Abstract
Background
Genetic studies of neuropsychiatric disease strongly suggest overlap in liability. There are growing efforts to characterize these diseases dimensionally rather than categorically, but the extent to which such dimensional models correspond to biology is unknown.
Methods
We applied a newly-developed natural language processing (NLP) method to extract five symptom dimensions, based on the NIMH Research Domain Criteria (RDoC) definitions, from narrative hospital discharge notes in a large biobank. We conducted a genome-wide association study to examine whether common variants were associated with each of these dimensions as quantitative traits.
Results
Among 4,687 individuals, loci in three of five domains exceeded a genome-wide threshold for statistical significance. These included a locus spanning the neocortical development genes RFPL3 and RFPL3S, for arousal (p=2.29e–8), and one spanning the FPR3 gene, for cognition (p=3.22e–8).
Discussion
NLP identifies dimensional phenotypes that may facilitate discovery of common genetic variation relevant to psychopathology.
Keywords: genetic, genomic, valence, arousal, social, transdiagnostic
Introduction
Family studies of psychiatric illnesses demonstrated decades ago the overlap in risk for these disorders, a finding now confirmed by genome-wide association.(1–3) Such overlap highlights the limitations of a nosologic system focused on categories of symptoms rather than dimensions. For this reason, recent initiatives emphasize the utility of identifying symptom domains that may better correspond to underlying neurobiology.(4, 5)
The rise of biobanks embedded in health care systems or national registries provides an opportunity to investigate the impact of genomic variation in a less biased fashion that traditional disease case-control designs. However, such biobanks typically capture primarily coded clinical data - i.e., categorical diagnoses. We have recently developed multiple methods to examine narrative clinical notes to extract symptom dimensions as a means of augmenting this coded data.(6, 30)†
We hypothesized that symptom dimensions based on expert-curated terms capturing NIMH Research Domain Criteria (RDoC) domains would be associated with common genomic variation and could thereby implicate novel sets of genes related to psychopathology. As proof of concept, we therefore applied a newly-described (30) natural language processing (NLP) method for extracting dimensional phenotypes to hospital discharge summaries drawn from the genomic biobank of an academic medical center, and used standard genome-wide association to investigate these novel phenotypes as quantitative traits.
Methods and Materials
Overview and Data Set Generation
We drew on three waves of participants in the Partners Biobank from the Brigham and Women's Hospital network as well as the Massachusetts General Hospital network, representing the first ~15,000 individuals genotyped as part of the Partners HealthCare Biobank initiative.(7) Narrative discharge summaries were extracted from the longitudinal electronic health record (EHR) of the Massachusetts General Hospital (MGH). We included any individuals age 18 or older with at least one hospitalization between 2010 and 2015.
A datamart containing all clinical data was generated with the i2b2 server software (i2b2 v1.6, Boston, MA, USA), a computational framework for managing human health data.(8–10) The Partners Institutional Review Board approved the both the study protocol, and the release of biobank data, which is collected after acquiring written informed consent from participants and explicitly allows identifiable data to be shared with qualified investigators.
Study Design and Analysis
Primary analyses utilized a cohort design with all patients admitted for any reason during the time period noted above. Discharge documentation was used to estimate dimensional psychopathology scores for one encounter per individual; where an individual was hospitalized on multiple occasions during the study period, a single hospitalization was selected at random to minimize bias resulting from other means of ascertainment. The derivation of dimensional psychopathology has been previously described †; in brief, it began with a set of seed terms for each of the five NIMH RDoC definitions drawn from NIMH workgroup statements, then expanded these term lists to include synonyms.(11) This second expansion step is important as it reduces potential bias introduced by a given specialty or set of providers who may use specific terminology to characterize symptoms, yielding a broader set of terms that should better generalize across providers and hospitals. Each note is assigned a score corresponding to a simple count of term appearance. We have developed simple code to facilitate dimension extraction in other data sets; please see (30) for this code.
Genotyping and quality control
DNA was extracted from buffy coat and genotyping was done using three versions of the Illumina Multi-Ethnic Global (MEG) array (MEGA n=4,927, MEGA EX n=5,353, and MEG n=4,784; mappable variants available for each were 1,411,334; 1,710,339; and 1,747,639 respectively). These common variant arrays all incorporate content from the 1000 Genomes Project Phase 3 (1000G Phase 3). SNP coordinates were remapped based on the TopGenomicSeq provided from IlluminaA; all rsID's correspond to build 142 of dbSNP. To determine the forward strand of the SNP, we aligned both SNP sequences (alleles A and B) to hg19 using BLAT with default parameters set by UCSC Genome Browser.(12)
Each cohort was cleaned, imputed, and analyzed separately to avoid batch effects. In each batch we included subjects with genotyping call rates exceeding 99%; no related individuals based on identity by descent (IBD) were included.(13) From these individuals, any genotyped SNP with call rate of at least 95% and Hardy-Weinberg equilibrium P value <1×10−6 was included. Imputation used the Michigan Imputation Server implementing Minimac3.(14–16) Imputation used all population subsets from 1000G Phase 3 v5 as reference panel; haplotype phasing was performed using SHAPEIT.(17)
For each batch, we applied principal components analysis (PCA) of a linkage-disequilibrium-pruned set of genotyped SNPs to characterize population structure, based on EIGENSTRAT as implemented in PLINK v1.9.(18) We then plotted these components with superimposition of HapMap samples to confirm location of Northern European individuals. The present analysis included only individuals of Northern European genomic ancestry in order to minimize risk for confounding by ancestry (i.e., population stratification), and because power to detect association in other ancestry groups would be limited.(19–21)
Analysis
We examined single-locus associations in each batch, then combined in inverse-variance-weighted fixed-effects meta-analysis. In all analyses, only bi-allelic SNPs with minor allele frequencies of at least 1% in all batches were retained. Tests for association used linear regression assuming an additive allelic effect, and examined each of the five dimensional measures as a quantitative trait, with adjustment for the first 10 principal components a priori. (In prior work analyses incorporating five or 20 components did not yield meaningfully different results.) Association results are presented in terms of independent loci after pruning using the clump command in PLINK 1.9, with a 250kb window and r2=0.2. Locus plots were generated using locuszoom.(18, 22)
Reported p-values are not adjusted for lambda or linkage disequilibrium (LD) scores; in prior work adjustment for lambda-1000 or LD score regression intercept did not meaningfully change relative results. Lambdas range from 0.998 to 1.003.(23)
Results
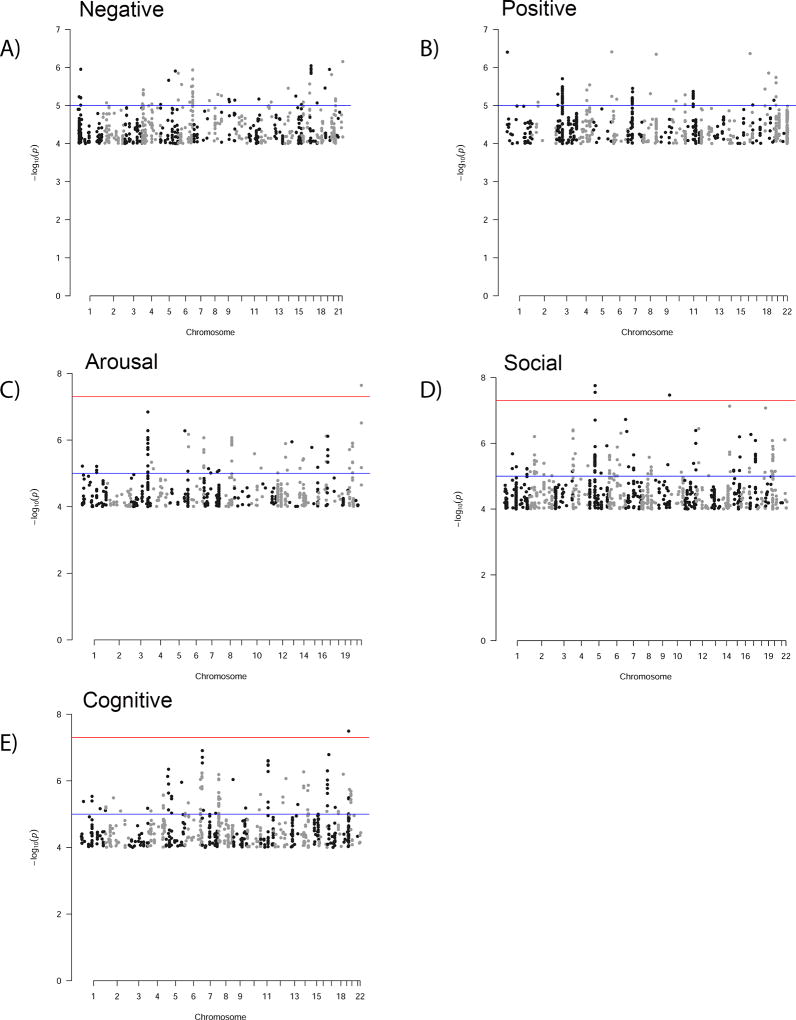
In total, we examined 4,687 individuals of Northern European ancestry across the 3 batches (wave 1, 1589; wave 2, 1547; wave 3, 1551), with meta-analysis of 893,900 SNPs with MAF of 0.01 or greater. The cohorts were 2,363/4,687 female (50.4%) and mean age was 64.3 (SD 14.9) years. Figure 1 (panels a–e) illustrates Manhattan plots for each of the five dimensional phenotypes (for Q-Q plots, see Supplemental Figure 1).
Figure 1.
Manhattan plots from genome-wide association for each of the five dimensions of psychopathology.
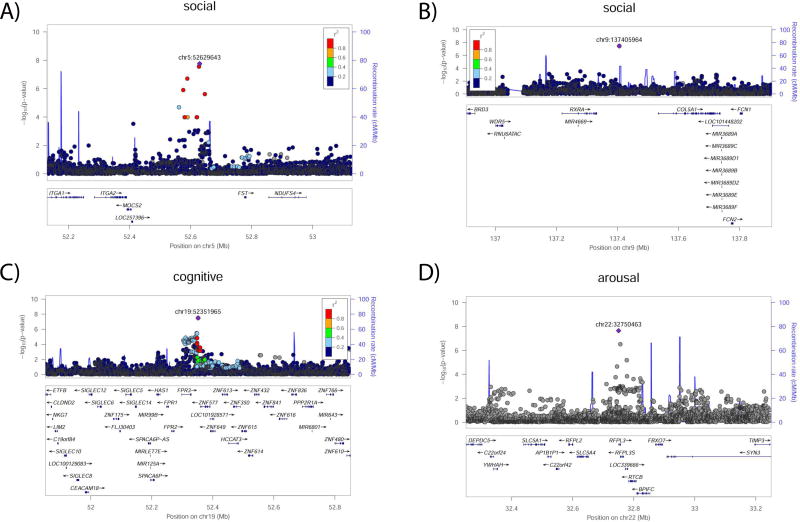
For each of the dimensions, the 10 independent loci with strongest evidence of association are described in Table 1. Overall, one locus was associated with arousal, two with social, and one with cognition at a standard genome-wide significance threshold (p<5×10−8); these four regions are depicted in Figure 2. Notably, for arousal, the associated locus spans Ret Finger Protein-Like -3 and -3S (RFPL3 and RFPL3S); this family of proteins has been suggested to be important in primate neocortical evolution.(24) For cognition, the associated locus spans Formyl Peptide Receptor 3 (FPR3), a chemoattractant (15623572) suggested to be relevant in immune response in Alzheimer's disease.(25)
Table 1.
Independent loci with strongest evidence of association for each dimension of psychopathology.
| CHR | SNP | P-value | N SNPs | Locus span | Locus size (kb) | Genes in locus | A1 | A2 | MAF | |
|---|---|---|---|---|---|---|---|---|---|---|
| Negative | ||||||||||
| 22 | 22:32750463 | 7.00E-07 | 8 | chr22:32738156..32800705 | 62.55 | [LOC339666,RFPL3,RFPL3S,RTCB] | A | C | 0.013 | |
| 17 | 17:14495883 | 9.03E-07 | 12 | chr17:14382314..14497857 | 115.544 | [] | A | C | 0.012 | |
| 1 | 1:27963259 | 1.12E-06 | 23 | chr1:27963259..27981314 | 18.056 | [] | T | A | 0.011 | |
| 19 | 19:51371390 | 1.13E-06 | 1 | chr19:51371390..51371390 | 0.001 | [] | T | C | 0.033 | |
| 6 | 6:155563548 | 1.16E-06 | 18 | chr6:155562206..155733894 | 171.689 | [CLDN20,NOX3,TFB1M,TIAM2] | G | A | 0.014 | |
| 5 | 5:155373160 | 1.24E-06 | 4 | chr5:155221281..155397446 | 176.166 | [] | T | C | 0.015 | |
| 6 | 6:5821150 | 1.42E-06 | 3 | chr6:5815134..5851189 | 36.056 | [] | G | T | 0.014 | |
| 20 | 20:15042429 | 1.54E-06 | 1 | chr20:15042429..15042429 | 0.001 | [MACROD2] | C | T | 0.013 | |
| 5 | 5:86039654 | 2.18E-06 | 4 | chr5:86037590..86040114 | 2.525 | [] | G | T | 0.010 | |
| 16 | 16:83664928 | 2.71E-06 | 38 | chr16:83656037..83753512 | 97.476 | [CDH13] | A | T | 0.084 | |
|
| ||||||||||
| Positive | ||||||||||
| 6 | 6:5821150 | 3.91E-07 | 3 | chr6:5815134..5851189 | 36.056 | [] | G | T | 0.014 | |
| 1 | 1:6039258 | 3.97E-07 | 3 | chr1:5953811..6039453 | 85.643 | [NPHP4] | T | C | 0.012 | |
| 16 | 16:56251428 | 4.32E-07 | 2 | chr16:56095547..56251428 | 155.882 | [DKFZP434H168,GNAO1,LOC283856] | G | A | 0.013 | |
| 8 | 8:132532229 | 4.51E-07 | 121 | chr8:132404887..132532936 | 128.05 | [] | A | G | 0.157 | |
| 18 | 18:77374268 | 1.40E-06 | 1 | chr18:77374268..77374268 | 0.001 | [] | G | C | 0.011 | |
| 20 | 20:15696084 | 1.83E-06 | 3 | chr20:15690854..15696084 | 5.231 | [MACROD2] | C | A | 0.093 | |
| 3 | 3:54508115 | 1.98E-06 | 71 | chr3:54488508..54575770 | 87.263 | [CACNA2D3] | T | C | 0.338 | |
| 20 | 20:16560345 | 2.54E-06 | 51 | chr20:16509803..16605627 | 95.825 | [KIF16B] | C | T | 0.167 | |
| 4 | 4:127370341 | 2.88E-06 | 102 | chr4:127360862..127402924 | 42.063 | [] | T | C | 0.100 | |
| 7 | 7:47324136 | 3.55E-06 | 3 | chr7:47324136..47328060 | 3.925 | [TNS3] | T | C | 0.081 | |
|
| ||||||||||
| Arousal | ||||||||||
| 22 | 22:32750463 | 2.29E-08 | 8 | chr22:32738156..32800705 | 62.55 | [LOC339666,RFPL3,RFPL3S,RTCB] | A | C | 0.013 | |
| 3 | 3:167741670 | 1.44E-07 | 81 | chr3:167544555..167741670 | 197.116 | [GOLIM4,LOC646168] | A | G | 0.016 | |
| 5 | 5:150327474 | 5.28E-07 | 4 | chr5:150115979..150327474 | 211.496 | [DCTN4,IRGM,SMIM3,ZNF300,ZNF300P1] | T | G | 0.057 | |
| 6 | 6:5821150 | 6.75E-07 | 2 | chr6:5821150..5851189 | 30.04 | [] | G | T | 0.014 | |
| 16 | 16:83664928 | 7.59E-07 | 49 | chr16:83656037..83753512 | 97.476 | [CDH13] | A | T | 0.084 | |
| 17 | 17:14496077 | 7.69E-07 | 11 | chr17:14495883..14497857 | 1.975 | [] | C | T | 0.012 | |
| 8 | 8:118469770 | 8.49E-07 | 86 | chr8:118379461..118588575 | 209.115 | [MED30] | C | T | 0.371 | |
| 6 | 6:155563548 | 8.55E-07 | 20 | chr6:155562206..155733894 | 171.689 | [CLDN20,NOX3,TFB1M,TIAM2] | G | A | 0.014 | |
| 13 | 13:43496853 | 1.13E-06 | 1 | chr13:43496853..43496853 | 0.001 | [EPSTI1] | C | T | 0.014 | |
| 20 | 20:45375674 | 1.24E-06 | 17 | chr20:45315786..45385268 | 69.483 | [SLC2A10,TP53RK] | T | A | 0.063 | |
|
| ||||||||||
| Social | ||||||||||
| 5 | 5:52629643 | 1.77E-08 | 22 | chr5:52564100..52661007 | 96.908 | [] | A | G | 0.012 | |
| 9 | 9:137405964 | 3.42E-08 | 3 | chr9:137341500..137405964 | 64.465 | [] | T | C | 0.021 | |
| 14 | 14:97095154 | 7.45E-08 | 4 | chr14:97087772..97117785 | 30.014 | [] | A | G | 0.043 | |
| 18 | 18:77374268 | 8.48E-08 | 5 | chr18:77365764..77396240 | 30.477 | [] | G | C | 0.011 | |
| 7 | 7:2472517 | 1.89E-07 | 2 | chr7:2230076..2472517 | 242.442 | [CHST12,EIF3B,FTSJ2,MAD1L1, MIR6836,NUDT1,SNX8] | T | C | 0.023 | |
| 12 | 12:27167220 | 3.60E-07 | 8 | chr12:27145587..27333632 | 188.046 | [C12orf71,MED21,TM7SF3] | A | G | 0.014 | |
| 4 | 4:2483900 | 3.94E-07 | 7 | chr4:2483900..2732557 | 248.658 | [FAM193A,RNF4] | A | C | 0.013 | |
| 11 | 11:125064877 | 4.10E-07 | 65 | chr11:125052718..125110079 | 57.362 | [PKNOX2] | T | A | 0.127 | |
| 7 | 7:14309510 | 4.37E-07 | 3 | chr7:14294006..14309510 | 15.505 | [DGKB] | T | C | 0.014 | |
| 6 | 6:125802803 | 4.93E-07 | 1 | chr6:125802803..125802803 | 0.001 | [] | A | G | 0.013 | |
|
| ||||||||||
| Cognitive | ||||||||||
| 19 | 19:52351965 | 3.22E-08 | 94 | chr19:52306547..52377699 | 71.153 | [FPR3,ZNF577] | T | C | 0.321 | |
| 7 | 7:3627391 | 1.24E-07 | 3 | chr7:3610381..3662960 | 52.58 | [SDK1] | G | A | 0.019 | |
| 17 | 17:13683929 | 1.63E-07 | 3 | chr17:13680505..13806459 | 125.955 | [] | A | G | 0.014 | |
| 11 | 11:73586112 | 2.49E-07 | 211 | chr11:73340835..73672187 | 331.353 | [COA4,DNAJB13,MRPL48,PAAF1, PLEKHB1,RAB6A] | G | C | 0.303 | |
| 5 | 5:22528391 | 4.49E-07 | 5 | chr5:22365713..22706775 | 341.063 | [CDH12] | T | G | 0.018 | |
| 17 | 17:167312 | 5.02E-07 | 11 | chr17:149460..172591 | 23.132 | [RPH3AL] | T | C | 0.023 | |
| 14 | 14:57183567 | 5.40E-07 | 6 | chr14:57182182..57194970 | 12.789 | [] | C | A | 0.018 | |
| 6 | 6:169616423 | 5.79E-07 | 31 | chr6:169596595..169622263 | 25.669 | [THBS2] | T | C | 0.257 | |
| 18 | 18:77374268 | 6.33E-07 | 1 | chr18:77374268..77374268 | 0.001 | [] | G | C | 0.011 | |
| 8 | 8:6034653 | 6.48E-07 | 43 | chr8:6021491..6061234 | 39.744 | [] | A | G | 0.363 | |
N SNPs, number of SNPs in LD block with nominal p<0.01; see text for details
MAF, minor allele frequency
Figure 2.
Region plots for four loci with genome-wide significance.
Discussion
In this analysis of 4,687 individuals drawn from a biobank spanning academic medical centers, we identified four loci associated with dimensional psychopathology at a standard genome-wide threshold based on NLP of narrative hospital discharge notes. Two of these span genes associated with neurodevelopment (RFPL3) or neurodegeneration (PFR3). While both of these are known to be brain-expressed, neither has previously been strongly associated with neuropsychiatric disease, suggesting the potential utility of the approach we describe in understanding brain function in a manner unbiased by traditional nosology.
While not achieving a genome-wide threshold for significance, we also note the observed association between the calcium channel subunit CACNA2D3 and positive valence. This locus has previously been associated with pain sensitivity, which may impact reward responsiveness, suggesting convergent validity (i.e., assay sensitivity).(26) This family of subunits represents the target for multiple anticonvulsants used to treat neuropathic pain, and has recently been shown to regulate accumulation of voltage gated calcium channels as well as exocytosis at the synapse.(27)
While these loci are promising as candidates for follow-up study, multiple limitations in this proof-of-concept study should be considered. First, while we exceed a standard threshold for genome-wide studies, replication will increase confidence in these results. (At a more stringent experiment-wide threshold, based upon correlation between these domains, one could also argue that a threshold of 2×10−8 would be appropriate). We elected to meta-analyze all data available to us, rather than holding out a replication set, and present these results in the hope that they will encourage other hospital-linked biobanks to consider our approach. Second, as with any common-variant study, none of these variants can be considered causal and biological studies will be required to characterize their effect.
More broadly, it is entirely possible - indeed, likely - that other dimensional features or extraction methods, as well as incorporation of other data types, would lead to identification of other loci. We adopted a new method for identifying dimensional psychopathology from narrative clinical notes based on seed terms extracted from RDoC workgroup statements, which we have recently described in more detail along with initial validation.(30) These scores do not yet address subdomains, sensitivity likely varies by domain, and indeed as with RDoC itself the presence of terms loading on a given domain does not necessarily represent psychopathology, and may capture normal or subsyndromal variation. We note that the present study represents an example of transfer learning: a model trained in one type of cohort (psychiatric hospitalizations) is applied to distinguish features of another (all-cause hospitalizations), but further investigations of portability will important. In particular, this approach complements rather than replacing analysis of more traditional curated phenotypes.(28, 29) Beyond investigating other strategies for concept extraction, it will be valuable to understand the extent to which incorporating other types of notes, or integrating these data with coded clinical data, improve identification of dimensions of psychopathology. (For further discussion of general methodologic considerations, please also see McCoy et al.(30))
With these caveats in mind, our results suggest an approach to identifying genes associated with psychopathology beyond traditional diagnostic categories, and demonstrate the feasibility and potential utility of this broad class of approaches, aiming to be both transparent and portable. Narrative clinical notes may contain a wealth of clinical detail relevant to developing dimensional representations of brain diseases. With increasing availability of biobanks and registries as a resource for genomic discovery and translation, NLP represents a way to amplify their utility for investigating complex phenotypes which avoids the constraint of traditional psychiatric nosology.
Supplementary Material
Acknowledgments
This study was funded by the National Human Genome Research Institute, the National Institute of Mental Health, and the Broad Institute. The sponsor had no role in study design, writing of the report, or data collection, analysis, or interpretation. The corresponding and senior authors had full access to all data and made the decision to submit for publication. The authors wish to acknowledge the participants and administrators of the Partners HealthCare Biobank for their contribution to this work.
Financial Disclosures:
RHP reports grants from the National Human Genome Research Institute and National Institute of Mental Health; serves on the scientific advisory board for Perfect Health, Genomind, and Psy Therapeutics; and consults to RID Ventures. THM reports grants from the Broad Institute and Brain and Behavior Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The other authors report no biomedical financial interests or potential conflicts of interest.
Please refer to our other submission to your journal, “High throughput phenotyping for dimensional psychopathology in electronic health records”.
MEGA_Consortium_v2_15070954_A2.csv
References and Notes
- 1.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cross-Disorder Group of the Psychiatric Genomics Consortium. Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilman SE, Ni MY, Dunn EC, Breslau J, McLaughlin KA, Smoller JW, et al. Contributions of the social environment to first-onset and recurrent mania. Mol Psychiatry. 2015;20:329–336. doi: 10.1038/mp.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167:748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 5.Sanislow CA, Pine DS, Quinn KJ, Kozak MJ, Garvey MA, Heinssen RK, et al. Developing constructs for psychopathology research: research domain criteria. J Abnorm Psychol. 2010;119:631–639. doi: 10.1037/a0020909. [DOI] [PubMed] [Google Scholar]
- 6.McCoy TH, Castro VM, Rosenfield HR, Cagan A, Kohane IS, Perlis RH. A clinical perspective on the relevance of research domain criteria in electronic health records. Am J Psychiatry. 2015;172:316–320. doi: 10.1176/appi.ajp.2014.14091177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gainer VS, Cagan A, Castro VM, Duey S, Ghosh B, Goodson AP, et al. The Biobank Portal for Partners Personalized Medicine: A Query Tool for Working with Consented Biobank Samples, Genotypes, and Phenotypes Using i2b2. Journal of personalized medicine. 2016;6:11. doi: 10.3390/jpm6010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murphy SN, Mendis M, Hackett K, Kuttan R, Pan W, Phillips LC, et al. Architecture of the open-source clinical research chart from Informatics for Integrating Biology and the Bedside. AMIA Annu Symp Proc. 2007:548–552. [PMC free article] [PubMed] [Google Scholar]
- 9.Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19:1675–1681. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17:124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Institute of Mental Health. RDoC Matrix. National Institute of Mental Health; 2017. [Google Scholar]
- 12.Wang C, Ward ME, Chen R, Liu K, Tracy TE, Chen X, et al. Scalable Production of iPSC-Derived Human Neurons to Identify Tau-Lowering Compounds by High-Content Screening. Stem cell reports. 2017;9:1221–1233. doi: 10.1016/j.stemcr.2017.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henn BM, Hon L, Macpherson JM, Eriksson N, Saxonov S, Pe'er I, et al. Cryptic distant relatives are common in both isolated and cosmopolitan genetic samples. PLoS One. 2012;7:e34267. doi: 10.1371/journal.pone.0034267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sala M, Lazzaretti M, De Vidovich G, Caverzasi E, Barale F, d'Allio G, et al. Electrophysiological changes of cardiac function during antidepressant treatment. Ther Adv Cardiovasc Dis. 2009;3:29–43. doi: 10.1177/1753944708096282. [DOI] [PubMed] [Google Scholar]
- 16.Wenger TL, Cohn JB, Bustrack J. Comparison of the effects of bupropion and amitriptyline on cardiac conduction in depressed patients. J Clin Psychiatry. 1983;44:174–175. [PubMed] [Google Scholar]
- 17.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nature methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S, Chang C. PLINK 1.90 beta. Cog Genomics 2013 [Google Scholar]
- 19.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 20.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:1. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nature genetics. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnefont J, Nikolaev SI, Perrier AL, Guo S, Cartier L, Sorce S, et al. Evolutionary forces shape the human RFPL1,2,3 genes toward a role in neocortex development. American journal of human genetics. 2008;83:208–218. doi: 10.1016/j.ajhg.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iribarren P, Zhou Y, Hu J, Le Y, Wang JM. Role of formyl peptide receptor-like 1 (FPRL1/FPR2) in mononuclear phagocyte responses in Alzheimer disease. Immunologic research. 2005;31:165–176. doi: 10.1385/IR:31:3:165. [DOI] [PubMed] [Google Scholar]
- 26.Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, et al. A genome-wide Drosophila screen for heat nociception identifies alpha2delta3 as an evolutionarily conserved pain gene. Cell. 2010;143:628–638. doi: 10.1016/j.cell.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoppa MB, Lana B, Margas W, Dolphin AC, Ryan TA. alpha2delta expression sets presynaptic calcium channel abundance and release probability. Nature. 2012;486:122–125. doi: 10.1038/nature11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Dushlaine C, Ripke S, Ruderfer DM, Hamilton SP, Fava M, Iosifescu DV, et al. Rare copy number variation in treatment-resistant major depressive disorder. Biol Psychiatry. 2014;76:536–541. doi: 10.1016/j.biopsych.2013.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castro VM, Minnier J, Murphy SN, Kohane I, Churchill SE, Gainer V, et al. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am J Psychiatry. 2015;172:363–372. doi: 10.1176/appi.ajp.2014.14030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy TH, Jr, Yu S, Hart KL, Castro VM, Brown HE, Rosenquist JN, et al. High throughput phenotyping for dimensional psychopathology in electronic health records. Biol Psychiatry. (in press) doi: 10.1016/j.biopsych.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.