Abstract
Background
The breast cancer (BC) epidemic is a multifactorial disease attributed to the early twenty-first century: about two million of new cases and half a million deaths are registered annually worldwide. New trends are emerging now: on the one hand, with respect to the geographical BC prevalence and, on the other hand, with respect to the age distribution. Recent statistics demonstrate that young populations are getting more and more affected by BC in both Eastern and Western countries. Therefore, the old rule “the older the age, the higher the BC risk” is getting relativised now. Accumulated evidence shows that young premenopausal women deal with particularly unpredictable subtypes of BC such as triple-negative BC, have lower survival rates and respond less to conventional chemotherapy compared to the majority of postmenopausal BC.
Working hypothesis
Here we hypothesised that a multi-level diagnostic approach may lead to the identification of a molecular signature highly specific for the premenopausal BC. A multi-omic approach using machine learning was considered as a potent tool for stratifying patients with benign breast alterations into well-defined risk groups, namely individuals at high versus low risk for breast cancer development.
Results and conclusions
The study resulted in identifying multi-omic signature specific for the premenopausal BC that can be used for stratifying patients with benign breast alterations. Our predictive model is capable of discriminating individually between high and low BC-risk with high confidence (>90%) and considered of potential clinical utility. Novel risk assessment approaches and advanced screening programmes—as the long-term target of this project—are of particular importance for predictive, preventive and personalised medicine as the medicine of the future, due to the expected health benefits for young subpopulations and the healthcare system as a whole.
Keywords: Predictive preventive personalised medicine, Breast cancer, Menopause, Patient stratification, Bioinformatics, Machine learning, Multi-level diagnostics, Biomarker panel, Laboratory medicine
Introduction
Breast cancer (BC) epidemic is attributed to the early twenty-first century as a multi-factorial disease: about two million of new cases and a half million of deaths are registered annually worldwide [1]. The highest incidence of BC has been recorded in Northern and Western Europe, North America, Australia and New Zealand [2]. Since a couple of years, the USA persistently demonstrate high incidence rates in BC with by 246,660 new BC cases and 40,450 BC-related deaths in 2016 [3], which corresponds to a lifetime risk for one in eight women. The persisting challenge is prevalent postmenopausal BC frequently linked to ageing, obesity and metabolic syndrome [4].
However, completely new trends are emerging now: on the one hand, with respect to the geographical BC prevalence and, on the other hand, with respect to the age distribution. More specifically, Eastern and African countries nowadays experience a dramatic increase in BC incidence and mortality rates [5, 6]. It has been reported that current BC incidence and related mortality rates for younger Chinese generations in Singapore and Taiwan are even higher than those in the USA [7]. Furthermore, recent statistics demonstrate that young populations are getting more and more affected by BC in both Eastern and Western countries, particularly women in the third and fourth decade of life [6, 8–11]. Johnson et al. have reported that the trend of increasing incidence was observed specifically in women aged 25–39 years without any significant increase in older subpopulations [9]. Therefore, the traditional rule “the older the age, the higher the BC risk” is getting revised now.
Noteworthy, BC occurring at younger age is particularly unpredictable comprising sporadic BC cases with strongly promoted metastatic spread to the life-important organs [12, 13]. Accumulated evidence shows that young premenopausal women deal with more aggressive subtypes of BC, have lower survival rates and respond less to conventional chemotherapy, when compared with postmenopausal women [9, 14–17]. Moreover, a great number of known BC risk factors are dependent on the menopausal status, which, however, is hardly considered in most risk assessment models [18]. Hence, the risks by abnormal (both decreased and increased) BMI are different for premenopausal and postmenopausal women and, further, modulate individual outcomes of the BC metastatic disease [1].
Many risk factors—both genetic and environmental—are currently considered by BC prediction models. However, the proportion of BC cases explained by these factors, particularly, if stratified by menopausal status, is unknown [19]. Exploratory breast proteomic assessment has demonstrated menopausal status-specific protein profiles and dietary (systemic) based modulation of breast cancer risk biomarkers involved in hormone and cytokine signalling pathways [20]. Metabolomic investigations (plasma folate, vitamin B6, vitamin B12, homocysteine) revealed alteration of breast cancer risks specifically in premenopausal women [21]. Finally, subcellular imaging of chromosomal DNA by the Comet Assay analysis has indicated differentiation of the “comet profiles” between younger and older breast cancer patients [22, 23].
Working hypothesis
Based on the above listed facts, we hypothesised that a multi-level diagnostic approach is crucial for identifying a molecular signature, which might be highly specific for the premenopausal BC (preBC). Consequently, the multi-omic modalities are considered as a potent tool for stratifying patients with benign breast alterations into well-defined risk groups, namely individuals at high versus low risk for breast cancer development. Further, particularly systemic alterations can be expected to underlie the pathomechanisms of preBC. Therefore, pathology-specific biomarker patterns in blood (but not in breast tissue) were the main target of the project, in order to create corresponding stratification algorithms. Finally, multi-parametric analysis using machine learning is essential for the predictive disease modelling and thus to generate high clinical utility. Novel risk assessment approaches and advanced screening programmes—as the long-term target of this project—are of particular importance from a healthcare economical point of view, due to the expected health benefits for young patients.
Materials and methods
Patient recruitment and stratification
Eighty-five premenopausal female patients were enrolled in the study. BC was diagnosed in 24 cases, and 61 patients demonstrated benign breast alterations (BCfree). All patients included into the current project were informed about the purposes of the study and have signed their “consent of the patient”. All investigations conformed to the principles outlined in the Declaration of Helsinki and were performed with the permission (Nr. 148/05) released by the responsible ethic committee of the Medical Faculty, Rheinische Friedrich-Wilhelms-University of Bonn.
Biobanking and biopreservation of blood samples
Venous blood draw was performed individually and prior to the core needle biopsy has been taken, in order to avoid any potential changes in molecular profiles related to the invasive approach of the breast tissue manipulation and drug application (anaesthesia, etc.). Blood samples (20 ml) were collected from the persons under investigation utilising anti-coagulation by heparin. One millilitre of the total blood was separated and centrifuged by a standard procedure described elsewhere for receiving blood plasma which was stored at − 80 °C until homocysteine measurements were performed. 19 ml of the fresh total blood were used for the isolation of peripheral leukocytes and, further, aliquoted for proteomic and Comet Assay analysis. All aliquots were stored at − 80 °C until corresponding experiments were performed.
Isolation of peripheral leukocytes
Peripheral leukocytes were separated using Ficoll-Histopaque gradients (Histopaque 1077, Sigma, USA) as described previously [24]. For that, individual samples were diluted with equal volumes of physiological buffer solution (PBS, Gibco™, USA). Then, 2 ml of histopaque were placed into 10-ml sterile centrifuge tubes and 5 ml of diluted blood samples were carefully layered onto each histopaque gradient. Gradients were centrifuged at 475 g and 20 °C for 15 min. The leukocytes bands were removed from the interface between plasma and histopaque layers of each tube and collected into one 50-ml tube. The total volume was brought to 50 ml with cold Dulbecco’s modified Eagle’s medium (DMEM, Gibco™, USA). The cell suspension was washed three times with DMEM and the total number of cells determined. Two equal aliquots of the isolated cells were prepared—one stored at − 80 °C as the dry pellet for consequent proteomic analyses. Another aliquot of the cells was re-suspended in PBS-DMSO solution, further aliquoted into Eppendorf tubes and stored at − 80 °C until subcellular imaging by Comet Assay analysis was performed.
Homocysteine measurements (metabolomics)
Total homocysteine was measured via automated HPLC with reversed phase separation and fluorescent detection [25]. Individual tHcy values were recorded for all the patients anonymously utilising the encoding system.
Clinical proteomics
Two-dimensional poly-acrylamide gel electrophoresis as the qualification approach
Two-dimensional poly-acrylamide gel electrophoresis (2D-PAGE) is routinely performed by the proteomic group at the Radiological Clinic, Breast Cancer Research Centre, Centre for Integrated Oncology, Rheinische Friedrich-Wilhelms-Universität Bonn, Germany. For the current project, altogether, 40 2D-PAGE-images were performed for the protein mapping and investigation of the expression patterns in peripheral leukocytes of patients with benign and malignant alterations in breast. Ten individual samples were used per corresponding group (malignant versus benign ones). Two parallel images were performed for each sample to confirm the reproducibility that doubled the final number of the gels analysed. A 200-μg aliquot of each protein sample was used for each 2D-PAGE analysis. First-dimensional separation was performed in immobilised pH gradient (IPG) strips (Bio-Rad, USA) in the range of IP 4-7, as recommended by the supplier. One hundred twenty-five-microlitre protein samples containing re-hydration buffer (8 M urea, 10 mM DTT, 1% CHAPS, 0.25% Bio-Lyte, pH 4–7) were loaded on the IPG-strips and subjected to 14 kVh overnight at 20 °C in a PROTEAN IEF Cell (Bio-Rad, USA). After the first-dimensional separation had been performed, the extruded IPG strips were equilibrated in gel equilibration buffer I (50 mM Tris-HCl, 6 M urea, 30% glycerol, 2% SDS, 1% DTT), followed by equilibration in buffer II (50 mM Tris-HCl, 6 M urea, 30% glycerol, 2% SDS and 260 mM iodacetamide) for 10 min before loading them onto poly-acrylamide gels (12% SDS-PAGE) for the second-dimensional resolution in Mini-PROTEAN 3 (Bio-Rad, USA). After the electrophoretic separation, resulting protein spots were visualised by silver staining (Silver Stain Plus™, Bio-Rad, USA). Differential gene expression was analysed using the task-dedicated software (Bio-Rad, USA).
MALDI-TOF
Selected spots were cut out from the gels (see the subchapter “Two-dimensional poly-acrylamide gel electrophoresis as the qualification approach”). Sample preparation for the MALDI-TOF performance was performed as described earlier [26]. The proteins localised within the individual spots were in-gel digested by incubating the samples with porcine trypsin (Promega, USA) at 37 °C overnight. The resulting peptide mixtures were then purified by ZipTip C18 according to the recommendations provided by the manufacturer (Millipore, USA). Elution was performed using 50% ACN/water solution saturated with CHCA. One microliter of each sample was spotted onto sample anchor and allowed to dry at room temperature. Afterwards, 0.7 μl of the re-crystallisation solution (ethanol/acetone/1%TFA in the ratio 60:30:10) were added. The follow-up analysis was performed using MALDI-TOF mass spectrometer (Bruker Daltonics) operated in positive ion reflector mode with an acceleration voltage of 25 kV. For internal calibration, the trypsin autolysis peptides were applied. Mass spectra were analysed automatically using Bruker software. The following search parameters were settled: (a) monoisotopic masses, (b) mass tolerance of 50 ppm, (c) one missing cleavage per peptide and (d) possible oxidation of methionine residues. No restrictions on Mr or pI were applied. A minimum of four matching peptides covering at least 15% of the overall sequence was required for a protein’s identification. Sequence similarity search was carried out using the BLASTP software.
Western blot analysis as the quantification approach
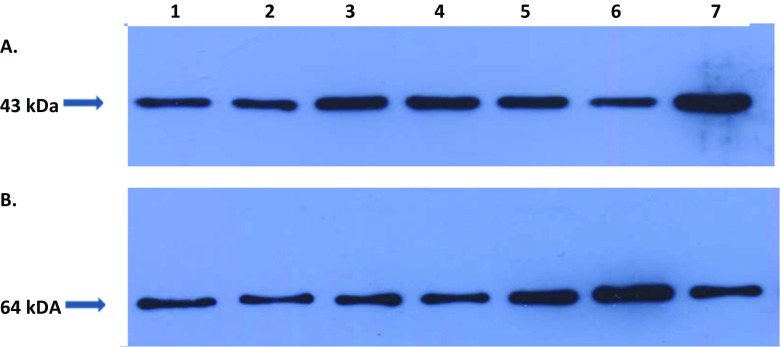
Actin and catalase were selected for quantification by 2D-PAGE followed by MALDI-TOF. The entire procedure for the Western blot analysis in the project has been described earlier [27]. Quantification of both protein targets was performed two times for each (blood) sample. The electrophoretic and blotting are standard procedures described elsewhere. Primary anti-body incubation was performed at room temperature using a 1:200 dilution of specific anti-bodies to human catalase 64 kDa (goat polyclonal IgG, recommended for detection of catalase of human origin by Western blotting, sc-34,282 Santa Cruz, USA) and to human actin 43 kDa (goat polyclonal IgG, recommended for detection of a broad range of actin isoforms of human origin, sc-1616 Santa Cruz, USA). The protein-specific signals were measured densitometrically using the “Quantity One®“ imaging system (Bio-Rad, USA)—see Fig. 1.
Fig. 1.
Western blot imaging of the expression rates for a actin and b catalase as demonstrated for the samples/patients numbered 1–7 which correspond to the patients 1 and 2 diagnosed with benign breast alterations, 3–7 breast cancer patients, whereby 3–6 are premenopausal BC and 7 is postmenopausal BC
Comet Assay analysis of the DNA patterns
DNA fragmentation assessment (Trevigen, Inc., Cat. No. 4250-050-K, USA) was performed for each blood sample (altogether 84) collected by evaluation of the DNA “comet” tail shape and specific migration patterns. Peripheral leukocytes were immobilised in a bed of low melting point agarose, on a Trevigen CometSlide™. The alkaline electrophoresis was applied to perform the most sensitive analysis of DNA breaks. After electrophoretic separation, staining with a fluorescent DNA intercalating dye (SYBR® Green I) was performed. The shape of individual comets was visualised by epifluorescence microscopy. The evaluation system developed by the authors has been already published earlier [28], and it was applied for the qualification and quantification of DNA fragmentation/damage in this project.
Bioinformatic analysis
NMF-based clustering of premenopausal patients with benign breast alterations
Sixty-one premenopausal patients with benign breast alterations were clustered into two groups using non-negative matrix factorisation (NMF) [29]. Briefly, NMF is a multi-variate, algebraic technique, which decomposes a non-negative data n × m matrix X into the product of two non-negative matrices W (n × m) and H (m × k). In our context, rows in X correspond to the molecular biomarkers measured in patients that are presented in different columns. Columns in W are sparse linear combinations of molecular markers in X, i.e. typically only a subset of biomarkers (biomarker panel) is used. The column vectors in W are also called meta-markers. Rows in H indicate, to which of the k clusters a particular patient can be assigned to. Consequently, NMF can be used to stratify patients into a predefined number of k clusters using a signature of multi-modal biomarkers. Notably, this signature can consist of a subset of available biomarkers (biomarker panel). To ensure robustness of our NMF based patient stratification, we repeated NMF clustering starting from random initial conditions 100 times and then looked at the consensus solution. That means that we have investigated how often each pair of patients would fall into the same cluster, yielding a consensus matrix. Based on the consensus matrix, a final and stable clustering result was then obtain via hierarchical clustering, as suggested in [29]. The whole analysis was done with the help of the implementation in R-package NMF [30].
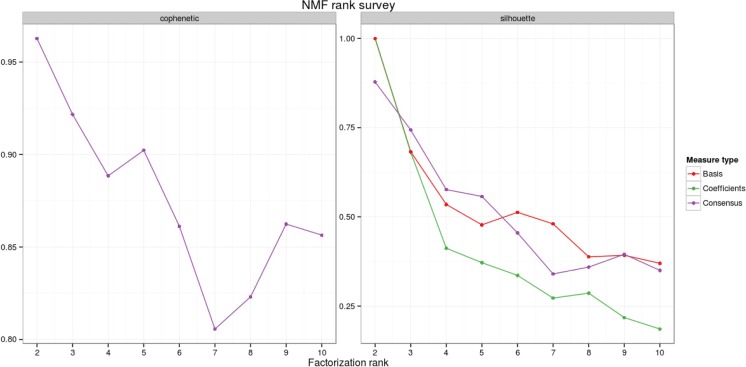
We varied the number k of clusters in our analysis from 2 to 10 and investigated the silhouette index [31] and the cophenetic correlation [32] as classical methods to evaluate clustering solutions. Both measures vary from − 1 to 1, where 1 indicates perfect agreement of the observed distance structure in the data to the proposed clustering. As indicated in Fig. 2, both measures favour a solution with two clusters.
Fig. 2.
Cophenetic correlation (left) and silhouette index (right) as a function of the number of NMF clusters. Both plots clearly favour a solution with two patient groups. The solution of a consensus clustering over 100 NMF runs (purple) is contrasted with the silhouette indices for clustering the markers (red) and the patients (green)
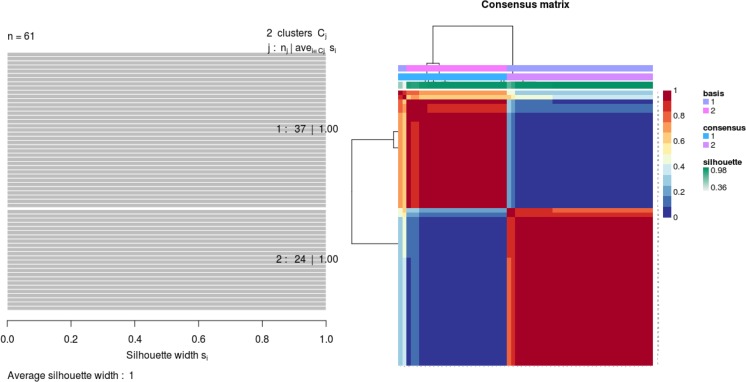
Investigating further the two-cluster solution, we depicted the consensus matrix (reflecting the frequency of each pair of patients falling into the same cluster), which shows a very clear block structure (Fig. 3). Further, Fig. 3 shows a full silhouette plot for the two-cluster solution. Both plots indicate a very clear separation of both clusters and high agreement with the observed distance structure of patient samples.
Fig. 3.
Left: silhouette plot of two patient subgroups with 37 and 24 patients, respectively. The x-axis shows the cluster silhouette for each patient on the y-axis. The cluster silhouette is a measure of how similar each patient is compared to patients in its own cluster and the closest patient from other clusters. The silhouette measure ranges from 0 to 1, where 1 indicates a perfect agreement of the assumed cluster assignment with patient distances. The average cluster silhouette for all patients in cluster 1 and 2 is shown. The overall average silhouette index was 1. Right: consensus matrix with super-imposed dendrogram of hierarchical clustering. The consensus matrix depicts the relative frequency of two patients falling into the same cluster across repeated NMF runs. A clear separation of two patient subgroups (red blocks) can be seen. Patients in these groups frequently fall into the same cluster.
Predicting patient subgroups via machine learning
To allow future patient stratification utilising the algorithms developed in the current study, Gradient Boosting Machine (GBM) as a supervised classification algorithm has been used [33]. A GBM classifier consists of an ensemble of decision trees of limited depth (here: at most five). Thanks to this aspect, GBMs is well suited for applications to multi-variate, heterogeneous data stemming from different measurement techniques and having different numerical scales and distributions—as in our situation. GBMs assign each decision tree a weight. During the iterative training procedure of GBMs, more and more decision trees are constructed and weighted. The optimal number of training (boosting) steps has a critical influence on the prediction performance of a GBM and can, e.g. be determined via a tenfold cross-validation. Notably, GBMs do not necessarily use all variables in the data for classification, but potentially only a subset. Hence, the resulting classifier can be sparse. Moreover, it is possible to extract a measure of importance of each individual variable for the classifier. This measure reflects the relative reduction of misfit to the training data (the relative loss reduction more precisely).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. Data on patients are available at a local database of the Radiological clinic, University of Bonn, Germany, that is not open for the public.
Results
Identification of pathology-specific biomarker patterns
NMF was applied as a modern data analytical technique to robustly identify clusters in multi-variate data (see “Bioinformatic analysis”). One of the features of NMF clustering is the selection of a suitable biomarker panel that induces the clustering of patients. In our case, we identified eight biomarkers, which resulted into a statistically clear separation of two patient groups with high silhouette indices (Figs. 2 and 3):
Hybridome: Homocystein plasma levels at the ratio of comet patterns CA IV/CA(I–III)
CA I patterns
CA II patterns
CA III patterns
CA IV patterns
Combined CA IV/ CA (I–III) patterns
Actin expression levels
Catalase expression levels
Reproducibility of the biomarker signature
We asked, how reproducible the identified biomarker panel was when repeating the NMF clustering on subsets of the patients. For this purpose, we applied a 10× repeated tenfold cross-validation procedure. That means, we randomly split all patients into tenfolds (subsets) and sequentially performed NMF clustering on 9/10 of these data while leaving out 1/10. This resulted into 100 NMF clustering solutions and corresponding biomarker sets. For all 100 NMF clustering solutions, two patient clusters were identified based on the silhouette index (see the description provided previously). Table 1 shows the frequency by which the previously identified 8 biomarkers were selected amongst these 100 clustering solutions, indicating specifically their high stability.
Table 1.
Frequency of NMF selected biomarkers within a 10× repeated tenfold cross-validation procedure
| Marker | Selection frequency |
|---|---|
| Hcy + CA I–IV | 94.00% |
| CA I | 100.00% |
| CA II | 89.00% |
| CA III | 68.00% |
| CA IV | 99.00% |
| CA I–IV | 99.00% |
| Actin | 98.00% |
| Catalase | 92.00% |
Algorithm to stratify future patients
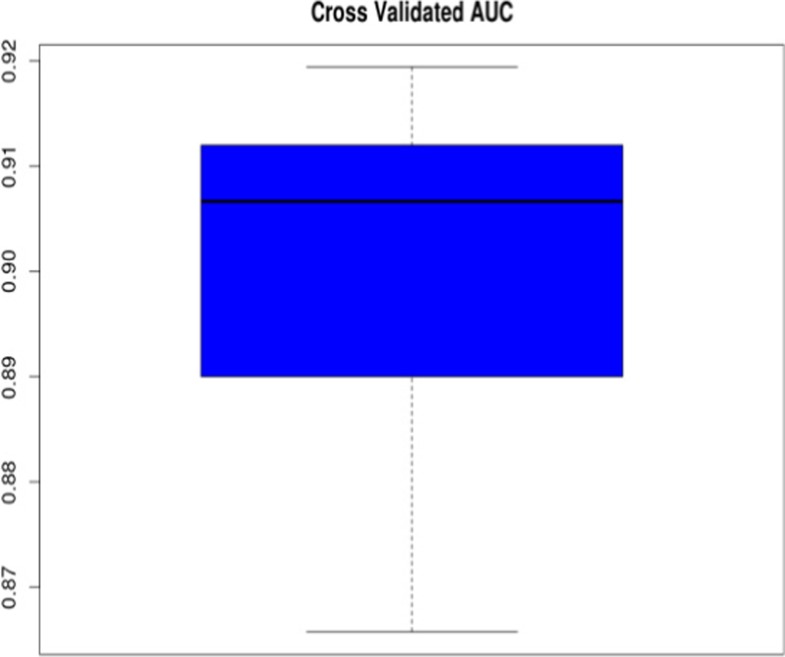
Based on the stratification of 61 patients into 2 clusters (classes), we evaluated the ability of a supervised machine learning classifier to stratify new patients into correct clusters this was done via a 10× repeated tenfold cross-validation procedure (see previous section). That means that a classifier was trained on 9/10 of 61 premenopausal patients with benign breast alterations and predictions of the patient cluster made for the remaining 1/10 of the patients. Prediction performance was measured via the area under ROC curve (AUC), indicating a highly accurate classification with ~ 91% AUC (Fig. 4). Notably, for this evaluation, we allowed classifiers to use all available variables rather than restricting them to the previously established signature based on the eight biomarkers chosen. This was done to reduce over-optimism, because the “8-biomarker” signature had previously been established on the entire dataset. Still, a prospective validation utilising external patient cohorts is required to gain a realistic picture of the generalisability of our results in future research.
Fig. 4.
Prediction of correct patient cluster assignment: The boxplot shows the distribution of 10 AUC values resulting from 10 repeats of a tenfold cross-validation procedure. Within each cross-validation loop, a GBM classifier was trained on 9/10 of the available patient data and tested on the hold out rest. A 50% AUC indicates chance level and a AUC of 100% a perfect prediction performance
Clinical interpretation of the results
We trained the final machine learning classifier with all 61 patients and 8 NMF selected biomarkers that resulted in the stratified patient clusters. Then, we applied this classifier to 24 premenopausal breast cancer patients. Fourteen of these patients were predicted to fall with high probability (> 90%) into one of our clusters. That means these 14 BC patients revealed a significant similarity to the 61 patients with benign breast alterations. Moreover, 11/14 (79%) patients fell into the cluster 2, indicating a higher similarity to patients in this subgroup. Hence, we interpreted patients in cluster 2 as the BC high-risk subgroup.
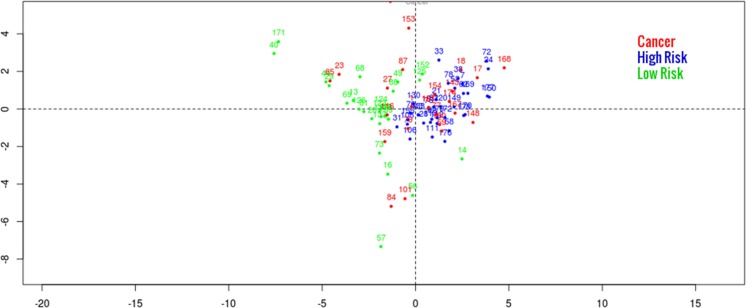
In agreement to this finding, Fig. 5 shows a principal component plot, which visualises two well-separated clusters (green and blue).Within the green cluster, there is clearly an enrichment of cancer samples (red) achieved.
Fig. 5.
Principal component plot depicting both identified patient subgroup and 14 breast cancer patients. Shown is the projection of patients (indicated by numbers) on the first two principal components of the biomarker signature space. The first two principal components explain 19.6 and 9.2% of the total variance
Relative influence of the individual biomarkers in the panel
We assessed the relative influence of each of the eight biomarkers that were found to have a power to stratify the patients between high- and low-risk clusters by our machine learning classifier (see Table 2). Table 2 shows, whether the corresponding biomarker increases or decreases the relative chance of a patient to be stratified into the high risk cluster. The latter was assessed by inspecting partial dependencies of variables [33].
Table 2.
Relative importance of NMF selected biomarkers in a GBM classifier distinguishing between two clusters as high- versus low-risk patients
| Marker | Relative importance | In high risk |
|---|---|---|
| CA I | 39.7973 | Up |
| Catalase | 19.4992 | Up |
| Hcy + CA I–IV | 13.6948 | Up |
| Actin | 12.6924 | Up |
| CA IV | 6.6395 | Down |
| CA III | 3.9636 | Down |
| CA II | 3.6177 | Down |
| CA I–IV | 0.0956 | Down |
Boxplots of individual features are shown in Fig. 6. Notably, each of the 8 biomarkers selected demonstrates univariately a clear difference between high- and low-risk patient groups that is statistically significant (false discovery rate < 0.05, Banjamini-Yekutieli method [34]) in all cases tested (Wilcoxon rank test). Further, Fig. 7 demonstrates individualised patient profiles relevant for their clinical utility.
Fig. 6.
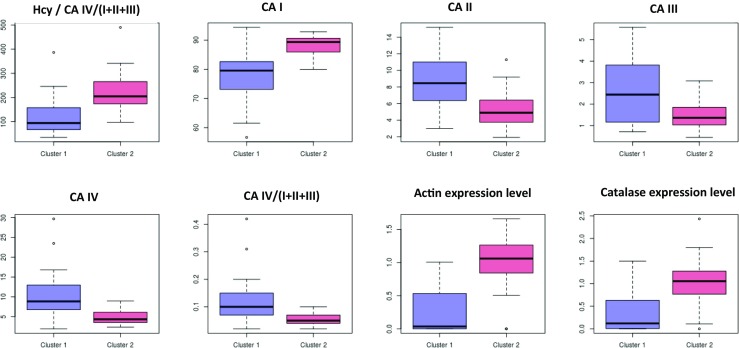
Boxplots depicting the distribution of individual variables in low risk (cluster 1) and high risk (cluster 2) patient subgroups. All markers show a statistically significant difference between high- and low-risk groups after multiple testing corrections
Fig. 7.
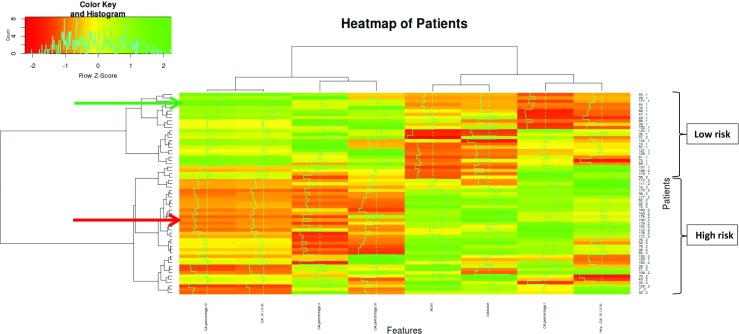
Heat map of patients—the x-axis displays individualised patient profiles (patients involved are listed on the y-axis); characteristic patterns of following biomarker are presented: CA IV, CA IV/I + II + III, CA II, CA III, actin expression, catalase expression, CA I, and hybridome Hcy/CA(I–IV). The colour code displays the row-wise normalised magnitude (e.g. expression levels for proteins) of each marker (z-score): green = high, red = low. Example: indicated with the red arrow patient is at high BC risk; in this case, low levels of CA IV, CA IV/I + II + III, CA II and CA III, but high levels of actin, catalase, CA I, and Hcy / CA (I–IV) have been demonstrated that is characteristic for the profile of the “high risk” cluster. The patient at low risk for BC indicated with the green arrow shows exactly opposite patterns
Discussion
Premenopausal breast cancer context
As stated in the “Introduction”, the management of premenopausal breast cancer (preBC) is challenging, due to increasing prevalence, no specialised screening programmes, underdeveloped predictive diagnostics and targeted prevention. The area attracts more and more attention that is reflected by the PubMed statistics demonstrating a permanently increasing annual number of the field-dedicated papers starting with only one paper in 1971 up to 270 PubMed-registered papers in 2016. However, altogether, there are currently only 366 and 60 papers which could be found as specifically dedicated to the “preBC risk assessment” and “preBC prediction”, respectively. This is an astonishing low amount of publications demonstrating evident deficits in the field-related research activities. Large-cohort studies continue to report on first-degree family history of BC (germline mutations resulting in familial BC comprising 5–10% of all BC cases), the extremely or heterogeneously dense breast tissue, anthropometric parameters, overweight/obesity, decreased physical activity, abnormal alcohol consumption, history of benign breast biopsy and disease-predisposing reproductive history as the main risk factors for both pre- and postmenopausal BC (postBC) with some more significance for one or another depending on the factor and population [1, 19, 35]. Far more clarity has been achieved for patient profiling of postBC, since the majority of BC patients comprises postBC in Western countries [4]. However, this finding does not hold true for some other world regions such as African countries: in Central Sudan, about 63% of all breast cancer cases are represented by preBC and 34% have been registered for women with five or more childbirths [36]. No clarity has been reached so far for patient profiling of preBC resulting in poorer prognosis and higher mortality rates typical for preBC compared to postBC. Moreover, the menopausal status is hardly considered in most currently applied risk assessment models [18]. Consequently, more effective diagnostic approaches, better adapted screening programmes and targeted treatments are of highest priority for research and medical services in the overall BC management. This, however, requires reliable preBC risk assessment based on multi-level diagnostics and comprehensive biomarker panels.
Advantages of the diagnostic approach proposed here
The diagnostic approach presented here is based on a multi-omic approach utilising blood samples, which underwent subcellular imaging by Comet Assay DNA anaylsis, disease-specific profiling of selected proteins and hybridome approach based on the comet patterns and homocysteine profiles. It was an intention of the authors to create the model based on a panel of biomarkers that fulfil two criteria:
They are complementary to each other with respect to their individual biological functions, which by differential patterns can be attributed to the tumour development and progression.
They represent possibly disease-specific and systemic biomarker patterns, which can be of particularly great clinical utility by creating risk assessment modalities based on blood tests correlated with tumour development in women predisposed to preBC before the clinically manifested disease.
Biological interpretation of the biomarkers selected
Oxidative stress resulting from an imbalanced production of reactive oxygen species (ROS) plays a key role in carcinogenesis. Several mechanisms underlie this functional link including an excessive damage to chromosomal DNA accompanied with ineffective repair [37], mitochondrial DNA damage and/or misguided repair [38], insufficient energy production and self-promotion of the “vicious circle” towards the formula “less energy = less repair but more ROS = more damage”. Catalase (Cat) is a primary antioxidant operating downstream in the SOD-Cat cascade detoxifying the most aggressive species, namely O2·− (superoxide radical) and H2O2 (hydrogen peroxide). Consequently, both significantly increased and/or decreased levels of Cat are strongly indicative for imbalanced production of ROS [39].
Although its role in BC pathology is inconsistent in the literature, homocysteine (Hcy) has been reported as a proliferation [40] and angiogenesis stimulating metabolite [41]. Moreover, Hcy induces metalloproteinase-2 in a dose-dependent manner [42], which is a prognostic biomarker for tumour aggressiveness and poor outcome in BC patients [43]. Furthermore, Hcy profiles in blood plasma can be regarded as readout of vitamin B12 and folate states, which are crucial for DNA synthesis. Contextually, DNA integrity has been assessed by the Comet Assay analysis. To this end, our previous studies demonstrated altered comet patterns as an attribute of BC in general [22]. Finally, the “hybridome” constructed as the ratio of the Hcy levels in blood plasma and the corresponding comet patterns in peripheral leukocytes has demonstrated statistically significant differences between low- and high-risk clusters (Fig. 6).
Finally, rearranged filamentous actin networks have been found as relevant for the cytoskeletal architecture of aggressive breast cancer cells which allows for cell migration and tumour invasion [44]. Altered expression rates of actin have been demonstrated in both breast cancer tissue and circulating leukocytes of BC patients [26]. Dysregulation of actin expression patterns might be genetically predisposed and could, therefore, be highly individual and systemic.
Systemic molecular patterns
Systemic metabolism has been shown to impact molecular patterns in breast tissue. However, breast tissue responsiveness is highly individual as demonstrated by energy restriction studies, which did not provide any interpretation for the control mechanisms decisive for the individual responsiveness [45]. Unfortunately, our knowledge regarding systemic interrelationship of individual risk factors is still rudimentary that strongly limits the predictive power of currently applied risk assessment models based on modifiable risk factors such as environment, dietary habits and behaviours predisposing to sporadic BCs. To this end, sporadic BCs comprise (over) 90% of all BC cases. Consequently, the greatest advantages of the preBC model presented here are (1) the use of blood samples reflecting systemic effects in the body and (2) correlation of the identified biomarker panels with the high- (disease similar) versus low (disease dissimilar)-risk clusters.
The main message
The approach seems to be highly promising for both positive and negative predictive diagnosis that allows, on the one hand, to trigger timely preventive measures and, on the other hand, to avoid unnecessary treatments such as biopsy, preventive chemotherapy and other procedures.
Concluding remarks
Using a multi-omic data approach the current project revealed two clearly and robustly separated clusters with high versus low BC-similarity in premenopausal, BC-free individuals. These clusters were induced by a highly reproducible subset of only eight biomarkers. Moreover, we developed a machine learning model, which is in principle able to predict any premenopausal woman as a member of either high or low BC-risk group based on the established biomarker panel.
As highlighted previously, the reported high and low BC-risk subgroups in premenopausal, BC-free patients require further validation via large-scale patient studies. We consider our present results as encouraging; however, the reproducibility level is currently difficult to estimate. Bringing our biomarker panel together with the developed stratification algorithm into clinical practice will require a prospective clinical trial. Such a study would have to demonstrate that the predicted high-risk individuals more frequently disease on BC during their lifetime compared to the low-risk individuals according to the evaluation system elaborated here. The ratio of lifetime risks in both clusters has then to be seen in relation to the stratification costs, in order to come to a final judgement with respect to the clinical utility of the proposed approach. Therefore, the presented work is to be considered as the first encouraging step made to benefit potentially affected patients, health care and society at large.
Acknowledgements
The authors thank Prof. Dr. H.J. Blom for measurements of homocysteine in blood plasma. The authors thank Dr. M. Fountoulakis and Dr. A. Papadopoulou for proteomic expertise strongly supported the project performance. Further, authors thank Ms. G. Windisch-Schuster for performing the Western blot analysis.
Abbreviations
- ACN
Acetonitrile
- AUC
Area under ROC (receiver operating characteristic) curve
- BC
Breast cancer
- CA
Comet Assay
- CA I, II, III
Comet classes I, II and III, respectively
- Cat
Catalase
- CHCA
α-cyano-4-hydroxycinnamic acid
- GBM
Gradient Boosting Machine
- Hcy
Homocysteine
- H2O2
Hydrogen peroxide
- MALDI-TOF
Matrix-assisted laser desorption/ionisation time-of-flight
- TFA
Trifluoroacetic acid
- NMF
Non-negative matrix factorisation
- preBC
Premenopausal breast cancer
- postBC
Postmenopausal breast cancer
- ROS
Reactive oxygen species
- SOD
Superoxide-dismutase
- O2·−
Superoxide radical
- 2D-PAGE
Two-dimensional poly-acrylamide gel electrophoresis
Authors’ contributions
OG created the concepts of the project and drafted the article. HF established the approach of machine learning applied to the project, and SP implemented the approach. KY carried out molecular biological investigations. HF and OG made the results interpretation. CK provided expertise in breast cancer. WK supervised the project with clinical expertise in breast cancer management.
Funding
The study funding has been performed by the Breast Cancer Research Centre, University of Bonn, Germany. KY has been awarded a fellowship by the European Association for Predictive, Preventive and Personalised Medicine (EPMA, Belgium).
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval
All the patient investigations conformed to the principles outlined in the Declaration of Helsinki and have been performed with the permission (Nr. 148/05) released by the responsible Ethic’s Committee of the Medical Faculty, Rheinische Friedrich-Wilhelms-University of Bonn. Human rights have been obligatory protected during the entire duration of the project according to the European standards. All the patients were informed about the purposes of the study and have signed their “consent of the patient”. This article does not contain any studies with animals performed by any of the authors.
References
- 1.Golubnitschaja O, Debald M, Yeghiazaryan K, Kuhn W, Pešta M, Costigliola V, Grech G. Breast cancer epidemic in the early 21st century: evaluation of risk factors, cumulative questionnaires and recommendations for preventive measures. Tumor Biol. 2016;37(10):12941–12957. doi: 10.1007/s13277-016-5168-x. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 4.Smokovski I, Risteski M, Polivka J, Jr, Zubor P, Konieczka K, Costigliola V, Golubnitschaja O. Postmenopausal breast cancer: European challenge and innovative concepts. EPMA J. 2017;8(2):159–169. doi: 10.1007/s13167-017-0094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American Cancer Society . Global cancer facts & figures. 2. Atlanta: American Cancer Society; 2011. [Google Scholar]
- 6.Ibrahim AS, Khaled HM, Mikhail NN, Baraka H, Kamel H. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014;2014:437971. doi: 10.1155/2014/437971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung H, Rosenberg PS, Chen WQ et al. Female breast cancer incidence among Asian and Western populations: more similar than expected. J Natl Cancer Inst. 2015;107. [DOI] [PMC free article] [PubMed]
- 8.Bouchardy C, Fioretta G, Verkooijen HM, Vlastos G, Schaefer P, Delaloye JF, Neyroud-Caspar I, Balmer Majno S, Wespi Y, Forni M, Chappuis P, Sappino AP, Rapiti E. Recent increase of breast cancer incidence among women under the age of forty. British J Cancer. 2007;96(11):1743–1746. doi: 10.1038/sj.bjc.6603783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson RH, Chien FL, Bleyer A. Incidence of breast cancer with distant involvement among women in the United States, 1976 to 2009. JAMA. 2013;309(8):800–805. doi: 10.1001/jama.2013.776. [DOI] [PubMed] [Google Scholar]
- 10.Merlo DF, Ceppi M, Filiberti R, Bocchini V, Znaor A, Gamulin M, Primic-Žakelj M, Bruzzi P, Bouchardy C, Fucic A, AIRTUM WG. Breast cancer incidence trends in European women aged 20–39 years at diagnosis. Breast Cancer Res Treat. 2012;134(1):363–370. doi: 10.1007/s10549-012-2031-7. [DOI] [PubMed] [Google Scholar]
- 11.Leclere B, Molinie F, Tretarre B, Stracci F, Daubisse-Marliac L, Colonna M. Trends in incidence of breast cancer among women under 40 in seven European countries: a GRELL cooperative study. Cancer Epidemiol. 2013;37(5):544–549. doi: 10.1016/j.canep.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Bubnov R, Polivka J Jr, Zubor P, Koniczka K, Golubnitschaja O. “Pre-metastatic niches” in breast cancer: are they created by or prior to the tumour onset? “Flammer Syndrome” relevance to address the question. EPMA J 2017;8(2):141–157. 10.1007/s13167-017-0092-8. [DOI] [PMC free article] [PubMed]
- 13.Polivka J, Jr, Kralickova M, Polivka J, Jr, Kaiser C, Kuhn W, Golubnitschaja O. Mystery of the brain metastatic disease in breast cancer patients: improved patient stratification, disease prediction and targeted prevention on the horizon? EPMA J. 2017;8(2):119–127. doi: 10.1007/s13167-017-0087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MCU, Nielsen TO, Moorman PG, Earp HS, Millikan RC. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 15.Anders CK, Fan C, Parker JS, Carey LA, Blackwell KL, Klauber-DeMore N, et al. Breast carcinomas arising at a young age: unique biology or a surrogate for aggressive intrinsic subtypes? J Clin Oncol. 2011;29(1):e18–e20. doi: 10.1200/JCO.2010.28.9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colleoni M, Rotmensz N, Peruzzotti G, Maisonneuve P, Orlando L, Ghisini R, Viale G, Pruneri G, Veronesi P, Luini A, Intra M, Cardillo A, Torrisi R, Rocca A, Goldhirsch A. Role of endocrine responsiveness and adjuvant therapy in very young women (below 35 years) with operable breast cancer and node negative disease. Ann Oncol. 2006;17(10):1497–1503. doi: 10.1093/annonc/mdl145. [DOI] [PubMed] [Google Scholar]
- 17.Ahn SH, Son BH, Kim SW, Kim SI, Jeong J, Ko SS, Han W, Korean Breast Cancer Society Poor outcome of hormone receptor-positive breast cancer at very young age is due to tamoxifen resistance: nationwide survival data in Korea—a report from the Korean Breast Cancer Society. J Clin Oncol. 2007;25(17):2360–2368. doi: 10.1200/JCO.2006.10.3754. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Dai J, Li M, Chan WC, Kwok CC, Leung SL, Wu C, Li W, Yu WC, Tsang KH, Law SH, Lee PM, Wong CK, Shen H, Wong SY, Yang XR, Tse LA. Risk assessment model for invasive breast cancer in Hong Kong women. Medicine (Baltimore) 2016;95(32):e4515. doi: 10.1097/MD.0000000000004515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engmann NJ, Golmakani MK, Miglioretti DL, Sprague BL, Kerlikowske K. Population-attributable risk proportion of clinical risk factors for breast cancer. Breast Cancer Surveillance Consortium. JAMA Oncol 2017. 10.1001/jamaoncol.2016.6326. [DOI] [PMC free article] [PubMed]
- 20.Fabian CJ, Kimler BF, Phillips TA, Box JA, Kreutzjans AL, Carlson SE, Hidaka BH, Metheny T, Zalles CM, Mills GB, Powers KR, Sullivan DK, Petroff BK, Hensing WL, Fridley BL, Hursting SD. Modulation of breast cancer risk biomarkers by high-dose omega-3 fatty acids: phase II pilot study in premenopausal women. Cancer Prev Res (Phila) 2015;8(10):912–921. doi: 10.1158/1940-6207.CAPR-14-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang SM, Willett WC, Selhub J, Hunter DJ, Giovannucci EL, Holmes MD, Colditz GA, Hankinson SE. Plasma folate, vitamin B6, vitamin B12, homocysteine, and risk of breast cancer. J Natl Cancer Inst. 2003;95(5):373–380. doi: 10.1093/jnci/95.5.373. [DOI] [PubMed] [Google Scholar]
- 22.Yeghiazaryan K, Cebioglu M, Braun M, Kuhn W, Schild HH, Golubnitschaja O. Noninvasive subcellular imaging in breast cancer risk assessment: construction of diagnostic windows. Personalized Med. 2011;8(3):321–330. doi: 10.2217/pme.11.17. [DOI] [PubMed] [Google Scholar]
- 23.Golubnitschaja O, Yeghiazaryan K, Costigliola V, Trog D, Braun M, Debald M, Kuhn W, Schild HH. Risk assessment, disease prevention and personalised treatments in breast cancer: is clinically qualified integrative approach in the horizon? EPMA J. 2013;4(1):6. doi: 10.1186/1878-5085-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golubnitschaja-Labudova O, Liu R, Decker C, Zhu P, Haefliger IO, Flammer J. Altered gene expression in lymphocytes of patients with normal-tension glaucoma. Curr Eye Res. 2000;21:867–876. doi: 10.1076/ceyr.21.5.867.5534. [DOI] [PubMed] [Google Scholar]
- 25.te Poele Pothoff MT, van den Berg M, Franken DG, Boers GH, Jakobs C, de Kroon IF, Eskes TK, Trijbels JM, Blom HJ. Three different methods for the determination of total homocysteine in plasma. Ann Clin Biochem. 1995;32:218–220. doi: 10.1177/000456329503200218. [DOI] [PubMed] [Google Scholar]
- 26.Braun M, Fountoulakis M, Papadopoulou A, Vougas K, Seidel I, Höller T, et al. Down-regulation of microfilamental network-associated proteins in leukocytes of breast cancer patients: potential application to predictive diagnostics. Cancer Genomics Proteomics. 2009;6:31–40. [PubMed] [Google Scholar]
- 27.Golubnitschaja O, Yeghiazaryan K, Abraham JA, Schild HH, Costigliola V, Debald D, Kuhn W. Breast cancer risk assessment: a non-invasive multiparametric approach to stratify patients by MMP-9 serum activity and RhoA expression patterns in circulating leucocytes. Amino Acids. 2017;49(2):273–281. doi: 10.1007/s00726-016-2357-2. [DOI] [PubMed] [Google Scholar]
- 28.Golubnitschaja O, Mönkemann H, Kim K, Mozaffari MS. Depletion of taurine induces DNA damage and expression of checkpoint genes p21WAF1/CIP1 and 14-3-3 σ in rat cardiomyocytes. Biochem Pharmacol 2003;66:511–517 [DOI] [PubMed]
- 29.Brunet J-P, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. PNAS. 2004;101(12):4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaujoux R, Seoighe C. A flexible R package for nonnegative matrix factorization. BMC Bioinformatics. 2010;11:367. doi: 10.1186/1471-2105-11-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comp Appl Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 32.Sokal RR, Rohlf FJ. The comparison of dendrograms by objective methods. Taxon. 1962;11(2):33–40. doi: 10.2307/1217208. [DOI] [Google Scholar]
- 33.Friedman JH. Stochastic gradient boosting. Comput Stat Data Anal Nonlinear Methods Data Mining. 2002;38(4):367–378. doi: 10.1016/S0167-9473(01)00065-2. [DOI] [Google Scholar]
- 34.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29:1164–1188. [Google Scholar]
- 35.Guldberg TL, Christensen S, Zachariae R, Jensen AB. Prognostic factors in early breast cancer associated with body mass index, physical functioning, physical activity, and comorbidity: data from a nationwide Danish cohort. Breast Cancer Res Treat. 2017;162(1):159–167. doi: 10.1007/s10549-016-4099-y. [DOI] [PubMed] [Google Scholar]
- 36.Awadelkarim KD, Aceto G, Veschi S, Elhaj A, Morgano A, Mohamedani AA, Eltayeb EA, Abuidris D, Di Gioacchino M, Battista P, Verginelli F, Cama A, Elwali NE, Mariani-Costantini R. BRCA1 and BRCA2 status in a Central Sudanese series of breast cancer patients: interactions with genetic, ethnic and reproductive factors. Breast Cancer Res Treat. 2007;102(2):189–199. doi: 10.1007/s10549-006-9303-z. [DOI] [PubMed] [Google Scholar]
- 37.Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, Rosebrock D, Livitz D, Kübler K, Mouw KW, Kamburov A, Maruvka YE, Leshchiner I, Lander ES, Golub TR, Zick A, Orthwein A, Lawrence MS, Batra RN, Caldas C, Haber DA, Laird PW, Shen H, Ellisen LW, D'Andrea AD, Chanock SJ, Foulkes WD, Getz G. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mencalha A, Victorino VJ, Cecchini R, Panis C. Mapping oxidative changes in breast cancer: understanding the basic to reach the clinics. Anticancer Res. 2014;34(3):1127–1140. [PubMed] [Google Scholar]
- 39.Ray G, Batra S, Shukla NK, Deo S, Raina V, Ashok S, Husain SA. Lipid peroxidation, free radical production and antioxidant status in breast cancer. Breast Cancer Res Treat. 2000;59(2):163–170. doi: 10.1023/A:1006357330486. [DOI] [PubMed] [Google Scholar]
- 40.Deng J, Lü S, Liu H, Liu B, Jiang C, Xu Q, Feng J, Wang X. Homocysteine activates B cells via regulating PKM2-dependent metabolic reprogramming. J Immunol. 2017;198(1):170–183. doi: 10.4049/jimmunol.1600613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YJ, Chiu CC, Ke CY, Tien N, Lin PK. Homocysteine facilitates prominent polygonal angiogenetic networks of a choroidal capillary sprouting model. Invest Ophthalmol Vis Sci. 2017;58(10):4052–4063. doi: 10.1167/iovs.17-22308. [DOI] [PubMed] [Google Scholar]
- 42.Wang ZS, Jin H, Wang DM. Influence of hydrogen sulfide on zymogen activation of homocysteine-induced matrix metalloproteinase-2 in H9C2 cardiocytes. Asian Pac J Trop Med. 2016;9(5):489–493. doi: 10.1016/j.apjtm.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 43.Ranogajec I, Jakić-Razumović J, Puzović V, Gabrilovac J. Prognostic value of matrix metalloproteinase-2 (MMP-2), matrix metalloproteinase-9 (MMP-9) and aminopeptidase N/CD13 in breast cancer patients. Med Oncol. 2012;29(2):561–569. doi: 10.1007/s12032-011-9984-y. [DOI] [PubMed] [Google Scholar]
- 44.Gari HH, DeGala GD, Ray R, Lucia MS, Lambert JR. PRL-3 engages the focal adhesion pathway in triple-negative breast cancer cells to alter actin structure and substrate adhesion properties critical for cell migration and invasion. Cancer Lett. 2016;380(2):505–512. doi: 10.1016/j.canlet.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harvie MN, Sims AH, Pegington M, Spence K, Mitchell A, Vaughan AA, Allwood JW, Xu Y, Rattray NJ, Goodacre R, Evans DG, Mitchell E, McMullen D, Clarke RB, Howell A. Intermittent energy restriction induces changes in breast gene expression and systemic metabolism. Breast Cancer Res. 2016;18(1):57. doi: 10.1186/s13058-016-0714-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article. Data on patients are available at a local database of the Radiological clinic, University of Bonn, Germany, that is not open for the public.