Short abstract
Objective
To elucidate the mechanism underlying secretion of human immunodeficiency virus type 1 (HIV-1) into the oral cavity, by examining the relationships between various oral and systemic factors and the viral load in saliva.
Methods
Plasma and saliva samples from HIV-1 infected patients were assayed using the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, version 1.0 and a Poisson distribution-based polymerase chain reaction (PCR) method for quantifying HIV-1 RNA and DNA.
Results
Forty-four pairs of samples were obtained from 18 patients. Salivary viral load was approximately 10% of the plasma viral load, but higher than the plasma load in two patients. The salivary viral DNA load was < 1% of the total HIV-1 nucleic acid load except in one patient who had more viral DNA than RNA. Multiple regression analysis showed that salivary viral load was significantly correlated with plasma viral load (partial correlation coefficient, 0.90) and the community periodontal index (–0.63).
Conclusions
The present results suggest that excretion through salivary glands, but not occult bleeding, may be a major pathway of HIV-1 into the oral cavity.
Keywords: HIV, quantification, plasma, saliva, Poisson distribution
Introduction
Although the annual number of new human immunodeficiency virus type 1 (HIV-1) infections is declining globally, the overall number of people living with HIV-1 has increased steadily.1 Recent advances in antiretroviral therapy have dramatically improved the prognosis of infected individuals, while HIV-1 infection is increasingly seen in the general practice setting.2 To improve early testing and medical care for infected individuals, it is important to learn about the oral manifestations of HIV infection and to understand the clinical significance of the HIV virus in the oral cavity.
HIV-1 is present in various body fluids including blood plasma, seminal plasma, breast milk, vaginal secretions and saliva.3 Blood, semen and vaginal secretions are potently infectious, while saliva is not considered infectious unless visibly bloody.4 The lack of HIV-1 transmission via saliva is thought to be due to factors including low viral load, the hypotonicity of saliva, and endogenous antiviral factors such as lysozyme, defensins, thrombospondin and secretory leukocyte protease inhibitor.5–9
Although many studies have quantified HIV-1 RNA in saliva and reported its correlation with the plasma viral load,3,10–13 little attention has been paid to HIV-1 DNA in infected cells, which may be more infectious than the free virus.14 Another shortcoming of these studies is that, to quantify the salivary viral load, they used commercial assays designed and approved to determine plasma viral load.
Thus, the aims of the present study were to develop a method for quantifying HIV-1 RNA and DNA in saliva based on the Poisson distribution of nested polymerase chain reaction (PCR) results, and to use this method to determine the salivary viral loads in HIV-1 infected individuals. Using the resulting data, the correlation between salivary viral loads and various systemic and oral parameters including cluster of differentiation (CD)4 cell count, patient age, plasma viral load, salivary occult bleeding, community periodontal index, salivary secretion rate, and the number of teeth was investigated to elucidate the mechanism of HIV-1 secretion into the oral cavity.
Patients and methods
Study population
This study included plasma and saliva samples from all HIV-1 infected patients who attended the Infection Control Department of Niigata University Medical and Dental Hospital between April 2008 and July 2009, and who provided written informed consent to participate in the study. There were no other inclusion or exclusion criteria for this study. Blood and saliva samples were collected at every visit (intervals ranged from 1 to 4 months, and were usually 2 months) for all patients during the study period unless the plasma viral loads were <40 copies/µl in consecutive tests. Seronegative participants, to provide negative control saliva samples, were recruited from the medical staff at the Division of Oral and Maxillofacial Surgery, Niigata University Graduate School of Medical and Dental Sciences. Approval was obtained from the Ethics Committee of the Niigata University School of Dentistry (19-R13-07-12).
Blood and saliva collection, and CD4 analysis
Approximately 5 ml of saliva was collected from each participant without stimulation, by expectoration after gargling with water and then swallowing the saliva secreted during the initial 1 min. Overall time to collect saliva was recorded, and the salivary secretion rate was obtained by dividing the volume by the sampling time. The saliva samples were stored at −20°C within 2 h of collection. Blood samples (8 ml) were collected using ethylenediaminetetra-acetic acid 2K (1.8 mg/ml) as an anticoagulant. The blood was centrifuged at 1 600 × g for 20 min to obtain plasma, and then the plasma was stored at –20°C prior to use. Cluster of differentiation (CD)4 cell count was obtained using CD4+ cell percentage, determined using the BD FACSCalibur™ automated flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA) and a fluorescein isothiocyanate (FITC)-conjugated anti-human CD4 antibody (T4-FITC; Beckman Coulter, Brea, CA, USA), and lymphocyte numbers per µl blood, obtained from complete cell count.
HIV-1 RNA
The 3rd World Health Organization (WHO) international standard for HIV-1 RNA (code 10/152) was purchased from the National Institute for Biological Standards and Control (Hertfordshire, UK) and was used as a source of HIV-1 RNA after purification using the QIAamp UltraSens Virus kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions.
Purification of nucleic acid from saliva
A 500 µl volume of saliva was mixed with 50 µl of 3.5 M sodium acetate (pH 5.2) and 1 ml of ethanol. The suspension was allowed to stand for 15 min and then centrifuged at 12 000 × g for 10 min. Nucleic acid was purified from the precipitate using the QIAamp UltraSens Virus kit. The recovery rate of HIV-1 RNA was determined by quantifying HIV-1 RNA after the purification of 500 µl of a blank saliva sample spiked with 100 copies of HIV-1 RNA.
In-house HIV-1 quantitative PCR assay
To amplify HIV-1 nucleic acid (RNA and DNA), purified nucleic acid was subjected to reverse transcription-nested PCR with primers designed in the HIV-1 group-specific antigen (gag) p24 region. The forward primer GA1FE, 5′-CAAGCAGCCATGCAAATGTTAA-3′ (nucleotides 1372–1393 of the HXB2 strain) and the reverse primer GA1RF, 5′-CATCCTATTTGTTCCTGAAGGGTACT-3′ (nucleotides 1510–1535) were used in the first round of PCR; the forward primer GA2FE, 5′-GAGGAAGCTGCAGAATGGGA-3′ (1408–1427) and the reverse primer GA2RE, 5′-GTTCTCTCATCTGGCCTGGTG-3′ (1460–1480) were used in the second round of PCR. Reverse transcription and the first round of PCR were performed in a 50-µl reaction mixture containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM of MgCl2, 0.2 mM of each dNTP, 0.2 µM of GA1FE and GA1RF primers, 4 units of RNasin® RNase Inhibitor, 20 units of SuperScript® III Reverse Transcriptase, and 1 unit of Platinum® Taq DNA Polymerase (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA). The second round of PCR was performed using 1 µl of the first-round PCR product in a 50-µl reaction mixture containing 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM of MgCl2, 0.2mM of each dNTP, 0.2 µM of GA2FE and GA2RE primers, and 1 unit of Platinum® Taq DNA Polymerase (Invitrogen). The cycling parameters for reverse transcription and the first round of PCR were 50°C for 10 min, then 94°C for 2 min, followed by 5 cycles of 94°C for 5 s, 48°C for 10 s, and 72°C for 15 s, then 25 cycles of 94°C for 5 s and 60°C for 15 s, followed by 72°C for 1 min, then held at 4°C. Conditions for the second round of PCR were 94°C for 2 min, then 5 cycles of 94°C for 5 s, 48°C for 10 s, and 72°C for 15 s, followed by 20 cycles of 94°C for 5 s and 60°C for 15 s, then 72°C for 1 min, followed by 4°C. The PCR products were analysed by agarose gel electrophoresis using 2% low electroendosmosis agarose (BMBio, Tokyo, Japan). To measure HIV-1 DNA only, the reverse transcription step was omitted. The concentration of HIV-1 nucleic acid was calculated using statistical analysis based on the Poisson distribution. Firstly, serial 10-fold dilutions of the extracted nucleic acid solution were assayed using nested PCR. Thereafter, the diluted solution that corresponded to the penultimate band was serially diluted two-fold and retested. Finally, 15 replicates of the two-fold dilution that produced the last positive signal were assayed. If the number of positive reactions was > 10 or < 5, further diluted or concentrated solutions, respectively, were used. When the original undiluted nucleic acid solution produced < 5 positive reactions in 15 replicates, this number was used for the calculation. The HIV-1 nucleic acid copy number in each reaction was calculated using the null class equation of the Poisson distribution.15
Commercial real-time PCR assay
The plasma and salivary viral RNA loads were determined by a reference laboratory (SRL, Tokyo, Japan) using the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, v1.0 (Roche Molecular Systems, Branchburg, NJ, USA), which is based on real-time PCR, according to the manufacturer’s instructions. Prior to assay, frozen saliva samples were thawed and mixed using end-over-end rotation to uniformly distribute the contents and food residues were removed by centrifugation at 400 × g for 2 min.
Quantification of salivary haemoglobin
The saliva blood content was evaluated by determining the ratio of haemoglobin concentration in saliva to that in blood, and referred to as the oral haemoglobin index. To determine the salivary haemoglobin levels, saliva was dripped onto a haemoglobin test strip (Salivaster, Showa Yakuhin Kako, Tokyo, Japan), incubated for 30 s and then fixed by a 10-s immersion in 30% hydrogen peroxide/ethanol (10:90). The strip was photographed with a digital camera, and the intensity of the developed colour was expressed numerically using ImageJ software (National Institutes of Health, Bethesda, MD, USA). A calibration curve was constructed using standard saliva obtained from a healthy volunteer (RI). The standard saliva was confirmed to be haemoglobin-free by comparing the colour reaction to physiological saline. Thereafter, the saliva samples for a calibration curve were prepared by spiking with various volumes of blood from the healthy volunteer to obtain seven different final haemoglobin concentrations ranging from 0.36 to 5.98 mg/dl. The haemoglobin concentration in the control blood was measured by automated haemocytometer in the clinical laboratory. If the sample results exceeded the limits of the calibration curve, the sample was diluted using the standard saliva.
Clinical evaluation of the oral cavity
The conditions of the oral cavity were examined by an oral surgeon (RT), including the number of teeth and HIV-associated lesions or diseases.16 Periodontal conditions were evaluated on the last day of sample collection using the community periodontal index.17
Statistical analyses
Data are presented as mean or individual values, depending on the number of samples obtained per patient, range, or n (%) sample prevalence. Single and multiple regression analyses were used to evaluate the associations between relevant parameters. The values of plasma and saliva viral loads were transformed to log10 prior to analysis. Data below the lower limit was treated as half the lower limit value. For example, in the COBAS assay, < 40 copies/ml was recorded as 20 copies/ml. Fisher’s exact test was used to analyse the differences in HIV-1 nucleic acid detection levels between the in-house Poisson quantitative PCR assay and commercial COBAS® TaqMan® PCR assay. Data were analysed using the statistical software package Statcel2 (OMS Publishing, Saitama, Japan). A P value <0.05 was considered statistically significant.
Results
Patient characteristics
A total of 18 patients were enrolled, 14 male and 4 female (Table 1), with a median age of 37 years (range, 26–60 years). The 18 patients provided 44 pairs of plasma and saliva samples, and the median number of samplings was 2 per patient (range, 1–6). The mean time required to collect saliva was 13 min (range, 4–40 min). Sixteen patients were undergoing antiretroviral therapy: 15 (83%) from the start of the study, and one patient, M, commenced antiretroviral therapy during the study period. Virologic suppression (<40 copies/ml) had been achieved with antiretroviral therapy in eight patients (44%). None of the patients complained of dry mouth or were diagnosed as having xerostomia during the study period.
Table 1.
Within-study characteristics of 18 patients with human immunodeficiency virus type 1 (HIV-1) infection
|
Patient characteristic | |||||||
|---|---|---|---|---|---|---|---|
| Patient ID | Sex | Age | Number of samplings | Antiretroviral therapy | CD4+cell count(cells/µl) | Viral load(copies/µl) | Community periodontal index |
| A | Male | 36 | 2 | TDF/FTC + ATV/r | 8–18 | 13000–50000 | 2.33–2.16 |
| B | Male | 48 | 3 | AZT/3TC + EFV | 481–536 | <40–110 | 0.00–0.67 |
| C | Male | 29 | 4 | ABC/3TC + ATV/r | 108–247 | <40–110 | 1.66–2.00 |
| D | Male | 36 | 1 | ABC/3TC + ATV/r | 466 | 45 | 2.00 |
| E | Male | 58 | 3 | ABC/3TC + LPV/r | 183–291 | <40–300 | 0.83–2.67 |
| F | Female | 26 | 1 | AZT/3TC + LPV/r | 446 | <40 | 0.83 |
| G | Female | 41 | 1 | AZT/3TC + LPV/r | 320 | <40 | 1.50 |
| H | Male | 41 | 5 | ABC/3TC + ATV/r | 258–406 | <40–300 | 0.67–1.67 |
| I | Male | 42 | 1 | ABC/3TC + LPV/r | 365 | <40 | 0.83 |
| J | Male | 48 | 2 | d4T + 3TC + LPV/r | 206–238 | <40–44 | 0.50–1.60 |
| K | Male | 39 | 4 | AZT + TDF + LPV/r | 300–505 | <40–50 | 0.00–2.17 |
| L | Female | 32 | 1 | TDF/FTC + LPV/r | 144 | 1300 | 0.67 |
| M | Male | 38 | 2 | TDF/FTC + EFV | 268–417 | <40–15000 | 2.50–2.70 |
| N | Female | 32 | 5 | TDF/FTC + ATV | 81–224 | Not detected to <40 | 1.00–2.00 |
| O | Male | 33 | 6 | Untreated | 286–388 | 1600–8600 | 0.00–1.00 |
| P | Male | 32 | 1 | ABC/3TC + LPV/r | 172 | <40 | 2.00 |
| Q | Male | 60 | 1 | TDF/FTC + ATV/r | 42 | 1600 | 2.80 |
| R | Male | 31 | 1 | Untreated | 651 | 7300 | 1.17 |
Data presented as value or range.
ID, patient study identification; AZT, zidovudine; d4T, stavudine; ABC, abacavir; 3TC, lamivudine; TDF, tenofovir; FTC, emtricitabine; EFV, efavirenz; ATV, atazanavir; LPV, lopinavir; r, ritonavir for boosting.
Accuracy of in-house quantification assay
The third WHO international standard for HIV-1 RNA was measured using the in-house Poisson quantitative PCR assay and its concentration was found to be 104 copies/µl. The HIV-1 RNA standard has been assigned a unitage of 5.27 log10 international units (IU)/ml,18 thus, the present results indicated that 1 IU was equivalent to 0.56 copies. This value was very close to previously reported conversion factors of 0.6 or 0.51 copy/IU for HIV-1 RNA,19 suggesting that the in-house quantification assay was capable of accurately determining the HIV-1 RNA copy number. Regarding specificity, no positive signals were detected in 50 runs of saliva samples from three seronegative individuals.
Recovery rate of HIV-1 RNA in saliva
The recovery rate of HIV-1 RNA in saliva following purification was determined using five replicates of HIV-1-negative saliva each spiked with 100 copies of HIV-1 RNA. The HIV-1 RNA quantities before and after purification were determined using the in-house Poisson quantitative PCR assay. The obtained recovery rate was 76% ± 30% (mean ± SD). Based on this result, the HIV-1 RNA concentration in saliva was corrected by dividing the raw data by 0.76.
In-house Poisson quantitative PCR versus COBAS® AmpliPrep/ COBAS® TaqMan® HIV-1 Test, v1.0, for salivary viral load
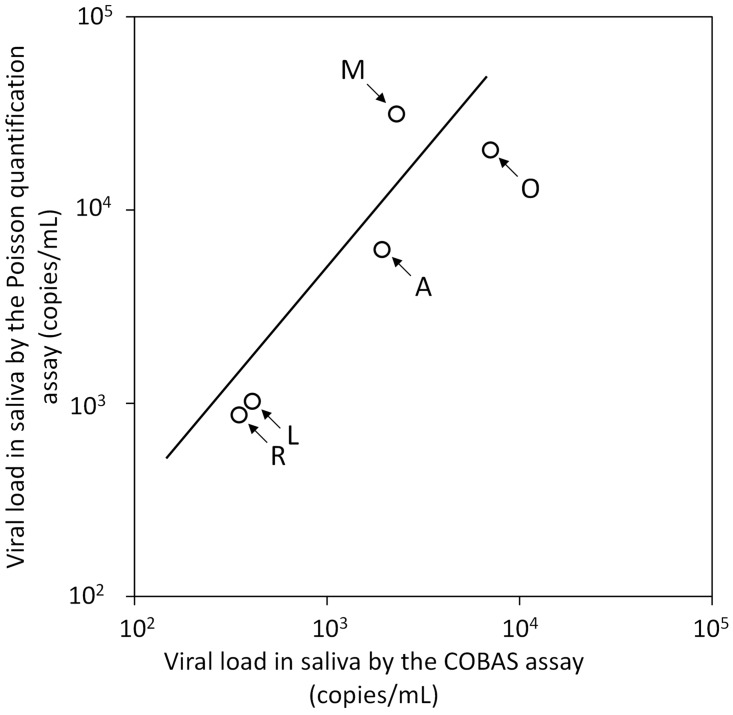
Viral nucleic acid (RNA and DNA) was detected in 73% (32/44) of saliva samples from the 18 participants using the Poisson quantitative PCR assay, and viral RNA was detected in 34% (15/44) of saliva samples using the COBAS assay (Table 2). Statistical analysis revealed that the Poisson quantitative PCR assay was significantly more sensitive than the COBAS assay in detecting HIV-1 nucleic acid (P = 0.027, Fisher’s exact test). Saliva samples were found to be positive using both assays in five patients (comprising 12 samples), and the mean viral load value for each patient was used to compare quantification performance between the two assays, which resulted in a good linear relationship between the two (P <0.05; r2 = 0.83; Figure 1). The mean value obtained from the Poisson quantitative PCR assay was 3.8 times higher than from the COBAS assay. Salivary viral loads of all samples that were determined using the Poisson quantitative PCR assay were used for further analysis.
Table 2.
Comparison between an in-house Poisson quantitative polymerase chain reaction (PCR) assay and the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, v1.0 for detecting human immunodeficiency virus type 1 (HIV-1) nucleic acid (DNA and RNA) in 44 saliva samples from 18 patients with HIV-1
|
In-house Poisson quantitative PCR assaya |
||||
|---|---|---|---|---|
| Detected | Undetected | Total | ||
| COBAS® TaqMan® assay | Detected | 14 | 1 | 15 |
| Undetected | 18 | 11 | 29 | |
| Total | 32 | 12 | 44 | |
Data presented as n sample prevalence.
aSignificantly more positive samples using the in-house Poisson quantitative PCR assay versus the COBAS® TaqMan® assay (P = 0.027; Fisher’s exact test).
Figure 1.
Regression analysis to compare salivary human immunodeficiency virus type 1 (HIV-1) viral loads in five patients with HIV-1 infections, measured using the COBAS® AmpliPrep/ COBAS® TaqMan® HIV-1 Test, v1.0 or an in-house Poisson quantitative polymerase chain reaction assay. Data presented as the mean of patient samples for each patient (each patient shown with an arrow). The solid diagonal line shows a single regression curve. A good linear relationship was seen between the results of the COBAS assay and the Poisson quantitative PCR assay (P <0.05; r2 = 0.83)
Plasma and salivary viral loads
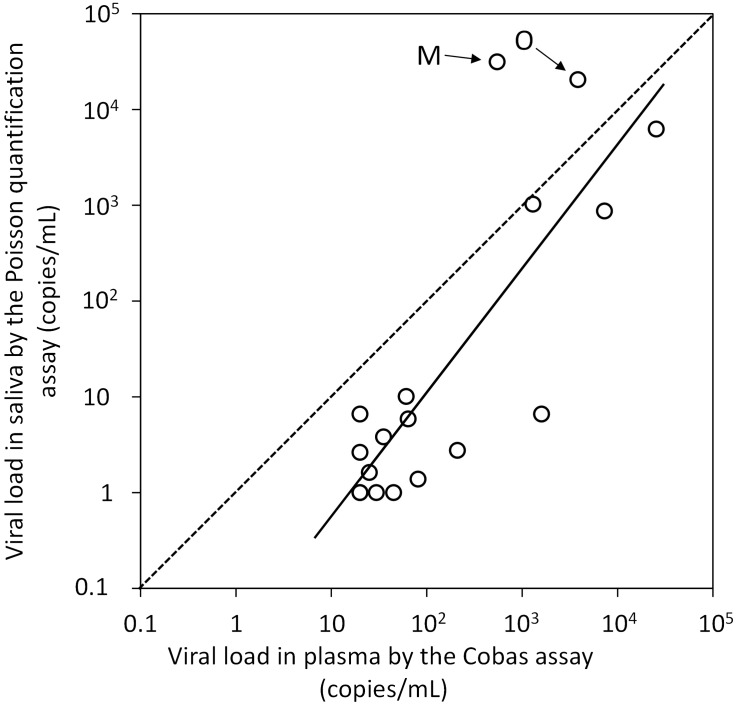
The relationship between plasma and salivary viral loads in 18 patients was studied using a single regression analysis of the mean sample values for each patient (Figure 2). Salivary viral load (DNA and RNA) was measured using the Poisson quantitative PCR assay and plasma viral load was measured using the COBAS assay. A good linearity was observed in terms of viral load between the two sample and measurement types (P <0.01; r2 = 0.75). The salivary viral load was found to be approximately 10% of the plasma viral load, expressed as geometric mean of all values. However, in two patients, M and O, the salivary viral loads were higher than the plasma viral loads (Figure 2).
Figure 2.
Regression analysis of the relationship between plasma human immunodeficiency virus type 1 (HIV-1) viral load (determined using the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, v1.0) and salivary HIV-1 viral load (determined using a Poisson quantitative polymerase chain reaction assay) in 18 patients with HIV-1. Two patients (M and O) were found to have salivary viral loads that were higher than the plasma viral loads (arrows). Data presented as the mean of all samples for each patient. The solid diagonal line represents the single regression curve for the data, with a good linear relationship between the saliva and plasma viral loads (P <0.01; r2 = 0.75). The dotted line represents equal viral loads in plasma and saliva (r2 = 1.0) for comparison
Viral DNA loads in saliva
Viral DNA was detected in 11 saliva samples from four of five patients who had salivary viral loads of ≥100 copies/ml. The mean plasma viral loads, salivary viral nucleic acid loads and salivary viral DNA loads of these five patients are compared in Table 3. Patient L had a viral DNA load accounting for 75.3% of the salivary nucleic acid load, while the proportions of viral DNA in the viral nucleic acid for the other four patients were ≤1%.
Table 3.
Comparison of mean plasma viral load, salivary viral load and salivary proviral DNA load in five patients with human immunodeficiency virus type 1 (HIV-1)
| Patient | Number of samples | Plasma HIV-1 viral load (copies/ml)a | Salivary HIV-1 viral load (copies/ml)b | Salivary HIV-1 DNA load (copies/ml)c |
|---|---|---|---|---|
| A | 2 | 31500 | 7630 | 63 (0.8) |
| L | 1 | 1300 | 1024 | 771 (75.3) |
| M | 1 | 15000 | 31345 | 97 (0.3) |
| O | 6 | 4600 | 22268 | 17 (0.1) |
| R | 1 | 7300 | 868 | 0 (0.0) |
Data presented as mean (% of salivary viral load).
aDetermined using the COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, v1.0. bDetermined using the Poisson quantitative polymerase chain reaction (PCR) assay with reverse transcription. cDetermined using the Poisson quantitative PCR assay without reverse transcription.
Factors influencing the salivary viral load
Multiple regression analysis was used to examine the relationships between salivary HIV-1 viral load and various systemic and oral parameters, comprising plasma viral load, community periodontal index, oral haemoglobin index, CD4+ cell count, salivary secretion rate, patient age, and the number of teeth (Table 4). As a result, salivary viral load (y) could be expressed in relation to two statistically significant predictors, namely, the plasma viral load (x1, P < 0.01) and community periodontal index (x2, P < 0.05): y = 1.13x1 – 0.60x2 (r2 = 0.92). Antiretroviral therapy was highly correlated with the plasma viral load but was not an independent predictor of the salivary viral load.
Table 4.
Multiple regression analysis to examine the relationship between salivary viral load, measured using the Poisson quantitative PCR assay, and various systemic and oral parameters in 18 patients with human immunodeficiency virus type 1
| Parameter | Regression coefficient(95% confidence interval) | Partial correlation coefficient | F | Statistical significance |
|---|---|---|---|---|
| Plasma viral load | 1.10 (0.76, 1.51) | 0.90 | 44.7 | P < 0.01 |
| Community periodontal index | –0.61 (–1.14, –0.08) | –0.63 | 6.53 | P < 0.05 |
| Oral haemoglobin index | 16033 (–2524, 34592) | 0.51 | 3.71 | NS |
| CD4 | 0.0005 (–0.002, 0.003) | 0.14 | 0.20 | NS |
| Salivary secretion rate | 0.29 (–0.82, 1.40) | 0.18 | 0.35 | NS |
| Patient age | –0.03 (–0.07, 0.02) | –0.36 | 1.45 | NS |
| Number of teeth | 0.06 (–0.04, 0.17) | 0.38 | 1.6 | NS |
NS, no statistically significant correlation (P > 0.05).
Discussion
The present study assessed the association between salivary HIV-1 loads and systemic and oral variables. Similar recent studies3,10–13 have differed from the present study in several ways: in the present study, salivary viral loads were determined using a Poisson distribution-based PCR method, which is considered to be an accurate and robust method for quantifying HIV-1;20,21 viral DNA loads were determined separately from viral RNA; and occult blood levels in saliva were quantitatively assessed by measuring the haemoglobin concentrations. These parameters together with salivary secretion rates and periodontal conditions, assessed using the community periodontal index, were used in multiple regression analysis to assess predictors of salivary viral load.
The in-house Poisson quantitative PCR assay was shown to quantify the WHO international HIV-1 RNA standard accurately, and was found to detect viral nucleic acid (RNA and DNA) more frequently in saliva samples, and yielded higher concentrations than the COBAS assay. Thus, data obtained using this assay was used for further analysis. One possible reason for the discrepancy between the two assays may be that the COBAS assay has been designed to measure viral loads in blood22 but not saliva.
Consistent with previous studies,3,10 multiple regression analysis in the present study showed that the plasma viral load was a significant predictor of the salivary viral load. Salivary viral load was not found to be correlated with saliva secretion rate. The present authors postulate that if the virus is excreted into the oral cavity from sites other than the salivary glands, a higher salivary secretion rate would reduce the salivary viral concentration. The lack of relationship between salivary secretion rate and salivary viral load in the present study suggests that the virus is mainly excreted along with saliva from the salivary glands. Higher levels of stimulated saliva flow rates have been associated with lower HIV-1 RNA shedding in the saliva.12 In the present study, unstimulated saliva samples were used, and the discrepancy between the previous12 and present results may be explained by the possibility that the excretion rate of HIV-1 from salivary grands into saliva is not significantly influenced by artificial stimulation of saliva secretion.
Interestingly, the present results showed that the community periodontal index contributed to salivary viral load in an inverse manner, suggesting that poor periodontal conditions may suppress viral excretion into the oral cavity. Epithelial attachments have wide intercellular spaces through which lymphocytes and antibodies can pass,23,24 and during the progression of periodontitis, epithelial attachments are gradually lost. If the virus is excreted more easily through epithelial attachments than connective tissue, healthy periodontal conditions may favour viral excretion from the gingival crevices. However, the number of patients in the present study was limited, and their community periodontal index ranged from 0.0 to 2.8, which represents mild periodontal inflammation. At more advanced stages of periodontal disease, the salivary viral load may increase.3
Viral hyper-excretion has been defined as a four-fold or higher viral load in saliva than in plasma.11 In the present study, one of three untreated patients was found to be a hyper-excreter, whereas there were no hyper-excreters among the 15 patients receiving antiretroviral therapy. The hyper-excreter did not have any distinctive oral findings or abnormal observations in the oral cavity, and the cause of the hyper-secretion remains unclear.
Among five patients in whom viral DNA was quantified, the salivary viral DNA load was less than 1% of the total viral nucleic acid in four patients, but viral DNA was more abundant than HIV-1 RNA in one patient (with viral DNA at 75.3% of total viral nucleic acid). Although infected milk cells have been argued to play a more important role in viral transmission via breast feeding than free virus,25 there is no clear evidence that cell-associated virus in saliva is more infectious than free virus. Regardless of whether this is true, standard precautions must be observed during dental procedures.
In conclusion, the present study showed that salivary HIV-1 load was correlated with plasma viral load and, to a lesser extent, with patient periodontal conditions. The study also suggested that HIV-1 virus may be mainly excreted together with saliva from the salivary glands. Measurement of HIV-1 viral load in saliva may be used as a noninvasive assay for estimating blood viral load to assess the effect of antiretroviral therapy in resource-poor settings.
Declaration of conflicting interests
The authors declare that there is no conflict of interest.
Funding
This study was performed with the support of a Category B Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
References
- 1.UNAIDS report on the global AIDS epidemic 2013, http://www.unaids.org/sites/default/files/en/media/unaids/contentassets/documents/epidemiology/2013/gr2013/UNAIDS_Global_Report_2013_en.pdf (2013, accessed 6 December 2015).
- 2.Rutland E, Foley E, O’Mahony C, et al. How normalised is HIV care in the UK? A survey of current practice and opinion. Sex Transm infect 2007; 83: 151–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shugars DC, Slade GD, Patton LL, et al. Oral and systemic factors associated with increased levels of human immunodeficiency virus type 1 RNA in saliva. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2000; 89: 432–440. [DOI] [PubMed] [Google Scholar]
- 4.Chapman LE, Sullivent EE, Grohskopf LA, et al. Recommendations for postexposure interventions to prevent infection with hepatitis B virus, hepatitis C virus, or human immunodeficiency virus, and tetanus in persons wounded during bombings and other mass-casualty events – United States, 2008: recommendations of the Centers for Disease Control and Prevention (CDC). MMWR Recomm Rep 2008; 57: 1–21. [PubMed] [Google Scholar]
- 5.Shugars DC. Endogenous mucosal antiviral factors of the oral cavity. J Infect Dis 1999; 179: S431–S435. [DOI] [PubMed] [Google Scholar]
- 6.Baron S Poast J andCloyd MW.. Why is HIV rarely transmitted by oral secretions? Saliva can disrupt orally shed, infected leukocytes. Arch Intern Med 1999; 159: 303–310. [DOI] [PubMed] [Google Scholar]
- 7.Shine N Konopka K andDuzgunes N.. The anti-HIV-1 activity associated with saliva. J Dent Res 1997; 76: 634–640. [DOI] [PubMed] [Google Scholar]
- 8.Shugars DC, Alexander AL, Fu K, et al. Endogenous salivary inhibitors of human immunodeficiency virus. Arch Oral Biol 1999; 44: 445–453. [DOI] [PubMed] [Google Scholar]
- 9.Kazmi SH, Naglik JR, Sweet SP, et al. Comparison of human immunodeficiency virus type 1-specific inhibitory activities in saliva and other human mucosal fluids. Clin Vaccine Immuno 2006; 13: 1111–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepard RN, Schock J, Robertson KJ, et al. Quantification of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol 2000; 38: 1414–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shugars DC, Patton LL, Freel SA, et al. Hyper-excretion of human immunodeficiency virus type 1 RNA in saliva. J Dent Res 2001; 80: 414–420. [DOI] [PubMed] [Google Scholar]
- 12.Navazesh M, Mulligan R, Kono N, et al. Oral and systemic health correlates of HIV-1 shedding in saliva. J Dent Res 2010; 89:1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kantor R, Bettendorf D, Bosch RJ, et al. HIV-1 RNA levels and antiretroviral drug resistance in blood and non-blood compartments from HIV-1-infected men and women enrolled in AIDS clinical trials group study A5077. PLoS One.2014; 9: e93537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costiniuk CT andJenabian MA.. Cell-to-cell transfer of HIV infection: implications for HIV viral persistence. J Gen Virol 2014; 95: 2346–2355. [DOI] [PubMed] [Google Scholar]
- 15.Kinai E Hanabusa H andKato S.. Prediction of the efficacy of antiviral therapy for hepatitis C virus infection by an ultrasensitive RT-PCR assay. J Med Virol 2007; 79: 1113–1119. [DOI] [PubMed] [Google Scholar]
- 16.EC-Clearinghouse on Oral Problems Related to HIV Infection and WHO Collaborating Centre on Oral Manifestations of the Immunodeficiency Virus Classification and diagnostic criteria for oral lesions in HIV infection. J Oral Pathol Med 1993; 22: 289–291. [PubMed] [Google Scholar]
- 17.World Health Organization Oral health surveys: basic methods. 4th ed. Geneva, Switzerland, 1997, pp.36–38.
- 18.Morris CL andHeath AB. International collaborative study to establish the 3rd WHO international standard for HIV-1 NAT assays. WHO ECBS report. Report no. WHO/BS/2011.2178, http://apps.who.int/iris/bitstream/10665/70784/1/WHO_BS_2011.2178_eng.pdf (2011, accessed 13 September 2017).
- 19.Glaubitz J, Sizmann D, Simon CO, et al. Accuracy to 2nd International HIV-1 RNA WHO Standard: assessment of three generations of quantitative HIV-1 RNA nucleic acid. J Clin Virol 2011; 50: 119–124. [DOI] [PubMed] [Google Scholar]
- 20.Kato S, Hanabusa H, Kaneko S, et al. Complete removal of HIV-1 RNA and proviral DNA from semen by the swim-up method: assisted reproduction technique using spermatozoa free from HIV-1. AIDS 2006; 20: 967–973. [DOI] [PubMed] [Google Scholar]
- 21.Kondo M, Sudo K, Tanaka R, et al. Quantitation of HIV-1 group M proviral DNA using TaqMan MGB real-time PCR. J Virol Methods 2009; 157: 141–146. [DOI] [PubMed] [Google Scholar]
- 22.Braun P, Ehret R, Wiesmann F, et al. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin Chem Lab Med 2007; 45: 93–99. [DOI] [PubMed] [Google Scholar]
- 23.Maticic M, Poljak M, Kramar B, et al. Proviral HIV-1 DNA in Gingival Crevicular Fluid of HIV-1-infected Patients in Various Stages of HIV Disease. J Dent Res 2000; 79: 1496–1501. [DOI] [PubMed] [Google Scholar]
- 24.O’Shea S, Cordery M, Barrett WY, et al . HIV excretion patterns and specific antibody responses in body fluids. J Med Virol 1990; 31: 291–296. [DOI] [PubMed] [Google Scholar]
- 25.Rousseau CM, Nduati RW, Richardson BA, et al. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J Infect Dis 2004; 190: 1880–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]