Abstract
Objective
To investigate the association of the CD11b single nucleotide polymorphism (SNP) rs1143679 with systemic lupus erythematosus (SLE) in Han Chinese patients, and to clarify this association with SLE clinical manifestations.
Methods
PCR–restriction fragment length polymorphism and direct sequencing of CD11b rs1143679 were conducted in 584 patients with SLE and 628 healthy controls in this case–control study to compare genotype and allele frequency distributions. Correlations between CD11b genotypes and clinical manifestations were also determined.
Results
The frequency of the CD11b rs1143679 GA genotype was 1.89% in Han Chinese patients with SLE, which was much lower than that of European and American populations, but close to the frequency observed in individuals from Hong Kong and Thailand. The CD11b rs1143679 GA genotype was also shown to confer susceptibility to SLE (odds ratio = 4.00, 95% confidence interval = 1.11–14.41). CD11b rs1143679 was found to be significantly associated with nephritis, but not with age of disease onset, arthritis, hematological involvement, or neural lesions.
Conclusion
CD11b rs1143679 appears to be associated with risk for SLE in the Han Chinese population, and may play an important role in the development of lupus nephritis.
Keywords: Systemic lupus erythematosus, CD11b, rs1143679, Lupus nephritis, Han Chinese, Polymorphism
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease characterized by a large number of aberrations in the immune system, typically resulting in autoantibody production and organ-specific tissue destruction. SLE primarily affects women and has a strong genetic basis. Several associations have been identified through genome-wide association studies, including the rs1143679 variant in exon 3 of the integrin subunit α-M gene (CD11b) also known as ITGAM, complement component receptor 3. This is robustly associated with SLE,1–3 and the rs1143679 A risk allele has also been linked to clinical manifestations including renal disease, discoid rash, and immunological manifestations that can be recorded on the immunologic index.4
This non-synonymous variant (R77H) of the CD11b protein converts the arginine at amino acid position 77 to a histidine. Together with CD18, CD11b forms complement receptor 3 (CR3, also known as Mac-1, CD11b/CD18, αMβ2).5 CR3/Mac-1 is a type I membrane glycoprotein and one of four members of the leucocyte-restricted β2-integrin family, which plays a crucial role in several immunological processes including leukocyte extravasation and phagocytosis.6 Increasing evidence has suggested that CR3 is a crucial regulator of immune tolerance.7,8 Additionally, Ding et al.9 found that integrin CD11b negatively regulates B cell receptor (BCR) signaling to mediate autoreactive B cell tolerance. The R77H mutation on CD11b destroys direct CD22–CD11b binding, which results in B cell proliferation and increased autoantibody production, leading to immunoglobulin (Ig) deposition in the kidneys.
Associations between rs1143679 and SLE have been reported in many populations, including those from European, African, and Amerindian descent, but no data are available about the prevalence of rs1143679 in the Han Chinese population. We therefore analyzed the mutation frequencies of CD11b in Han Chinese patients with SLE using PCR–restriction fragment length polymorphism (RFLP) and direct sequencing in a case–control study.
Methods
Patients and controls
A total of 584 SLE patients were recruited from two Chinese hospitals: Nanjing First Hospital and Nanjing Pukou Central Hospital. All patients were diagnosed with SLE according to American College of Rheumatology classification criteria. A total of 628 unrelated healthy volunteer donors were recruited from the same ethnic background and geographic area. The 584 SLE patients included 38 men and 546 women with a mean age of 37.14 ± 12.35 years; the 628 healthy subjects comprised 27 men and 601 women with a mean age of 36.63 ± 11.27 years. There were no significant differences in sex or age between cases and controls. All clinical data were documented in an Excel database by trained, professional rheumatologists according to clinical rules. Lupus nephritis was diagnosed if patients met at least one of these three criteria: 1) permanent proteinuria >0.5 g/day, 2) persistent hematuria, and 3) pathological biopsy defined as nephritis. This study was reviewed and approved by Ethics Committees at the institutions, and all subjects provided written informed consent.
Isolation of genomic DNA
Genomic DNA was isolated from EDTA-anticoagulated peripheral blood using an E.Z.N.A.™ Blood DNA kit (Omega Bio-Tek, Norcross, GA, USA).
PCR–RFLP and direct sequencing
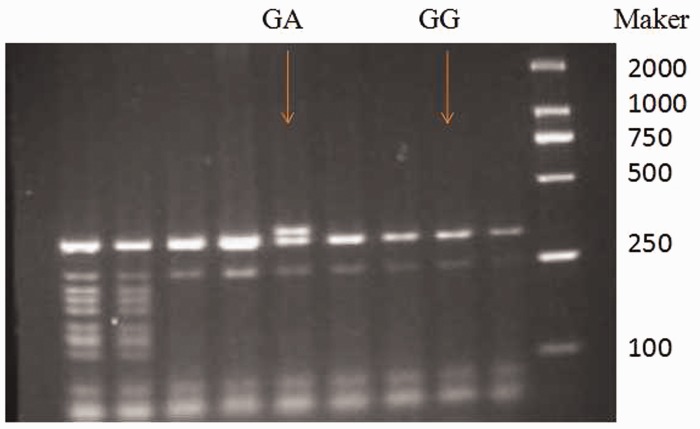
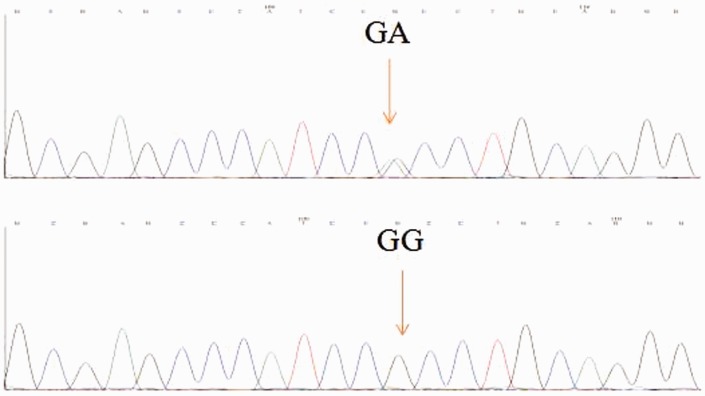
Primers were designed using Primer Express Software (Thermo Fisher Scientific, Inc., Rockford, IL, USA) to recognize specific sequences, and were obtained from Invitrogen Corp. (Carlsbad, CA, USA): forward primer, 5′-CTGGTTTTTGTGTCATTCTTAGG-3′; reverse primer: 5′-CAATCCCAGTCCCAGCCCG-3′. PCR amplifications were performed using 1 µl of each primer, 12.5 µl 2 × Taq PCR MasterMix (Tiangen, Beijing, China), 1 µl DNA template, and 9.5 µl ddH2O in a total volume of 25 µl. The following program was used for amplification: 95℃ for 30 s, then 35 cycles of 95℃ for 30 s, 61℃ for 30 s, 68℃ for 30 s, and 68℃ for 5 min. Fragments of 350 bp were obtained for each allele after amplification, which were then purified and 16 µl PCR product was digested with 2 µl Aci I (Ssi I) (Thermo Fisher Scientific, Inc.) and 2 µl 10 × FastDigest buffer (Thermo Fisher Scientific, Inc.) for 2 h at 37℃ (Figure 1). R77H genotyping was also confirmed using direct sequencing carried out by Yinjun Biological Engineering Technology Company (Shanghai, China; Figure 2).
Figure 1.
Confirmation of genotyping by PCR-restriction fragment length polymorphism.
Figure 2.
Confirmation of genotyping by direct sequencing.
Statistical analysis
Quantitative variables are presented as mean ± SD, and differences between variables were analyzed by the t-test. Genotype frequencies in patients and control subjects were examined on the basis of the Hardy–Weinberg equilibrium (HWE) using Genepop 4.1 software (http://kimura.univ-montp2.fr/∼rousset/Genepop.htm). Allele and genotype frequencies were compared between patients and control subjects by Fisher’s exact test. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated to estimate the effect of the rs1143679 SNP on SLE. P-values were two-sided and considered statistically significant if <0.05. All analyses were conducted using IBM SPSS statistics V.21 software (Chicago, IL, USA).
Results
Gene frequencies
Table 1 shows allelic and genotypic frequencies of the CD11b rs1143679 polymorphism in SLE patients with or without nephritis and controls. Genotype frequencies in both SLE patients and controls were in agreement with HWE expectations. The rs1143679 AA genotype was found to be absent from both SLE patients and controls. The frequency of the rs1143679 GA genotype was 1.89% in the SLE group, which was significantly higher than in the control group (0.48%; P = 0.02). The GA genotype also conferred susceptibility to SLE (OR = 4.00, 95% confidence interval: 1.11–14.41), whereas the homozygous GG genotype conferred protection against SLE.
Table 1.
Allelic and genotypic frequencies of CD11b rs1143679 in systemic lupus erythematosus (SLE) patients with and without nephritis and controls.
| Genotypes |
Alleles |
||||
|---|---|---|---|---|---|
| rs1143679 | GG, n (%) | GA, n (%) | AA, n (%) | G, n (%) | A, n (%) |
| Controls (n = 628) | 625 (99.52) | 3 (0.48) | NA | 1253 (99.76) | 3 (0.24) |
| SLE patients (n = 584) | 573 (98.11) | 11 (1.89) | NA | 1157 (99.06) | 11 (0.94) |
| P valuea | reference | 0.02 | NA | reference | 0.02 |
| OR [95%CI] | 4.00 [1.11–14.41] | NA | 3.97 [1.11–14.28] | ||
| SLE with nephritis (n = 284) | 275 (96.83) | 9 (3.17) | NA | 559 (98.42) | 9 (0.58) |
| P valuea | reference | 0.001 | NA | reference | 0.005 |
| OR [95%CI] | 6.82 [1.83–25.38] | NA | 6.28 [1.70–23.26] | ||
| SLE without nephritis (n = 300) | 298 (99.33) | 2 (0.67) | NA | 598 (99.67) | 2 (0.33) |
| P valuea | reference | 0.599 | NA | reference | 0.661 |
| OR [95%CI] | 2.10 [0.35–12.68] | NA | 1.397 [0.23–8.38] | ||
NA, no data; OR, odds ratio; 95%CI, 95% confidence interval; aFisher’s exact test.
The allelic distribution between SLE patients and controls was significantly different (P = 0.02). The minor allele (A) was shown to confer susceptibility to SLE (OR = 3.97) when compared with the major allele (G). The A allele also showed a significant association with nephritis (OR = 6.28) but only in patients with renal nephritis, not in those without, compared with controls (P = 0.005). An association with SLE and nephritis was also observed for the CD11b rs1143679 GA genotype.
Clinical characteristics of SLE patients
Table 2 shows the clinical characteristics of SLE patients with different CD11b rs1143679 genotypes. This SNP was found to be significantly associated with nephritis (P = 0.01), but not with age of disease onset, arthritis, hematological involvement, skin involvement, or neural lesions.
Table 2.
Clinical characteristics of systemic lupus erythematosus and genotype frequency distributions.
| GG (n = 573) | GA (n = 11) | χ2/t | P value | |
|---|---|---|---|---|
| Age at disease onset (years) | 27.23 ± 7.56 | 26.49 ± 9.17 | 0.32 | 0.74 |
| Arthritis, n (%) | 329 (57.4) | 7 (63.4) | 0.01 | 0.92 |
| Hematological involvement, n (%) | 346 (60.4) | 8 (72.7) | 0.27 | 0.60 |
| Neural involvement, n (%) | 262 (45.7) | 4 (36.4) | 0.10 | 0.76 |
| Nephritis, n (%) | 275 (48.0) | 9 (81.8) | 6.55 | 0.01 |
Discussion
SLE is an autoimmune disease mainly affecting childbearing women that is characterized by a loss of tolerance to self-antigens and dysregulated immune responses. Chinese subjects have a higher SLE disease prevalence and more renal involvement than Caucasians.10,11 Population differences in term of susceptibility genes are well documented; for example, the A risk allele of rs1143679 has a frequency of 9%–11% in European, Hispanic, Brazilian, and African–American populations, and shows a significant association with SLE. However, this minor risk allele has a lower frequency in Asian populations of Thailand and Hong Kong (<2%).12 Our study also demonstrated an association with the minor A allele and SLE in the Han Chinese population, despite its low frequency (1.98%), which is similar to that of other Asian populations, and the low power of the study on assessing rare SNPs. We also found that the heterozygous GA genotype was associated with a higher risk for SLE (OR = 4.00) compared with the GG genotype. The AA genotype was absent in both SLE patients and controls, perhaps because of the low prevalence of the homozygous mutation.
The A risk allele has been found to reduce adhesion and phagocytosis in human monocytes and monocyte-derived macrophages, without affecting the cell surface expression of CD11b.13 Fossati-Jimack et al.14 also suggested that this allele was responsible for impaired phagocytosis, but did not affect neutrophil adhesion or transmigration in vivo. A reduction in CR3-mediated phagocytosis might lead to increased tissue damage and inflammation, providing a plausible explanation for the genetic linkage identified by genome-wide association studies. It is also possible that a decrease in “waste disposal” contributes to disease, perhaps through reduced clearance of apoptotic cells, thereby leading to the activation of dendritic cells, inflammatory cytokine production, the presentation of nuclear antigens to T cells, and subsequent B cell activation.15 CR3 is an important receptor that mediates phagocytosis for opsonized particles such as apoptotic cells and immune complexes.16 The R77H variant is therefore thought to reduce phagocytosis of complement opsonized particles through influencing the ability of integrin to bind CD11b ligands such as iC3b, ICAM-1, and ICAM-2.6 CR3 may also interact with Fc receptors on the surface of neutrophils/macrophages, thus modulating the effect of immune complex binding; however, this requires further analysis.17
We found that the GA genotype was significantly associated with nephritis (P = 0.01), which agrees with studies of other populations; however, we found no association with age of disease onset, arthritis, hematological involvement, or neural lesions. It is conceivable that presence of the A allele predisposes patients to nephritis.. Overall, our data indicated that individuals with the CD11b GA genotype are more likely to develop lupus nephritis, and may have a more serious impaired renal function. Similar results were observed in mice; for example, a study by Alexander et al.18 showed that CD11b was protective in complement-mediated immune complex glomerulonephritis. CD11b−/− chimeras had a significantly higher number of M1 macrophages and CD4+T cells in their kidneys and functional renal insufficiency. Hence, CD11b expression on mononuclear cells appears to be instrumental in generating an anti-inflammatory response. Additionally, the engagement of BCR in CD11b-deficent mice was shown to increase autoantibody production and kidney Ig deposition.9
Our study is the first to demonstrate the distribution of CD11b rs1143679 allele and genotype frequencies in the Han Chinese SLE population. We showed that this SNP is associated with SLE susceptibility and the development of lupus nephritis. However, considering that a low minor allele frequency can introduce a wider confidence interval and a higher risk of random false positive associations, the sample size be increased in future work to confirm these findings. Moreover, further studies should investigate the role of B1 cell CD11b expression on the control of immune responses and inflammation.19,20 Previous in vitro work revealed that the suppressive function of CD11b was mediated by an impairment of T cell antigen receptor signaling, while increased B cell CD11b expression was regulated by interleukin 10.21 Moreover, CD11b was shown to negatively regulate BCR signaling to maintain autoreactive B cell tolerance. Identifying differences in genetic susceptibility to SLE could aid our understanding of disease mechanisms and have implications on its clinical intervention.
Acknowledgements
We thank Dr Bingwei Shi for help with English language support.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the National Natural Science Foundation of China (Grant #: 81228018); the Nanjing Health Youth Talent Project, Nanjing Department of Health (Grant #: QRX11026); and the Medical Science and Technology Development Foundation, Nanjing Department of Health (Grant #: YKK13207).
References
- 1.Nath SK, Han S, Kim-Howard X, et al. A nonsynonymous functional variant in integrin-alpha (M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet 2008; 40: 152–154. [DOI] [PubMed] [Google Scholar]
- 2.Harley JB, Alarcón-Riquelme ME, et al. International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN). Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008; 40: 204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez E, Webb RD, Rasmussen A, et al. Genetically determined Amerindian ancestry correlates with increased frequency of risk alleles for systemic lupus erythematosus. Arthritis Rheum 2010; 62: 3722–3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim-Howard X, Maiti AK, Anaya JM, et al. ITGAM coding variant (rs1143679) influences the risk of renal disease, discoid rash and immunological manifestations in patients with systemic lupus erythematosus with European ancestry. Ann Rheum Dis 2010; 69: 1329–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross GD. Role of the lectin domain of Mac-1/CR3 (CD11b/CD18) in regulating intercellular adhesion. Immunol Res 2002; 25: 219–227. [DOI] [PubMed] [Google Scholar]
- 6.MacPherson M, Lek HS, Prescott A, et al. A systemic lupus erythematosus-associated R77H substitution in the CD11b chain of the Mac-1 integrin compromises leukocyte adhesion and phagocytosis. J Biol Chem 2011; 286: 17303–17310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling GS, Bennett J, Woollard KJ, et al. Integrin CD11b positively regulates TLR4-induced signalling pathways in dendritic cells but not in macrophages. Nat Commun 2014; 5: 3039–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehirchiou D, Xiong Y, Xu G, et al. CD11b facilitates the development of peripheral tolerance by suppressing Th17 differentiation. J Exp Med 2007; 204: 1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding C, Ma Y, Chen X, et al. Integrin CD11b negatively regulates BCR signalling to maintain autoreactive B cell tolerance. Nat Commun 2013; 4: 2813–2813. [DOI] [PubMed] [Google Scholar]
- 10.Mok CC, Tang SS. Incidence and predictors of renal disease in Chinese patients with systemic lupus erythematosus. Am J Med 2004; 117: 791–795. [DOI] [PubMed] [Google Scholar]
- 11.Seligman VA, Lum RF, Olson JL, et al. Demographic differences in the development of lupus nephritis: a retrospective analysis. Am J Med 2002; 112: 726–729. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Zhao M, Hirankarn N, et al. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Hum Mol Genet 2009; 18: 2063–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes B, Fürnrohr BG, Roberts AL, et al. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann Rheum Dis 2012; 71: 2028–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fossati-Jimack L, Ling GS, Cortini A, et al. Phagocytosis is the main CR3-mediated function affected by the lupus-associated variant of CD11b in human myeloid cells. PLoS One 2013; 8: e57082–e57082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fagerholm SC, MacPherson M, James MJ, et al. The CD11b-integrin (ITGAM) and systemic lupus erythematosus. Lupus 2013; 22: 657–663. [DOI] [PubMed] [Google Scholar]
- 16.Morelli AE, Larrengina AT, Shufesky WJ, et al. Internalization of circulating apoptotic cells by splenic marginal zone dendritic cells: dependence on complement receptors and effect on cytokine production. Blood 2003; 101: 611–620. [DOI] [PubMed] [Google Scholar]
- 17.van Spriel AB, Leusen JH, van Egmond M, et al. Mac-1 (CD11b/CD18) is essential for Fc receptor-mediated neutrophil cytotoxicity and immunologic synapse formation. Blood 2001; 97: 2478–2486. [DOI] [PubMed] [Google Scholar]
- 18.Alexander JJ, Chaves LD, Chang A, et al. CD11b is protective in complement-mediated immune complex glomerulonephritis. Kidney Int 2015; 87: 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin DO, Rothstein TL. Human “orchestrator” CD11b (+) B1 cells spontaneously secrete interleukin-10 and regulate T-cell activity. Mol Med 2012; 18: 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai K, Tsuno NH, Matsuhashi M, et al. CD11b-mediated migratory property of peripheral blood B cells. J Allergy Clin Immunol 2005; 116: 192–197. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Jiang X, Liu R, et al. B cells expressing CD11b effectively inhibit CD4+ T-cell responses and ameliorate experimental autoimmune hepatitis in mice. Hepatology 2015; 62: 1563–1575. [DOI] [PubMed] [Google Scholar]