Abstract
High levels of fetal hemoglobin (HbF) reduce sickle cell anemia (SCA) morbidity and mortality. HbF levels vary considerably and there is a strong genetic component that influences HbF production. Genetic polymorphisms at three quantitative trait loci (QTL): Xmn1-HBG2, HMIP-2 and BCL11A, have been shown to influence HbF levels and disease severity in SCA. Hydroxyurea (HU) is a drug that increases HbF. We investigated the influence of single nucleotide polymorphisms (SNPs) at the Xmn1-HBG2 (rs7482144); BCL11A (rs1427407, rs4671393 and rs11886868); and HMIP-2 (rs9399137 and rs9402686) loci on baseline and HU-induced HbF levels in 111 HbSS patients. We found that both BCL11A and HMIP-2 were associated with increased endogenous levels of HbF. Interestingly, we also found that BCL11A was associated with higher induction of HbF with HU. This effect was independent of the effect of BCL11A on baseline HbF levels. Additional studies will be needed to validate these findings and explain the ample inter-individual variations in HbF levels at baseline and HU-induced in patients with SCA.
Keywords: Sickle cell anemia, Single nucleotide polymorphism, BCL11A, HMIP-2
1. Introduction
Although sickle cell anemia is a monogenic Mendelian disorder caused by a single point mutation, the clinical manifestations of the disease include a striking phenotypic heterogeneity. The interplay of many genetic and environmental factors results in a remarkable diversity of disease severity. Increased levels of fetal hemoglobin (HbF) are known to reduce SCA morbidity and mortality due to the ability of HbF to inhibit sickle Hb (HbS) polymerization and overall reduction in the mean corpuscular HbS concentration [1,2].
Genome wide association studies have identified 3 major quantitative trait loci (QTL) with robust and reproducible association with levels of HbF. These loci are: the Xmn1-HBG2 polymorphism on chromosome 11p15; the HMIP-2 intergenic region on chromosome 6q23; and the BCL11A locus on chromosome 2p16 [3]. These three QTL account for approximately 20–50% of the variation in HbF levels in patients with sickle cell anemia (SCA), β-thalassemia, and even in healthy adults [4–9]. The drug hydroxyurea (HU) has been shown to be an effective agent to stimulate increased levels of HbF in patients with SCD. There is limited information available whether the three main QTL associated with endogenous HbF levels also affect the induced levels of HbF in SCA patients treated with HU.
Although HU reduces morbidity and mortality in adults and pediatric patients with SCA, almost one third of patients do not respond with a clinically beneficial increase in HbF. This suggests there is an underlying spectrum of quantitative and/or qualitative response to HU induced HbF in these patients [10]. Elucidation of specific genetic modifiers associated with HU induction of HbF may detect reasons for this phenotypic variability and avoid unnecessary exposure to this toxic drug for patients that are predicted to not respond well to the drug in advance.
2. Methods
We enrolled patients with SCA (SS) who received regular treatment with HU for at least 6 months at the Sickle Cell Center of the Hospital de Clínicas de Porto Alegre, Brazil. Patients were excluded if they were being treated with any other drugs that stimulate the synthesis of HbF (e.g. erythropoietin) or if they had received blood transfusion within 3 months prior to the study. The study was approved by the local Institutional Review Board under the number 100585 and a written informed consent was signed by each patient or their parents.
Of 121 patients enrolled, 111 (56% female) fulfilled the eligibility criteria: age N 3 years old and been on HU therapy for at least 6 months. The patient ages ranged from 4 to 54 years of age (mean 21 ± 14). Maximum tolerated doses of HU ranged from 8.6–42.8 mg/kg/day (mean 23 ± 7.6 mg/kg/day), duration of treatment with HU ranged from 6 to 254 months (mean 102 ± 67 months). HbF was measured by capillary electrophoresis using Sebia CAPILLARYS 2 Flex Piercing™ equipment (Cap 2FP; Sebia, Lisses, France). We obtained HbF measurements for patients at commencement of HU treatment (BL HbF) and after each patient had achieved their maximum tolerated dose (MTD HbF). We calculated the change in HbF for each patient from baseline to MTD (delta HbF).
Six widely-known and validated genetic variants were selected from the 3 main HbF modifier loci: locus Xmn1-HBG2 (rs7482144); rs1427407, rs4671393 and rs11886868 at the BCL11A; rs9399137 and rs9402686 at HMIP-2 [4–9]. These variants have been genotyped previously and we performed comparable genotyping methods using Taq-Man™ procedure (Applied Biosystems, Foster City, CA).
2.1. Statistical analysis
The Shapiro-Wilk test was applied to check for normal distribution of the data and the Bartlett’s test to check for equal variances across samples. None of the three HbF phenotypes passed tests for normality; the Box-Cox method [T(Y) = (Yˆlambda − 1)/lambda] was used to transform the baseline HbF (lambda = −0.01), MTD HbF (lambda = 0.67), and delta HbF (lambda = 0.46) data to allow linear regression analysis.
Differences in quantitative data were analyzed with Student’s t-test and the values were given as mean ± standard deviation (SD). Non parametric values were compared with Wilcoxon signed-rank test. Correlations (R) between variables were tested using Pearson (parametric). The analysis was performed using the software R version 3.2.2 (www.r-project.org) with the packages “genetics” version 1.3.8.1 and “haplo.stats” version 1.7.6. Linear regression was performed to analyze the association of the outcome with the BCL11A and HMIP-2 genotypes (multilevel variables). The regression model was fitted at a genotype level assuming an additive model and corrected for age and sex. For analysis of MTD HbF and delta HbF, the model also included covariate correction for ANC levels at MTD. The strength of the correlation is reported as a beta coefficient value that is the average change in %HbF per allele copy of the respective SNP. A P-value < 0.05 was considered statistically significant.
3. Results
Similar to previous reports of HU treatment, we observed statistically significant improvements in several hematological parameters following HU exposure (Table 1) in the 111 patients analyzed. We did not observe any differences in HbF between males (N = 49) and females (N = 62) at either baseline (6.0 ± 5.1 vs. 7.2 ± 6.4, P = 0.29) or MTD (18.3 ± 8.1 vs. 18.0 ± 9.4, P = 0.87). There was a correlation of baseline HbF with MTD HbF following hydroxyurea treatment (R = 0.40, P = 0.0004), indicating patients with higher baseline HbF tended to have higher MTD HbF values. Interestingly, there was an inverse correlation of delta HbF with HbF at baseline (R = −0.22, P = 0.026). All correlations were corrected for sex and age.
Table 1.
Hematological parameters and comparison between HbF at baseline and MTD.
| Baseline
|
MTD
|
|||||
|---|---|---|---|---|---|---|
| Mean | SD | 25th, median, 75th | Mean | SD | 25th, median, 75th | |
| WBC | 14.3 | 5.7 | 10.0, 13.4, 17.3 | 8.1 | 3.1 | 6.0, 7.4, 9.4 |
| ANC | 7734 | 4375 | 4900, 6840, 9430 | 3956 | 2352 | 2422, 3455, 4990 |
| Hb | 7.6 | 1.3 | 6.7, 7.7, 8.5 | 8.5 | 1.3 | 7.6, 8.4, 9.4 |
| ARC | 343 | 146 | 236, 311, 455 | 211 | 105 | 125, 199, 267 |
| MCV | 86.6 | 10.9 | 80.5, 86.9, 93.6 | 100.5 | 11.7 | 92.7, 100.2, 107.8 |
| MCHC | 32.2 | 1.7 | 31.2, 32, 32.7 | 34.9 | 1.4 | 34.0, 35.1, 35.9 |
| HbF | 6.5 | 6.0 | 2.1, 4.4, 9.6 | 17.3 | 8.8 | 10.4, 16.3, 22.8 |
The hematological parameters for 111 patients with SCD are shown. ANC, absolute neutrophil count; ARC, absolute reticulocyte count; HbF, fetal hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MTD, maximum tolerated dose; MCV, mean corpuscular volume; WBC, white blood cells. All parameters were statistically significant between Baseline and MTD values (p-values < 0.0001).
3.1. Single nucleotide polymorphism (SNP) analysis
The SNP HBG2-Xmn1-rs7482144 had a very low allele frequency in our Brazilian cohort (1 heterozygote; minor allele frequency = 0.45%) and was excluded from our study. The five other candidate SNPs were all in agreement with Hardy-Weinberg Equilibrium and their characteristics are described in Table 2. We used regression analysis to independently test association of these five SNPs with baseline HbF, MTD HbF, and delta HbF. The regression model for baseline HbF included covariate correction for patient age at start of HU treatment and sex. The regression model for MTD and delta incorporated covariate correction for age at start of HU, sex and ANC at MTD. For each SNP, the wild-type homozygote genotype was used as the reference in the regression models. In a separate analysis, we did not detect any correlation between the 5 SNPs studied on any of the other measured hematological parameters before or after exposure to HU (data not shown).
Table 2.
Analysis of association between SNPs and HbF levels at baseline, MTD, and with delta HbF following HU treatment.
| Gene | SNP | Genotype | MAF | Baseline HbF
|
MTD HbF
|
Delta HbF
|
|||
|---|---|---|---|---|---|---|---|---|---|
| β | P-value | β | P-value | β | P-value | ||||
| BCL11A | rs1427407 | G>T | 0.273 | 1.84 | 0.033 | 3.30 | 0.004 | 2.55 | 0.021 |
| rs4671393 | G>A | 0.282 | 2.09 | 0.006 | 3.16 | 0.009 | 2.02 | 0.069 | |
| rs11886868 | T>C | 0.409 | 1.58 | 0.056 | 2.87 | 0.005 | 2.31 | 0.026 | |
| HMIP-2 | rs9399137 | T>C | 0.145 | 4.57 | 0.00078 | 0.93 | 0.239 | −0.607 | 0.797 |
| rs9402686 | G>A | 0.150 | 5.18 | 0.00005 | 1.00 | 0.185 | −0.621 | 0.842 | |
HbF, fetal hemoglobin; SNP, single nucleotide polymorphisms; MTD, maximum tolerated dose; MAF, Minor Allele Frequency; β is the beta coefficient that represents the average change in %HbF per allele copy of each individual SNP (effect size). The P-values were calculated using linear regression testing of each HbF phenotype. The presented P-values for the MTD HbF analysis include covariate correction for baseline HbF.
For the three BCL11A SNPs, rs1427407 and rs4671393 were both associated with increased baseline and MTD HbF (Fig. 1). When we included covariate correction for baseline HbF, the association of the SNPs with higher MTD HbF was diminished but still statistically significant (P = 0.004 and P = 0.009 for the rs1427407 and rs4671393, respectively). In addition, the two SNPs were associated with increased delta HbF (Table 2). The BCL11A rs11886868 only approached statistical significance for association with baseline HbF in our cohort but did show association with MTD and delta HbF (Table 2). To analyze the combinatorial effect of the BCL11A SNPs, we performed a haplotype analysis of the three SNPs. We computed the haplotypes using the expectation-maximization algorithm implemented in the haplo.stats R package. The haplotype with the highest frequency had all three BCL11A SNPs with the wild-type reference allele (G-G-T, frequency = 59%) and this haplotype was used as reference for the analysis. Compared to this reference G-G-T haplotype, patients with at least one haplotype carrying all three BCL11A SNPs (T-A-C, frequency = 28%) had higher baseline and MTD HbF levels (effect size = 0.15, P = 0.014 and effect size = 0.28, P = 0.001, respectively). As in the single SNP analysis outlined above, covariate correction for baseline HbF diminished the association of the T-A-C haplotype with MTD HbF but was still statistically significant (P = 0.016). The T-A-C haplotype was also associated with increased delta HbF (effect size = 0.25, P = 0.005).
Fig. 1.
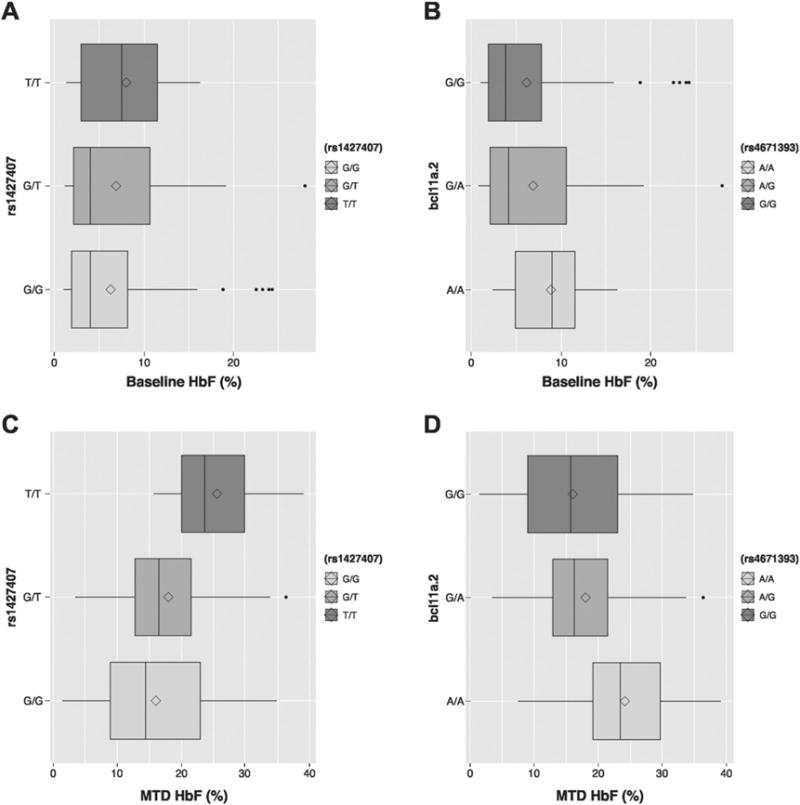
Effect of BCL11A SNPs on fetal hemoglobin (HbF) levels at baseline and maximum tolerated dose of hydroxyurea (MTD). A) Effect of rs1427407 on baseline HbF. B) Effect of rs4671393 on baseline HbF. C) Effect of rs11886868 on baseline HbF. D) Effect of rs1427407 on HbF at MTD. E) Effect of rs4671393 on HbF at MTD. F) Effect of rs11886868 on HbF at MTD. The line represents the median and the diamond the mean of the untransformed HbF (%). n.s., not statistically significant. *P < 0.05; **P < 0.01.
For HMIP-2, both rs9399137 and rs9402686 SNPs were associated with increased baseline HbF and MTD HbF (Fig. 2). Covariate correction for baseline HbF eliminated association of the HMIP-2 rs9399137 and rs9402686 SNPs with MTD HbF (P = 0.239 and P = 0.185, respectively) and there was no association of either SNP with delta HbF (Table 2). Linkage disequilibrium testing showed that the two HMIP-2 SNPs are in strong linkage (D′ = 0.9 and r2 = 0.90). Haplotype analysis showed that there were three haplotypes present in our Brazilian cohort. The haplotype with the highest frequency contained the wild-type alleles for both SNPs (T-G, frequency = 84.2%). Compared to this reference T-G haplotype, patients with the C-A or T-A haplotypes (frequency = 13.1% and 1.7%, respectively) had higher baseline HbF levels (effect size = 0.28, P = 0.0006 and effect size = 0.10, P = 0.0075, respectively). Similar to our observations of the individual HMIP-2 SNP analysis, the association of the C-A haplotype with MTD HbF lost significance when corrected for baseline HbF and there was no association of any HMIP-2 haplotype with delta HbF.
Fig. 2.
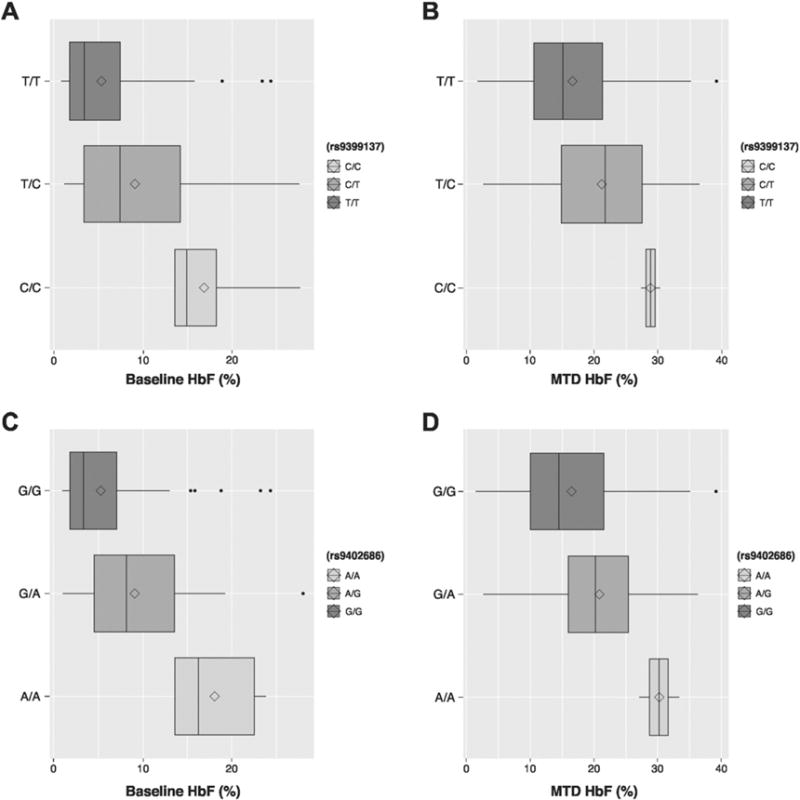
Effect of HMIP-2 SNPs on HbF at baseline and hydroxyurea MTD. A) Effect of rs9399137 on baseline HbF. B) Effect of rs9402686 on baseline HbF. C. Effect of rs9399137 on HbF at MTD. D. Effect of rs9402686 on HbF at MTD. Each plot shows the untransformed HbF levels of the three genotypes. The line represents the median and the diamond the mean of the HbF (%). *P < 0.05; **P < 0.01; ***P < 0.001.
4. Discussion
This study examined the role of two well-known QTL associated with HbF levels in a large cohort of SCA patients in Brazil. Similar to previous studies, we found that the BCL11A and HMIP-2 are associated with endogenous levels of HbF in patients with SCA [8,11]. In our cohort of patients, the frequency of the HMIP-2 SNPs was higher than other studies looking at patients with SCA [12]. In contrast, the frequency of the BCL11A SNPs was lower in our cohort and the Xmn1-HBG2 SNP was almost absent. This likely reflects the distinct genetic ancestry of our SCA patients based in Porto Alegre, where there may have potentially been more admixture of the population with individuals of European ancestry compared to African-American or African patients with SCA. Despite these frequency differences, we still found that these SNPs were associated with increased levels of HbF. The strength of this study was that we also examined whether these same SNPs also affect the induction of HbF by hydroxyurea. Our study had >70% power to detect genetic associations with baseline HbF and >60% power to detect associations with either MTD or delta HbF.
Hydroxyurea is a well-established agent capable of inducing increased HbF production. There is little known about the factors that contribute to the wide variability observed in individual response to hydroxyurea, even when patients are of similar ages and receiving similar dosages of the drug. Previous studies found that individual MTD HbF responses are correlated with baseline hematocrit, WBC, ARC and HbF levels as well as time of exposure to hydroxyurea [13]. In our cohort, only baseline HbF levels were associated with final HbF levels (Fig. 3). We observed an inverse relationship between baseline HbF and delta HbF values in response to hydroxyurea treatment. This suggested that, although patients with higher baseline HbF tended to have higher absolute final HbF values, the absolute drug-induced change tends to be smaller in patients with higher baseline HbF values. In our SNP analysis, the three BCL11A SNPs showed association with MTD HbF (even when corrected for baseline HbF levels) and with delta HbF. Furthermore, BCL11A may also contribute to hydroxyurea induction of HbF, independently of its effect on endogenous HbF levels.
Fig. 3.
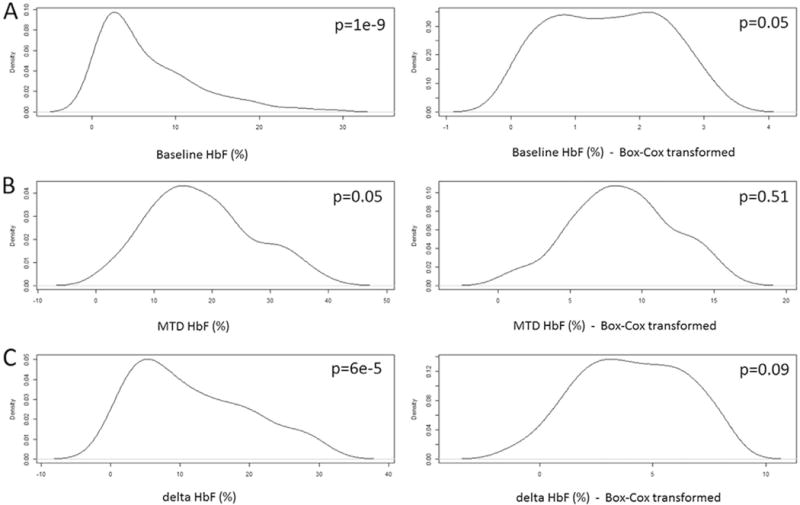
Distribution of fetal hemoglobin (HbF). Data is shown untransformed and transformed using Box-Cox method. A) Distribution of HbF at baseline. B) Distribution of HbF at maximum tolerated dose (MTD). C) Distribution of delta HbF (HbF at MTD – HbF at baseline). P: P-value of the Shapiro-Wilk test.
BCL11A is a negative regulator of HbF expression. Individuals with any of the established BCL11A SNPs are known to have decreased expression of BCL11A, which results in increased HbF production [8,11,14]. In patients treated with hydroxyurea, BCL11A expression has been shown to be reduced after patients have reached their maximum tolerated dose [15]. This effect was most evident in patients without the BCL11A SNPs but it seemed that BCL11A had a limited role in hydroxyurea induction of HbF. In these same patients, there was no association of the BCL11A SNPs with MTD HbF or delta HbF [16]. In contrast, we have found that, in our Brazilian cohort, patients with the BCL11A SNPs have an improved response to hydroxyurea. With no phenotype data correction or data transformation, the average delta HbF was 13.3 ± 7.9% versus 10.6 ± 8.3% for patients with and without the BCL11A variants, respectively. Although this effect size is relatively small, it is an interesting finding that warrants further validation.
In conclusion, we have shown that BCL11A and HMIP-2 are potent QTL associated with endogenous HbF levels in our Brazilian cohort. We also showed that patients with the BCL11A SNPs have a favorable probability of producing more HbF in response to hydroxyurea treatment. Genetic studies examining larger populations with SCA on HU are needed to better assess the specific variants in the SNPs we studied and other genetic modifiers related to HbF expression and variability in HU response.
Acknowledgments
The authors thank all of the patients who participated in this study and their families.
Financial disclosure
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Abbreviations
- HbF
fetal hemoglobin
- SCA
sickle cell anemia
- QTL
quantitative trait loci
- HU
hydroxyurea
- SNP
single nucleotide polymorphism
- MTD
maximum tolerated dose
Footnotes
Conflict of interest
The authors have no conflicts of interest to disclose.
Authors’ contributions
Concept, patient follow-up, data collection, data analysis and manuscript writing: João Ricardo Friedrisch.
Genetic polymorphism analysis, data analysis and manuscript writing: Vivien Sheehan.
Genetic polymorphism laboratory analysis, data analysis and manuscript writing: Jonathan M. Flanagan, Alessandro Baldan, Carly C. Ginter Summarell.
Patient assessment and clinical and laboratory data collection: Christina Matzembacher Bittar, Bruno Kras Friedrisch, Ianaê Indiara Wilke, Camila Blos Ribeiro, Liane Esteves Daudt.
Research advising and supervision and manuscript writing supervision: Lucia Mariano da Rocha Silla.
All authors have approved the final article.
References
- 1.Silla LM. Sickle cell disease: a serious and unknown problem of public health in Brazil. J Pediatr (Rio J) 1999;75:145–146. doi: 10.2223/jped.288. [DOI] [PubMed] [Google Scholar]
- 2.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH, Klug PP. Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med. 1994;330:1639–1644. doi: 10.1056/NEJM199406093302303. [DOI] [PubMed] [Google Scholar]
- 3.Sankaran VG, Weiss MJ. Anemia: progress in molecular mechanisms and therapies. Nat Med. 2015;21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galarneau G, Palmer CD, Sankaran VG, Orkin SH, Hirschhorn JN, Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat Genet. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thein SL, Menzel S, Lathrop M, Garner C. Control of fetal hemoglobin: new insights emerging from genomics and clinical implications. Hum Mol Genet. 2009;18:R216–R223. doi: 10.1093/hmg/ddp401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, Voit RA, Yuan GC, Porteus MH, Stamatoyannopoulos JA, Lettre G, Orkin SH. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzel S, Garner C, Gut I, Matsuda F, Yamaguchi M, Heath S, Foglio M, Zelenika D, Boland A, Rooks H, Best S, Spector TD, Farrall M, Lathrop M, Thein SL. A QTL influencing F cell production maps to a gene encoding a zinc-finger protein on chromosome 2p15. Nat Genet. 2007;39:1197–1199. doi: 10.1038/ng2108. [DOI] [PubMed] [Google Scholar]
- 8.Lettre G, Sankaran VG, Bezerra MA, Araujo AS, Uda M, Sanna S, Cao A, Schlessinger D, Costa FF, Hirschhorn JN, Orkin SH. DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci U S A. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae HT, Baldwin CT, Sebastiani P, Telen MJ, Ashley-Koch A, Garrett M, Hooper WC, Bean CJ, Debaun MR, Arking DE, Bhatnagar P, Casella JF, Keefer JR, Barron-Casella E, Gordeuk V, Kato GJ, Minniti C, Taylor J, Campbell A, Luchtman-Jones L, Hoppe C, Gladwin MT, Zhang Y, Steinberg MH. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood. 2012;120:1961–1962. doi: 10.1182/blood-2012-06-432849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ware RE. How I use hydroxyurea to treat young patients with sickle cell anemia. Blood. 2010;115:5300–5311. doi: 10.1182/blood-2009-04-146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, Lenhard B, Rooks H, Best S, Menzel S, Grosveld F, Thein SL, Soler E. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menzel S, Rooks H, Zelenika D, Mtatiro SN, Gnanakulasekaran A, Drasar E, Cox S, Liu L, Masood M, Silver N, Garner C, Vasavda N, Howard J, Makani J, Adekile A, Pace B, Spector T, Farrall M, Lathrop M, Thein SL. Global genetic architecture of an erythroid quantitative trait locus, HMIP-2. Ann Hum Genet. 2014;78:434–451. doi: 10.1111/ahg.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ware RE, Eggleston B, Redding-Lallinger R, Wang WC, Smith-Whitley K, Daeschner C, Gee B, Styles LA, Helms RW, Kinney TR, Ohene-Frempong K. Predictors of fetal hemoglobin response in children with sickle cell anemia receiving hydroxyurea therapy. Blood. 2002;99:10–14. doi: 10.1182/blood.v99.1.10. [DOI] [PubMed] [Google Scholar]
- 14.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin SH, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flanagan JM, Steward S, Howard TA, Mortier NA, Kimble AC, Aygun B, Hankins JS, Neale GA, Ware RE. Hydroxycarbamide alters erythroid gene expression in children with sickle cell anaemia. Br J Haematol. 2012;157:240–248. doi: 10.1111/j.1365-2141.2012.09061.x. [DOI] [PubMed] [Google Scholar]
- 16.Sheehan VA, Crosby JR, Sabo A, Mortier NA, Howard TA, Muzny DM, Dugan-Perez S, Aygun B, Nottage KA, Boerwinkle E, Gibbs RA, Ware RE, Flanagan JM. Whole exome sequencing identifies novel genes for fetal hemoglobin response to hydroxyurea in children with sickle cell anemia. PLoS One. 2014;9:e110740. doi: 10.1371/journal.pone.0110740. [DOI] [PMC free article] [PubMed] [Google Scholar]


