Abstract
Background
Probiotics have tremendous potential to develop healthy diets, treatment, and prevention. Investigation of in vitro cultural properties of health-promoting microorganisms like lactic acid bacteria (LAB) and bifidobacteria is crucial to select probiotic strains for treatments based on gut microbiota modulation to justify individualized and personalized approach for nutrition and prevention of variety of diseases. The aim was to study the biological properties of LAB and bifidobacteria probiotic strains, namely adhesive properties; resistance to antibiotics; and biological fluids (gastric juice, bile, pancreatic enzymes), and to overview the literature in the field.
Materials and methods
We studied six LAB strains (Lactobacillus acidophilus ІМV В-7279, L. casei ІМV В-7280, L. delbrueckii subsp. bulgaricus ІМV В-7281, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, L. plantarum LM VK7), and two bifidobacteria strains (Bifidobacterium animalis VKL, B. animalis VKB). We characterized tinctorial, culturally morphological, physiological, and biochemical properties of probiotic strains of LAB and bifidobacteria by commonly used research methods. Determination of the resistance to antibiotics was carried out using disc-diffusion method. The effects of gastric juice, bile, and pancreatin on the viability of LAB and bifidobacteria were evaluated. Adhesive properties of LAB and bifidobacteria to epithelial cells were assessed calculating three indicators: average adhesion rate (AAR), participation rate of epithelial cells (PRE), and adhesiveness index of microorganisms (AIM). Electron microscopy of LAB and bifidobacteria cells was conducted.
Results
The studied strains of LAB and bifidobacteria did not form spores, were positively stained by Gram, grow on medium in a wide range of pH (1.0–9.0, optimum pH 5.5–6.5), were sensitive to a wide range of antibiotics; and showed different resistance to gastric juice, bile, and pancreatic enzymes. The most resistant to antibiotics were L. rhamnosus LB-3 VK6 and L. delbrueckii LE VK8 strains. The most susceptible to gastric juice was L. plantarum LM VK7, which stopped its growth at 8% of gastric juice; L. acidophilus IMV B-7279, B. animalis VKL, and B. animalis VKB strains were resistant even in the 100% concentration. Strains L. acidophilus IMV В-7279, L. casei IMV В-7280, B. animalis VKL, B. animalis VKB, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. delbrueckii subsp. bulgaricus IMV В-7281 were resistant to pancreatic enzymes. Adhesive properties of the strains according to AIM index were high in L. casei IMV В-7280, B. animalis VKL, and B. animalis VKB; were moderate in L. delbrueckii subsp. bulgaricus IMV В-7281; and were low in L. acidophilus IMV В-7279, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. plantarum LM VK7.
Conclusion
We recognized strain-dependent properties of studied LAB and bifidobacteria probiotic strains (adhesive ability, resistance to antibiotics, and gut biological fluids) and discussed potential for most effective individualized treatment for gut and distant sites microbiome modulation.
Keywords: Predictive preventive personalized medicine, Lactobacillus, Bifidobacterium, Probiotics, Gut microbiota, Antibiotics, Gastric juice, Bile, Pancreatic enzymes, Adhesive properties, Pili, Patient phenotype, Individualized medicine
Overview
Relevance of in vitro research to support strains stratification for effective personalized probiotic interventions
Intestinal microbial population largely represented by Bacteroidetes and Firmicutes, has been proven to impact on human health and maintaining homeostasis [1].
The definition of a probiotic as “live microorganisms which when administered in adequate amounts confer a health benefit on the host” was determined by Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) in 2001 [2] and confirmed in 2014 by the International Scientific Association for Probiotics and Prebiotics (ISAPP) experts [3] and later remain unchanged being agreed in the broad expert communities. Probiotics have tremendous potential to develop healthy diets and integrated approach for immunity-related disease treatment and prevention [3–12] and are effective actors in the gut and in distant sites [12] with strong potential for applications in personalized medicine and nutrition [13, 14]. Modification of the gut microbiota in chronic diseases and metabolic syndrome [14, 15] is among the leading tasks of microbiome research and needs for clinical use of probiotics [16].
However, evidence-supported knowledge on probiotics contribution to disease pathophysiology and applicability to clinical care is not yet sufficient [17], excluding very few aspects. Thus, in cases of antibiotic- and Clostridium difficile-associated diarrhea and respiratory tract infections, the effects of probiotics are considered “evidence-based” [18].
Studies conducted in vitro and in vivo, including probiotics mechanism of function, gut microbiota composition ecology, and metabolomic researches in regards to screening strains for clinical application are needed to implement personalized probiotic treatment to the clinical care and set relevant designs of clinical trials of particular strains [8, 19, 20]. Since clinical studies are very complicated to design and conduct, in vitro [21] and animal models [22] research still can provide a high quality data in this matter.
Recently Koch’s original postulates have been adapted to identify microorganisms that contribute to human health and formulated necessary requirements for a microorganism to be considered a probiotic as follows [23]:
The strain of the commensal microorganism is associated with the health of the host, which is regularly manifested in healthy hosts, but less common in patients with disease.
The strain of the commensal microorganism can be identified as pure culture and cultivated in the laboratory.
A strain of a commensal microorganism improves or alleviate the disease when introduced into a new host organism.
The strain of the commensal microorganism can be detected after its introduction into a host to which health was restored.
One of the principle mechanisms of probiotics’ role in the pathogenesis of a number of diseases is the ability of bacterial strains to contribute to immune response, prevent colonization of pathogen microorganisms, matabolize nutrients and process toxic metabolites, and regulate energy balance [1, 2]. Probiotics demonstrate antagonistic relations with pathogenic and opportunistic bacteria due to the synthesis of a number of organic acids, which leads to pH lowering, hydrogen peroxide, lysozyme, and bacteriocins—polypeptide antibiotic-like substances, which differ in the strength and spectrum of antibiotic action [2, 3, 24].
In addition, probiotic strains are able to synthesize digestive enzymes (amylase, lipase, proteases, pectinases, and endoglucanases) and vitamins A, B, PP, E, C, K, and others [25, 26] and produce metabolites such as short-chain fatty acids and histamine [27]. Due to the effects of these substances, probiotic strains are able to restore microbiota; participate in the metabolism of glucose, cholesterol, bilirubin, choline, bile, and fatty acids; as well as indirectly influence on the metabolism of iron and calcium and to demonstrate immunomodulatory and antitoxic properties [28]. Also, due to the adhesion to the mucosa and epithelium, inhibiting pathogen adhesion and/or growth [28] probiotic strains can prevent lesions of tissues by pathogenic and opportunistic microorganisms.
On the other hand, a broad use of antibiotics alters host phenotypes and gut microbiota and glucose metabolism [29]. In recent decades, uncontrolled antibiotic therapy led to the formation of associations of microorganisms with increased virulence, in particular the so-called hospital strains. Gut microbiota is known to be a potential reservoir of antimicrobial resistance (AMR) genes, demonstrating the ability for the horizontal transfer to potential pathogenic bacteria within this ecosystem [30]. Microorganisms AMR are extensively studied over the past decades as so-called “resistome.” Antibiotic susceptibility of probiotic strain can be an important specific indicator; antimicrobial resistance was studied for LAB and bifidobacterium strains [31].
Thus, each individual strain may demonstrate multiple mechanisms of action and have specific phenotype/genotype accordingly; these relevant markers has not been formulated yet for traditional probiotic strains [10] and a comprehensive understanding of these mechanisms full discovered.
Thus, insufficient viability and survival of probiotic bacteria remain a problem in commercial products of fermented food [32]. Therefore, cultural properties of strains are crucial for stratification before inclusion of probiotic bacteria as therapeutic fermented products by selecting best functional probiotic strains and improvement methods of increasing survival, using appropriate prebiotics and the development optimal combination of probiotics and prebiotics [4] (synbiotics) an increasing viable strains delivery of bacteria is essential task [32].
Despite extensive agenda of genomic research in microbiome, metagenomic studies have been recently revived in particular as used to the human gut [33–35]. Browne et al. suggested that the gut microbiota is largely culturable and the majority of strains can be cultured using a single bacteriological medium [36]. Such “microbial culturomics” approach [33] opens new insights for phenotypic analysis of the human gut microbiota and probiotic strains.
The tests conducted in vitro have widely demonstrated the strain-dependent immunomodulation potential of bifidobacteria [37, 38]. In vitro models have important limitations but they enable the preliminary screening of the effects that bacterial cells or fractions might have on different components of the immune response [39, 40], e.g., in vitro models using immune cells as macrophage-like cell lines for bifidobacteria and lactic acid bacteria (LAB) [39] and cells isolated from the gut-associated lymphoid tissues (GALT) [40]. In vitro properties like the cell wall parameters [39, 40] play an essential role in many aspects of modulating beneficial immune response and EPS-producing phenotype can depend on a single gene (e.g., Balat_1410 for Bifidobacterium animalis subsp. lactis) [41]. Such established crosslinks between phenotype-genotype can warrant to stratify strains on their modulatory activity on innate immunity to justify individualized and personalized approach for nutrition and prevention.
Lactobacillus and bifidobacterium strains are the most commonly used probiotics [42]; although their cultural properties have not been widely used to examine the immunomodulation potential to select probiotic strains for individualized treatments based on gut microbiota modulation in close regards to patient’s phenotype analysis.
The aim was to investigate the biological properties of Lactobacillus acidophilus ІМV В-7279, L. casei ІМV В-7280, L. delbrueckii subsp. bulgaricus ІМV В-7281, L. rhamnosus LB-3 VK6, L. plantarum LM VK7, L. delbrueckii LE VK8, B. animalis VKL, and B. animalis VKB strains, namely adhesive properties, resistance to antibiotics and biological fluids (gastric juice, bile, pancreatic enzymes); to overview the literature in the field and discuss the potential for individualized use in adherence with patient’s needs according to host phenotype to obtain the best effect possible.
Materials and methods
Вacterial strain, media, and growth conditions
The objects of the study were as follows:
Six LAB strains (L. acidophilus ІМV В-7279, L. casei ІМV В-7280, L. delbrueckii subsp. bulgaricus ІМV В-7281, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, L. plantarum LM VK7)
and two strains of the Bifidobacterium genus (B. animalis VKL, B. animalis VKB).
Strains used in our study are deposited in the Ukrainian collection of microorganisms (Zabolotny Institute of Microbiology and Virology of NAS of Ukraine, Kyiv, Ukraine).
No human participant were included and no animals used in this study.
Studies were performed using microorganisms, freeze-dried in a Cuddon Freeze Dryer FD1500 (New Zealand). Before each experiment, the viability of the probiotic strains was tested by monitoring their growth on the Man-Rogosa-Sharpe (MRS) agar or bifidum agar (BA) in aerobic and anaerobic conditions, respectively, at 37 °C for 24–48 h. The same cultural media without agar adding were used to determine the growth curve of bacterial cultures.
Determination of the resistance to antibiotics of different groups was carried out on MRS agar (MRSA) and BA mediums by disc-diffusion method [19, 22]. Standardized discs with antibiotics (Obolensk, Russia) were used. The degree of bacteria sensitivity to antibiotics was determined by the size of growth inhibition zone: less than 10 mm was evaluated as resistant (R); 10–20 mm as medium resistant (M); and over 20 mm as sensitive (S) [31].
The effects of gastric juice, bile, and pancreatin on the viability of LAB and bifidobacteria were determined. Daily bacterial strains were grown in liquid media at 37 °C with gastric juice in a concentration of 1, 2, 5, 8, 10, 20, 30, 50, 75, and 100% (for 2.5 h) or in bile in a concentration of 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4, and 5% (for 5 h) or pancreatin at a concentration of 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4, and 5% (for 15 h). After this, LAB and bifidobacteria were sown to MRSА and BA, respectively. After 24–48 h cultivation at 37 °C with high content of carbon dioxide in the air (5%), the number of colony-forming units (CFU) was counted. Complex effect of gastric juice, bile, and pancreatin on the activity of LAB and bifidobacteria was evaluated. For this purpose, the strains were grown in MRS and bifidum media, respectively, with the gradual addition of 2% of gastric juice (for 2.5 h), 1% of bile (for 5 h), and 1% of pancreatin (for 15 h) [43, 44]. Determination of adhesive properties of probiotic strains was performed by standard method as suggested in [45].
Adhesive properties of LAB and bifidobacteria were assessed by three indicators: the average adhesion rate (AAR; the average number of microorganisms that have attached to the surface of one epitheliocyte in case of counting no less than 100 epithelial cells, given no more than five in a single field of view); the participation rate of epithelial cells (PRE; percent of epithelial cells, having on its surface-adhered microorganisms), the adhesiveness index of microorganisms (AIM; the average number of microorganisms on one epitheliocyte that participate in the adhesive process). AIM is calculated by the formula [45]:
It was considered that microorganisms had no adhesive activity at AIM index ≤ 1.75; had low adhesive activity at AIM 1.75–2.5, average adhesive activity at AIM 2.51–4.0, and high adhesive activity at AIM > 4.0 [29].
For electron microscopy, daily culture of lactobacilli and bifidobacteria cells were twice washed after cultivation on MRSA or BA media by centrifugation at 2000 rpm and resuspended in a physiological solution. The concentration of microorganisms was adjusted to 1 × 106 cells/ml. The electron microscopy of the samples was carried out by the generally accepted method [46] using the JEM-1400 electron microscope (Zabolotny Institute of Microbiology and Virology, NAS of Ukraine) at 80 kV.
Statistical analysis
Statistical analysis was performed by using one-way analysis of variance (ANOVA) using Epi Info software (USA, version 8.0) and Microsoft Office Excel. Numeric data were presented as mean arithmetic values and their standard deviations (M ± m). For single comparisons, values of P were determined using Student’s t test. Differences between groups were defined significant at P < 0.05.
Results
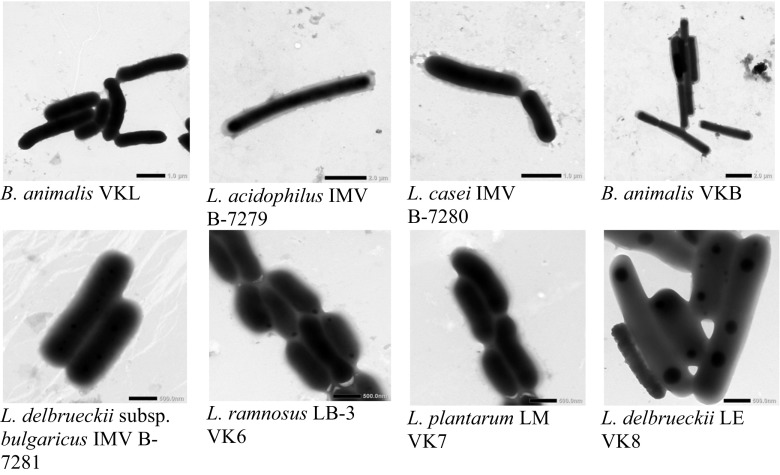
We established tinctorial, culturally morphological, physiological, and biochemical properties of probiotic strains of LAB and bifidobacteria by commonly used research methods. L. acidophilus ІМV В-7279, L. casei ІМV В-7280, L. delbrueckii subsp. bulgaricus ІМV В-7281, L. rhamnosus LB-3 VK6, L. plantarum LM VK7, L. delbrueckii LE VK8, B. animalis VKL, and B. animalis VKB strains were previously obtained from fermented biological material of the intestines of clinically healthy adults. All studied strains were motionless. LAB had a rod-shaped form. Bifidobacteria were characterized by cell polymorphism when the cell’s shape depended on the stage of development of the culture and varied from the rod-shaped to the pin-shaped, spindle-shaped, amorphous, and Y- or X-shaped cells (Fig. 1).
Fig. 1.
Subcellular imaging (electronic microscopy) of the strains B. animalis VKL, B. animalis VKВ, L. acidophilus ІМV В-7279, L. casei ІМV В-7280, L. delbruеckii subsp. bulgaricus ІМV В-7281, L. rhamnosus LB-3 VK6 (ІМV В-7038), L. plantarum LM VK7, and L. delbrueckii LE VK8. The scale is shown in the images
The studied strains of LAB and bifidobacteria did not form spores, were positively stained by Gram and grow on medium in a wide range of pH (1.0–9.0, optimum pH 5.5–6.5).
For further investigation of biological properties of L. acidophilus ІМV В-7279, L. casei ІМV В-7280, L. delbrueckii subsp. bulgaricus ІМV В-7281, L. rhamnosus LB-3 VK6, L. plantarum LM VK7, L. delbrueckii LE VK8, B. animalis VKL, and B. animalis VKB, we studied the dynamics of their growth in the liquid media. Based on the obtained results, we constructed the growth curves of each culture and determined the beginning of the stationary phase of growth in each particular case. The stationary growth phase of L. delbrueckii subsp. bulgaricus IMV V-7281, L. delbrueckii LE VK8, and L. plantarum LM VK7 after inoculation in MRS medium and incubation at 37 °C occurred only at 11–12 h. It was found that during growth in the MRS medium, the stationary growth phase in L. acidophilus IMV B-7279, B. animalis VKL, and B. animalis VKB occurred approximately 8 h after the inoculation of these strains. For L. casei IMV В-7280 and L. rhamnosus LB-3 VK6 strains, the stationary growth phase began only at 10 h after strains inoculation.
Thus, it has been determined that the beginning of the stationary phase of growth in all strains studied was 7–12 h from the beginning of the growth of culture and was strain-dependent. In the studied bifidobacteria, it occurred a little earlier than in LAB strains.
Resistance to antibiotics
As a result of our study, it was determined the resistance of probiotic strains of LAB and bifidobacteria to antibiotics of different groups with the aim of developing recommendations about the advisability of their further use during antibiotic treatment. The obtained data are provided in Table. 1.
Table 1.
The resistance of investigated strains of LAB and bifidobacteria to antibiotics
| Mechanism of action | Group of antibiotic | Strain of bacteria | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Generation | Name | B. animalis VKL | B. animalis VKB | L. acidophilus ІМV В-7279 | L. casei ІМV В-7280 | L. delbrueckii subsp. bulgaricus ІМV В-7281 | L. rhamnosus LB-3 VK6 | L. plantarum LM VK7 | L. delbrueckii LE VK8 | |
| Antibiotic resistance/diameter of growth inhibition zone | ||||||||||
| Inhibition of cell wall synthesis | Penicillins | |||||||||
| I | Penicillin | M | M | S | S | M | R | S | M | |
| II | Oxacillin | R | R | R | R | R | R | M | R | |
| Ampicillin | S | S | S | S | M | R | M | M | ||
| Carbenicillin | S | S | S | S | S | R | S | S | ||
| Azlocillin | S | S | S | S | S | R | S | M | ||
| Piperacillin | M | S | S | S | S | R | M | M | ||
| Amoxicillin | S | S | S | S | S | R | S | M | ||
| Amoxicillin/clavulanic acid | S | S | S | S | S | R | S | M | ||
| Ticarcillin | M | S | S | S | S | R | S | M | ||
| Ticarcillin/clavulanic acid | M | S | S | S | S | M | S | S | ||
| Cephalosporins | ||||||||||
| I | Cefazolin | R | M | M | M | M | R | M | R | |
| Cefalexin | M | M | M | M | R | M | M | R | ||
| II | Cefuroxime | M | R | M | M | R | R | M | M | |
| Cefaclor | M | S | M | M | M | M | R | M | ||
| III | Cefotaxime | R | S | S | M | S | R | S | R | |
| Ceftriaxone | M | M | S | M | S | R | M | M | ||
| Cefoperazone | M | M | S | M | S | R | M | M | ||
| Ceftazidime | R | M | S | R | R | R | R | R | ||
| Cefamandole | M | R | S | S | S | S | R | M | ||
| Ceftibuten | R | R | R | M | R | R | R | R | ||
| IV | Cefepime | R | M | M | M | R | R | R | R | |
| Glycopeptides | ||||||||||
| Vancomycin | R | R | R | R | R | R | R | M | ||
| Teicoplanin | R | R | R | R | R | R | R | M | ||
| Carbapenems | ||||||||||
| Imipenem | M | S | S | M | M | M | S | M | ||
| Meropenem | S | S | S | M | M | M | M | M | ||
| Inhibition of protein synthesis | Aminoglycosides | |||||||||
| I | Streptomycin | M | M | S | S | M | R | M | M | |
| Neomycin | S | M | S | M | R | M | M | M | ||
| Kanamycin | R | R | S | M | R | R | R | M | ||
| II | Garamycin | S | S | S | M | M | M | S | M | |
| Tobramycin | R | S | S | M | M | M | S | M | ||
| Sisomycin | M | M | S | S | M | M | S | S | ||
| Amikacin | M | M | S | M | R | M | R | R | ||
| III | Netilmicin | M | M | S | M | M | R | M | M | |
| Macrolides | ||||||||||
| I | Oleandomycin | S | S | S | S | M | R | S | M | |
| Erythromycin | M | S | S | S | M | R | S | S | ||
| II | Azithromycin | R | S | S | S | M | R | S | S | |
| Clarithromycin | S | R | S | S | S | R | S | S | ||
| Roxithromycin | M | M | S | S | S | R | S | M | ||
| Lincosamides | ||||||||||
| I | Lincomycin | M | S | S | S | M | M | S | S | |
| II | Clindamycin | M | S | S | S | S | R | S | M | |
| Tetracyclines | ||||||||||
| I | Tetracycline | M | S | S | M | M | S | S | M | |
| I | Chlortetrazyklin | M | R | S | S | S | S | S | S | |
| II | Doxycycline | M | S | S | M | S | R | S | M | |
| Violations of the synthesis of respiratory enzymes and biosynthesis of cell membrane proteins | Nitrofurans | |||||||||
| Nitrofurantoin | R | M | S | R | S | R | M | M | ||
| Furazolidone | R | M | S | R | M | R | R | R | ||
| Fusidin | M | M | S | R | M | R | R | R | ||
| Nitroxoline (oxyquinolines) | S | M | S | S | S | M | S | S | ||
| Rifampicin | M | S | S | S | S | R | S | S | ||
| Levomycetin | S | S | S | S | S | R | M | M | ||
| Violation of synthesis of nucleic acids | Fluoroquinolones | |||||||||
| I | Nalidixic acid | R | R | R | R | R | R | R | R | |
| II | Ofloxacin | R | M | M | M | R | R | M | M | |
| Ciprofloxacin | R | R | S | M | R | R | M | M | ||
| Norfloxacin | R | M | R | M | M | R | M | R | ||
| III | Levofloxacin | S | R | R | M | R | R | M | M | |
| Sparfloxacin | S | S | S | R | R | R | M | M | ||
| II | Pefloxacin | M | M | M | M | R | R | M | M | |
S sensitive, M medium resistant, R resistant
We have established that the following strains were sensitive for most of the investigated antibiotics which effects associated with the blocking of peptidoglycan synthesis: L. acidophilus IMV B-7279 (except oxacillin); L. casei IMV B-7280; or B. animalis VKB (except penicillin, oxacillin, meropenem, and imipenem). L. rhamnosus LB-VK6 3 strain was resistant to most inhibitors of peptidoglycan synthesis, while L. delbrueckii LE VK8 strain was mostly moderately sensitive to these antibiotics. At the same time, all studied strains were resistant to vancomycin, teicoplanin, and oxacillin. Studied strains had different sensitivities for antibiotics of cephalosporin group, in which mechanism of action is also inhibition of cell wall synthesis. Highly sensitive to the effects of most cephalosporins was L. acidophilus IMV B-7279, to a lesser extent—L. delbrueckii subsp. bulgaricus IMV B-7281. Other strains were resistant or moderately resistant.
Highly sensitive to the effect of antibiotics that are inhibitors of protein synthesis were strains L. plantarum LM VK7, L. acidophilus IMV B-7279, L. casei IMV B-7280 and B. animalis VKB (except clarithromycin, roxithromycin, chlortetracycline). Highly sensitive to the action of aminoglycosides, which mechanism of action is also inhibition of protein synthesis, was L. acidophilus IMV B-7279 strain only. Other strains were mostly moderately resistant to aminoglycosides.
Nitrofurans had different effects on the studied probiotic strains. Thus, L. acidophilus IMV B-7279 strain was highly sensitive, B. animalis VKB was moderately resistant, and L. casei IMV B-7280 and L. rhamnosus LB-3 VK6 strains were resistant. L. delbrueckii subsp. bulgaricus IMV B-7281 strain was sensitive to furadonin. L. delbrueckii LE VK8 and L. plantarum LM VK7 strains were resistant to the action of fusidin and furazolidone, but moderately resistant to furadonin.
Inhibitors of nucleic acid synthesis also had different degrees of effect on the studied strains. All strains were resistant to nalidixic acid. L. delbrueckii LE VK8, L. plantarum LM VK7, and L. casei IMV B-7280 strains were mostly moderately resistant to the action of inhibitors of nucleic acids synthesis, whereas L. rhamnosus LB-3 VK6 strain was resistant. B. animalis VKB was highly sensitive to sparfloxacin, L. acidophilus IMV B-7279 to sparfloxacin and ciprofloxacin, and B. animalis VKL to sparfloxacin and levofloxacin.
Thus, the investigated strains of LAB and bifidobacteria were sensitive to a wide range of antibiotics. The most resistant to the studied drugs were L. rhamnosus LB-3 VK6 and L. delbrueckii LE VK8 strains. Therefore, the sensitivity of the studied strains to antibiotics was strain-depended by nature and did not depend on the genus and species of microorganism.
Resistance to biological agents
The next step was to study the resistance of lactobacilli and bifidobacteria to the action of gastric juice, bile, pancreatin, as well as their complex effect.
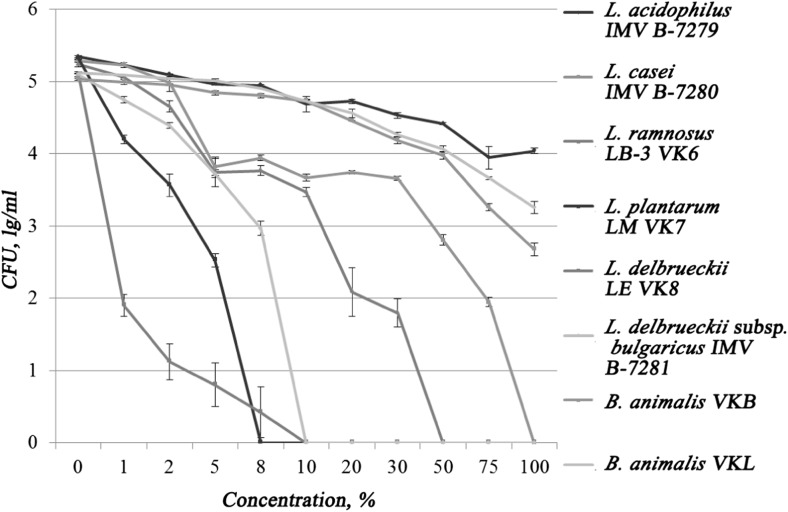
In accordance with Fig. 2, all tested strains showed different resistance to gastric juice. The most susceptible was L. plantarum LM VK7, which stopped its growth at 8% of gastric juice. The next gastric juice-sensitive strain was L. delbrueckii LE VK8, which growth was completely inhibited by gastric juice at a concentration of 10%, with significant growth inhibition already observed at 1% concentration. Complete inhibition of L. delbrueckii subsp. bulgaricus IMV-7281 growth was observed at a 10% gastric juice concentration in a nutrient medium.
Fig. 2.
Resistance of investigated strains to gastric juice
The average resistance to gastric juice had L. rhamnosus LB-3 VK6 and L. casei IMV B-7280 strains. Growth of L. rhamnosus strain LB-3 VK6 was completely suppressed at 50% gastric juice concentration. Growth of the L. casei IMV B-7280 strain was not observed after its cultivation in a 100% gastric juice. Strains L. acidophilus IMV B-7279, B. animalis VKL, and B. animalis VKB were resistant to gastric juice even in the 100% concentration.
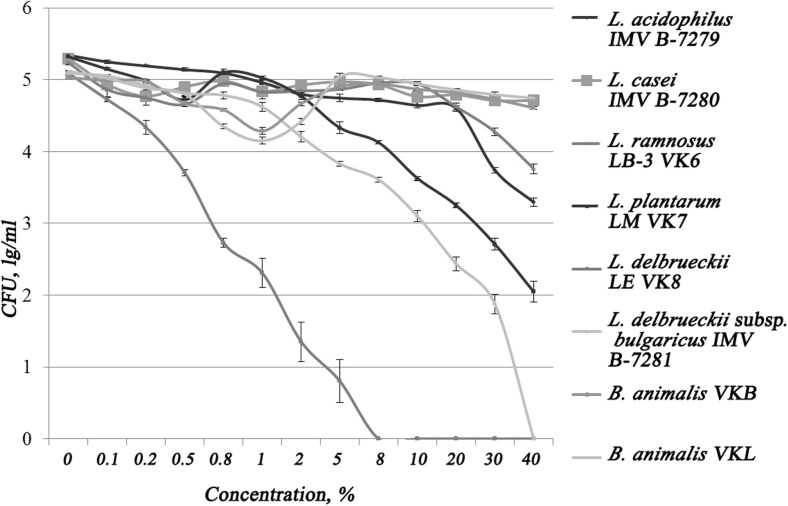
Probiotic strains of LAB and bifidobacteria have shown different resistance to bile (Fig. 3). The most sensitive was strain L. delbrueckii LE VK8, its complete inhibition was observed at a concentration of 8% bile.
Fig. 3.
Strains resistance to bile acids
Lactobacillus delbrueckii subsp. bulgaricus IMV B-7281 and L. plantarum LM VK7 strains had average resistance to bile. L. delbrueckii subsp. bulgaricus IMV B-7281 strain lost its viability after adding bile into medium at a concentration of 40%. The number of live cells of L. plantarum LM VK7 was significantly reduced after strain cultivation in a physiological saline solution with addition of bile in concentrations higher than 10%. The most resistant to bile were L. acidophilus IMV B-7279, L. casei IMV B-7280, L. rhamnosus LB-3 VK6, B. animalis VKL, and B. animalis VKB; their viability was not completely suppressed under the influence of bile in concentrations up to 40% inclusive.
Importantly, that according to our previous studies (unpublished data), B. animalis VKL and B. animalis VKB strains began to lose their viability after culturing in a medium with bile adding at concentrations of 75 and 50%, respectively.
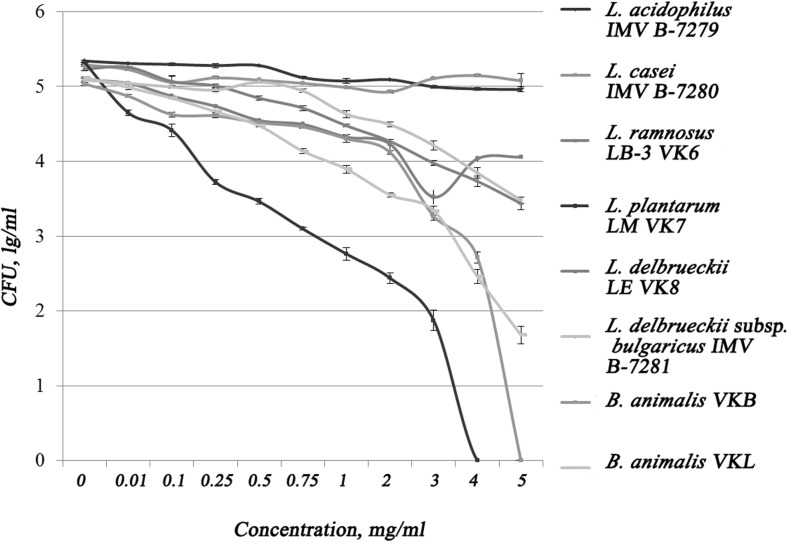
The results of the study of the resistance of probiotic strains of LAB and bifidobacteria to the action of pancreatin are shown in Fig. 4. All strains tested showed different resistance to pancreatin as demonstrated on Fig. 4. The most susceptible were strains L. plantarum LM VK7 and B. animalis VKB.
Fig. 4.
Resistance of strains to the action of proteolytic enzymes
Complete inhibition of L. plantarum LM VK7 was observed at a concentration of 4% proteolytic enzymes. B. animalis VKB strain lost its vitality at 5% proteolytic enzymes. It should be noted that the growth of L. acidophilus IMV B-7279, L. casei IMV B-7280, B. animalis VKL, B. animalis VKB, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. delbrueckii subsp. bulgaricus IMV B-7281 was not suppressed completely under the influence of pancreatin in concentrations up to 5% (inclusively).
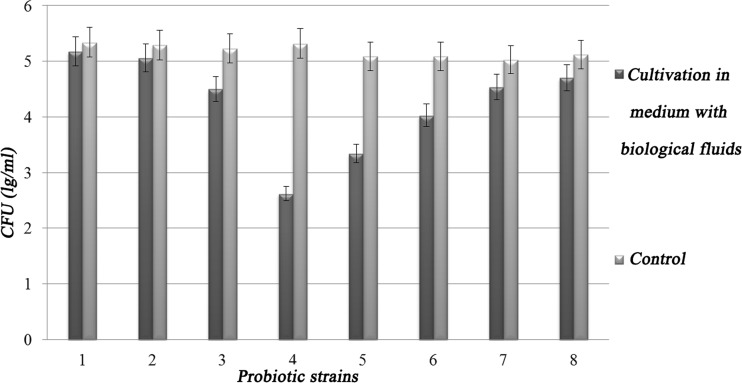
Since in a living organism probiotics must withstand the effect of these biological fluids in stages, we have investigated their complex effect on the viability of probiotic strains of LAB and bifidobacteria. Since the physiological concentrations of biological fluids are constantly changing and often depend on many factors ranging from food content to human health, we have investigated the phased effects of 2% gastric juice, 1% bile, and 1% proteolytic enzymes.
As presented in Fig. 5, investigated probiotic strains have shown different resistance to the complex influences of biological fluids of the gastrointestinal tract. Thus, the most resistant strains L. acidophilus IMV В-7279, L. casei IMV В-7280, B. animalis VKB, and B. animalis VKL, which survival was more than 90% compared to control (96.96, 95.65, 90.16, 91.9, respectively). Moderate sensitivity to the complex action of biological fluids in the gastrointestinal tract was noted for strains of L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. delbrueckii subsp. bulgaricus IMV В-7281, the survival rate of which was, respectively, 50.00, 86.02, 65.64, and 79.16% compared to control. L. plantarum LM VK7 was more sensitive to the complex effects of biological fluids, as its survival rate was 49.25% compared to control.
Fig. 5.
Stability of probiotic strains to the complex action of biological fluids in the gastrointestinal tract, where 1 = L. acidophilus IMV В-7279, 2 = L. casei IMV В-7280, 3 = L. rhamnosus LB-3 VK6, 4 = L. plantarum LM VK7, 5 = L. delbrueckii LE VK8, 6 = L. delbrueckii subsp. bulgaricus IMV В-7281, 7 = B. animalis VKB, and 8 = B. animalis VKL
The ability of LAB and bifidobacteria strains to adhere to epithelial cells in vitro is one of the most important criteria for the selection of potentially probiotic strains for intravaginal use, since it indicates their ability to attach and colonize the vaginal surfaces [18].
Results of study of adhesion of LAB strains L. acidophilus IMV В-7279, L. casei IMV В-7280, L. delbrueckii subsp. bulgaricus IMV В-7281, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, L. plantarum LM VK7, B. animalis VKL, and B. animalis VKB to the buccal epithelium cells are shown in Table. 2.
Table 2.
Adhesive properties of LAB and bifidobacteria strains, M ± m
| Strain | Parameter of adhesion | ||
|---|---|---|---|
| AAR, units | PRE, % | AIM, units | |
| L. acidophilus IMV В-7279 | 2.25 ± 0.10 | 91.82 ± 2.24 | 2.45 ± 0.11 |
| L. delbrueckii subsp. bulgaricus IMV В-7281 | 1.98 ± 0.06 | 75.21 ± 4.57 | 2.64 ± 0.16 |
| L. casei IMV В-7280 | 6.83 ± 0.27 | 87.45 ± 3.29 | 7.81 ± 0.86 |
| B. animalis VKВ | 4.82 ± 0.42 | 93.77 ± 2.69 | 5.14 ± 0.48 |
| B. animalis VKL | 4.02 ± 0.53 | 85.16 ± 4.45 | 4.72 ± 0.54 |
| L. rhamnosus VK6 | 1.96 ± 0.17 | 88.00 ± 4.00 | 2.23 ± 0.29 |
| L. plantarum VK7 | 1.88 ± 0.12 | 88.00 ± 5.00 | 2.14 ± 0.34 |
| L. delbrueckii LE VK8 | 2.12 ± 0.19 | 92.00 ± 3.00 | 2.30 ± 0.24 |
According to AIM: 0–1.75 unit = nonadhesive strains, 1.76–2.5 units = low adhesive properties strains, 2.51–4.0 units = moderate adhesive properties, ≥ 4.0 units = high adhesive properties
The obtained data showed that adhesive properties of the strains according to AIM index were high in L. casei IMV В-7280, B. animalis VKL, and B. animalis VKB; were moderate in L. delbrueckii subsp. bulgaricus IMV В-7281; and were low in L. acidophilus IMV В-7279, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. plantarum LM VK7.
According to the AIM, the strains were distributed as follows:
L. casei IMV В-7280 ≥ B. animalis VKВ ≥ B. animalis VKL ≥ L. delbrueckii subsp. bulgaricus IMV В-7281 ≥ L. acidophilus IMV В-7279 ≥ L. delbrueckii LE VK8 ≥ L. rhamnosus VK6 ≥ L. plantarum VK7.
Consequently, the investigated LAB and bifidobacteria strains can be used in the production of probiotics for oral use, since they have eliminated the resistance to gastric juice, bile, pancreatic enzymes in physiological concentrations, had the ability to adhere to buccal epithelium cells, and also had different sensitivity to antibiotics of different groups.
Discussion
Resistance to antibiotics
Thus, the data received on the antibiotic resistance of individual strains of LAB and bifidobacteria partially coincide with the data of other researchers [30, 31, 47–52]. It has been shown that antibiotic resistant bacteria via food chain could be transmitted from animals to humans [48] and most LAB isolated from humans and farm animals are susceptible to amikacin, ampicillin, first generation of cephalosporins, erythromycin, gentamicin, imipinem, oxacillin, and penicillin. Teuber et al. [47] reported that bifidobacteria have sensitivity to ampicillin, penicillin, cephalosporin, erythromycin, and tetracycline and resistance to vancomycin, gentamicin, and streptomycin. It was also reported that B. animalis, L. delbrueckii subsp. bulgaricus, L. casei, and L. acidophilus were susceptible to ampicillin, bacitracin, clindamycin, dicloxacillin, erytromycin, novobiocin, and penicillin G and strain-dependent susceptible to cephalothin, chloramphenicol, gentamicin, lincomycin, metronidazole, neomycin, paromomycin, streptomycin, tetracycline, and vancomycin [31]. We have determined that B. animalis VKB was susceptible to doxycycline and erythromycin, moderately sensitive to penicillin and canamicin resistant, but it was moderately resistant to ampicillin, penicillin, and tetracycline, and resistant to ofloxacin. Similar patterns were observed with respect to other strains examined. These results have some discrepancies with literature data, since all biological properties are strongly strain-dependent and may have certain differences even among similar strains. Current data have much importance for individualized application of probiotics.
A matter of concern to use resistant to antibiotics strains is that antibiotic resistance per se is not a parameter of safety due to the risk of resistance transfer to pathogenic strains [49–51]. Antibiotic resistant determinants have been previously demonstrated to have the ability to transfer from one Lactobacillus to another and, also from Lactobacilli to other species including pathogens such as Staphylococcus [52, 53].
The studied strains meet such important selection criteria as antibiotic resistance according to international guidelines for probiotics like the FAO and WHO [2] and European Food Safety Authority (EFSA) [54, 55].
Resistance to gastric juice
Probiotic strains should have ability to survive the gastrointestinal tract of the host. Such properties as stress response mechanisms and adhesion and colonization factors, as well as by taking advantage of specific energy recruitment pathways increase survival and facilitate to carry out their functional activities [56]. Investigated strains of LAB and bifidobacteria have shown resistance to gastric juice, bile, and pancreatin separately and to their complex action, which is consistent with the results obtained by other researchers. Liu et al. reported that the bifidobacteria strains are resistant to the conditions of the gastrointestinal tract [43]. Chou et al. reported similar results on L. acidophilus [44] and noted that their stability depends on the acidity of bile and gastric juice and cultivation time. Consequently, the studied strains tested meet the requirements for probiotics according to this parameter [24, 54, 55].
Resistance to bile acids
Bile tolerance is one of the most crucial properties for probiotic bacteria and among others as it determines its ability to survive in the small intestine and consequently, its capacity to play its functional role as a probiotic [57]. It is not commonly detected, therefore Hassanzadazar et al. reported that among 27 isolated strains, only one, namely L. casei, could tolerate acid and bile salt and had antibacterial activity [58].
Recently, the novel modalities for therapeutics intervention targeting the gut microbiome or plasma BAs were reported [59–61] based on the discovery of metabolic significance of BA signaling. Gut microbiota can alters BA composition contributing to the biotransformation of primary BAs to secondary BAs and to the activation or inhibition of FXR [60].
Gu et al. [61] suggested that gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment via so-called acarbose-gut microbiota-BA axis and distinguished two microbiome clusters (Bacteroides and Prevotella clusters) interacting with BA metabolism.
Cholesterol metabolism via bile acids interplay with microbiome
Since cholesterol metabolism has strong crosslinks with bile acids circulation according to the “bile salt hydrolase hypothesis” (BSH) and the potential associations between immune modulatory and hypocholesterolemic activity of probiotic strains formulated [8], finding on bile resistance might have a strong input on management patients with cholestasis, atherosclerosis, and associated conditions.
Some strains of LAB and bifidobacteria had effective hypocholesterolemic activity in various models of metabolic diseases and in vitro associated with enzymatic degradation of bile acids, direct binding of cholesterol by cell walls of bacteria changes in expression of several genes involved in lipid metabolism [58, 62, 63]. Almost all bifidobacteria species showed BSH activity, while this activity was detected only in a few species of LAB [62]. In addition, many of the bacterial species of the phylum Firmicutes produce butyrate, and a decreased abundance of these bacteria was observed in patients with colorectal cancer [63]. L. casei IMV B-7280 (separately) and composition of B. animalis VKL/B. animalis VKB/L. casei IMV B-7280 are effective to decrease cholesterol level, beneficially modulate gut microbiome, and recover the liver morphology in high calorie-induced obesity [8].
Resistance to pancreatic juice
All the tested strains are sensitive to artificial and human pancreatic juice depending on time contact while bifidobacterium strains were found in the study by Del Piano [64] to be more sensitive than LAB strains in particular at higher time contact. No significant difference reported between sensitivity to simulated and human pancreatic juice was reported. Artificial pancreatic fluid is recommended as a standardized, easier, and less costly procedure to test probiotics activity [65]. The prolonged exposures to acid stress is suggested to improve the stability of probiotic strains in the human gut indicating strategy for the production of robust probiotic strains [65].
Adhesiveness, pili
Gram-positive strains of genus Lactobacillus have bacterial pili for strengthening adhesion to mucus and protection against stresses of the environment. However, molecular mechanism of mucosal mechanical properties of bacterial pili and their role in immune interaction are still largely unknown [66–69]. Gene cluster spaCBA, and the pilus-associated SpaC pilin makes possible exertion both long distance and intimate contact with tissue of the host and provides mucus-binding [69]. Gram-positive pili are often modified by sortase-specific cleavage reactions. Glycosylation as a modification of sortase-dependent pili has been reported to play in the immunomodulation of the host by glycoproteins of beneficial [69]. These findings altered vision of underappreciated role of glycoconjugates in bacteria-host interplay. Modification of the complex heterotrimeric pili is of importance for the functional interaction with the host immune lectin receptor DC-SIGN on human dendritic cells [69]. Interaction between pili and whey proteins depends on pH; at acidic pH, pili are immobilized in the collapsed EPS layer; and at neutral pH, pili are easily accessible for interaction with whey proteins [70]. Assessment of probiotic bacteria wall using imaging and molecular modalities is important to evaluate or predict role in adhesion properties and immunomodulatory potential of the strain [39, 71–73]. Growing knowledge on nanomechanics of pili can help to design nanomaterials (potential prebiotics) capable of promote adhesive properties for benefits of bacterial-host interactions [74].
Probiotic gut colonization and biofilm formation and microbiome profiles
Microorganisms are usually studied either in isolation as monoclonal model populations that we manage to grow in the laboratory or highly complex natural communities with influence of number unpredictable factors [75, 76]. Probiotics are able to form complex communities, known as biofilms producing an extracellular polysaccharide matrix, expanding colonization in gut and using adhesion properties of bacteria [77, 78]. This helps to prolong and stabilize their residence in the epithelium and triggering the immune response of the host cell [77]. This facilitate probiotic strains to show their antimicrobial potential against human pathogens [79]. Collectively, the disease protection activity of LAB is correlated with their spatial distribution in the intestinal tissue, with strains showing a balanced distribution (hybrid type) more efficient in protection [80]. Probiotic combinations that inhibit and displace pathogens may be excellent candidates to use in fermented milk products [81]. Using new encapsulation technologies based on ability to biofilms formation allow development of fourth-generation probiotics [77].
Gut microbiota being proved with body of evidence to be key players in the regulation of host energy homoeostasis and in the pathogenesis of obesity remain limited to be properly studied in clinical set due to effects of number unpredictable and poorly controllable factors in humans [75, 76, 82]. Thus, genetic and environmental factors (diet, drug assumption, lifestyle, and the complex interactions within the complexity of microbiome as a whole till inadequately investigated and can evoke significant inconsistency in the results observed in such trials [8, 82].
Searching for reliable phenotype markers of microbiome relevant for longitudinal observation and reproducible in large population is an essential task for microbiome and probiotic research in clinicо.
Recently, mycobiome was suggested to have the protective benefits via intestinal colonization by commensal fungi [83, 84] that functionally replace intestinal bacteria in alleviating tissue injury and positively activate protective CD8 T cells. Thus, commensal gut fungi protect local and systemic immunity reactivity by providing tonic microbial stimulation that can functionally replace intestinal bacteria.
Microbial diversity is an important parameter of intestinal health. Thus, lower richness of gut microbiota compositions was found in Western diet consumers shapes the microbial ecosystem [15] and in the populations under the burden of obesity and metabolic disease [85, 86]. Individuals with higher diversity were reported to have a healthier dietary pattern [87, 88]. In a subset from the lower diversity was associated with greater abdominal adiposity according to the TwinsUK cohort study [89]. Variety of metabolites are modulated by the action of gut microbiota richness, number of recently discovered crosslinks between gut microbes, and different circulating metabolites with high predictive and diagnostic potential have been recently identified [90].
Pathologic microbiomes in gut and in distant sites—beneficial modulation by probiotics
In the current research, the probiotic bacteria of LAB and bifidobacteria demonstrated different strain-dependent properties (ability for adhesion and resistance to antibiotics and biological fluids) strongly relevant for their clinical application for effective modulation of microbiota in the gut and in distant sites (e.g., oral, skin, vaginal, and other sites microbiota). This supposes their potential secondary beneficial effects for individualized use in adherence with patient’s needs.
Thus, according to their adhesive properties according to the AIM, the strains were distributed in the sequence as follows: L. casei IMV В-7280 ≥ B. animalis VKВ ≥ B. animalis VKL ≥ L. delbrueckii subsp. bulgaricus IMV В-7281 ≥ L. acidophilus IMV В-7279 ≥ L. delbrueckii LE VK8 ≥ L. rhamnosus VK 6 ≥ L. plantarum VK 7. This property provide an opportunity for the bacteria to be fixed on the mucosa and effectively modulate oral, vaginal microbiome and initiate complex immune response [12], and enhance therapeutic potential for conditions, in which treatment by probiotics is still not supported with significant evidence, like bacterial vaginosis, urinary tract infections, periodontitis, and wound healing, etc. Adhesive properties of strains favor biofilm formation that alter interstrain interaction; therefore, synergic activity of probiotic strains appear rather speculative. In our previous works and in the literature, only few strain compositions demonstrate specifically higher activity [8] and comparative was not assessed properly up to date.
We hypothesize that composition prescription if necessary should be carefully selected for individual case and recommend to always pick one optimal strain or include as less strains as possible. Effects of separate or combined use with prebiotics should be studied.
The level tolerance to antibiotics has importance for probiotic safety considering AMR genes transfer and the concept of “resistome” and also has a potential to stratify strains according to the antibiotic use history during patient’s life, and pertinence for infections prevention.
Tolerance to gastric juice and to pancreatic enzymes can provide insights for probiotic pharmacokinetics and development of microencapsulation for probiotic preparations [64], nanomaterials, and potential prebiotics [4] to enhance probiotic function [74], consider conditions like gastritis, hyperacidity on the probiotic efficacy; and on other hand, can suppose treatment modalities for gut diseases like dyspepsia, IBS, IBD, postoperative gut, personalized malnutrition correction, and open prospects for development ferment-guided personalized diets.
These studied basic bacterial characteristics can contribute to many secondary properties of probiotic species. Thus, relatively neutral to trigger biological effects properties like elasticity of bacterial wall can be measured using subcellular imaging [39, 71–73]; in newly discovered strains to predict their probiotic potential and justify stratification strains on potential for immunomodulatory activity [39].
Immunomodulatory properties [5, 7, 39] are hypothesized to be a pillar of clinical effect of beneficial microbes. Development the concept and the term of “immunobiotics”—a probiotics with pronounced immunomodilative activity is a promising avenue for future research [7]. This associated with anti-inflammatory properties [5, 7], treatment broad immune-related pathological conditions; enhancement efficacy of immunotherapy [91], and use of vaccines.
It has been hypothesized that immunomodulatory and anti-inflammatory properties [5] likely correlate with secondary clinically relevant effects, in particular the ability to modulate metabolic conditions and to demonstrate antiobesogenic, hypocholesterolemic, and liver protective properties [8]; hypouricemic activity has been reported [92] and should be further studied to provide treatment agenda for patients with gout, also using prebiotics [93].
All probiotic strains demonstrate their primary native antibacterial [94], antiviral [95, 96], and antifungal properties [83, 84, 97], and have a clear perspective to be a routine alternative for antibiotics.
Oxygen tolerance of probiotic strains is another essential parameter to be studied in the future. Only a few studies have been done in the field [98, 99]; LAB can potentiate intestinal hypoxia-inducible factor [100]. These data can open new perspectives to manage cases associated with hypoxia-associated conditions and stress [100]; develop individualized treatment to patients demonstrating Flammer syndrome phenotype [101]; consider patient stratification on important gut marker as hypoxia signaling, mesenteric ischemia in patients with atherosclerosis [102], and ischemic niches in cancer genesis [101].
Potential of probiotics for enhancement safety and efficacy of regenerative therapy [103], transplantation stem cells is intriguing challenge to develop hybrid biological therapies.
Finally, many mentioned properties should be implemented to cancer case management as supportive therapy [18] and to facilitate symptoms, associated with treatment [104, 105]. Age and gender aspects are essential issues for selection probiotic species for individual use.
Recommendation for individualized clinical use of probiotics
Product quality;
Effectiveness should be proven on the basis of evidence-based medicine for routine use in the clinical setting;
Personalized (or individualized) approach needed in prescribing probiotic according to the disease, clinical case, and phenotype of the patient;
Using live microorganisms is essential for therapeutic effect;
Selecting the “best” strain for particular case (for example, the L casei strain has strongest properties in most characteristics);
The higher effectiveness of multiprobiotics has not been proved;
The dose should be at least 109 microbial bodies per 1 ml;
Use of “prebiotic” substances;
The appropriate route of delivering a probiotic drug (capsule, gel, novel encapsulation technologies);
Crucially important is combination with an appropriate diet.
Summarizing data of properties of individual probiotic strains (those studied and augmented with literature data) for translating to the human microbiome working as a whole resulting on host’s health are presented in the Table 3.
Table 3.
LAB and bifidobacteria probiotic strains properties and potential secondary effects for beneficial individualized use in adherence with patient’s needs
| Strain property | Strains with rather low activity | Strains with rather high activity | Host phenotype, clinical condition, potential application |
|---|---|---|---|
| Adhesive properties | According to the AIM, the strains were distributed as follows: L. casei IMV В-7280 ≥ B. animalis VKВ ≥ B. animalis VKL ≥ L. delbrueckii subsp. bulgaricus IMV В-7281 ≥ L. acidophilus IMV В-7279 ≥ L. delbrueckii LE VK8 ≥ L. rhamnosus VK6 ≥ L. plantarum VK7 |
Distant sites [12] use: oral, vaginal, skin microbiome, wound healing; microencapsulation for probiotic preparations [64]; development of nanomaterials (potential prebiotic [4]) to enhance probiotic function [74]; using subcellular imaging [39, 71–73]; microbiome phenotype (diversity), biofilms | |
| Tolerance to antibiotics | Highly sensitive to the effect of antibiotics that are inhibitors of protein synthesis were strains L. plantarum LM VK7, L. acidophilus IMV B-7279, L. casei IMV B-7280 and B. animalis VKB | – | Probiotic use safety, stratification strains according to antibiotic use history during patient’s life, adhere to infection prevention; AMR genes, “resistome” |
| Tolerance to gastric juice | L. plantarum LM VK7 | L. acidophilus IMV В-7279, L. casei IMV В-7280, B. animalis VKB, B. animalis VKL | Gastritis, microencapsulation is the future of probiotic preparations [64] |
| Tolerance to bile | The most susceptible were strains L. plantarum LM VK7 and B. animalis VKB. Complete growth inhibition of L. plantarum LM VK7 was observed at a concentration of 4% proteolytic enzymes. B. animalis VKB strain lost its vitality at 5% proteolytic enzymes | The growth of L. acidophilus IMV B-7279, L. casei IMV B-7280, B. animalis VKL, B. animalis VKB, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. delbrueckii subsp. bulgaricus IMV B-7281 was not suppressed completely under the influence of pancreatin in concentrations up to 5% (inclusive) | Cholestasis; BA-associated diseases, cholesterol metabolism |
| Tolerance to pancreatic enzymes | Bifidobacterium strains was reported to be more sensitive than LAB strains [64] | Strains L. acidophilus IMV В-7279, L. casei IMV В-7280, B. animalis VKL, B. animalis VKB, L. rhamnosus LB-3 VK6, L. delbrueckii LE VK8, and L. delbrueckii subsp. bulgaricus IMV В-7281 were resistant to pancreatic enzymes | Dyspepsia, IBS, IBD, postoperative gut; personalized malnutrition correction; prospects for development ferment-guided personalized diets |
| Elasticity of wall [39] | The rigidity of the cell walls among LAB was distributed as follows: L. acidophilus IMV B-7279 > L. casei IMV B-7280 > L. delbrueckii subsp. bulgaricus IMV B-7281; among the strains of bifidobacteria: B. animalis VKB > B. animalis VKL | Stratification strains on potential for immunomodulatory activity | |
| Immunomodulatory properties [5, 7, 39] |
L. delbruеckii subsp. bulgaricus IMV B-7281, B. animalis VKL, and B. animalis VKB |
L. acidophilus IMV B-7279 or L. casei IMV B-7280 | Development immunobiotics [7] broad immune-related pathological states; enhance immunotherapy [91] |
| Anti-inflammatory properties [5] | Variety of bifidobacterium and LAB strains | Clostridial infection, respiratory infections (evidence-supported), vaginal infection, dysbiosis, visceral pain, cancer, etc. | |
| Antiobesogenic [8] | B. animalis VKB/B. animalis VKL | L. casei IMV B-7280, L. delbrueckii subsp. bulgaricus IMV B-7281, B. аnimalis VKB, B. аnimalis VKL (separately) or B. animalis VKL/B. animalis VKB/L. casei IMV B-7280, and L. casei IMV B-7280/L. delbrueckii subsp. bulgaricus IMV B-7281 | Metabolic syndrome (MetS) management |
| Hypocholesterolemic properties [8] | B. animalіs VKL and B. animalіs VKB | L. casei IMV B-7280 and L. delbrueckii subsp. bulgaricus. Nearly all bifidobacteria strains but only few species of LAB showed BSH activity [62]. | Cardio-vascular diseases, MetS |
| Liver protective properties [8] | B. animalіs VKB or L. delbrueckii subsp. bulgaricus IMV B-7281 (separately) and B. animalіs VKL/B. animalіs VKB | L. delbrueckii subsp. bulgaricus IMV B-7281, B. аnimalis VKB | Liver fibrosis, cancer |
| Hypouricemic properties [92]* | B. longum 51A | Gout; use of prebiotics (phenugreek [93]) | |
| Effects with prebiotics | To be studied—concept has been modified [4] | Create strain-dependent synbiotics and disease-specific for particular conditions (reproductive function [106]), kidney involvement [93]) | |
| Antibacterial [94], antiviral [95, 96] properties | L. casei IMV B-7280, L. acidophilus IMV B-7279, B. animalis VKL, and B. animalis VKB have antibacterial and LAB have antiviral properties | Various infections, cancer | |
| Antifungal properties [83, 84, 97]* | L. plantarum [84] | Promising to stratify host’s phenotype (to be studied) | |
| Vaccine efficacy enhancement [107, 108]* | L. casei IMV B-7280 –VHB vaccine [107]; B. longum BL999 L. rhamnosus LPR (Hep B vaccine); L. casei GG (LGG)—oral rotavirus vaccine; L. rhamnosus GG—vaccine against H1N1 influenza [108] | Prevention and treatment infectious diseases in particular groups of patients | |
| Oxygen tolerance [98, 99]* | To be studied. Only a few studies have been conducted on the oxygen tolerance of probiotic bacteria. Most of these studies have focused on Bifidobacterium spp. Little is known about the effect of oxygen on the physiology of L. acidophilus L. rhamnosus GG can potentiate intestinal hypoxia-inducible factor [100] |
Relevance to hypoxic-conditions, stress; hypoxia-inducible factor [100]; Flammer syndrome phenotype; metastatic cancer [101]; mesenteric ischemia [102]; regenerative treatments [102] | |
| Regenerative therapy [103]* | To be studied | Potential for enhancement safety and efficacy of regenerative therapy [103], transplantation stem cells | |
*Augmented with literature data
Conclusion
We recognized strain-dependent properties of studied LAB and bifidobacteria probiotic strains (adhesive ability, resistance to antibiotics and gut biological fluids) and discussed potential for most effective individualized treatment for gut and distant sites microbiome modulation in adherence with patient’s needs according to host phenotype to obtain the best effect possible. These strains correspond to the probiotic characteristics according to probiotic guidelines, are safe and can be recommended for creation of probiotic preparations for application in humans.
Highlights and recommendations
Current research demonstrated that necessary requirements for the inclusion of studied strains into the composition of probiotic preparations were fully met [2, 54, 55, 65].
Current research confirms the safety of biological properties of potentially probiotic strains of lactobacilli (LAB) and bifidobacteria;
Studied strains adhere requirement to probiotic guidelines [2, 54, 55];
Biological properties of studied bacteria are strain-dependent;
Individual characteristics provide relevant data for clinical diagnosis and stratification to facilitate individualized/personalized use;
Other phenotypic changes that require an individual characterization should be studied [65];
Patient’s phenotype stratification is needed for individualized probiotics use.
We believe that a comprehensive approach for evaluating properties of bacteria to select the “best” probiotic strains for development effective probiotic drug and follow up with further preclinical and clinical studies. Host’s phenotype should be clearly analyzed to suggest a panel of biomarkers for individualized use of probiotics and prebiotics. Some properties demonstrate clear associations among each other with significant correlations [8, 39] and this interplay has high importance for further research. Secondary clinically relevant properties of probiotic treatment can be considered like anti-aging or gut-brain axis effects. Mathematical approach using a variety of algorithmic calculation would provide preparing evidence-supported guideline for prevention and treatment. Considering anticipated rapid microbiome and probiotics research progress within paradigm of predictive preventive personalized medicine, the unification of multidisciplinary approaches is a demand. Thus, as a completion of this new knowledge, open potential road in this direction of creating the consolidated model of medical application of probiotics for PPPM as follows.
Personalized and/or individualized approach
“Individual” phenotype of bacterial characteristics considered for most effective individualized treatment via gut, oral, and vaginal and other sites microbiome modulation according to phenotype of the patient is excellent example of individualized medicine, while using broad panel of molecular biomarkers and genomic approach (including microbiome obtained in the microbiome-wide association studies) these data will be easily self-translated to “personalized” medical approach for probiotics. Clarification terms “personalized”/“individualized” for probiotics use is important question [13] and also to establish correlations and associations between both approaches.
On the other hand, raising the question for future innovative research needed in direction of cumulating evidence, since, according to Cochrane reviews, the effects of probiotics are considered “evidence-based” only in cases of antibiotic- and C. difficile-associated diarrhea and respiratory tract infections [18], the incomplete evidence is a possibly biased by the heterogeneity of trials conducted. Therefore novel PPPM-oriented protocols for a randomized controlled trial using the microbiome-wide association studies on the largest cohorts possible should be developed; e.g., recently protocol was suggested for type 2 diabetes [109]. Probiotic interstrain reciprocity and study of growing panel of potential prebiotics is a great challenge for near future.
Microbiome phenotypes like microbial diversity, fungal (mycobiome) component, and many others are underestimated parameters of predictive medicine to recognize predispositions and evaluate treatment responses almost to any existing disease and number of phenotype markers to be effectively considered during microbiome modulation.
Strong and well-documented preventive potential of probiotics includes evidence-supported prevention of respiratory infections and gut disorders [18] and prevention and treatment infectious diseases via enhancement of vaccine use [107, 108].
A clear message sounds from the research to avoid overusing antibiotics and broadly propagate the knowledge in this concern. This can reduce the incidence of hospital infections and largely preserve public health.
Acknowledgements
The study was conducted with the support of the State Agency on Science, Innovations, and Informatization of Ukraine.
Authors’ contributions
RVB suggested the idea and design of the study, participated in experiments, did the study analysis, prepared discussion, formulated future outlooks, and performed article drafting. LML and LPB prepared the first draft of manuscript, did the literature analysis, interpreted the results, and performed the statistical analysis. LPB and LML, performed the experiments on animals and participated analysis of the study. VVM performed the experiments. MYS did the organization, revision manuscript, and data interpretation, and contributed to the overall development of the studied topic. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Rostyslav V. Bubnov, Email: rostbubnov@gmail.com
Lidiia P. Babenko, Email: babenkolidiia@gmail.com
Liudmyla M. Lazarenko, Email: lazarenkolm@gmail.com
Victoria V. Mokrozub, Email: viktoriiamokrozub@gmail.com
Mykola Ya. Spivak, Email: n.spivak@ukr.net
References
- 1.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol. 2015;6:e91. doi: 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO/FAO scientific document. http://who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf. Accessed 11 Feb 2018.
- 3.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;8:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 4.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14(8):491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 5.Bubnov RV, Spivak MY, Lazarenko LM, Bomba A, Boyko NV. Probiotics and immunity: provisional role for personalized diets and disease prevention. EPMA J. 2015;6:14. doi: 10.1186/s13167-015-0036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aron-Wisnewsky J, Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol. 2016 Mar;12(3):169-81. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed]
- 7.Lazarenko LM, Babenko LP, Bubnov RV, Demchenko OM, Zotsenko VM, Boyko NV, et al. Imunobiotics are the novel biotech drugs with antibacterial and immunomodulatory properties. Mikrobiol Z. 2017;79(1):66–75. doi: 10.15407/microbiolj79.01.066. [DOI] [Google Scholar]
- 8.Bubnov RV, Babenko LP, Lazarenko LM, Mokrozub VV, Demchenko OA, Nechypurenko OV, et al. Comparative study of probiotic effects of lactobacillus and bifidobacteria strains on cholesterol levels, liver morphology and the gut microbiota in obese mice. EPMA J. 2017;8(4):357–376. doi: 10.1007/s13167-017-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jobin C. Precision medicine using microbiota. Science. 2018;359(6371):32–34. doi: 10.1126/science.aar2946. [DOI] [PubMed] [Google Scholar]
- 10.Lebeer S, Bron PA, Marco ML, VanPijkeren JP, O'Connell Motherway M, Hill C, et al. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol. 2017;49:217–23. 10.1016/j.copbio.2017.10.007. [DOI] [PubMed]
- 11.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reid G, Abrahamsson T, Bailey M, Bindels LB, Bubnov R, Ganguli K, et al. How do probiotics and prebiotics function at distant sites? Benef Microbes. 2017;8(4):521–533. doi: 10.3920/BM2016.0222. [DOI] [PubMed] [Google Scholar]
- 13.Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation—EPMA position paper 2016. EPMA J. 2016;7:23. doi: 10.1186/s13167-016-0072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro H, Suez J, Elinav E. Personalized microbiome-based approaches to metabolic syndrome management and prevention. J Diabetes. 2017;9(3):226–36. 10.1111/1753-0407.12501. Review. [DOI] [PubMed]
- 15.Dao MC, Clément K. Gut microbiota and obesity: Concepts relevant to clinical care. Eur J Intern Med. 2018;48:18-24. 10.1016/j.ejim.2017.10.005. [DOI] [PubMed]
- 16.van den Nieuwboer M, Browne PD, Claassen E. Patient needs and research priorities in probiotics: a quantitative KOL prioritization analysis with emphasis on infants and children. Pharma Nutrition. 2016;4(1):19–28. doi: 10.1016/j.phanu.2015.09.004. [DOI] [Google Scholar]
- 17.Park S, Bae JH. Probiotics for weight loss: a systematic review and meta-analysis. Nutr Res. 2015;35:566–575. doi: 10.1016/j.nutres.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Rondanelli M, Faliva MA, Perna S, Giacosa A, Peroni G, Castellazzi AM. Using probiotics in clinical practice: where are we now? A review ofexisting meta-analyses. Gut Microbes. 2017;8(6):521–543. doi: 10.1080/19490976.2017.1345414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Papadimitriou K, Zoumpopoulou G, Foligné B, et al. Discovering probiotic microorganisms: in vitro, in vivo, genetic and omics approaches. Front Microbiol. 2015;6:58. doi: 10.3389/fmicb.2015.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fijan S. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health. 2014;11(5):4745–4767. doi: 10.3390/ijerph110504745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen TL, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. 2015;8(1):1–16. doi: 10.1242/dmm.017400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neville BA, Forster SC, Lawley TD. Commensal Koch’s postulates: establishing causation in human microbiota research. CurrOpinMicrobiol. 2017;42:47–52. doi: 10.1016/j.mib.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Aimmo MR, Mattarelli P, Biavati B, Carlsson NG, Andlid T. The potential of bifidobacteria as a source of natural folate. J Appl Microbiol. 2012;112(5):975–84. 10.1111/j.1365-2672.2012.05261.x. [DOI] [PubMed]
- 26.Lilly DM, Stillwell RH. Growth promoting factors produced by probiotics. Science. 1965;147:747–748. doi: 10.1126/science.147.3659.747. [DOI] [PubMed] [Google Scholar]
- 27.Gao C, Ganesh BP, Shi Z, Shah RR, Fultz R, Major A, et al. Gut microbe-mediated suppression of inflammation-associated colon carcinogenesis by luminal histamine production. Am J Pathol. 2017 Oct;187(10):2323–36. 10.1016/j.ajpath.2017.06.011. [DOI] [PMC free article] [PubMed]
- 28.Bermudez-Brito M, Plaza-Díaz J, Muñoz-Quezada S, Gómez-Llorente C, Gil A. Probiotic mechanisms of action. Ann Nutr Metab. 2012;61(2):160–74. 10.1159/000342079. [DOI] [PubMed]
- 29.Rodrigues RR, Greer RL, Dong X, DSouza KN, Gurung M, Wu JY, et al. Antibiotic-induced alterations in gut microbiota are associated with changes in glucose metabolism in healthy mice. Front Microbiol. 2017;8:2306. 10.3389/fmicb.2017.02306. [DOI] [PMC free article] [PubMed]
- 30.Penders J, Stobberingh EE, Savelkoul PHM, Wolffs PFG. The human microbiome as a reservoir of antimicrobial resistance. Front Microbiol. 2013;4:87. doi: 10.3389/fmicb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Aimmo MR, Modesto M, Biavati B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int J Food Microbiol. 2007;115(1):35–42. doi: 10.1016/j.ijfoodmicro.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 32.Avdeeva LV, Osadchaia AI, Kharkhota MA. Influence of lactitol and lactulose on adhesion properties of Bacillus subtilis probiotic strains. Mikrobiol Z. 2012;74(5):22–5. [PubMed]
- 33.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18:1185–93. [DOI] [PubMed]
- 34.Lagier JC, Hugon P, Khelaifia S, Fournier PE, la Scola B, Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015;28:237–264. doi: 10.1128/CMR.00014-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH et al.: Culture of previously uncultured members of the human gut microbiota by culturomics. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Retracted]
- 36.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, et al. Culturing of “unculturable” human microbiota reveals novel taxa and extensive sporulation. Nature. 2016 May 26;533(7604):543–6. 10.1038/nature17645. [DOI] [PMC free article] [PubMed]
- 37.Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. Bifidobacteria and their molecular communication with the immune system. Front Microbiol. 2017;8:2345. doi: 10.3389/fmicb.2017.02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi H, Kanmani P, Ishizuka T, Miyazaki A, Soma J, Albarracin L, et al. Development of an in vitro immunobiotic evaluation system against rotavirus infection in bovine intestinal epitheliocytes. Benef Microbes. 2017;8:309–21. 10.3920/BM2016.0155. [DOI] [PubMed]
- 39.Mokrozub VV, Lazarenko LM, Sichel LM, Bubnov RV, Spivak MY. The role of beneficial bacteria wall elasticity in regulating innate immune response. EPMA J. 2015;6:13. doi: 10.1186/s13167-015-0035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hidalgo-Cantabrana C, Sánchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. ApplEnvironMicrobiol. 2014;80(1):9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hidalgo-Cantabrana C, Sánchez B, Álvarez-Martín P, López P, Martínez-Álvarez N, Delley M, et al. A single mutation in the gene responsible for the mucoid phenotype of Bifidobacterium animalis subsp. lactis confers surface and functional characteristics. ApplEnvironMicrobiol. 2015;81(23):7960–8. 10.1128/AEM.02095-15. [DOI] [PMC free article] [PubMed]
- 42.Kailasapathy K, Chin J. Survival and therapeutic potential of probiotic organisms with reference to lactobacillus acidophilus and Bifidobacterium spp. Immunol Cell Biol. 2000;78(1):80–8. [DOI] [PubMed]
- 43.Liu Z, Jiang Z, Zhou K, Li P, Liu G, Zhang B. Screening of bifidobacteria with acquired tolerance to human gastrointestinal tract. Anaerobe. 2007;13(5–6):215–219. doi: 10.1016/j.anaerobe.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Chou LS, Weimer B. Isolation and characterization of acid- and bile-tolerant isolates from strains of lactobacillus acidophilus. J Dairy Sci. 1999;82(1):23–31. doi: 10.3168/jds.S0022-0302(99)75204-5. [DOI] [PubMed] [Google Scholar]
- 45.Bernet MF, Brassart D, Neeser JR, Servin AL. Adhesion of human bifidobacterial strains to cultured human intestinal epithelial cells and inhibition of enteropathogen-cell interactions. Appl Environ Microbiol. 1993;59(12):4121–4128. doi: 10.1128/aem.59.12.4121-4128.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Golding CG, Lamboo LL, Beniac DR, Booth TF. The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci Rep. 2016;6:26516. doi: 10.1038/srep26516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teuber M, Meile L, Schwarz F. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek. 1999;76(1–4):115–137. doi: 10.1023/A:1002035622988. [DOI] [PubMed] [Google Scholar]
- 48.Singer RS, Finch R, Wegener HC, Bywater R, Walters J, Lipsitch M. Antibiotic resistance—the interplay between antibiotic use in animals and human beings. Lancet Infect Dis. 2003;3(1):47–51. doi: 10.1016/S1473-3099(03)00490-0. [DOI] [PubMed] [Google Scholar]
- 49.Gueimonde M, Sánchez B, de los Reyes-Gavilán CG, Margolles A. Antibiotic resistance in probiotic bacteria. Front Microbiol. 2013;4:202. doi: 10.3389/fmicb.2013.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng M, Zhang R, Tian X, Zhou X, Pan X, Wong A. Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front Microbiol. 2017;8:908. doi: 10.3389/fmicb.2017.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R. Antibiotic resistance among commercially available probiotics. Food Res Int. 2014;57:176–195. doi: 10.1016/j.foodres.2014.01.025. [DOI] [Google Scholar]
- 52.Tannock GW, Luchansky JB, Miller L, Connell H, Thode-Andersen S, Mercer AA, et al. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-63. Plasmid. 1994;31(1):60–71. [DOI] [PubMed]
- 53.Mater DD, Langella P, Corthier G, Flores MJ. A probiotic lactobacillus strain can acquire vancomycin resistance during digestive transit in mice. J Mol Microbiol Biotechnol. 2008;14(1–3):123–127. doi: 10.1159/000106091. [DOI] [PubMed] [Google Scholar]
- 54.EFSA NDA Panel (EFSA Panel on Dietetic Products, Nutrition and Allergies) General scientific guidance for stakeholders on health claim applications. EFSA Journal. 2016;14(1):4367. doi: 10.2903/j.efsa.2016.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.European Food Safety Authority (EFSA) Technical guidance—update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J. 2008;732:1–15. doi: 10.2903/j.efsa.2008.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.González-Rodríguez I, Ruiz L, Gueimonde M, Margolles A, Sánchez B. Factors involved in the colonization and survival of bifidobacteria in the gastrointestinal tract. FEMS Microbiol Lett. 2013;340(1):1–10. doi: 10.1111/1574-6968.12056. [DOI] [PubMed] [Google Scholar]
- 57.Hassanzadazar H, Ehsani A, Mardani K, Hesari J. Investigation of antibacterial, acid and bile tolerance properties of lactobacilli isolated from Koozeh cheese. Veterinary research Forum. 2012;3(3):181–185. [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz L, Margolles A, Sánchez B. Bile resistance mechanisms in lactobacillus and bifidobacterium. Front Microbiol. 2013;4:396. doi: 10.3389/fmicb.2013.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nie YF, Hu J, Yan XH. Cross-talk between bile acids and intestinal microbiota in host metabolism and health. J Zhejiang Univ Sci B. 2015 Jun; 16(6):436-46. doi: 10.1631/jzus.B1400327 [DOI] [PMC free article] [PubMed]
- 60.Park MY, Kim SJ, Ko EK, Ahn SH, Seo H, Sung MK. Gut microbiota-associated bile acid deconjugation accelerates hepatic steatosis in Ob/Ob mice. J Appl Microbiol. 2016;121(3):800–810. doi: 10.1111/jam.13158. [DOI] [PubMed] [Google Scholar]
- 61.Gu Y, Wang X, Li J, Zhang Y, Zhong H, Liu R, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8(1):1785. 10.1038/s41467-017-01682-2. [DOI] [PMC free article] [PubMed]
- 62.Tanaka H, Doesburg K, Iwasaki T, Mierau I. Screening of lactic acid bacteria for bile salt hydrolase activity. J Dairy Sci. 1999;82:2530–2535. doi: 10.3168/jds.S0022-0302(99)75506-2. [DOI] [PubMed] [Google Scholar]
- 63.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–9. [DOI] [PMC free article] [PubMed]
- 64.Del Piano M, Strozzi P, Barba M, Allesina S, Deidda F, Lorenzini P, et al. In vitro sensitivity of probiotics to human pancreatic juice. J Clin Gastroenterol. 2008;42(Suppl 3 Pt 2):S170–3. 10.1097/MCG.0b013e3181815976. [DOI] [PubMed]
- 65.Collado MC, Sanz Y. Induction of acid resistance in bifidobacterium: a mechanism for improving desirable traits of potentially probiotic strains. J Appl Microbiol. 2007;103(4):1147–1157. doi: 10.1111/j.1365-2672.2007.03342.x. [DOI] [PubMed] [Google Scholar]
- 66.Lebeer S, Vanderleyden J, De Keersmaecker SC. Genes and molecules of lactobacilli supporting probiotic action. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Microbiol Mol Biol Rev. 2008;72(4):728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, et al. Functional analysis of lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78(1):185–93. 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed]
- 68.von Ossowski I, Reunanen J, Satokari R, Vesterlund S, Kankainen M, Huhtinen H, et al. Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl Environ Microbiol. 2010;76(7):2049–57. 10.1128/AEM.01958-09. [DOI] [PMC free article] [PubMed]
- 69.Tytgat HL, van Teijlingen NH, Sullan RM, Douillard FP, Rasinkangas P, Messing M, et al. Probiotic gut microbiota isolate interacts with dendritic cells via glycosylated heterotrimeric pili. PLoS One. 2016;11(3):e0151824. 10.1371/journal.pone.0151824. eCollection 2016. [DOI] [PMC free article] [PubMed]
- 70.Burgain J, Scher J, Lebeer S, Vanderleyden J, Corgneau M, Guerin J, et al. Impacts of pH-mediated EPS structure on probiotic bacterial pili-whey proteins interactions. Colloids Surf B Biointerfaces. 2015;134:332–8. 10.1016/j.colsurfb.2015.06.068. Epub 2015 Jul 15 [DOI] [PubMed]
- 71.Burgain J, Gaiani C, Francius G, Revol-Junelles AM, Cailliez-Grimal C, Lebeer S, et al. In vitro interactions between probiotic bacteria and milk proteins probed by atomic force microscopy. Colloids Surf B Biointerfaces. 2013;104:153–62. 10.1016/j.colsurfb.2012.11.032. [DOI] [PubMed]
- 72.Tytgat HL, Schoofs G, Vanderleyden J, Van Damme EJ, Wattiez R, Lebeer S, et al. Systematic exploration of the glycoproteome of the beneficial gut isolate Lactobacillus rhamnosus GG. J Mol Microbiol Biotechnol. 2016;26(5):345–58. 10.1159/000447091. [DOI] [PubMed]
- 73.Guerin J, Burgain J, Borges F, Bhandari B, Desobry S, Scher J, et al. Use of imaging techniques to identify efficient controlled release systems of lactobacillus rhamnosus GG during in vitro digestion. Food Funct. 2017;8(4):1587–98. 10.1039/c6fo01737a. [DOI] [PubMed]
- 74.Tripathi P, Beaussart A, Alsteens D, Dupres V, Claes I, von Ossowski I, et al. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano. 2013;7(4):3685–97. 10.1021/nn400705u. [DOI] [PubMed]
- 75.Garcia SL, Buck M, McMahon KD, Grossart HP, Eiler A, Auxotrophy WF. Intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol Ecol. 2015;24(17):4449–59. 10.1111/mec.13319. [DOI] [PubMed]
- 76.Garcia SL, Stevens SLR, Crary B, Martinez-Garcia M, Stepanauskas R, Woyke T, et al. Contrasting patterns of genome-level diversity across distinct co-occurring bacterial populations. ISME J. 2018;12(3):745-55. doi: 10.1038/s41396-017-0001-0 [DOI] [PMC free article] [PubMed]
- 77.Salas-Jara MJ, Ilabaca A, Vega M, García A. Biofilm forming Lactobacillus: new challenges for the development of probiotics. Microorganisms. 2016;4(3). 10.3390/microorganisms4030035. [DOI] [PMC free article] [PubMed]
- 78.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, et al. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A. 2011 Jul 5;108(27):11217–22. 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed]
- 79.Shokryazdan P, Sieo CC, Kalavathy R, Liang JB, Alitheen NB, Faseleh Jahromi M, et al. Probiotic potential of lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int. 2014;2014:927268. 10.1155/2014/927268. [DOI] [PMC free article] [PubMed]
- 80.He S, Ran C, Qin C, Li S, Zhang H, de Vos WM, et al. Anti-infective effect of adhesive probiotic lactobacillus in fish is correlated with their spatial distribution in the intestinal tissue. Sci Rep. 2017;7:13195. 10.1038/s41598-017-13466-1. [DOI] [PMC free article] [PubMed]
- 81.Collado MC, Jalonen L, Meriluoto J, Salminen S. Protection mechanism of probiotic combination against human pathogens: in vitro adhesion to human intestinal mucus. Asia Pac J Clin Nutr. 2006;15(4):570–575. [PubMed] [Google Scholar]
- 82.Compare D, Rocco A, Zamparelli MS, Nardone G. The gut bacteria-driven obesity development. Dig Dis. 2016;34(3):221–229. doi: 10.1159/000443356. [DOI] [PubMed] [Google Scholar]
- 83.Jiang TT, Shao TY, Ang WXG, Kinder JM, Turner LH, Pham G, et al. Commensal Fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe. 2017;22(6):809–816.e4. 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed]
- 84.Ilavenil S, Park HS, Vijayakumar M, Arasu MV, Kim DH, Ravikumar S, et al. Probiotic potential of lactobacillus strains with antifungal activity isolated from animal manure. ScientificWorldJournal. 2015;2015:802570. 10.1155/2015/802570. [DOI] [PMC free article] [PubMed]
- 85.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed]
- 86.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong LC, Holmes BA, Cotillard A, Habi-Rachedi F, Brazeilles R, Gougis S, et al. Dietary patterns differently associate with inflammation and gut microbiota in overweight and obese subjects. PLoS One. 2014;9:e109434. 10.1371/journal.pone.0109434. [DOI] [PMC free article] [PubMed]
- 88.Cotillard A, Kennedy SP, Kong LC, Prifti E, Pons N, Le Chatelier E, et al. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 89.Beaumont M, Goodrich JK, Jackson MA, Yet I, Davenport ER, Vieira-Silva S, et al. Heritable components of the human fecal microbiome are associated with visceral fat. Genome Biol. 2016;17:189. 10.1186/s13059-016-1052-7. [DOI] [PMC free article] [PubMed]
- 90.Org E, Blum Y, Kasela S, Mehrabian M, Kuusisto J, Kangas AJ, et al. Relationships between gut microbiota, plasma metabolites, and metabolic syndrome traits in the METSIM cohort. Genome Biol. 2017;18:70. 10.1186/s13059-017-1194-2. [DOI] [PMC free article] [PubMed]
- 91.Liu J, Chen FH, Qiu SQ, Yang LT, Zhang HP, Liu JQ, et al. Probiotics enhance the effect of allergy immunotherapy on regulating antigen specific B cell activity in asthma patients. Am J Transl Res. 2016 Dec 15;8(12):5256–70. [PMC free article] [PubMed]
- 92.Vieira AT, Galvão I, Amaral FA, Teixeira MM, Nicoli JR, Martins FS. Oral treatment with Bifidobacterium longum 51A reduced inflammation in a murine experimental model of gout. Benef Microbes. 2015;6(6):799–806. doi: 10.3920/BM2015.0015. [DOI] [PubMed] [Google Scholar]
- 93.Konopelniuk VV, Goloborodko II, Ishchuk TV, Synelnyk TB, Ostapchenko LI, Spivak MY, et al. Efficacy of fenugreek-based bionanocomposite on renal dysfunction and endogenous intoxication in high-calorie diet-induced obesity rat model-comparative study. EPMA J. 2017;8(4):377–90. 10.1007/s13167-017-0098-2. [DOI] [PMC free article] [PubMed]
- 94.Babenko LP, Lazarenko LM, Shynkarenko LM, Mokrozub VV, Pidgorskyi VS, Spivak MY. The effect of lacto- and bifidobacteria compositions on the vaginal microflora in cases of intravaginal staphylococcosis. Mikrobiol Z. 2012;74(6):80–89. [PubMed] [Google Scholar]
- 95.Al Kassaa I, Hober D, Hamze M, Chihib NE, Drider D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob Proteins. 2014;6(3–4):177–185. doi: 10.1007/s12602-014-9162-6. [DOI] [PubMed] [Google Scholar]
- 96.Dillon SM, Frank DN, Wilson CC. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS. 2016;30(18):2737–51. [DOI] [PMC free article] [PubMed]
- 97.Hager CL, Ghannoum MA. The mycobiome: role in health and disease, and as a potential probiotic target in gastrointestinal disease. Dig Liver Dis. 2017;49(11):1171–1176. doi: 10.1016/j.dld.2017.08.025. [DOI] [PubMed] [Google Scholar]
- 98.Talwalkar A, Kailasapathy K. Metabolic and biochemical responses of probiotic bacteria to oxygen. J Dairy Sci. 2003;86(8):2537–2546. doi: 10.3168/jds.S0022-0302(03)73848-X. [DOI] [PubMed] [Google Scholar]
- 99.Talwalkar A, Kailasapathy K. The role of oxygen in the viability of probiotic bacteria with reference to L. acidophilus and Bifidobacterium spp. Curr Issues Intest Microbiol. 2004;5(1):1–8. [PubMed] [Google Scholar]
- 100.Wang Y, Kirpich I, Liu Y, Ma Z, Barve S, McClain CJ, et al. Lactobacillus rhamnosus GG treatment potentiates intestinal hypoxia-inducible factor, promotes intestinal integrity and ameliorates alcohol-induced liver injury. Am J Pathol. 2011;179(6):2866–75. 10.1016/j.ajpath.2011.08.039. [DOI] [PMC free article] [PubMed]
- 101.Bubnov R, Jr PJ, Zubor P, Koniczka K, Golubnitschaja O. Pre-metastatic niches in breast cancer: are they created by or prior to the tumour onset? “Flammer syndrome” relevance to address the question. EPMA J. 2017;8:141–157. doi: 10.1007/s13167-017-0092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bubnov RV. Ultrasonography diagnostic capability for mesenteric vascular disorders. Gut. 2011;60(Suppl 3):A104. [Google Scholar]
- 103.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124(7):1174–82. [DOI] [PMC free article] [PubMed]
- 104.Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, et al. Position paper in cancer: current overview and future perspectives. EPMA J. 2015;6(1):9. 10.1186/s13167-015-0030-6. [DOI] [PMC free article] [PubMed]
- 105.York A. Microbiome: gut microbiota sways response to cancer immunotherapy. Nat Rev Microbiol. 2018;16(3):121. doi: 10.1038/nrmicro.2018.12. [DOI] [PubMed] [Google Scholar]
- 106.Kobyliak NM, Falalyeyeva TM, Kuryk OG, Beregova TV, Bodnar PM, Zholobak NM, et al. Antioxidative effects of cerium dioxide nanoparticles ameliorate age-related male infertility: optimistic results in rats and the review of clinical clues for integrative concept of men health and fertility. EPMA J. 2015 Jun 10;6(1):12. 10.1186/s13167-015-0034-2. [DOI] [PMC free article] [PubMed]
- 107.Babenko L, Bubnov R, Lazarenko L, Mokrozub V, Ganova L Kiseleva E, Shevchuk V, Spivak M. Use of probiotic bacteria increase effectiveness of vactination against hepatitis B in experimental studies Reviews in antiviral therapy infectious diseases (abstract book of the 3rd CEE meeting on viral hepatitis and co-infection with HIV, 27-28 September 2017, Ljubliana, Slovenia) 2017, 9:35.
- 108.Vitetta L, Saltzman ET, Thomsen M, Nikov T, Hall S. Adjuvant probiotics and the intestinal microbiome: enhancing vaccines and immunotherapy outcomes. Vaccines (Basel). 2017;5 (4). doi: 10.3390/vaccines5040050. [DOI] [PMC free article] [PubMed]
- 109.Kassaian N, Aminorroaya A, Feizi A, Jafari P, Amini M. The effects of probiotic and synbiotic supplementation on metabolic syndrome indices in adults at risk of type 2 diabetes: study protocol for a randomized controlled trial. Trials. 2017;18(1):148. 10.1186/s13063-017-1885-8. [DOI] [PMC free article] [PubMed]