Summary
Mammalian genomes are promiscuously transcribed, yielding protein-coding and non-coding products. Many transcripts are short lived due to their nuclear degradation by the ribonucleolytic RNA exosome. Here, we show that abolished nuclear exosome function causes the formation of distinct nuclear foci, containing polyadenylated (pA+) RNA secluded from nucleocytoplasmic export. We asked whether exosome co-factors could serve such nuclear retention. Co-localization studies revealed the enrichment of pA+ RNA foci with “pA-tail exosome targeting (PAXT) connection” components MTR4, ZFC3H1, and PABPN1 but no overlap with known nuclear structures such as Cajal bodies, speckles, paraspeckles, or nucleoli. Interestingly, ZFC3H1 is required for foci formation, and in its absence, selected pA+ RNAs, including coding and non-coding transcripts, are exported to the cytoplasm in a process dependent on the mRNA export factor AlyREF. Our results establish ZFC3H1 as a central nuclear pA+ RNA retention factor, counteracting nuclear export activity.
Keywords: nuclear RNA decay, nuclear export, nuclear RNA retention, ZFC3H1, AlyREF, RNA exosome, RNA aggregates, MTR4
Graphical Abstract
Highlights
-
•
Abolished RNA exosome function leads to pA+ RNA accumulation in nuclear foci
-
•
pA+ RNA foci are enriched with various transcripts and exosome adaptor proteins
-
•
The exosome adaptor protein ZFC3H1 is required for pA+ RNA foci formation
-
•
ZFC3H1 functionally counteracts the mRNA export factor AlyREF
Silla et al. report that the RNA exosome adaptor protein ZFC3H1 acts as a nuclear RNA retention factor. In the absence of ZFC3H1, exosome targets are exported to the cytoplasm in a AlyREF-dependent manner. The discovery establishes ZFC3H1 as a central factor in the retention and degradation of polyadenylated RNA.
Introduction
Mammalian genomes transcribe a large repertoire of RNAs, which experience markedly different fates. This includes (1) their transport to nuclear compartments for functioning or for the passage of processing steps, (2) their nuclear export, or (3) their rapid nuclear decay. Nuclear export of RNA polymerase II (Pol II) transcripts is tightly coupled to their 5′ m7G cap formation, splicing, and 3′polyadenylation. mRNAs serve as the prototypic example of such flow from transcription to cytoplasmic translation (recently reviewed in Williams et al. [2018]). Some long non-coding RNAs (lncRNAs) are processed similarly to mRNAs (Kung et al., 2013). However, many are also rapidly degraded in the nucleus without being extensively spliced and where the polyadenylation status is unclear. This includes divergently transcribed enhancer RNAs (eRNAs) (Andersson et al., 2014, Djebali et al., 2012) and so-called promoter upstream transcripts (PROMPTs)/upstream antisense (UA) RNAs (Preker et al., 2008, Seila et al., 2008). Despite of all these RNAs being Pol II products and displaying structural similarity, at least at their 5′ ends, lncRNAs appear to be more concentrated in the nucleus and to be more labile than their mRNA counterparts (Djebali et al., 2012, Preker et al., 2011). Thus, mechanisms must exist that “sort” Pol II-derived transcripts for their differential nuclear and cytoplasmic destinies.
RNA-processing steps largely take place co-transcriptionally but, nevertheless, may occur in specialized nuclear areas; nucleoli, speckles, Cajal bodies (CBs), and paraspeckles (Sleeman and Trinkle-Mulcahy, 2014). Commonly, such nuclear bodies are self-organizing and dynamic structures that harbor specific substrates and enzymes and where enzymatic and RNP assembly reactions might be accelerated. With relevance for Pol II-derived transcripts, CBs contain a variety of proteins and RNAs that are involved in the assembly and modification of small nuclear ribonucleoproteins/small nucleolar ribonucleoproteins (snRNPs/snoRNPs) (Nizami et al., 2010). After their maturation, snRNPs travel to speckles, which may host multiple steps of gene expression, such as Pol II transcription, pre-mRNA splicing, and other mRNP maturation events in preparation for nuclear export (recently reviewed in Galganski et al. [2017]). Paraspeckles, which are proximal to speckles, form around the NEAT1 lncRNA and are enriched for specific RNA-binding proteins (Naganuma and Hirose, 2013). Their function is still debated, but recent reports suggest that paraspeckle components are involved in gene expression regulation (West et al., 2014), immune responses (Morchikh et al., 2017), and microRNA processing (Jiang et al., 2017). The so-called “Mmi foci” are another group of nuclear bodies of interest. These foci, which, so far, appear specific for S. pombe cells, are enriched with proteins involved in nuclear RNA decay, and it has been proposed that Mmi1 foci are degradation sites for meiosis-specific transcripts during vegetative growth (Harigaya et al., 2006, Sugiyama and Sugioka-Sugiyama, 2011, Yamanaka et al., 2010, Yamashita et al., 2013). Where and whether similar degradation centers exist in mammalian cell nuclei are still open questions.
The best described and major, nuclear RNA decay machinery is the highly conserved 3′–5′ exo- and endo-nucleolytic RNA exosome (Kilchert et al., 2016). This multi-subunit complex is present in the cytoplasm as well as in the nucleus, where it deals with most RNA biotypes. The human nucleoplasmic exosome is composed of a nine-subunit catalytically inactive core (known as EXO9) that achieves its activity via interaction with the exonuclease RRP6 and the exo- and endonuclease RRP44 (also known as DIS3) (Kilchert et al., 2016, Zinder and Lima, 2017). RNA exosome function critically depends on the RNA helicase MTR4 (also called SKIV2L2) to unwind exosome substrates, hereby facilitating their entry into the central channel of the exosome (Johnson and Jackson, 2013, Schneider and Tollervey, 2014). Critically, MTR4 also serves as a platform for recruiting adaptor proteins, providing RNA substrate binding and specificity (Meola and Jensen, 2017). In human nuclei, three such MTR4-containing adaptor complexes have been described. The human hTRAMP complex, harboring the non-canonical poly(A) (pA) polymerase TRF4-2 and the zinc-knuckle protein ZCCHC7, is exclusively nucleolar and predominantly involved in rRNA processing and decay (Lubas et al., 2011). Two other MTR4-containing complexes, the nuclear-exosome-targeting (NEXT) complex (Lubas et al., 2011) and the pA-tail-exosome-targeting (PAXT) connection (Meola et al., 2016), both have nucleoplasmic localizations. In the NEXT complex, MTR4 interacts with the zinc-knuckle protein ZCCHC8 and the promiscuous RNA-binding protein RBM7 (Giacometti et al., 2017, Lubas et al., 2011), which facilitate exosome decay of, e.g., PROMPTs, eRNAs, and 3′ extended products of snRNAs and snoRNAs (Andersen et al., 2013, Hrossova et al., 2015, Lubas et al., 2011, Lubas et al., 2015, Meola et al., 2016). In case of the PAXT connection, another zinc-finger protein, ZFC3H1, mediates an interaction of MTR4 with the nuclear pA-binding protein PABPN1, in turn, targeting polyadenylated RNAs to the exosome (Meola et al., 2016). Thus, although substrate overlap exists, PAXT generally promotes the degradation of longer and more extensively polyadenylated RNAs, whereas NEXT targets shorter and more immature transcripts (Meola et al., 2016).
Adding an m7G cap to the 5′ end of the ∼20-nt-long nascent transcripts is considered to be a hallmark of successful Pol II transcription initiation (Ramanathan et al., 2016). Shortly after its capping, the nascent RNA is bound by the cap-binding proteins, CBP20 and CBP80, forming the cap-binding complex (CBC) (Izaurralde et al., 1994, Moteki and Price, 2002, Visa et al., 1996). Interestingly, both PAXT and NEXT components can connect to the CBC via bridging proteins ZC3H18 and ARS2 (Andersen et al., 2013, Hallais et al., 2013, Meola et al., 2016). ARS2 directly interacts with the CBC to form the CBC-ARS2 (CBCA) complex, which behaves as a general suppressor of pervasive transcription (Andersen et al., 2013, Iasillo et al., 2017), whereas ZC3H18 connects the CBCA with NEXT for immediate substrate degradation (Andersen et al., 2013). The CBC also plays a role in intra-nuclear transcript transport as well as nuclear RNA export (Boulon et al., 2004, Cheng et al., 2006). For small nuclear RNAs (snRNAs), the transport adaptor protein PHAX binds the CBCA complex to promote the nuclear export of capped precursor transcripts (Boulon et al., 2004, Ohno et al., 2000). Sorting these RNAs between nuclear decay and export is suggested to occur in a competition between ZC3H18/NEXT and PHAX for CBCA binding (Giacometti et al., 2017). For most polyadenylated RNAs, like mRNAs, nuclear export is promoted in a process involving the AlyREF protein, which interacts with CBP80 to recruit the transcription/export (TREX) machinery to mRNA 5′ ends (Masuda et al., 2005). How the interplay between RNA export and degradation factors leads to a final decision of RNA fate remains unresolved.
In this study, we demonstrate that abolishing RNA exosome activity, by depleting its core component RRP40 (also known as EXOSC3), leads to the sequestration of pA+ RNAs in distinct nuclear foci (coined pA+ RNA foci). We find that MTR4 and PAXT, but not NEXT, components, are enriched in pA+ RNA foci, and we identify ZFC3H1 as a critical factor for foci formation. We surmise that ZFC3H1 is a nuclear retention factor for its bound pA+ RNA, and we demonstrate that it functionally competes with the pA+ RNA export factor AlyREF.
Results
RNA Exosome Depletion Leads to the Accumulation of pA+ RNA in Distinct Nuclear Foci
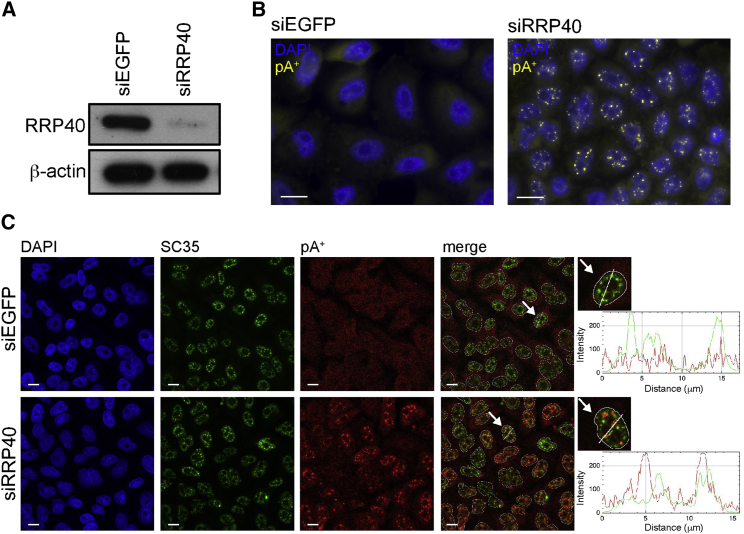
While both NEXT and PAXT pathways act on nuclear transcripts, it is unclear where targeting occurs and which mechanism(s) might ensure the nuclear retention of substrates prior to their decay. To begin to shed light on these questions, we first used small interfering RNAs (siRNAs) to deplete the exosome core protein RRP40 in HeLa cells (Figure 1A) and visualized pA+ RNA with a fluorescently labeled oligo(dT) probe. By removing an exosome core component, all exosome targets are impacted, regardless of the utilized adaptor complex. Moreover, it has been demonstrated that, in the absence of the exosome, at least some of its targets undergo poly- (Preker et al., 2011) or hyper-adenylation (Bresson and Conrad, 2013). RRP40 depletion triggered the appearance of distinct nuclear pA+ RNA foci only vaguely visible in control cells (Figure 1B). To ascertain whether these foci were related to known nuclear bodies, we performed co-localization analysis using SC35, Coilin, and PSF protein as markers for speckles, paraspeckles, and Cajal bodies, respectively. These analyses did not disclose any exclusive overlap with pA+ RNA foci in exosome-depleted conditions (Figures 1C, S1A, and S1B), although we noted an occasional co-localization with SC35 (Figure 1C, right line scan) and PSF (Figure S1B, right line scan) and a frequent close proximity to Coilin (Figure S1A, right line scan). In addition, we did not observe pA+ RNA localization in nucleoli, constituting condensed regions poorly stained with DAPI. We conclude that at least some exosome targets are nuclear retained and largely aggregate outside of known nuclear compartments when prevented from getting degraded.
Figure 1.
RNA Exosome Depletion Leads to the Formation of Distinct Nuclear pA+ RNA Foci
(A) Western blotting analysis of RRP40 levels in HeLa cells treated with control siRNA (siEGFP) or siRNA targeting RRP40 (siRRP40). β-actin was used as a loading control.
(B) RNA-FISH analysis of pA+ RNA in control (siEGFP) or RRP40-depleted (siRRP40) HeLa cells. pA+ RNA, detected with an oligo(dT)-LNA probe (Thomsen et al., 2005), is shown in yellow, and nuclear DAPI stain is shown in blue.
(C) Co-localization analysis of pA+ RNA foci with the speckle marker SC35 using cells from (B). Confocal images of SC35 immunofluorescence (IF) signal in green, pA+ RNA-FISH signal in red, and DAPI stain in blue. SC35 IF and RNA-FISH signals were merged (merge). Arrows point to cells that were used for line scan analyses, and zoom-ins of the relevant cells are shown at the right of the corresponding panel. Line scan profiles represent IF and RNA-FISH signal intensities along the drawn line. Dashed lines represent outlines of nuclei. pA+ RNA was detected as in (B).
Scale bars, 10 μm.
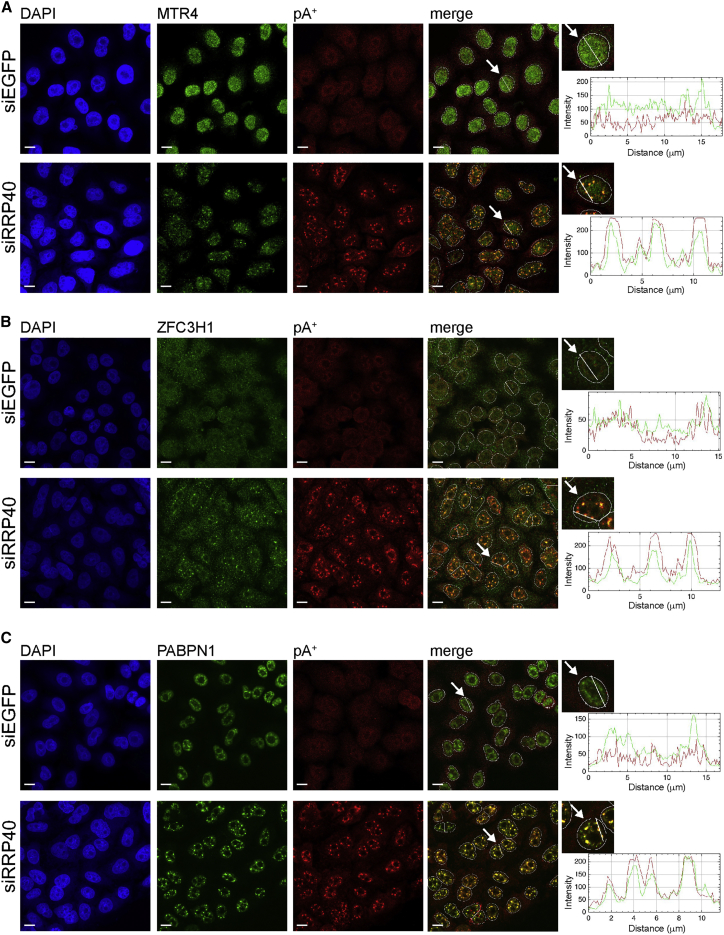
PAXT Components Are Selectively Enriched in pA+ RNA Foci
Besides RNA, nuclear bodies harbor high concentrations of protein, potentially involved in their biological function. Therefore, we asked whether pA+ RNA foci contain proteins involved in nuclear RNA decay. Immunolocalization (IL) analysis of MTR4, involved in both NEXT and PAXT pathways, displayed a clear punctate accumulation in RRP40-depleted cells, whereas the protein showed an even nuclear distribution in control cells (Figure 2A). Importantly, dual MTR4 IL/pA+ RNA fluorescence in situ hybridization (FISH) analysis demonstrated that MTR4 punctae coincide with pA+ RNA foci (Figure 2A, right bottom line scan).
Figure 2.
PAXT Complex Components Localize to pA+ RNA Foci
(A–C) Co-localization analysis of PAXT components MTR4 (A), ZFC3H1 (B), and PABPN1 (C) with pA+ RNA in control (siEGFP) and RRP40-depleted (siRRP40) HeLa cells as in Figure 1C. Scale bars, 10 μm.
To inquire about PAXT and/or NEXT enrichment in the pA+ RNA foci, we repeated such co-localization analysis using antibodies toward the PAXT components PABPN1 and ZFC3H1, as well as the NEXT components RBM7 and ZCCHC8. This revealed a strong overlap between PABPN1 and ZFC3H1 with the pA+ RNA signal upon exosome inactivation (Figures 2B and 2C, right bottom line scans), whereas neither of the NEXT components ZCCHC8 and RBM7 exhibited any noticeable localization change between RRP40-depleted and control cells (Figures S2A and S2B). Interestingly, we could also detect an enrichment of the ARS2 protein in pA+ RNA foci (Figure S2C). As ARS2 is part of the CBCA complex that binds capped RNA, this suggests that accumulating RNAs have a proper cap structure. Thus, PAXT components MTR4, ZFC3H1, and PABPN1 accumulate on nuclear exosome targets, of which at least some are capped and polyadenylated, when turnover is prevented.
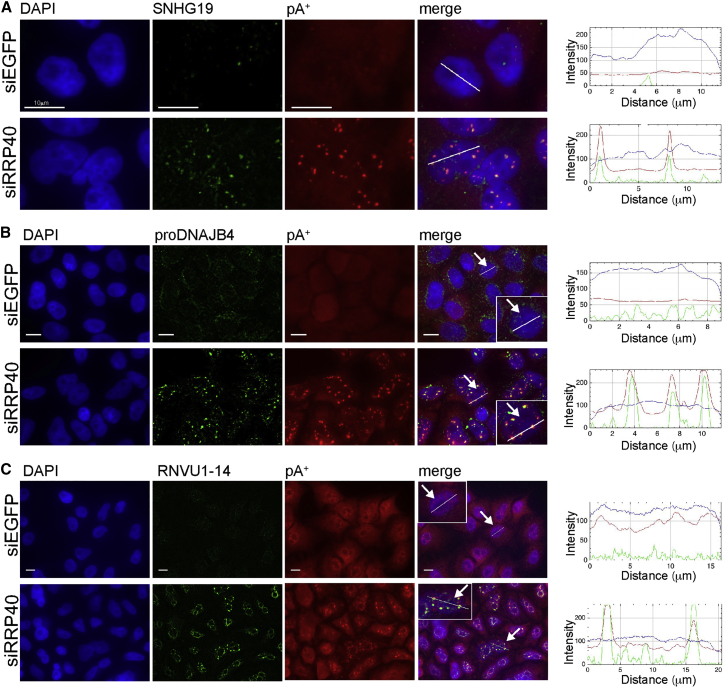
Diverse RNAs Accumulate in pA+ RNA Foci upon Exosome Inactivation
Having established an enrichment of PAXT proteins in pA+ RNA foci, we next addressed whether known RNA substrates of the PAXT pathway were also present. Hence, we performed specific RNA-FISH analysis of the highly expressed and spliced SNHG19 RNA previously established as a PAXT pathway target (Meola et al., 2016) (Figure S3A). FISH analysis using probes spanning the SNHG19 RNA exon-exon junction demonstrated its co-accumulation with polyadenylated RNA in pA+ RNA foci (Figure 3A). To determine any pathway specificity of such accumulation, we next localized NEXT pathway targets. These are generally short and lowly expressed, making RNA-FISH challenging; therefore, we chose the highly expressed DNAJB4 PROMPT (Figure S3B) and the 3′ extended form of U1 snRNA (RNVU1-14) (Figure S3C) for our analysis. Both these transcripts accumulated in pA+ RNA foci upon RNA exosome depletion (Figures 3B and 3C). Finally, we analyzed RNA expressed from an artificial PROMPT (proPOGZ) locus, stably integrated in the HeLa genome between a CMV promoter and a BGH pA signal (Ntini et al., 2013). qRT-PCR analysis showed this transcript to be stabilized upon RNA exosome depletion, although neither ZCCHC8 (NEXT) nor ZFC3H1 (PAXT) depletion affected its levels (Figure S3D). Yet, RNA-FISH analysis still revealed a clear accumulation of this transcript in pA+ RNA foci (Figure S3E). Thus, even though the nuclear pA+ foci specifically concentrate PAXT components, they contain both PAXT and non-PAXT RNA substrates. It is plausible that non-PAXT targets become polyadenylated in exosome-depletion conditions (Preker et al., 2011) and that longer pA tails trigger PAXT loading and pA+ RNA foci localization (see also Discussion).
Figure 3.
Diverse RNAs Accumulate in pA+ RNA Foci
(A–C) RNA-FISH co-localization analyses of pA+ RNA with SNHG19 (A), proDNAJB4 (B), and RNVU1-14 3′ extension (C) in control (siEGFP) or RRP40-depleted (siRRP40) HeLa cells. pA+ RNA was detected with an oligo(dT) 50-mer probe. Line scans are indicated as in Figure 1C, including DAPI channel. Scale bars, 10 μm.
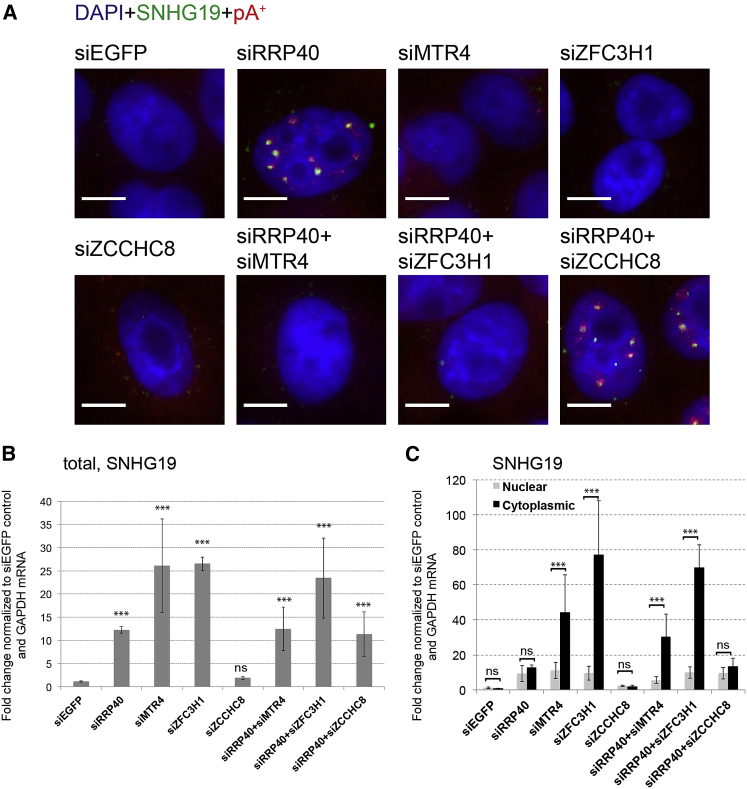
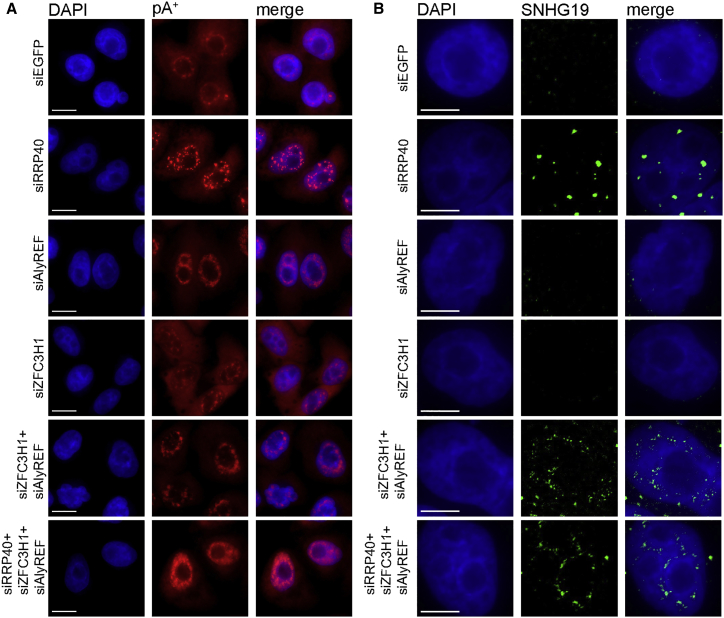
The ZFC3H1 Protein Is Required for Formation of pA+ RNA Foci
Given the massive condensation of nuclear RNA exosome substrates into pA+ RNA foci, we next wondered whether exosome adapters NEXT or PAXT might be involved in foci formation. To examine this question, we depleted HeLa cells of RRP40, MTR4, ZFC3H1, or ZCCHC8 (Figure S4A) and followed pA+ RNA foci and SNHG19 RNA localization by RNA-FISH, as described earlier. As previously reported (Lubas et al., 2011, Ogami et al., 2017), MTR4 depletion also reduced ZCCHC8, ZFC3H1, and, to some extent, RRP40 protein levels (Figure S4A). Still, neither MTR4 nor ZFC3H1 and ZCCHC8 depletions resulted in pA+ or SNHG19 RNA accumulation in nuclear foci (Figures 4A and S4B). At the same time, parallel qRT-PCR analysis showed the robust accumulation of SNHG19 RNA upon both MTR4 and ZFC3H1 depletions (Figure 4B; see also Meola et al. [2016]).
Figure 4.
The ZFC3H1 Protein Is Required for RNA Foci Formation
(A) Dual SNHG19 and pA+ RNA-FISH analysis in the indicated factor-depleted HeLa cells. Merged images of DAPI (blue), SNHG19 (green), and pA+ RNA (red) are shown. Images from separate channels are shown in Figure S4B. pA+ RNA was detected with an oligo(dT) 50-mer probe.
(B and C) qRT-PCR analysis of SNHG19 RNA using total (B) or cytoplasmic and nuclear (C) fractions harvested from the indicated factor-depleted HeLa cells. In (C), 1 μg RNA from the respective fractions was used for reverse transcriptase reactions. Data are displayed as mean values, with error bars denoting SD (n = 4 biological replicates, except for siZCCHC8: n = 3). ∗∗∗p < 0.01, Student’s t test; ns, not significant. Scale bars, 10 μm.
See also Figure S4B.
A possible reconciliation of these data could be that the MTR4 and/or ZFC3H1 proteins themselves are, apart for their roles as exosome co-factors, required for the creation of pA+ RNA foci. To test this notion, we depleted cells for RRP40 (to form pA+ RNA foci) and combined this with depletion of MTR4, ZFC3H1, or ZCCHC8, yielding largely comparable levels of SNHG19 RNA as upon single depletion of RRP40 (Figure 4B). Parallel RNA-FISH analysis showed that cells subjected to RRP40/MTR4 and RRP40/ZFC3H1 co-depletions were devoid of any discernable pA+ and SNHG19 RNA accumulation (Figure 4A). In contrast, cells subjected to RRP40/ZCCHC8 co-depletion displayed similar focal patterns of pA+ and SNHG19 RNA, as observed for cells depleted only for RRP40. Given the presence of MTR4 protein in RRP40/ZFC3H1-depleted cells and the co-depletion of MTR4 and ZFC3H1 in RRP40/MTR4-depleted cells (Figure S4A), these results, therefore, implicate the ZFC3H1 protein as an important contributor to pA+ RNA foci formation in exosome-depleted cells. If so, exogenous ZFC3H1 expression in RRP40/ZFC3H1 co-depleted cells should re-install pA+ foci formation. To test this, we used ZFC3H1 5′ and 3′ UTR-specific siRNAs, which both efficiently deplete endogenous ZFC3H1 protein but do not target a plasmid-expressed FLAG-tagged ZFC3H1 version (Figure S4C). Indeed, parallel IL/pA+ RNA-FISH analysis of the plasmid-transfected cells demonstrated that pA+ foci formation was rescued by exogenous FLAG-tagged ZFC3H1 expression (Figure S4D). We conclude that ZFC3H1 is required for pA+ RNA foci formation.
What then is the fate of exosome substrates, e.g., SNHG19 RNA, in ZFC3H1-depleted cells, where the transcript is stabilized but does not accumulate in nuclear foci? If ZFC3H1 activity generally retains exosome substrates in cell nuclei, SNHG19 transcripts escaping such retention would be likely to enter the mRNA export pathway. To explore this possibility, we performed subcellular fractionation of HeLa cells from Figure 4A and analyzed the abundance of SNHG19 RNA in nuclear and cytoplasmic fractions. qRT-PCR analysis showed a clear accumulation of SNHG19 transcripts in the cytoplasmic fractions of cells subjected to factor depletion combinations; MTR4, ZFC3H1, RRP40/MTR4, and RRP40/ZFC3H1, which all yielded low ZFC3H1 levels (Figures 4C and S4A). Consistently, ZCCHC8 depletion alone, or in combination with RRP40 depletion, did not result in elevated cytoplasmic SNHG19 RNA levels (Figure 4C). These results all point toward ZFC3H1 being responsible for retaining RNA exosome targets in the cell nucleus.
The mRNA Export Factor AlyREF Functionally Competes with ZFC3H1
Our results so far suggest that ZFC3H1 might compete with RNA export activity to prevent nuclear RNA exosome substrates from entering the cytoplasm. IL analysis of mRNA export factors AlyREF (Figures S5A and S5B) and NXF1 (data not shown) did not reveal any accumulation in pA+ RNA foci, suggesting mutual exclusive binding of export and decay factors. Therefore, the observed export of exosome substrates upon ZFC3H1 depletion may be caused by recruitment of the export machinery to substrates normally blocked by decay co-factors. To test this prediction, we depleted AlyREF alone or in ZFC3H1- and RRP40/ZFC3H1-depletion conditions (Figure S5C). As expected, depletion of AlyREF alone resulted in nuclear accumulation of pA+ RNA (Figure 5A) but did not lead to nuclear accumulation of SNHG19 RNA (Figure 5B), which is most likely degraded by the exosome in this condition. When co-depleted with ZFC3H1 or RRP40/ZFC3H1, AlyREF depletion resulted in re-established nuclear accumulation of SNGH19 and pA+ RNA (Figures 5A and 5B). However, as opposed to the distinct foci of RRP40-depleted cells, pA+ and SNHG19 RNA now appeared more diffusely in the nucleoplasm (Figures 5A and 5B; compare panel “siRRP40” to panels “siZFC3H1/siAlyREF” and “siRRP40/siZFC3H1/siAlyREF”). These results reiterate a role of ZFC3H1 in pA+ RNA aggregation and suggest that the protein might “shield” RNA exosome targets from the mRNA export machinery. In the absence of ZFC3H1, however, even exosome targets with no normal cytoplasmic business, like SNHG19 RNA, will get exported from the nucleus.
Figure 5.
ZFC3H1 Counteracts AlyREF Function
(A and B) RNA-FISH analysis of pA+ RNA (A) and SNHG19 RNA (B) in factor-depleted (indicated to the left side of each image panel) HeLa cells. Image display is as shown in Figure 3B. pA+ RNA was detected with an oligo(dT) 50-mer probe. Scale bars, 10 μm.
Full-Length mRNAs Accumulate upon Exosome Depletion
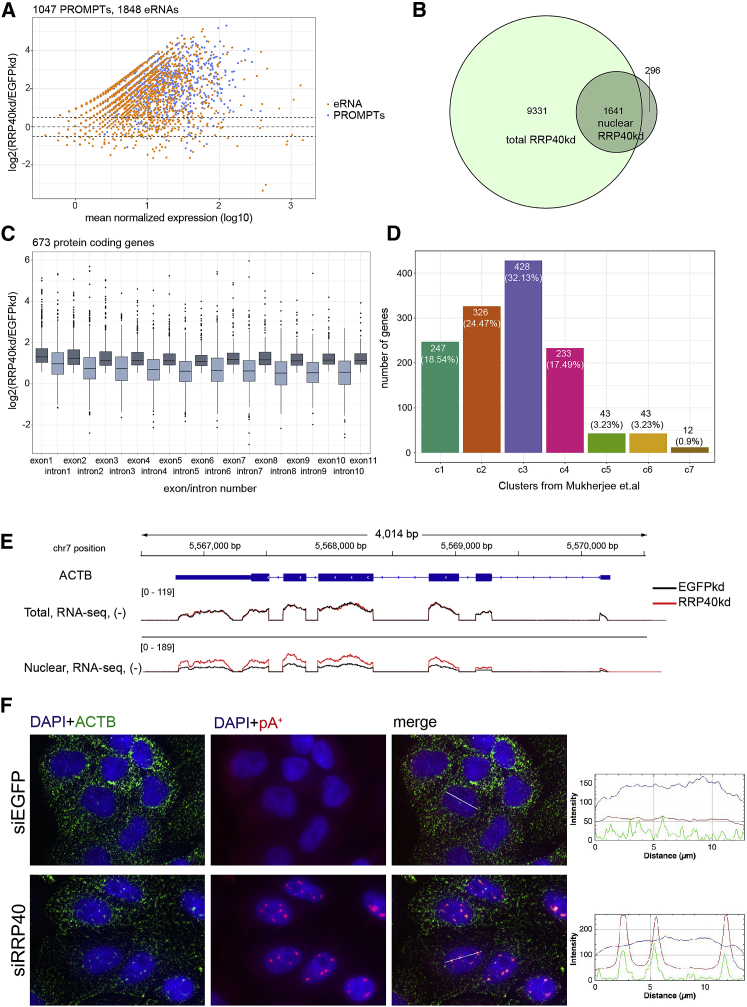
To further examine which transcripts get turned over by the nuclear exosome and might accumulate in pA+ RNA foci in its absence, we performed high-throughput RNA-sequencing (RNA-seq) analysis of pA+ RNA from nuclear fractions of cells subjected to RRP40 depletion (“siRRP40”) or treated with control EGFP siRNAs (“siEGFP”) (Figure S6A). Both types of libraries were produced either in duplicates (EGFPkd) or triplicates (RRP40kd), which generally displayed good reproducibility between replicates (Figure S6B). Nuclear exosome targets were revealed by calculating fold changes between the RRP40kd and the control sample, using DESeq2, and using all GENCODE-annotated RNAs expressed in the samples as well as PROMPTs and eRNAs as previously described (Meola et al., 2016). As expected PROMPTs and eRNAs were generally upregulated in the RRP40-depletion condition (Figure 6A), consistent with the notion that a large share of these transcripts are adenylated (Preker et al., 2011). More surprisingly, we noticed that RNA stemming from a significant number of annotated protein-coding genes were either up- or downregulated upon RRP40 depletion (Figure S6C), which was also observed in previously published RRP40kd libraries of total RNA preparations (Meola et al., 2016) (Figure S6D). However, aberrant products, e.g., those constituting premature transcription termination events, as well as intragenic eRNAs are stabilized upon RRP40 depletion (Iasillo et al., 2017). Therefore, we intersected the nuclear RNA-seq analysis with our previously published total RRP40kd RNA-seq data (Meola et al., 2016) and also interrogated the extent to which the upregulated signals constituted full-length and/or spliced mRNA. To this end, we selected 1,641 protein-coding genes that were significantly upregulated (log2[RRP40kd/EGFPkd] > 0, and adjusted p value [padj] < 0.05) in the nuclear samples and were upregulated/unaffected (log2[RRP40kd/EGFPkd] > −0.5) in the total RNA datasets (Figure 6B). Log2 fold changes (RRP40kd/EGFPkd) were then calculated for each exon and intron, and genes were selected that had their exons upregulated in the nuclear RNA samples, resulting in a set of 673. On average, introns were less upregulated than exons, indicating that mature spliced RNAs were being stabilized by exosome depletion (Figure 6C). Moreover, upregulation occurred across all exons, strongly suggesting that full-length mRNAs were affected. We note that our analysis cannot discriminate which of these are direct targets of the exosome and which accumulate due to indirect effects.
Figure 6.
Full-Length mRNAs Can Be Upregulated upon Exosome Depletion
(A) Log2 fold changes (y axis) between PROMPTs and eRNAs in nuclear RRP40kd versus EGFPkd libraries plotted against their mean normalized expression (log10 scale, x axis).
(B) Venn diagram representing the overlap between significantly upregulated protein-coding genes (log2[RRP40kd/EGFPkd] > 0, padj < 0.05) in the nuclear RNA libraries and the upregulated and unaffected protein-coding genes (log2[RRP40kd/EGFPkd] > −0.5) in the total RNA libraries.
(C) Boxplots of log2 fold changes (RRP40kd/EGFPkd) of each individual exon and intron of the selected protein coding genes in ascending order up to the 11th exon.
(D) Bar plots representing the classification of 1,641 protein-coding genes—overlap from (B)—to previously described clusters (Mukherjee et al., 2017). Note the enrichment of genes belonging to cluster c3 (Pearson’s chi-square test, p = 6.12e−10; calculated against all expressed genes in nuclear samples represented in Figure S6E).
(E) Genome browser view of the ACTB locus, showing tracks of normalized total and nuclear RNA-seq data from control (EGFPkd, black line) and RRP40-depleted (red line) HeLa cells. All tracks represent an average of three biological replicates, except the nuclear EGFPkd sample, which represents average of two biological replicates.
(F) Dual ACTB and pA+ RNA-FISH analysis in the indicated factor-depleted HeLa cells. Merged images of DAPI (blue), ACTB (green), and pA+ RNA (red) are shown. Image display is as in Figure 1C.
See also Figure S6E.
To characterize exosome-sensitive mRNAs, we used a published classification system for human coding and non-coding genes (Mukherjee et al., 2017). In relation to a background set (Figure S6E), our set of 1,641 upregulated protein-coding genes was significantly enriched for cluster c3 (p = 6.12e−10) from Mukherjee et al. (2017) (Figure 6D). A significant enrichment (p = 3.207e−07) was obtained for the same cluster when analyzing only those 366 genes giving rise to mRNAs upregulated (log2[RRP40kd/EGFPkd] > 0, and padj < 0.05) in both total and nuclear samples (Figures S6F and S6G). Interestingly, cluster c3 is enriched for transcription-factor-encoding mRNAs, that are more nuclear than the average, are well synthesized, and exhibit high degradation rates (Mukherjee et al., 2017).
Finally, we asked whether exosome-sensitive mRNAs might also localize to pA+ RNA foci upon RRP40 depletion. The ACTB and NFKB2 mRNAs were selected for analysis, as both transcripts are relatively abundant, and their full-length mature RNA species displayed elevated nuclear expression upon RRP40 depletion (Figures 6E, S6H, and S7A). As expected, RNA-FISH experiments revealed a predominant cytoplasmic localization for both RNAs, which did not differ between control and RRP40-depleted cells (Figures 6F and S7B). However, some ACTB mRNA could be detected in nuclear pA+ RNA foci upon RRP40 depletion (Figure 6F), while a noticeable accumulation of NFKB2 RNA was not observed (Figure S7B). Collectively, these results suggest that a fraction of spliced, full-length mRNA is subject to nuclear exosomal decay.
Discussion
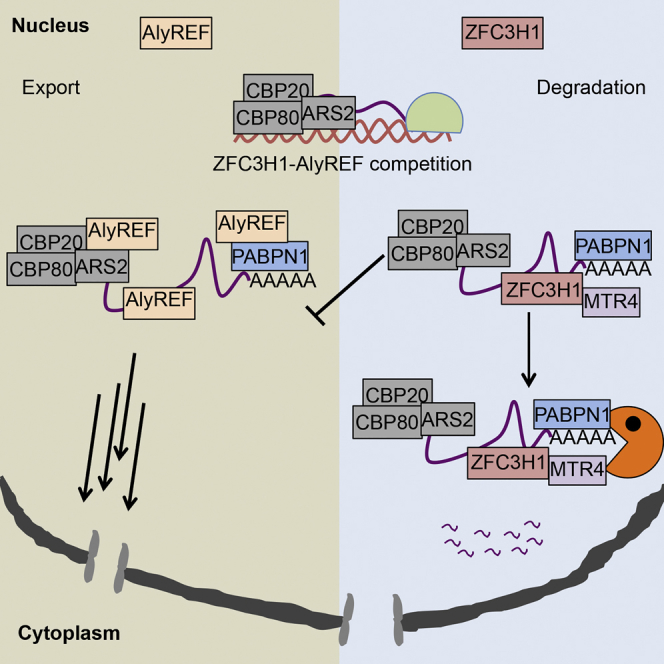
The vast majority of the human genome is transcribed, and commonly from non-genic regions, giving rise to a broad variety of ncRNA. These are often transcribed by Pol II and, therefore, pass some processing steps characteristic of mRNA. Yet, unlike most mRNA, a large fraction of ncRNA is rapidly turned over by the nuclear RNA exosome. How such molecular decision is enabled, and with what means nuclear export of these ncRNA is prevented, is not well understood. Here, we demonstrate that precluding the degradation of RNA exosome targets, including some mRNA, does not automatically lead to their nuclear export. Instead, they form distinct nuclear pA+ RNA foci, requiring retention that counteracts export. We unveil one aspect of such mechanism(s) by demonstrating that the PAXT component ZFC3H1 is required for establishing the pA+ RNA foci phenotype. Moreover, we suggest that this occurs in functional competition with AlyREF-mediated pA+ RNA nuclear export.
Cell biological data presented here echo results from mitotically growing S. pombe cells, where unwanted meiosis-specific transcripts are cleared by the nuclear exosome (Chen et al., 2011, Harigaya et al., 2006, Hiriart et al., 2012, Yamashita et al., 2012, Zofall et al., 2012). These transcripts are specifically recognized by the Mmi1 protein (Harigaya et al., 2006) in the context of MTREC, a multisubunit complex that also contains Red1 and Pab2, which are S. pombe homologs of ZFC3H1 and PAPBN1, respectively (Marayati et al., 2016, Zhou et al., 2015). Both these proteins are required for the decay of meiotic transcripts, in a process that also requires the polyadenylation activity of the canonical pA polymerase Pla1 (St-André et al., 2010, Sugiyama and Sugioka-Sugiyama, 2011, Yamanaka et al., 2010). Interestingly, Mmi1, Red1, Pab2, polyadenylation factors, and the nuclear exosome subunit Rrp6 accumulate in distinct nuclear dots, suggested to be transcript degradation sites (Harigaya et al., 2006, Sugiyama and Sugioka-Sugiyama, 2011, Yamanaka et al., 2010). Given the obvious similarity to our results, it is possible that such RNA decay centers also exist in mammalian cells. However, unlike in S. pombe, where “Mmi1 foci” are also visible in wild-type cells, we do not measure discernable condensates of exosome adaptor proteins in normally growing HeLa cells. Although we cannot fully exclude that RNA foci are pre-existing in normal cells as condensates of non-adenylated RNA, which becomes adenylated after RNA exosome depletion, the fact that neither adaptor proteins nor targets of the exosome accumulate in foci in the control condition is at odds with this possibility. Nuclear pA+ RNA foci are also observed in S. cerevisiae cells carrying a mutant allele of the MTR4 gene (Kadowaki et al., 1994). It is, therefore, an outstanding task, across different species, to discern whether macromolecular interactions exist, that are difficult to detect by conventional microscopy, and which coalesce into detectable foci only when the nuclear concentration of RNA increases upon exosome inactivation. Additionally, we do not have evidence to discriminate whether active RNA decay takes place in such putative RNP assemblies. Finally, S. cerevisiae strains debilitated in mRNA nuclear export also retain pA+ RNA in nuclear focal structures (Hilleren et al., 2001, Jensen et al., 2001a, Jensen et al., 2001b, Libri et al., 2002). Curiously, this retention is alleviated upon inactivation of the S. cerevisiae nuclear exosome, revealing that decay factors can also, in some situations, control nuclear RNA retention. Clearly, much still has to be learned about the relationships between nuclear RNA metabolism, localization, and export.
Although the mechanisms underlying the formation of RNA-containing foci are generally also not understood, a number of recent studies have demonstrated that non-membranous cellular compartments can form by so-called liquid-liquid phase transitions (Shin and Brangwynne, 2017). Their formation is often driven by sequence-specific physicochemical properties of RNA, facilitating multivalent protein-RNA and protein-protein interactions enriched with amino acid sequences of low complexity (Jain and Vale, 2017, Jiang et al., 2015, Kato et al., 2012, Uversky, 2017). The N-terminal part of ZFC3H1 harbors several low-complexity regions and is highly disordered (data not shown; using the low-complexity-predicting program PONDR-FIT; Xue et al., 2010), with particularly serine- and proline-rich stretches predicted to be important for phase separation (Shin and Brangwynne, 2017). Thus, it is likely that this property of ZFC3H1 drives formation of pA+ RNA foci.
Consistent with their ZFC3H1 dependency, we found PAXT components, and the bona fide PAXT target SNHG19, to be enriched in pA+ RNA foci. However, despite the absence of NEXT components ZCCHC8 and RBM7, we somewhat surprisingly also found the accumulation of NEXT substrates. Since PAXT complex loading onto target RNA presumably is pA-tail dependent (Meola et al., 2016), it is possible that NEXT substrates, stabilized in RNA exosome-depleted conditions, achieve pA-tails long enough to accommodate PAXT binding. Indeed, recent RNA-seq data (Meola et al., 2016) showed a partial stabilization of tested NEXT targets in ZFC3H1-depleted HeLa cells (see also Figures S3A and S3B), indicating that a fraction of transcripts can be degraded by both pathways. Moreover, NEXT targets tested in this study are highly expressed compared to bulk NEXT substrates and may, therefore, be especially prone to experiencing such pA tailing and subsequent loading of PAXT components. This suggestion is consistent with our previous observation that RBM7 binds to newly synthetized and unprocessed Pol II transcripts, including pre-mRNAs, and with the proposition that RBM7/NEXT recruits the exosome to trigger RNA degradation when unprotected RNA 3′ ends are available (Lubas et al., 2015). It is, therefore, possible that NEXT components, which are not present in pA+ RNA foci, have already been discarded from these more processed/polyadenylated RNAs.
Our data imply that ZFC3H1-mediated nuclear retention of pA+ RNA might occur in functional competition with its nuclear export by, e.g., AlyREF. Interestingly, AlyREF is loaded onto RNA in a 5′ cap-dependent manner (Cheng et al., 2006, Shi et al., 2017) and also binds mRNA 3′ ends PABPN1 dependently (Shi et al., 2017). It is conceivable that these interactions underlie aspects of transcript sorting between nuclear export and retention/degradation, with ZFC3H1 and AlyREF competing for transcript binding via a common interaction with PABPN1 as well as MTR4 competing with AlyREF for association with the CBC-bound ARS2 protein (Fan et al., 2017). Our analysis of AlyREF localization in exosome-depleted cells did not reveal any enrichment of this protein in pA+ RNA foci. In contrast, ARS2, MTR4, and ZFC3H1 were all clearly enriched, indicating that accumulated transcripts are not immediately prone for export. It was proposed that formation of ARS2-MTR4 versus ARS2-AlyREF links is decisive in whether transcript gets degraded or exported, respectively (Fan et al., 2017). However, as previously reported (Andersen et al., 2013, Ogami et al., 2017), and as recapitulated here, MTR4 depletion in HeLa cells negatively affects the levels of ZFC3H1, ZCCHC8, and RRP40. Hence, it is difficult to address any direct impact of MTR4 on pA+ RNA retention. Yet, RNA exosome targets escape the nucleus upon ZFC3H1 depletion (data in the present paper; Ogami et al., 2017), where MTR4 levels are unchanged, arguing against a direct role of MTR4 in transcript retention. It thus appears likely that PABPN1-dependent AlyREF versus ZFC3H1 loading on transcript 3′ends is a major RNA-sorting determinant, which also makes intuitive sense, since pA-tailing is a final step in RNA production, warranting an ultimate decision of whether to undergo export or decay. Moreover, it is possible that protein interactions with transcript 5′ and 3′ ends are cooperative so that any ZFC3H1-AlyREF 3′ end competition might affect ARS2-MTR4 5′ cap interactions and vice versa. Such cooperative factor loading was recently demonstrated by the depletion of CBP80, which impacts the 3′ end loading of AlyREF, and the depletion of the 3′ end processing factor CstF64, which disturbs 5′ cap association of AlyREF (Shi et al., 2017).
Our results are in agreement with a recent study reporting that the absence of ZFC3H1 results in the association of different types of pervasive transcripts with active ribosomes, causing global translation repression (Ogami et al., 2017). This positions ZFC3H1 as a key factor in preventing the nuclear export of RNAs that are seemingly unwanted in the cytoplasm. However, we also found that a fraction of fully spliced and full-length mRNAs was subject to exosomal decay. This suggests that bona fide translation substrates might also be targeted for nuclear turnover, echoing a recent demonstration that different cell types and tissues harbor 13%–30% of nuclear retained mRNAs (Halpern et al., 2015). Moreover, using annotation-agnostic classification of RNAs, Mukherjee et al. also found that certain transcription-factor-encoding transcripts were nuclear labile, and it was proposed that these RNAs may emanate from transcriptional bursts and that their nuclear retention might help buffer cytoplasmic transcript levels (Mukherjee et al., 2017). Consistently, this class of mRNA was upregulated upon RRP40 depletion (Figures 6D and S6G). Taken together, these data argue that nuclear turnover of full-length mRNA occurs. However, it remains an open question whether ZFC3H1-bound RNAs are committed to nuclear decay or whether the RNP assemblies, which we visualize as pA+ RNA foci by inactivating the RNA exosome, might also serve a storage function capable of releasing, e.g., retained mRNA, in resemblance to cytoplasmic processing bodies (P-bodies) (Buchan, 2014). Previously reported nuclear retained mRNAs were localized in nuclear speckles (Halpern et al., 2015), the function of which is connected to active transcription (Galganski et al., 2017). Since pA+ RNA foci do not co-localize with known nuclear bodies, including nuclear speckles, we suggest that these represent a post-transcriptional event, which is further supported by our observation that full-length spliced mRNA may appear in pA+ RNA foci. Future research will be focused on determining how sorting between productive export and retention/decay is orchestrated and which additional factors contribute to this process.
Experimental Procedures
Cell Culture and Transfections
HeLa cells were grown in DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C and 5% CO2. Cells were seeded at low confluence (approximately 44 cells per square millimeter) 1 day before siRNA transfection. The following day, DMEM was replaced with DMEM without penicillin/streptomycin, and transfections were performed using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions, with a final concentration of 20 nM of each siRNA (Table S1) and Lipofectamine 2000 (final dilution, 1:1,000) in RPMI 1640 medium. Two days after siRNA transfection, the medium was changed to new DMEM without penicillin/streptomycin, and the transfection procedure was repeated. For our complementation experiment, the ZFC3H1 coding construct pcDNA5-ZFC3H1-3xFLAG (Meola et al., 2016) or its empty vector pcDNA5-3xFLAG was transfected (using Lipofectamine 2000) into cells 1 day after the first siRNA transfection, and cells were harvested 1 day after the second siRNA transfection.
RNA-FISH Analysis
Cells were grown on microscope coverslips placed in tissue culture dishes. After siRNA treatment, medium was removed, and cells were washed twice with PBS prior to fixation with 4% formaldehyde in PBS for 20 min at room temperature. After fixation, cells were washed twice with PBS and permeabilized overnight at −20°C in 70% ethanol. Subsequently, cells were washed twice for 5 min with 2× SSC and blocked for 15 min in 2× SSC/30% formamide solution. The relevant FISH probe was added to hybridization mixture 1 (30% formamide, 2× SSC, 1 μg/μL E. coli tRNA), heated for 5 min at 95°C, and then cooled on ice. Thereafter, hybridization mixture 1 was combined with hybridization mixture 2 (10% dextran, 0.2 mg/mL BSA, 40 U RiboLock RNase inhibitors [Thermo Fisher Scientific]) and placed on a 37°C thermostat. 20 μL drops (per 12-mm coverslip) of combined hybridization mixtures 1 and 2 were pipetted onto a parafilm-coated box placed on wet filter paper, and coverslips were placed on the hybridization mixtures (cells facing the drops). The box was sealed with thin household film, and hybridization was performed overnight at 37°C. On the next day, coverslips were washed twice for 30 min each at 37°C in 2× SSC/30% formamide and twice for 5 min with PBS at room temperature. Cells were mounted with ProLong Gold Antifade Mountant with DAPI (Thermo Fisher Scientific).
ACTB, NFKB2, proDNAJB4, and RNVU1-14 3′ extension probes were designed using the R script Oligostan (Tsanov et al., 2016), and RNA-FISH was performed according to the smiFISH protocol described in Tsanov et al. (2016). All RNA-FISH probes used are listed in Table S2, except the oligo(dT)-LNA probe described in Thomsen et al., (2005).
Sequential RNA-FISH and IL Analysis
For co-staining of RNA and protein, RNA-FISH reactions were followed by protein IF staining. Briefly, a primary antibody dilution in 2% BSA was added to cells for 1 hr at room temperature, followed by three short washes with PBS. Thereafter, Alexa-Fluor-488-conjugated secondary antibody (Thermo Fisher Scientific) in 2% BSA was added for 1 hr at room temperature, followed by three short washes with PBS. Cells were mounted with ProLong Gold Antifade Mountant with DAPI. All primary antibodies and applied concentrations are listed in Table S3.
Microscopy and Image Analysis
Non-confocal images were obtained using a Zeiss Axiovert 200M epifluorescent microscope equipped with a coolSNAPHQ camera (Photometrics) and 60× (1.4) and 100× (1.3) objectives. Confocal images were obtained with a Zeiss LSM 710 confocal microscope and a 60× (1.4) objective. All images within the same experiment were taken with same excitation power and exposure time and processed similarly using the publicly available Fiji software (Schindelin et al., 2012). For all transcript-specific RNA-FISH images, background subtraction was performed using the Fiji Subtract Background plugin, with a rolling ball radius of 10 pixels. When image color balance adjustment was needed, all comparable images were processed in the same manner.
Western Blotting Analysis
Western blotting was performed by standard procedures (see Supplemental Experimental Procedures).
RNA Isolation, qRT-PCR Analysis, and Cellular Fractionation
RNA was purified using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions, and RNA was analyzed by qRT-PCR using standard procedures (see Supplemental Experimental Procedures).
RNA-Seq Library Preparation, Data Processing, and Computational Analysis
RNA-seq libraries were produced, and reads were quality controlled and mapped using standard procedures (see Supplemental Experimental Procedures).
Statistical Analysis
Values of biological replicates, p values, and statistical tests are reported in the figure legends. qRT-PCR data are shown as means ± SDs, except in Figure S6H where data are shown as means ± SEM. Statistical tests were done in the environment of the R Project for Statistical Computing (https://www.r-project.org).
Acknowledgments
This work was supported by the ERC (grant 339953), the Lundbeck Foundation, and the Novo Nordisk Foundation. We thank Marie Vestergård and Claudia Scheffler for technical support and Manfred Schmid for critical comments.
Author Contributions
T.S. and T.H.J. conceived the project. T.S. designed, conducted, and analyzed the experiments. E.K. performed the bioinformatics analysis. M.L. and D.M. contributed to the RNA-FISH analysis. T.H.J. supervised the project. T.S. and T.H.J. wrote the manuscript.
Declaration of Interests
The authors declare that they have no competing interests.
Published: May 15, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.04.061.
Data and Software Availability
The accession number for the RNA-seq data reported in this paper is GEO: GSE108197.
Supplemental Information
List of FISH and smiFISH probes.
References
- Andersen P.R., Domanski M., Kristiansen M.S., Storvall H., Ntini E., Verheggen C., Schein A., Bunkenborg J., Poser I., Hallais M. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol. 2013;20:1367–1376. doi: 10.1038/nsmb.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonné R., Bertrand E. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol. Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Bresson S.M., Conrad N.K. The human nuclear poly(A)-binding protein promotes RNA hyperadenylation and decay. PLoS Genet. 2013;9:e1003893. doi: 10.1371/journal.pgen.1003893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014;11:1019–1030. doi: 10.4161/15476286.2014.972208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-M., Futcher B., Leatherwood J. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS ONE. 2011;6:e26804–e26815. doi: 10.1371/journal.pone.0026804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Dufu K., Lee C.-S., Hsu J.L., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Kuai B., Wu G., Wu X., Chi B., Wang L., Wang K., Shi Z., Zhang H., Chen S. Exosome cofactor hMTR4 competes with export adaptor ALYREF to ensure balanced nuclear RNA pools for degradation and export. EMBO J. 2017;36:2870–2886. doi: 10.15252/embj.201696139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galganski L., Urbanek M.O., Krzyzosiak W.J. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017;45:10350–10368. doi: 10.1093/nar/gkx759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti S., Benbahouche N.E.H., Domanski M., Robert M.C., Meola N., Lubas M., Bukenborg J., Andersen J.S., Schulze W.M., Verheggen C. Mutually exclusive CBC-containing complexes contribute to RNA fate. Cell Rep. 2017;18:2635–2650. doi: 10.1016/j.celrep.2017.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallais M., Pontvianne F., Andersen P.R., Clerici M., Lener D., Benbahouche Nel.H., Gostan T., Vandermoere F., Robert M.C., Cusack S. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat. Struct. Mol. Biol. 2013;20:1358–1366. doi: 10.1038/nsmb.2720. [DOI] [PubMed] [Google Scholar]
- Halpern K.B., Caspi I., Lemze D., Levy M., Landen S., Elinav E., Ulitsky I., Itzkovitz S. Nuclear retention of mRNA in mammalian tissues. Cell Rep. 2015;13:1–11. doi: 10.1016/j.celrep.2015.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y., Tanaka H., Yamanaka S., Tanaka K., Watanabe Y., Tsutsumi C., Chikashige Y., Hiraoka Y., Yamashita A., Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- Hilleren P., McCarthy T., Rosbash M., Parker R., Jensen T.H. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Hiriart E., Vavasseur A., Touat-Todeschini L., Yamashita A., Gilquin B., Lambert E., Perot J., Shichino Y., Nazaret N., Boyault C. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012;31:2296–2308. doi: 10.1038/emboj.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrossova D., Sikorsky T., Potesil D., Bartosovic M., Pasulka J., Zdrahal Z., Stefl R., Vanacova S. RBM7 subunit of the NEXT complex binds U-rich sequences and targets 3′-end extended forms of snRNAs. Nucleic Acids Res. 2015;43:4236–4248. doi: 10.1093/nar/gkv240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iasillo C., Schmid M., Yahia Y., Maqbool M.A., Descostes N., Karadoulama E., Bertrand E., Andrau J.-C., Jensen T.H. ARS2 is a general suppressor of pervasive transcription. Nucleic Acids Res. 2017;45:10229–10241. doi: 10.1093/nar/gkx647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E., Lewis J., McGuigan C., Jankowska M., Darzynkiewicz E., Mattaj I.W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Jain A., Vale R.D. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.H., Boulay J., Rosbash M., Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 2001;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- Jensen T.H., Patricio K., McCarthy T., Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Jiang H., Wang S., Huang Y., He X., Cui H., Zhu X., Zheng Y. Phase transition of spindle-associated protein regulate spindle apparatus assembly. Cell. 2015;163:108–122. doi: 10.1016/j.cell.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Shao C., Wu Q.-J., Chen G., Zhou J., Yang B., Li H., Gou L.-T., Zhang Y., Wang Y. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017;24:816–824. doi: 10.1038/nsmb.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.J., Jackson R.N. Ski2-like RNA helicase structures: common themes and complex assemblies. RNA Biol. 2013;10:33–43. doi: 10.4161/rna.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T., Chen S., Hitomi M., Jacobs E., Kumagai C., Liang S., Schneiter R., Singleton D., Wisniewska J., Tartakoff A.M. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilchert C., Wittmann S., Vasiljeva L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 2016;17:227–239. doi: 10.1038/nrm.2015.15. [DOI] [PubMed] [Google Scholar]
- Kung J.T.Y., Colognori D., Lee J.T. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas M., Christensen M.S., Kristiansen M.S., Domanski M., Falkenby L.G., Lykke-Andersen S., Andersen J.S., Dziembowski A., Jensen T.H. Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- Lubas M., Andersen P.R., Schein A., Dziembowski A., Kudla G., Jensen T.H. The human nuclear exosome targeting complex is loaded onto newly synthesized RNA to direct early ribonucleolysis. Cell Rep. 2015;10:178–192. doi: 10.1016/j.celrep.2014.12.026. [DOI] [PubMed] [Google Scholar]
- Marayati B.F., Hoskins V., Boger R.W., Tucker J.F., Fishman E.S., Bray A.S., Zhang K. The fission yeast MTREC and EJC orthologs ensure the maturation of meiotic transcripts during meiosis. RNA. 2016;22:1349–1359. doi: 10.1261/rna.055608.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N., Jensen T.H. Targeting the nuclear RNA exosome: poly(A) binding proteins enter the stage. RNA Biol. 2017;14:820–826. doi: 10.1080/15476286.2017.1312227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N., Domanski M., Karadoulama E., Chen Y., Gentil C., Pultz D., Vitting-Seerup K., Lykke-Andersen S., Andersen J.S., Sandelin A., Jensen T.H. Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell. 2016;64:520–533. doi: 10.1016/j.molcel.2016.09.025. [DOI] [PubMed] [Google Scholar]
- Morchikh M., Cribier A., Raffel R., Amraoui S., Cau J., Severac D., Dubois E., Schwartz O., Bennasser Y., Benkirane M. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell. 2017;67:387–399.e5. doi: 10.1016/j.molcel.2017.06.020. [DOI] [PubMed] [Google Scholar]
- Moteki S., Price D. Functional coupling of capping and transcription of mRNA. Mol. Cell. 2002;10:599–609. doi: 10.1016/s1097-2765(02)00660-3. [DOI] [PubMed] [Google Scholar]
- Mukherjee N., Calviello L., Hirsekorn A., de Pretis S., Pelizzola M., Ohler U. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat. Struct. Mol. Biol. 2017;24:86–96. doi: 10.1038/nsmb.3325. [DOI] [PubMed] [Google Scholar]
- Naganuma T., Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami Z., Deryusheva S., Gall J.G. The Cajal body and histone locus body. Cold Spring Harb. Perspect. Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntini E., Järvelin A.I., Bornholdt J., Chen Y., Boyd M., Jørgensen M., Andersson R., Hoof I., Schein A., Andersen P.R. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–928. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- Ogami K., Richard P., Chen Y., Hoque M., Li W., Moresco J.J., Yates J.R., 3rd, Tian B., Manley J.L. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017;31:1257–1271. doi: 10.1101/gad.302604.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Segref A., Bachi A., Wilm M., Mattaj I.W. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell. 2000;101:187–198. doi: 10.1016/S0092-8674(00)80829-6. [DOI] [PubMed] [Google Scholar]
- Preker P., Nielsen J., Kammler S., Lykke-Andersen S., Christensen M.S., Mapendano C.K., Schierup M.H., Jensen T.H. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Preker P., Almvig K., Christensen M.S., Valen E., Mapendano C.K., Sandelin A., Jensen T.H. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A., Robb G.B., Chan S.-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016;44:7511–7526. doi: 10.1093/nar/gkw551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Tollervey D. Looking into the barrel of the RNA exosome. Nat. Struct. Mol. Biol. 2014;21:17–18. doi: 10.1038/nsmb.2750. [DOI] [PubMed] [Google Scholar]
- Seila A.C., Calabrese J.M., Levine S.S., Yeo G.W., Rahl P.B., Flynn R.A., Young R.A., Sharp P.A. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Zhang H., Wu X., He Z., Wang L., Yin S., Tian B., Li G., Cheng H. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 2017;45:9640–9653. doi: 10.1093/nar/gkx597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Sleeman J.E., Trinkle-Mulcahy L. Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr. Opin. Cell Biol. 2014;28:76–83. doi: 10.1016/j.ceb.2014.03.004. [DOI] [PubMed] [Google Scholar]
- St-André O., Lemieux C., Perreault A., Lackner D.H., Bähler J., Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(A)-binding protein in fission yeast. J. Biol. Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Sugioka-Sugiyama R. Red1 promotes the elimination of meiosis-specific mRNAs in vegetatively growing fission yeast. EMBO J. 2011;30:1027–1039. doi: 10.1038/emboj.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R., Nielsen P.S., Jensen T.H. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA. 2005;11:1745–1748. doi: 10.1261/rna.2139705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov N., Samacoits A., Chouaib R., Traboulsi A.-M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016;44:e165. doi: 10.1093/nar/gkw784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- Visa N., Izaurralde E., Ferreira J., Daneholt B., Mattaj I.W. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J. Cell Biol. 1996;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J.A., Davis C.P., Sunwoo H., Simon M.D., Sadreyev R.I., Wang P.I., Tolstorukov M.Y., Kingston R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell. 2014;55:791–802. doi: 10.1016/j.molcel.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T., Ngo L.H., Wickramasinghe V.O. Nuclear export of RNA: Different sizes, shapes and functions. Semin. Cell Dev. Biol. 2018;75:70–77. doi: 10.1016/j.semcdb.2017.08.054. [DOI] [PubMed] [Google Scholar]
- Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Yamashita A., Harigaya Y., Iwata R., Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Shichino Y., Tanaka H., Hiriart E., Touat-Todeschini L., Vavasseur A., Ding D.Q., Hiraoka Y., Verdel A., Yamamoto M. Hexanucleotide motifs mediate recruitment of the RNA elimination machinery to silent meiotic genes. Open Biol. 2012;2:120014. doi: 10.1098/rsob.120014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Takayama T., Iwata R., Yamamoto M. A novel factor Iss10 regulates Mmi1-mediated selective elimination of meiotic transcripts. Nucleic Acids Res. 2013;41:9680–9687. doi: 10.1093/nar/gkt763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu J., Schermann G., Ohle C., Bendrin K., Sugioka-Sugiyama R., Sugiyama T., Fischer T. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat. Commun. 2015;6:7050. doi: 10.1038/ncomms8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder J.C., Lima C.D. Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev. 2017;31:88–100. doi: 10.1101/gad.294769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M., Yamanaka S., Reyes-Turcu F.E., Zhang K., Rubin C., Grewal S.I.S. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of FISH and smiFISH probes.