI] [PubMed] [Google Scholar]
- 25.Kurian L, Aguirre A, Sancho-Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA, Rodriguez Esteban C, Campistol JM, Yeo GW, Izpisua Belmonte JC. Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation. 2015;131(14):1278–90. doi: 10.1161/CIRCULATIONAHA.114.013303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casero D, Sandoval S, Seet CS, Scholes J, Zhu Y, Ha VL, Luong A, Parekh C, Crooks GM. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nature immunology. 2015;16(12):1282–91. doi: 10.1038/ni.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez Y, Huarte M. Long non-coding RNAs: challenges for diagnosis and therapies. Nucleic acid therapeutics. 2013;23(1):15–20. doi: 10.1089/nat.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Challagundla KB, Fanini F, Vannini I, Wise P, Murtadha M, Malinconico L, Cimmino A, Fabbri M. microRNAs in the tumor microenvironment: solving the riddle for a better diagnostics. Expert Rev Mol Diagn. 2014;14(5):565–74. doi: 10.1586/14737159.2014.922879. [DOI] [PubMed] [Google Scholar]
- 29.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 30.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148(6):1172–87. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11(3):228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 33.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. International journal of cancer. 2017;140(9):1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 34.Bottai G, Pasculli B, Calin GA, Santarpia L. Targeting the microRNA-regulating DNA damage/repair pathways in cancer. Expert Opin Biol Ther. 2014;14(11):1667–83. doi: 10.1517/14712598.2014.950650. [DOI] [PubMed] [Google Scholar]
- 35.De Mattos-Arruda L, Bottai G, Nuciforo PG, Di Tommaso L, Giovannetti E, Peg V, Losurdo A, Perez-Garcia J, Masci G, Corsi F, Cortes J, Seoane J, Calin GA, Santarpia L. MicroRNA-21 links epithelial-to-mesenchymal transition and inflammatory signals to confer resistance to neoadjuvant trastuzumab and chemotherapy in HER2-positive breast cancer patients. Oncotarget. 2015;6(35):37269–80. doi: 10.18632/oncotarget.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotillo E, Thomas-Tikhonenko A. Shielding the messenger (RNA): microRNA-based anticancer therapies. Pharmacol Ther. 2011;131(1):18–32. doi: 10.1016/j.pharmthera.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang X, Mei H, Zhao X, Pu J, Li D, Qu H, Jiao W, Zhao J, Huang K, Zheng L, Tong Q. miRNA-337-3p suppresses neuroblastoma progression by repressing the transcription of matrix metalloproteinase 14. Oncotarget. 2015;6(26):22452–66. doi: 10.18632/oncotarget.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drake CG, Jaffee E, Pardoll DM. Mechanisms of immune evasion by tumors. Adv Immunol. 2006;90:51–81. doi: 10.1016/S0065-2776(06)90002-9. [DOI] [PubMed] [Google Scholar]
- 39.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldberg MV, Drake CG. LAG-3 in Cancer Immunotherapy. Curr Top Microbiol Immunol. 2011;344:269–78. doi: 10.1007/82_2010_114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang D, Lotze MT. Tumor immunity times out: TIM-3 and HMGB1. Nat Immunol. 2012;13(9):808–10. doi: 10.1038/ni.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, Murphy TL, Russell JH, Allison JP, Murphy KM. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4(7):670–9. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- 43.Smolle MA, Calin HN, Pichler M, Calin GA. Noncoding RNAs and immune checkpoints-clinical implications as cancer therapeutics. FEBS J. 2017;284(13):1952–1966. doi: 10.1111/febs.14030. [DOI] [PubMed] [Google Scholar]
- 44.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11(11):3887–95. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ceeraz S, Nowak EC, Noelle RJ. B7 family checkpoint regulators in immune regulation and disease. Trends Immunol. 2013;34(11):556–63. doi: 10.1016/j.it.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206(13):3015–29. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5(12):1365–9. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 48.Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, Iwai Y, Long AJ, Brown JA, Nunes R, Greenfield EA, Bourque K, Boussiotis VA, Carter LL, Carreno BM, Malenkovich N, Nishimura H, Okazaki T, Honjo T, Sharpe AH, Freeman GJ. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2(3):261–8. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 49.Tseng SY, Otsuji M, Gorski K, Huang X, Slansky JE, Pai SI, Shalabi A, Shin T, Pardoll DM, Tsuchiya H. B7-DC, a new dendritic cell molecule with potent costimulatory properties for T cells. J Exp Med. 2001;193(7):839–46. doi: 10.1084/jem.193.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, Rodig SJ, Chapuy B, Ligon AH, Zhu L, Grosso JF, Kim SY, Timmerman JM, Shipp MA, Armand P. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372(4):311–9. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou MX, Peng AB, Lv GH, Wang XB, Li J, She XL, Jiang Y. Expression of programmed death-1 ligand (PD-L1) in tumor-infiltrating lymphocytes is associated with favorable spinal chordoma prognosis. Am J Transl Res. 2016;8(7):3274–87. [PMC free article] [PubMed] [Google Scholar]
- 52.Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51(3):221–8. doi: 10.1016/j.oraloncology.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 53.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, Chen L. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li CW, Lim SO, Xia W, Lee HH, Chan LC, Kuo CW, Khoo KH, Chang SS, Cha JH, Kim T, Hsu JL, Wu Y, Hsu JM, Yamaguchi H, Ding Q, Wang Y, Yao J, Lee CC, Wu HJ, Sahin AA, Allison JP, Yu D, Hortobagyi GN, Hung MC. Glycosylation and stabilization of programmed death ligand-1 suppresses T-cell activity. Nat Commun. 2016;7:12632. doi: 10.1038/ncomms12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim SO, Li CW, Xia W, Cha JH, Chan LC, Wu Y, Chang SS, Lin WC, Hsu JM, Hsu YH, Kim T, Chang WC, Hsu JL, Yamaguchi H, Ding Q, Wang Y, Yang Y, Chen CH, Sahin AA, Yu D, Hortobagyi GN, Hung MC. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 2016;30(6):925–939. doi: 10.1016/j.ccell.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crane CA, Panner A, Murray JC, Wilson SP, Xu H, Chen L, Simko JP, Waldman FM, Pieper RO, Parsa AT. PI(3) kinase is associated with a mechanism of immunoresistance in breast and prostate cancer. Oncogene. 2009;28(2):306–12. doi: 10.1038/onc.2008.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, Mischel PS, Stokoe D, Pieper RO. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84–8. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 58.Majzner RG, Simon JS, Grosso JF, Martinez D, Pawel BR, Santi M, Merchant MS, Geoerger B, Hezam I, Marty V, Vielh P, Daugaard M, Sorensen PH, Mackall CL, Maris JM. Assessment of programmed death-ligand 1 expression and tumor-associated immune cells in pediatric cancer tissues. Cancer. 2017 doi: 10.1002/cncr.30724. [DOI] [PubMed] [Google Scholar]
- 59.Cheng JM, Hiemstra JL, Schneider SS, Naumova A, Cheung NK, Cohn SL, Diller L, Sapienza C, Brodeur GM. Preferential amplification of the paternal allele of the N-myc gene in human neuroblastomas. Nature genetics. 1993;4(2):191–4. doi: 10.1038/ng0693-191. [DOI] [PubMed] [Google Scholar]
- 60.Emanuel BS, Balaban G, Boyd JP, Grossman A, Negishi M, Parmiter A, Glick MC. N-myc amplification in multiple homogeneously staining regions in two human neuroblastomas. Proc Natl Acad Sci U S A. 1985;82(11):3736–40. doi: 10.1073/pnas.82.11.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224(4653):1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 62.Casinelli G, LaRosa J, Sharma M, Cherok E, Banerjee S, Branca M, Edmunds L, Wang Y, Sims-Lucas S, Churley L, Kelly S, Sun M, Stolz D, Graves JA. N-Myc overexpression increases cisplatin resistance in neuroblastoma via deregulation of mitochondrial dynamics. Cell Death Discov. 2016;2:16082. doi: 10.1038/cddiscovery.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kohl NE, Kanda N, Schreck RR, Bruns G, Latt SA, Gilbert F, Alt FW. Transposition and amplification of oncogene-related sequences in human neuroblastomas. Cell. 1983;35(2 Pt 1):359–67. doi: 10.1016/0092-8674(83)90169-1. [DOI] [PubMed] [Google Scholar]
- 64.Melaiu O, Mina M, Chierici M, Boldrini R, Jurman G, Romania P, D'Alicandro V, Benedetti MC, Castellano A, Liu T, Furlanello C, Locatelli F, Fruci D. PD-L1 is a therapeutic target of the Bromodomain inhibitor JQ1 and, combined with HLA class I, a promising prognostic biomarker in neuroblastoma. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-2601. [DOI] [PubMed] [Google Scholar]
- 65.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352(6282):227–31. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chuang JH, Chuang HC, Huang CC, Wu CL, Du YY, Kung ML, Chen CH, Chen SC, Tai MH. Differential toll-like receptor 3 (TLR3) expression and apoptotic response to TLR3 agonist in human neuroblastoma cells. J Biomed Sci. 2011;18:65. doi: 10.1186/1423-0127-18-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hsu WM, Huang CC, Wu PY, Lee H, Huang MC, Tai MH, Chuang JH. Toll-like receptor 3 expression inhibits cell invasion and migration and predicts a favorable prognosis in neuroblastoma. Cancer Lett. 2013;336(2):338–46. doi: 10.1016/j.canlet.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 68.van Dam LS, de Zwart VM, Meyer-Wentrup FA. The role of programmed cell death-1 (PD-1) and its ligands in pediatric cancer. Pediatr Blood Cancer. 2015;62(2):190–197. doi: 10.1002/pbc.25284. [DOI] [PubMed] [Google Scholar]
- 69.Dondero A, Pastorino F, Della Chiesa M, Corrias MV, Morandi F, Pistoia V, Olive D, Bellora F, Locatelli F, Castellano A, Moretta L, Moretta A, Bottino C, Castriconi R. PD-L1 expression in metastatic neuroblastoma as an additional mechanism for limiting immune surveillance. Oncoimmunology. 2016;5(1):e1064578. doi: 10.1080/2162402X.2015.1064578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mina M, Boldrini R, Citti A, Romania P, D'Alicandro V, De Ioris M, Castellano A, Furlanello C, Locatelli F, Fruci D. Tumor-infiltrating T lymphocytes improve clinical outcome of therapy-resistant neuroblastoma. Oncoimmunology. 2015;4(9):e1019981. doi: 10.1080/2162402X.2015.1019981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wong RM, Scotland RR, Lau RL, Wang C, Korman AJ, Kast WM, Weber JS. Programmed death-1 blockade enhances expansion and functional capacity of human melanoma antigen-specific CTLs. Int Immunol. 2007;19(10):1223–34. doi: 10.1093/intimm/dxm091. [DOI] [PubMed] [Google Scholar]
- 72.Yu AL, Gilman AL, Ozkaynak MF, London WB, Kreissman SG, Chen HX, Smith M, Anderson B, Villablanca JG, Matthay KK, Shimada H, Grupp SA, Seeger R, Reynolds CP, Buxton A, Reisfeld RA, Gillies SD, Cohn SL, Maris JM, Sondel PM, Children's Oncology G. Anti-GD2 antibody with GM-CSF, interleukin-2, and isotretinoin for neuroblastoma. N Engl J Med. 2010;363(14):1324–34. doi: 10.1056/NEJMoa0911123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kanold J, Paillard C, Tchirkov A, Lang P, Kelly A, Halle P, Isfan F, Merlin E, Marabelle A, Rochette E, Demeocq F. NK cell immunotherapy for high-risk neuroblastoma relapse after haploidentical HSCT. Pediatric blood & cancer. 2012;59(4):739–42. doi: 10.1002/pbc.24030. [DOI] [PubMed] [Google Scholar]
- 74.Eissler N, Mao Y, Brodin D, Reutersward P, Andersson Svahn H, Johnsen JI, Kiessling R, Kogner P. Regulation of myeloid cells by activated T cells determines the efficacy of PD-1 blockade. Oncoimmunology. 2016;5(12):e1232222. doi: 10.1080/2162402X.2016.1232222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao Y, Eissler N, Blanc KL, Johnsen JI, Kogner P, Kiessling R. Targeting Suppressive Myeloid Cells Potentiates Checkpoint Inhibitors to Control Spontaneous Neuroblastoma. Clin Cancer Res. 2016;22(15):3849–59. doi: 10.1158/1078-0432.CCR-15-1912. [DOI] [PubMed] [Google Scholar]
- 76.Boes M, Meyer-Wentrup F. TLR3 triggering regulates PD-L1 (CD274) expression in human neuroblastoma cells. Cancer Lett. 2015;361(1):49–56. doi: 10.1016/j.canlet.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 77.Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, Majem M, Fidler MJ, de Castro G, Jr, Garrido M, Lubiniecki GM, Shentu Y, Im E, Dolled-Filhart M, Garon EB. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540–50. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 78.Kerr KM, Tsao MS, Nicholson AG, Yatabe, Wistuba Y, II, Hirsch FR, Committee IP. Programmed Death-Ligand 1 Immunohistochemistry in Lung Cancer: In what state is this art? J Thorac Oncol. 2015;10(7):985–9. doi: 10.1097/JTO.0000000000000526. [DOI] [PubMed] [Google Scholar]
- 79.Massi D, Brusa D, Merelli B, Falcone C, Xue G, Carobbio A, Nassini R, Baroni G, Tamborini E, Cattaneo L, Audrito V, Deaglio S, Mandala M. The status of PD-L1 and tumor-infiltrating immune cells predict resistance and poor prognosis in BRAFi-treated melanoma patients harboring mutant BRAFV600. Ann Oncol. 2015;26(9):1980–7. doi: 10.1093/annonc/mdv255. [DOI] [PubMed] [Google Scholar]
- 80.Taube JM, Klein A, Brahmer JR, Xu H, Pan X, Kim JH, Chen L, Pardoll DM, Topalian SL, Anders RA. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064–74. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin EY, Li JF, Gnatovskiy L, Deng Y, Zhu L, Grzesik DA, Qian H, Xue XN, Pollard JW. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66(23):11238–46. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 82.Zheng Y, Cai Z, Wang S, Zhang X, Qian J, Hong S, Li H, Wang M, Yang J, Yi Q. Macrophages are an abundant component of myeloma microenvironment and protect myeloma cells from chemotherapy drug-induced apoptosis. Blood. 2009;114(17):3625–8. doi: 10.1182/blood-2009-05-220285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jubinsky PT, Dickens DS, Short MK. New roles for mononuclear phagocytes in cancer biology. J Pediatr Hematol Oncol. 2008;30(8):584–91. doi: 10.1097/MPH.0b013e31816e2358. [DOI] [PubMed] [Google Scholar]
- 84.Sica A, Larghi P, Mancino A, Rubino L, Porta C, Totaro MG, Rimoldi M, Biswas SK, Allavena P, Mantovani A. Macrophage polarization in tumour progression. Semin Cancer Biol. 2008;18(5):349–55. doi: 10.1016/j.semcancer.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 85.Laoui D, Van Overmeire E, Movahedi K, Van den Bossche J, Schouppe E, Mommer C, Nikolaou A, Morias Y, De Baetselier P, Van Ginderachter JA. Mononuclear phagocyte heterogeneity in cancer: different subsets and activation states reaching out at the tumor site. Immunobiology. 2011;216(11):1192–202. doi: 10.1016/j.imbio.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 86.Asgharzadeh S, Salo JA, Ji L, Oberthuer A, Fischer M, Berthold F, Hadjidaniel M, Liu CW, Metelitsa LS, Pique-Regi R, Wakamatsu P, Villablanca JG, Kreissman SG, Matthay KK, Shimada H, London WB, Sposto R, Seeger RC. Clinical significance of tumor-associated inflammatory cells in metastatic neuroblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2012;30(28):3525–32. doi: 10.1200/JCO.2011.40.9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Song L, Asgharzadeh S, Salo J, Engell K, Wu HW, Sposto R, Ara T, Silverman AM, DeClerck YA, Seeger RC, Metelitsa LS. Valpha24-invariant NKT cells mediate antitumor activity via killing of tumor-associated macrophages. J Clin Invest. 2009;119(6):1524–36. doi: 10.1172/JCI37869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ara T, Song L, Shimada H, Keshelava N, Russell HV, Metelitsa LS, Groshen SG, Seeger RC, DeClerck YA. Interleukin-6 in the bone marrow microenvironment promotes the growth and survival of neuroblastoma cells. Cancer Res. 2009;69(1):329–37. doi: 10.1158/0008-5472.CAN-08-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5(3):219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 90.Zhang H, Qi M, Li S, Qi T, Mei H, Huang K, Zheng L, Tong Q. microRNA-9 targets matrix metalloproteinase 14 to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells. Mol Cancer Ther. 2012;11(7):1454–66. doi: 10.1158/1535-7163.MCT-12-0001. [DOI] [PubMed] [Google Scholar]
- 91.Franklin RA, Liao W, Sarkar A, Kim MV, Bivona MR, Liu K, Pamer EG, Li MO. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–5. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Colvin EK. Tumor-associated macrophages contribute to tumor progression in ovarian cancer. Front Oncol. 2014;4:137. doi: 10.3389/fonc.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu P, Fu YX. Tumor-infiltrating T lymphocytes: friends or foes? Lab Invest. 2006;86(3):231–45. doi: 10.1038/labinvest.3700389. [DOI] [PubMed] [Google Scholar]
- 94.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV, Gonzalez-Ericsson PI, Sanders M, Solomon B, Solinas C, Van den Eynden G, Allory Y, Preusser M, Hainfellner J, Pruneri G, Vingiani A, Demaria S, Symmans F, Nuciforo P, Comerma L, Thompson EA, Lakhani S, Kim SR, Schnitt S, Colpaert C, Sotiriou C, Scherer SJ, Ignatiadis M, Badve S, Pierce RH, Viale G, Sirtaine N, Penault-Llorca F, Sugie T, Fineberg S, Paik S, Srinivasan A, Richardson A, Wang Y, Chmielik E, Brock J, Johnson DB, Balko J, Wienert S, Bossuyt V, Michiels S, Ternes N, Burchardi N, Luen SJ, Savas P, Klauschen F, Watson PH, Nelson BH, Criscitiello C, O'Toole S, Larsimont D, de Wind R, Curigliano G, Andre F, Lacroix-Triki M, van de Vijver M, Rojo F, Floris G, Bedri S, Sparano J, Rimm D, Nielsen T, Kos Z, Hewitt S, Singh B, Farshid G, Loibl S, Allison KH, Tung N, Adams S, Willard-Gallo K, Horlings HM, Gandhi L, Moreira A, Hirsch F, Dieci MV, Urbanowicz M, Brcic I, Korski K, Gaire F, Koeppen H, Lo A, Giltnane J, Rebelatto MC, Steele KE, Zha J, Emancipator K, Juco JW, Denkert C, Reis-Filho J, Loi S, Fox SB. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol. 2017;24(6):311–335. doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV, Gonzalez-Ericsson PI, Sanders M, Solomon B, Solinas C, Van den Eynden G, Allory Y, Preusser M, Hainfellner J, Pruneri G, Vingiani A, Demaria S, Symmans F, Nuciforo P, Comerma L, Thompson EA, Lakhani S, Kim SR, Schnitt S, Colpaert C, Sotiriou C, Scherer SJ, Ignatiadis M, Badve S, Pierce RH, Viale G, Sirtaine N, Penault-Llorca F, Sugie T, Fineberg S, Paik S, Srinivasan A, Richardson A, Wang Y, Chmielik E, Brock J, Johnson DB, Balko J, Wienert S, Bossuyt V, Michiels S, Ternes N, Burchardi N, Luen SJ, Savas P, Klauschen F, Watson PH, Nelson BH, Criscitiello C, O'Toole S, Larsimont D, de Wind R, Curigliano G, Andre F, Lacroix-Triki M, van de Vijver M, Rojo F, Floris G, Bedri S, Sparano J, Rimm D, Nielsen T, Kos Z, Hewitt S, Singh B, Farshid G, Loibl S, Allison KH, Tung N, Adams S, Willard-Gallo K, Horlings HM, Gandhi L, Moreira A, Hirsch F, Dieci MV, Urbanowicz M, Brcic I, Korski K, Gaire F, Koeppen H, Lo A, Giltnane J, Rebelatto MC, Steele KE, Zha J, Emancipator K, Juco JW, Denkert C, Reis-Filho J, Loi S, Fox SB. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv Anat Pathol. 2017;24(5):235–251. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Turk MJ, Guevara-Patino JA, Rizzuto GA, Engelhorn ME, Sakaguchi S, Houghton AN. Concomitant tumor immunity to a poorly immunogenic melanoma is prevented by regulatory T cells. J Exp Med. 2004;200(6):771–82. doi: 10.1084/jem.20041130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hendry S, Salgado R, Gevaert T, Russell PA, John T, Thapa B, Christie M, van de Vijver K, Estrada MV, Gonzalez-Ericsson PI, Sanders M, Solomon B, Solinas C, Van den Eynden G, Allory Y, Preusser M, Hainfellner J, Pruneri G, Vingiani A, Demaria S, Symmans F, Nuciforo P, Comerma L, Thompson EA, Lakhani S, Kim SR, Schnitt S, Colpaert C, Sotiriou C, Scherer SJ, Ignatiadis M, Badve S, Pierce RH, Viale G, Sirtaine N, Penault-Llorca F, Sugie T, Fineberg S, Paik S, Srinivasan A, Richardson A, Wang Y, Chmielik E, Brock J, Johnson DB, Balko J, Wienert S, Bossuyt V, Michiels S, Ternes N, Burchardi N, Luen SJ, Savas P, Klauschen F, Watson PH, Nelson BH, Criscitiello C, O'Toole S, Larsimont D, de Wind R, Curigliano G, Andre F, Lacroix-Triki M, van de Vijver M, Rojo F, Floris G, Bedri S, Sparano J, Rimm D, Nielsen T, Kos Z, Hewitt S, Singh B, Farshid G, Loibl S, Allison KH, Tung N, Adams S, Willard-Gallo K, Horlings HM, Gandhi L, Moreira A, Hirsch F, Dieci MV, Urbanowicz M, Brcic I, Korski K, Gaire F, Koeppen H, Lo A, Giltnane J, Rebelatto MC, Steele KE, Zha J, Emancipator K, Juco JW, Denkert C, Reis-Filho J, Loi S, Fox SB. Assessing Tumor-Infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method from the International Immuno-Oncology Biomarkers Working Group: Part 2: TILs in Melanoma, Gastrointestinal Tract Carcinomas, Non-Small Cell Lung Carcinoma and Mesothelioma, Endometrial and Ovarian Carcinomas, Squamous Cell Carcinoma of the Head and Neck, Genitourinary Carcinomas, and Primary Brain Tumors. Adv Anat Pathol. 2017 doi: 10.1097/PAP.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314(5796):126–9. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 100.Bergfeld SA, DeClerck YA. Bone marrow-derived mesenchymal stem cells and the tumor microenvironment. Cancer metastasis reviews. 2010;29(2):249–61. doi: 10.1007/s10555-010-9222-7. [DOI] [PubMed] [Google Scholar]
- 101.Lee NK, Park SE, Kwon SJ, Shim S, Byeon Y, Kim JH, Na DL, Chang JW. Agouti Related Peptide Secreted Via Human Mesenchymal Stem Cells Upregulates Proteasome Activity in an Alzheimer's Disease Model. Scientific reports. 2017;7:39340. doi: 10.1038/srep39340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Borriello L, Nakata R, Sheard MA, Fernandez GE, Sposto R, Malvar J, Blavier L, Shimada H, Asgharzadeh S, Seeger RC, DeClerck YA. Cancer-Associated Fibroblasts Share Characteristics and Pro-tumorigenic Activity with Mesenchymal Stromal Cells. Cancer research. 2017 doi: 10.1158/0008-5472.CAN-16-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hashimoto O, Yoshida M, Koma Y, Yanai T, Hasegawa D, Kosaka Y, Nishimura N, Yokozaki H. Collaboration of cancer-associated fibroblasts and tumour-associated macrophages for neuroblastoma development. The Journal of pathology. 2016;240(2):211–23. doi: 10.1002/path.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kimura K, Kishida T, Wakao J, Tanaka T, Higashi M, Fumino S, Aoi S, Furukawa T, Mazda O, Tajiri T. Tumor-homing effect of human mesenchymal stem cells in a TH-MYCN mouse model of neuroblastoma. J Pediatr Surg. 2016;51(12):2068–2073. doi: 10.1016/j.jpedsurg.2016.09.041. [DOI] [PubMed] [Google Scholar]
- 105.Rincon E, Cejalvo T, Kanojia D, Alfranca A, Rodriguez-Milla MA, Hoyos RAG, Han Y, Zhang L, Alemany R, Lesniak MS, Garcia-Castro J. Mesenchymal stem cell carriers enhance antitumor efficacy of oncolytic adenoviruses in an immunocompetent mouse model. Oncotarget. 2017 doi: 10.18632/oncotarget.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Conroy H, Mawhinney L, Donnelly SC. Inflammation and cancer: macrophage migration inhibitory factor (MIF)--the potential missing link. QJM. 2010;103(11):831–6. doi: 10.1093/qjmed/hcq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bach JP, Rinn B, Meyer B, Dodel R, Bacher M. Role of MIF in inflammation and tumorigenesis. Oncology. 2008;75(3–4):127–33. doi: 10.1159/000155223. [DOI] [PubMed] [Google Scholar]
- 108.Ren Y, Chan HM, Li Z, Lin C, Nicholls J, Chen CF, Lee PY, Lui V, Bacher M, Tam PK. Upregulation of macrophage migration inhibitory factor contributes to induced N-Myc expression by the activation of ERK signaling pathway and increased expression of interleukin-8 and VEGF in neuroblastoma. Oncogene. 2004;23(23):4146–54. doi: 10.1038/sj.onc.1207490. [DOI] [PubMed] [Google Scholar]
- 109.Russell HV, Hicks J, Okcu MF, Nuchtern JG. CXCR4 expression in neuroblastoma primary tumors is associated with clinical presentation of bone and bone marrow metastases. J Pediatr Surg. 2004;39(10):1506–11. doi: 10.1016/j.jpedsurg.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 110.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, Poellinger L, Pahlman S. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10(5):413–23. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 111.Hamidian A, von Stedingk K, Munksgaard Thoren M, Mohlin S, Pahlman S. Differential regulation of HIF-1alpha and HIF-2alpha in neuroblastoma: Estrogen-related receptor alpha (ERRalpha) regulates HIF2A transcription and correlates to poor outcome. Biochem Biophys Res Commun. 2015;461(3):560–7. doi: 10.1016/j.bbrc.2015.04.083. [DOI] [PubMed] [Google Scholar]
- 112.Gupta SC, Sundaram C, Reuter S, Aggarwal BB. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim Biophys Acta. 2010;1799(10–12):775–87. doi: 10.1016/j.bbagrm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bian X, Opipari AW, Jr, Ratanaproeksa AB, Boitano AE, Lucas PC, Castle VP. Constitutively active NFkappa B is required for the survival of S-type neuroblastoma. J Biol Chem. 2002;277(44):42144–50. doi: 10.1074/jbc.M203891200. [DOI] [PubMed] [Google Scholar]
- 114.Mantovani A. Molecular pathways linking inflammation and cancer. Curr Mol Med. 2010;10(4):369–73. doi: 10.2174/156652410791316968. [DOI] [PubMed] [Google Scholar]
- 115.Tafani M, Pucci B, Russo A, Schito L, Pellegrini L, Perrone GA, Villanova L, Salvatori L, Ravenna L, Petrangeli E, Russo MA. Modulators of HIF1alpha and NFkB in Cancer Treatment: Is it a Rational Approach for Controlling Malignant Progression? Front Pharmacol. 2013;4:13. doi: 10.3389/fphar.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Challagundla KB, Wise PM, Neviani P, Chava H, Murtadha M, Xu T, Kennedy R, Ivan C, Zhang X, Vannini I, Fanini F, Amadori D, Calin GA, Hadjidaniel M, Shimada H, Jong A, Seeger RC, Asgharzadeh S, Goldkorn A, Fabbri M. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107(7) doi: 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhi F, Wang R, Wang Q, Xue L, Deng D, Wang S, Yang Y. MicroRNAs in neuroblastoma: small-sized players with a large impact. Neurochem Res. 2014;39(4):613–23. doi: 10.1007/s11064-014-1247-9. [DOI] [PubMed] [Google Scholar]
- 118.Zhao D, Tian Y, Li P, Wang L, Xiao A, Zhang M, Shi T. MicroRNA-203 inhibits the malignant progression of neuroblastoma by targeting Sam68. Mol Med Rep. 2015;12(4):5554–60. doi: 10.3892/mmr.2015.4013. [DOI] [PubMed] [Google Scholar]
- 119.Xiang X, Mei H, Qu H, Zhao X, Li D, Song H, Jiao W, Pu J, Huang K, Zheng L, Tong Q. miRNA-584-5p exerts tumor suppressive functions in human neuroblastoma through repressing transcription of matrix metalloproteinase 14. Biochim Biophys Acta. 2015;1852(9):1743–54. doi: 10.1016/j.bbadis.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 120.Qu H, Zheng L, Pu J, Mei H, Xiang X, Zhao X, Li D, Li S, Mao L, Huang K, Tong Q. miRNA-558 promotes tumorigenesis and aggressiveness of neuroblastoma cells through activating the transcription of heparanase. Hum Mol Genet. 2015;24(9):2539–51. doi: 10.1093/hmg/ddv018. [DOI] [PubMed] [Google Scholar]
- 121.Mestdagh P, Fredlund E, Pattyn F, Schulte JH, Muth D, Vermeulen J, Kumps C, Schlierf S, De Preter K, Van Roy N, Noguera R, Laureys G, Schramm A, Eggert A, Westermann F, Speleman F, Vandesompele J. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2010;29(9):1394–404. doi: 10.1038/onc.2009.429. [DOI] [PubMed] [Google Scholar]
- 122.Ziyan W, Yang L. MicroRNA-21 regulates the sensitivity to cisplatin in a human osteosarcoma cell line. Ir J Med Sci. 2016;185(1):85–91. doi: 10.1007/s11845-014-1225-x. [DOI] [PubMed] [Google Scholar]
- 123.Xin C, Buhe B, Hongting L, Chuanmin Y, Xiwei H, Hong Z, Lulu H, Qian D, Renjie W. MicroRNA-15a promotes neuroblastoma migration by targeting reversion-inducing cysteine-rich protein with Kazal motifs (RECK) and regulating matrix metalloproteinase-9 expression. FEBS J. 2013;280(3):855–66. doi: 10.1111/febs.12074. [DOI] [PubMed] [Google Scholar]
- 124.Chen Y, Stallings RL. Differential patterns of microRNA expression in neuroblastoma are correlated with prognosis, differentiation, and apoptosis. Cancer Res. 2007;67(3):976–83. doi: 10.1158/0008-5472.CAN-06-3667. [DOI] [PubMed] [Google Scholar]
- 125.De Preter K, Mestdagh P, Vermeulen J, Zeka F, Naranjo A, Bray I, Castel V, Chen C, Drozynska E, Eggert A, Hogarty MD, Izycka-Swieszewska E, London WB, Noguera R, Piqueras M, Bryan K, Schowe B, van Sluis P, Molenaar JJ, Schramm A, Schulte JH, Stallings RL, Versteeg R, Laureys G, Van Roy N, Speleman F, Vandesompele J. miRNA expression profiling enables risk stratification in archived and fresh neuroblastoma tumor samples. Clin Cancer Res. 2011;17(24):7684–92. doi: 10.1158/1078-0432.CCR-11-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shalaby T, Fiaschetti G, Baumgartner M, Grotzer MA. Significance and therapeutic value of miRNAs in embryonal neural tumors. Molecules. 2014;19(5):5821–62. doi: 10.3390/molecules19055821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26(34):5017–22. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 128.Swarbrick A, Woods SL, Shaw A, Balakrishnan A, Phua Y, Nguyen A, Chanthery Y, Lim L, Ashton LJ, Judson RL, Huskey N, Blelloch R, Haber M, Norris MD, Lengyel P, Hackett CS, Preiss T, Chetcuti A, Sullivan CS, Marcusson EG, Weiss W, L'Etoile N, Goga A. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med. 2010;16(10):1134–40. doi: 10.1038/nm.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Le MT, Xie H, Zhou B, Chia PH, Rizk P, Um M, Udolph G, Yang H, Lim B, Lodish HF. MicroRNA-125b promotes neuronal differentiation in human cells by repressing multiple targets. Mol Cell Biol. 2009;29(19):5290–305. doi: 10.1128/MCB.01694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Foley NH, Bray I, Watters KM, Das S, Bryan K, Bernas T, Prehn JH, Stallings RL. MicroRNAs 10a and 10b are potent inducers of neuroblastoma cell differentiation through targeting of nuclear receptor corepressor 2. Cell Death Differ. 2011;18(7):1089–98. doi: 10.1038/cdd.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Das S, Bryan K, Buckley PG, Piskareva O, Bray IM, Foley N, Ryan J, Lynch J, Creevey L, Fay J, Prenter S, Koster J, van Sluis P, Versteeg R, Eggert A, Schulte JH, Schramm A, Mestdagh P, Vandesompele J, Speleman F, Stallings RL. Modulation of neuroblastoma disease pathogenesis by an extensive network of epigenetically regulated microRNAs. Oncogene. 2013;32(24):2927–36. doi: 10.1038/onc.2012.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Pahlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci U S A. 2010;107(4):1553–8. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Buechner J, Henriksen JR, Haug BH, Tomte E, Flaegstad T, Einvik C. Inhibition of mir-21, which is up-regulated during MYCN knockdown-mediated differentiation, does not prevent differentiation of neuroblastoma cells. Differentiation. 2011;81(1):25–34. doi: 10.1016/j.diff.2010.09.184. [DOI] [PubMed] [Google Scholar]
- 134.Tivnan A, Orr WS, Gubala V, Nooney R, Williams DE, McDonagh C, Prenter S, Harvey H, Domingo-Fernandez R, Bray IM, Piskareva O, Ng CY, Lode HN, Davidoff AM, Stallings RL. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One. 2012;7(5):e38129. doi: 10.1371/journal.pone.0038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, Huang K, Zheng L, Tong Q. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2 alpha. Oncogene. 2014;33(3):387–97. doi: 10.1038/onc.2012.574. [DOI] [PubMed] [Google Scholar]
- 136.Tivnan A, Foley NH, Tracey L, Davidoff AM, Stallings RL. MicroRNA-184-mediated inhibition of tumour growth in an orthotopic murine model of neuroblastoma. Anticancer Res. 2010;30(11):4391–5. [PMC free article] [PubMed] [Google Scholar]
- 137.Chakrabarti M, Khandkar M, Banik NL, Ray SK. Alterations in expression of specific microRNAs by combination of 4-HPR and EGCG inhibited growth of human malignant neuroblastoma cells. Brain Res. 2012;1454:1–13. doi: 10.1016/j.brainres.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes & development. 2002;16(20):2699–712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Buechner J, Einvik C. N-myc and noncoding RNAs in neuroblastoma. Molecular cancer research : MCR. 2012;10(10):1243–53. doi: 10.1158/1541-7786.MCR-12-0244. [DOI] [PubMed] [Google Scholar]
- 140.Atmadibrata B, Liu PY, Sokolowski N, Zhang L, Wong M, Tee AE, Marshall GM, Liu T. The novel long noncoding RNA linc00467 promotes cell survival but is down-regulated by N-Myc. PloS one. 2014;9(2):e88112. doi: 10.1371/journal.pone.0088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sahu D, Hsu CL, Lin CC, Yang TW, Hsu WM, Ho SY, Juan HF, Huang HC. Co-expression analysis identifies long noncoding RNA SNHG1 as a novel predictor for event-free survival in neuroblastoma. Oncotarget. 2016;7(36):58022–58037. doi: 10.18632/oncotarget.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mazar J, Rosado A, Shelley J, Marchica J, Westmoreland TJ. The long non-coding RNA GAS5 differentially regulates cell cycle arrest and apoptosis through activation of BRCA1 and p53 in human neuroblastoma. Oncotarget. 2017;8(4):6589–6607. doi: 10.18632/oncotarget.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, Pasquier E, Haber M, Norris MD, Suzuki T, Marshall GM, Liu T. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014;5(7):1793–804. doi: 10.18632/oncotarget.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tee AE, Liu B, Song R, Li J, Pasquier E, Cheung BB, Jiang C, Marshall GM, Haber M, Norris MD, Fletcher JI, Dinger ME, Liu T. The long noncoding RNA MALAT1 promotes tumor-driven angiogenesis by up-regulating pro-angiogenic gene expression. Oncotarget. 2016;7(8):8663–75. doi: 10.18632/oncotarget.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bi S, Wang C, Li Y, Zhang W, Zhang J, Lv Z, Wang J. LncRNA-MALAT1-mediated Axl promotes cell invasion and migration in human neuroblastoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39(5) doi: 10.1177/1010428317699796. 1010428317699796. [DOI] [PubMed] [Google Scholar]
- 146.Chen L, Feng P, Zhu X, He S, Duan J, Zhou D. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. Journal of cellular and molecular medicine. 2016;20(11):2102–2110. doi: 10.1111/jcmm.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tang W, Dong K, Li K, Dong R, Zheng S. MEG3, HCN3 and linc01105 influence the proliferation and apoptosis of neuroblastoma cells via the HIF-1alpha and p53 pathways. Scientific reports. 2016;6:36268. doi: 10.1038/srep36268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Pandey GK, Kanduri C. Fighting Neuroblastomas with NBAT1. Oncoscience. 2015;2(2):79–80. doi: 10.18632/oncoscience.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Maris JM, Mosse YP, Bradfield JP, Hou C, Monni S, Scott RH, Asgharzadeh S, Attiyeh EF, Diskin SJ, Laudenslager M, Winter C, Cole KA, Glessner JT, Kim C, Frackelton EC, Casalunovo T, Eckert AW, Capasso M, Rappaport EF, McConville C, London WB, Seeger RC, Rahman N, Devoto M, Grant SF, Li H, Hakonarson H. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. The New England journal of medicine. 2008;358(24):2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Russell MR, Penikis A, Oldridge DA, Alvarez-Dominguez JR, McDaniel L, Diamond M, Padovan O, Raman P, Li Y, Wei JS, Zhang S, Gnanchandran J, Seeger R, Asgharzadeh S, Khan J, Diskin SJ, Maris JM, Cole KA. CASC15-S Is a Tumor Suppressor lncRNA at the 6p22 Neuroblastoma Susceptibility Locus. Cancer research. 2015;75(15):3155–66. doi: 10.1158/0008-5472.CAN-14-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yu M, Ohira M, Li Y, Niizuma H, Oo ML, Zhu Y, Ozaki T, Isogai E, Nakamura Y, Koda T, Oba S, Yu B, Nakagawara A. High expression of ncRAN, a novel non-coding RNA mapped to chromosome 17q25.1, is associated with poor prognosis in neuroblastoma. International journal of oncology. 2009;34(4):931–8. doi: 10.3892/ijo_00000219. [DOI] [PubMed] [Google Scholar]
- 152.Chalei V, Sansom SN, Kong L, Lee S, Montiel JF, Vance KW, Ponting CP. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. eLife. 2014;3:e04530. doi: 10.7554/eLife.04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, Feo S. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. Journal of cellular physiology. 2007;213(2):511–8. doi: 10.1002/jcp.21133. [DOI] [PubMed] [Google Scholar]
- 154.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12(3):215–29. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 155.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 156.Scaruffi P. The transcribed-ultraconserved regions: a novel class of long noncoding RNAs involved in cancer susceptibility. TheScientificWorldJournal. 2011;11:340–52. doi: 10.1100/tsw.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mestdagh P, Fredlund E, Pattyn F, Rihani A, Van Maerken T, Vermeulen J, Kumps C, Menten B, De Preter K, Schramm A, Schulte J, Noguera R, Schleiermacher G, Janoueix-Lerosey I, Laureys G, Powel R, Nittner D, Marine JC, Ringner M, Speleman F, Vandesompele J. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene. 2010;29(24):3583–92. doi: 10.1038/onc.2010.106. [DOI] [PubMed] [Google Scholar]
- 158.Scaruffi P, Stigliani S, Moretti S, Coco S, De Vecchi C, Valdora F, Garaventa A, Bonassi S, Tonini GP. Transcribed-Ultra Conserved Region expression is associated with outcome in high-risk neuroblastoma. BMC cancer. 2009;9:441. doi: 10.1186/1471-2407-9-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ishizuka A, Hasegawa Y, Ishida K, Yanaka K, Nakagawa S. Formation of nuclear bodies by the lncRNA Gomafu-associating proteins Celf3 and SF1. Genes to cells : devoted to molecular & cellular mechanisms. 2014;19(9):704–21. doi: 10.1111/gtc.12169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wada M, Seeger RC, Mizoguchi H, Koeffler HP. Maintenance of normal imprinting of H19 and IGF2 genes in neuroblastoma. Cancer research. 1995;55(15):3386–8. [PubMed] [Google Scholar]
- 161.Barnhill LM, Williams RT, Cohen O, Kim Y, Batova A, Mielke JA, Messer K, Pu M, Bao L, Yu AL, Diccianni MB. High expression of CAI2, a 9p21-embedded long noncoding RNA, contributes to advanced-stage neuroblastoma. Cancer research. 2014;74(14):3753–63. doi: 10.1158/0008-5472.CAN-13-3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu PY, Erriquez D, Marshall GM, Tee AE, Polly P, Wong M, Liu B, Bell JL, Zhang XD, Milazzo G, Cheung BB, Fox A, Swarbrick A, Huttelmaier S, Kavallaris M, Perini G, Mattick JS, Dinger ME, Liu T. Effects of a novel long noncoding RNA, lncUSMycN, on N-Myc expression and neuroblastoma progression. J Natl Cancer Inst. 2014;106(7) doi: 10.1093/jnci/dju113. [DOI] [PubMed] [Google Scholar]
- 163.Liu PY, Atmadibrata B, Mondal S, Tee AE, Liu T. NCYM is upregulated by lncUSMycN and modulates N-Myc expression. International journal of oncology. 2016;49(6):2464–2470. doi: 10.3892/ijo.2016.3730. [DOI] [PubMed] [Google Scholar]
- 164.Yarmishyn AA, Batagov AO, Tan JZ, Sundaram GM, Sampath P, Kuznetsov VA, Kurochkin IV. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC genomics. 2014;15(Suppl 9):S7. doi: 10.1186/1471-2164-15-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang S, Zhang X, Guo Y, Rong H, Liu T. The long noncoding RNA HOTAIR promotes Parkinson's disease by upregulating LRRK2 expression. Oncotarget. 2017;8(15):24449–24456. doi: 10.18632/oncotarget.15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP. The long non-coding RNA Paupar regulates the expression of both local and distal genes. The EMBO journal. 2014;33(4):296–311. doi: 10.1002/embj.201386225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Le Novere N. The long journey to a Systems Biology of neuronal function. BMC Syst Biol. 2007;1:28. doi: 10.1186/1752-0509-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–44. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Scianna M, Munaron L. Computational Approaches for Translational Oncology: Concepts and Patents. Recent Pat Anticancer Drug Discov. 2016;11(4):384–392. doi: 10.2174/1574892811666161003111543. [DOI] [PubMed] [Google Scholar]
- 170.Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, Kamburov A, Ghiassian SD, Yang X, Ghamsari L, Balcha D, Begg BE, Braun P, Brehme M, Broly MP, Carvunis AR, Convery-Zupan D, Corominas R, Coulombe-Huntington J, Dann E, Dreze M, Dricot A, Fan C, Franzosa E, Gebreab F, Gutierrez BJ, Hardy MF, Jin M, Kang S, Kiros R, Lin GN, Luck K, MacWilliams A, Menche J, Murray RR, Palagi A, Poulin MM, Rambout X, Rasla J, Reichert P, Romero V, Ruyssinck E, Sahalie JM, Scholz A, Shah AA, Sharma A, Shen Y, Spirohn K, Tam S, Tejeda AO, Trigg SA, Twizere JC, Vega K, Walsh J, Cusick ME, Xia Y, Barabasi AL, Iakoucheva LM, Aloy P, De Las Rivas J, Tavernier J, Calderwood MA, Hill DE, Hao T, Roth FP, Vidal M. A proteome-scale map of the human interactome network. Cell. 2014;159(5):1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18(3):507–22. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 172.Szklarczyk D, Jensen LJ. Protein-protein interaction databases. Methods Mol Biol. 2015;1278:39–56. doi: 10.1007/978-1-4939-2425-7_3. [DOI] [PubMed] [Google Scholar]
- 173.Garcia-Alcalde F, Garcia-Lopez F, Dopazo J, Conesa A. Paintomics: a web based tool for the joint visualization of transcriptomics and metabolomics data. Bioinformatics. 2011;27(1):137–9. doi: 10.1093/bioinformatics/btq594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Marbach D, Costello JC, Kuffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, Consortium D, Kellis M, Collins JJ, Stolovitzky G. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9(8):796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Margolin AA, Nemenman I, Basso K, Wiggins C, Stolovitzky G, Dalla Favera R, Califano A. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mestdagh P, Bostrom AK, Impens F, Fredlund E, Van Peer G, De Antonellis P, von Stedingk K, Ghesquiere B, Schulte S, Dews M, Thomas-Tikhonenko A, Schulte JH, Zollo M, Schramm A, Gevaert K, Axelson H, Speleman F, Vandesompele J. The miR-17-92 microRNA cluster regulates multiple components of the TGF-beta pathway in neuroblastoma. Molecular cell. 2010;40(5):762–73. doi: 10.1016/j.molcel.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lynch J, Fay J, Meehan M, Bryan K, Watters KM, Murphy DM, Stallings RL. MiRNA-335 suppresses neuroblastoma cell invasiveness by direct targeting of multiple genes from the non-canonical TGF-beta signalling pathway. Carcinogenesis. 2012;33(5):976–85. doi: 10.1093/carcin/bgs114. [DOI] [PMC free article] [PubMed] [Google Scholar]