Short abstract
Objective
To evaluate the clinical value of human papillomavirus (HPV) E6 and E7 oncoprotein (HPV E6/E7) detection in the early screening of cervical cancer.
Methods
This prospective study evaluated all patients with suspected cervical intraepithelial neoplasia (CIN) as identified by the presence of at least one positive indicator from a ThinPrep cytologic test (TCT) and/or a Hybrid Capture 2 (HC2) HPV DNA test. The levels of E6/E7 oncoproteins were determined using Western blot analysis. The diagnostic value of the HPV E6/E7 protein assay was compared with the clinical diagnosis from TCT, HC2 and the gold standard of cervical biopsy histology.
Results
A total of 450 patients were enrolled in the study and based on histological findings, 102 patients were diagnosed with CIN1 (22.7%), 241 with CIN2 (53.6%), 96 with CIN3 (21.3%) and 11 with squamous cell carcinoma (2.4%). For a diagnosis of CIN2+, although the sensitivity of the HPV E6/E7 assay was lower than HC2 (65.5% versus 96.6%, respectively), the specificity was higher (38.2% versus 5.9%, respectively). The sensitivity of the HPV E6/E7 assay was higher than TCT (65.5% versus 36.2%, respectively).
Conclusion
Measuring HPV E6/E7 oncoprotein levels is a potential new biomarker for HPV type 16.
Keywords: E6/E7 oncoproteins, HPV DNA, HPV mRNA, prognostic detection
Introduction
Cervical cancer (CaCx) is closely associated with human papillomavirus (HPV) infection. More than half a million new cases of CaCx are diagnosed each year worldwide and the incidence in developing countries is much higher compared with developed countries.1,2 More importantly, around half of all new cases are in China and India.3 More than one-third of women of reproductive age in China have been infected with HPV, with a 5%–10% rate of continuous infection.4 Among those with long-term HPV infections, approximately one-quarter will develop cervical intraepithelial neoplasia (CIN) and less than 1% will develop CaCx.5 The technical developments in molecular biology and virology have enabled researchers to determine that integration of the viral HPV DNA into the host cell genome is the key step in the development of CaCx.6 Two high-risk HPV E6/E7 oncoproteins, which are found in nearly all HPV-positive cases, are expressed from HPV DNA after it has inserted into the host cell genome.7,8
The current mainstream methods for screening for CaCx in clinical trials include the Papanicolaou test, ThinPrep cytological test (TCT), HPV DNA test (e.g. Hybrid Capture 2 [HC2]), HPV mRNA test (e.g. PreTect HPV-Proofer), colposcopy, and visual inspection with acetic acid. Women with abnormal results from cytological testing and/or HPV DNA detection will undergo a subsequent pathological biopsy to check for the existence and staging of CIN during long-term follow-up. Histology is recognized as the gold standard for diagnosing the pathological progress of CaCx development, while CIN2+ (CIN2, CIN3 or cancer) is the cut-off for intervention in clinical practice.9 However, it is unknown which kind of lesions will finally develop into infiltrative cancers. In developed countries, primary screening based on cytological testing can prevent more than 80% of CaCx.10 However, abnormal diseases are often missed or misdiagnosed due to the limitations of the testing sensitivity and sampling techniques.11 Owing to the low sensitivity of the cytological test, females at a high risk are required to have regular retests to confirm the accuracy of the negative result.9 Among those patients with a negative cytology result but a positive HPV DNA test, only a small proportion will eventually develop CaCx.12 For the clinical management of these particular patients who are TCT–/HPV+, it would be useful to have new biomarkers to improve the prognostic abilities of the current screening methods.
The aim of this study was to investigate two new biomarkers, the E6/E7 oncoproteins, which are thought to be the main inducing factors for pre-CaCx lesions,13 as prognostic indicators for CaCx development.
Patients and methods
Patient population
This prospective study recruited patients with at least one positive indicator of cervical abnormalities either from a TCT or a HC2 HPV DNA test in the Department of Laboratory Medicine, Henan Province Chinese Medicine Hospital, Zhengzhou, Henan Province, China between October 2010 and September 2014. The patients had not undergone any treatment, hysterectomy, radiotherapy or chemotherapy. Each cervical cytology sample was analysed by TCT, HC2 HPV DNA test, Western blot analysis of HPV E6/E7 oncoproteins and a histological diagnosis to determine the stage of CIN as described below. The performance of the HPV E6/E7 assay was compared with the clinical diagnosis determined using the TCT and HC2 assays, as well as the histological diagnosis to determine the stage of CIN. Women attending for routine gynaecological examinations who had a negative HPV DNA test were included as healthy control subjects.
This study was approved by the Ethics Committee of Henan Province Chinese Medicine Hospital, Zhengzhou, Henan Province, China (no. YC2010309316). Patients or their legal representative provided written informed consent prior to study enrolment.
Preparation and preservation of samples
Cervical cytology samples were prepared using PreservCyt® Solution (ThinPrep Pap Test; Hologic, Marlborough, MA, USA). Cervical cytology specimens (2 ml; approximately 75 µl of cell pellet per 1 ml of solution) were collected into vials containing 20 ml PreservCyt® Solution as specimens for TCT as described below. All samples were tested within 2 weeks after collection and stored at –80°C until tested.
Cervical cytology specimens with abnormal cytological findings (approximately 75 µl of cell pellet per 1 ml of solution) were rinsed in Specimen Transport Medium (Qiagen, Valencia, CA, USA) in preparation for the detection of HPV DNA by HC2 (Qiagen) according to manufacturer's instructions for PreservCyt® specimens as described below. Specimens were stored at 4°C and remained viable for analysis within 3 months of initial specimen collection. The threshold for positive HPV DNA detection was a relative light unit/cut-off ratio of ≥1.0. Any sample containing < 5 µl of cell precipitate was discarded.
TCT testing
ThinPrep cytological testing was undertaken using a commercial kit according to the manufacturer’s instructions (Beijing TCT Medical Technology, Beijing, China). Results of the cytological screening were classified into five grades according to The Bethesda System:14 within normal limits (normal); atypical squamous cells with undetermined significance (ASC-US); low-grade squamous intraepithelial lesion (LSIL); high-grade squamous intraepithelial lesion (HSIL); atypical squamous cells–cannot exclude HSIL (ASC-H). TCT could also identify squamous cell carcinoma (SCC).
HC2 HPV DNA analysis
The digene HC2 HPV DNA Test (Qiagen) was used to quantitate the level of HPV DNA using a standardized test that has been approved by the US Food and Drug Administration.15 Residual cervical cytological samples remaining after the TCT test were denatured to get single-stranded DNA, which was mixed and reacted with a cocktail of 13 full-length RNA probes designed to capture the following oncogenic HPV types: 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68. All samples were repeated in triplicate.
Western blot analysis of E6/E7 protein levels
Cervical cytology samples were collected and frozen for protein extraction, which was performed by adding RIPA rapid cell lysis buffer (Jierdun Biotechnology, Shanghai, China) containing protease and phosphatase inhibitors (Thermo Fisher Scientific, Rockford, IL, USA) as described previously.16 The protein concentration of the cell lysate was determined using a BCA Protein Assay kit according to the manufacturer’s instructions (Thermo Fisher Scientific). Total protein extracts (25 μg) were separated using 15% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA, USA) according to the molecular weight of the E6/E7 oncoproteins (17 kDa of HPV16 E6/E7) as described previously.17 Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control to confirm equal loading of cell lysates. After electrophoresis, the samples were transferred to polyvinylidene fluoride membranes (Thermo Fisher Scientific). The membranes were blocked with 5% skimmed milk powder in Tris-buffered saline Tween-20 buffer (TBST; pH 8.6; 20 mM Tris-HCl, 150 mM NaCl and 0.1 % Tween 20) for 1 h at room temperature. Mouse anti-human papillomavirus 16 (E7) monoclonal antibody (1:1500 dilution: Abcam, Cambridge, UK) and goat anti-human HPV16 (E6) polyclonal antibody (1:200 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were incubated with the membranes overnight at 4 °C. The primary and secondary antibodies against GAPDH were mixed with the antibody solutions for the target proteins and incubated under the same conditions (primary antibody: rabbit anti-human GAPDH polyclonal antibody, 1:5000 dilution, Abcam; secondary antibody: goat anti-rabbit IgG H&L horseradish peroxidase [HRP], 1:10 000 dilution, Abcam). After incubation with primary antibodies, the membranes were washed in TBST (pH 8.6) four times for 10 min each wash. The membranes were then incubated for 1 h at room temperature with the secondary antibodies, which were rabbit anti-mouse IgG H&L (HRP) (Abcam) for HPV16 E7 protein (1:10 000 dilution) and rabbit anti-goat IgG H&L (HRP) (Abcam) for HPV16 E6 protein (1:10 000 dilution). The membranes were washed in TBST (pH 8.6) four times for 10 min each wash. The protein bands on the membranes were detected using an Immun-Star™ HRP enhanced chemiluminescence kit (Bio-Rad) for 1 min according to the manufacturer’s instructions and exposed to X-ray film (Beyotime Institute of Biotechnology, Shanghai, China). The extent of the grey value of the protein bands was determined using a Personal Densitometer SI (Amersham Pharmacia, Piscataway, NJ, USA) and densitometric analysis was taken using ImageJ Software.18
Histological diagnosis
Histological examination of cervical biopsy samples is deemed the gold standard for the diagnosis of CaCx and it is used to clarify the staging of CIN. The main steps have been reported previously.19 Cervical biopsies were only feasible to those patients with abnormal colposcopy. A cervical biopsy would usually be undertaken when collecting the cervical cytology specimen as part of standard care. Under certain conditions, biopsies were performed during the subsequent follow-up; and in this situation, histological diagnosis of biopsies taken within 1 year were included. The samples were randomly and blindly evaluated by at least two pathologists (X.Y.Z. and H.Y.W.) and were recognized as the end-point of this study.
Statistical analyses
All statistical analyses were performed using the SPSS® statistical package, version 16.0 (SPSS Inc., Chicago, IL, USA) for Windows®. Statistical significance was evaluated by paired Student’s t-test or one-way analysis of variance followed by a Student–Newman–Keuls post-hoc test when a statistically significant improvement of outcome was observed. A P-value < 0.05 was considered statistically significant.
Results
A total of 450 patients had at least one positive indicator of cervical abnormalities and were recruited to the study. All patients were Asian women from China aged 20–45 years. According to the histological findings, 102 patients were diagnosed with CIN1 (22.7%), 241 with CIN2 (53.6%), 96 with CIN3 (21.3%) and 11 with SCC (2.4%). As shown in Table 1, there was no significant difference between the CIN2+ and CIN2– groups in terms of the proportion of E6/E7 oncoprotein-positive patients. Similar findings were observed in the high-risk HC2-positive patients, although the sensitivity of HC2 was higher than that of the E6/E7 oncoprotein test, with a value of 96.6% in the CIN2+ group. When the patients were stratified by age, CIN2+ cases were mainly younger patients aged 20 to <30 years. In contrast, the majority of CIN2– cases were patients aged ≥40 years.
Table 1.
Characteristics of female patients (n = 450) with cervical intraepithelial neoplasia (CIN) stratified according to the histological diagnosis.
| CIN2+ groupn = 348 | CIN2– groupn = 102 | Statistical significancea | |
|---|---|---|---|
| Age, years | 39.7 ± 8.9 | 42.2 ± 9.7 | NS |
| 20 to <30 | 207 (59.5%) | 14 (13.7%) | P < 0.001 |
| 30 to <40 | 84 (24.1%) | 22 (21.6%) | NS |
| ≥40 | 57 (16.4%) | 66 (64.7%) | P < 0.001 |
| HPV E6/E7 oncoproteins | |||
| Positive | 228 (65.5%) | 63 (61.8%) | NS |
| Negative | 120 (34.5%) | 39 (38.2%) | |
| High-risk HC2 | |||
| Positive | 336 (96.6%) | 96 (94.1%) | NS |
| Negative | 12 (3.4%) | 6 (5.9%) | |
| TCT | |||
| HSIL+ | 126 (36.2%) | 12 (11.8%) | P < 0.001 |
| LSIL | 108 (31.0%) | 39 (38.2%) | |
| ASC-H | 48 (13.8%) | 6 (5.9%) | |
| ASC-US | 45 (12.9%) | 21 (20.6%) | |
| Normal | 21 (6.0%) | 24 (23.5%) | |
Data presented as mean ± SD or n of patients (%).
Statistical significance was evaluated by paired Student’s t-test or one-way analysis of variance followed by a Student–Newman–Keuls post-hoc test.
HPV, human papillomavirus; HC2, Hybrid Capture 2; TCT, ThinPrep cytological test; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; ASC-H, atypical squamous cells–cannot exclude HSIL; ASC-US, atypical squamous cells with undetermined significance; NS, no significant between-group difference (P ≥ 0.05).
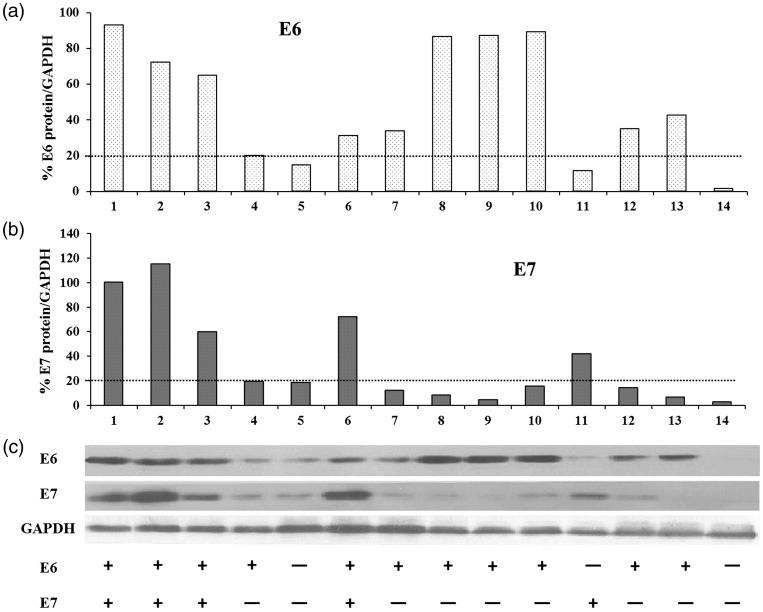
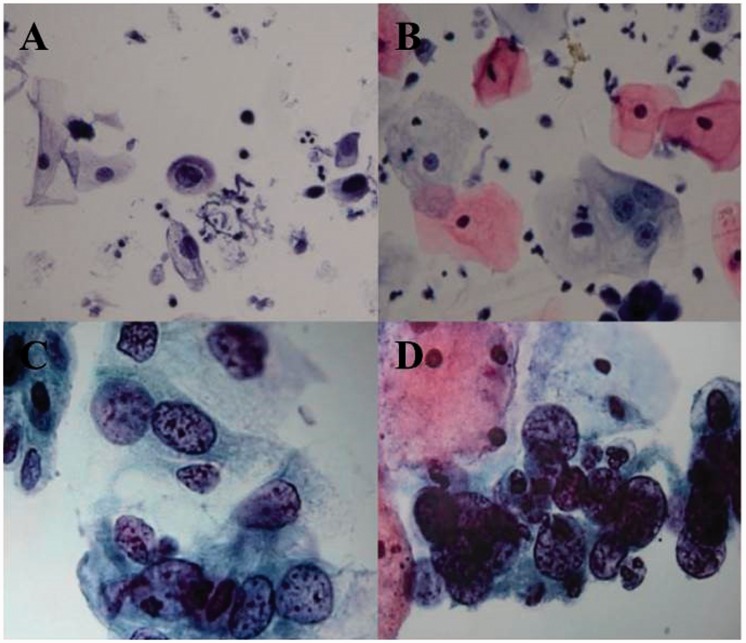
The levels of E6/E7 oncoproteins in all cytological samples were measured using Western blot analysis (Figure 1). According to the densitometric analysis, patients were considered to be positive for the HPV oncoproteins if the ratio of E6 or E7 to GAPDH was greater than the positive threshold value of 20%. Cytological samples from healthy volunteers were used as negative controls. Representative photomicrographs of the typical appearance of four subtypes of cervical epithelial lesions as they appear during the TCT are shown in Figure 2. The prevalence of abnormal cytological findings ≥ ASC-US in CIN2+ patients was 94.0% (327/348) compared with 76.5% (78/102) in CIN2– patients. In total, 45 HC2+ patients tested as normal in the TCT.
Figure 1.
Densitometric analysis of human papillomavirus (HPV) E6/E7 oncoproteins was performed on Western blots to quantify the E6/E7 oncoprotein-positivity in cervical cytological specimens using a positive threshold value of 20% (ratio of E6 or E7 to glyceraldehyde 3-phosphate dehydrogenase [GAPDH]). (A) Percentage value of E6/GAPDH; (B) percentage value of E7/GAPDH; (C) Western blot analysis of E6/E7 oncoproteins in representative samples with definition of protein expression (+ or –). 1–13: samples of HPV-infected patients, 14: negative control sample from a healthy volunteer.
Figure 2.
Representative photomicrographs of the typical appearance of four subtypes of cervical epithelial lesions as they appear during the ThinPrep cytological test: (A) atypical squamous cells with undetermined significance; (B) low-grade squamous intraepithelial lesion; (C) high-grade squamous intraepithelial lesion; (D) squamous cell carcinoma. The colour version of this figure is available at: http://imr.sagepub.com. Scale bar 10 µm.
For the diagnosis of CIN2+, although the sensitivity of the E6/E7 oncoprotein test was lower than HC2, the specificity was greater (Table 2). Compared with TCT, the sensitivity of the E6/E7 oncoprotein test was much higher, but had lower specificity.
Table 2.
Diagnostic value of the human papillomavirus (HPV) E6/E7 oncoprotein test, Hybrid Capture 2 (HC2) HPV DNA test, and ThinPrep cytological test (TCT) for high-grade cervical lesions (CIN2+).
| Diagnostic tests | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|
| HPV E6/E7 | 65.5 (60.5, 70.5) | 38.2 (28.6, 47.8) | 78.4 (73.6, 83.1) | 24.5 (17.8, 31.3) |
| HC2 | 96.6 (94.6, 98.5) | 5.9 (1.2, 10.5) | 77.8 (73.8, 81.7) | 33.3 (9.2, 57.5) |
| TCT | 36.2 (31.1, 41.3) | 88.2 (81.2, 94.6) | 91.3 (86.5, 96.1) | 28.8 (23.8, 33.9) |
Data presented as percentage (95% confidence interval).
PPV, positive predictive value; NPV, negative predictive value.
Sensitivity = true positive/(true positive + false negative); specificity = true negative/(true negative + false positive); PPV = true positive/(true positive + false positive); NPV = true negative/(true negative + false negative).
Table 3 presents the sensitivity and specificity data for the E6/E7 oncoprotein test and the HC2 HPV DNA test for different cervical lesions based on TCT. The specificity of the E6/E7 oncoprotein test was higher than the HC2 test for patients with ASC-US, LSIL or HSIL+.
Table 3.
Diagnostic value of the human papillomavirus (HPV) E6/E7 oncoprotein test and Hybrid Capture 2 (HC2) HPV DNA test for different cervical lesions.
| Sensitivity | Specificity | PPV | NPV | |
|---|---|---|---|---|
| ASC-US, n = 66 | ||||
| HPV E6/E7 | 48.4 (38.0, 58.7) | 55.6 (35.5, 75.6) | 78.9 (68.0, 89.9) | 23.8 (13.0, 34.6) |
| HC2 | 93.5 (88.5, 98.6) | 11.1 (–1.6, 23.8) | 78.4 (70.6, 86.2) | 33.3 (–5.1, 71.8) |
| LSIL, n = 147 | ||||
| HPV E6/E7 | 77.8 (69.8, 85.8) | 15.4 (3.5, 27.2) | 71.2 (63.5, 80.1) | 20 (4.8, 35.2) |
| HC2 | 97.2 (94.1, 100) | 7.7 (–1.1, 16.4) | 74.5 (67.2, 81.8) | 7.7 (–1.1, 16.4) |
| HSIL+, n = 138 | ||||
| HPV E6/E7 | 88.1 (83.6, 91.9) | 66.7 (48.9, 86.2) | 83.3 (74.8, 92.9) | 27.3 (18.4, 36.7) |
| HC2 | 98.4 (96.9, 100) | 6.3 (–1.0, 14.2) | 72.7 (65.6, 80.1) | 6.9 (–1.4, 13.8) |
Data presented as percentage (95% confidence interval).
PPV, positive predictive value; NPV, negative predictive value; ASC-US, atypical squamous cells with undetermined significance; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion.
Sensitivity = true positive/(true positive + false negative); specificity = true negative/(true negative + false positive); PPV = true positive/(true positive + false positive); NPV = true negative/(true negative + false negative).
Discussion
Human papillomavirus E6/E7 oncoproteins are overexpressed after HPV invasion into the host cervical cells in the form of episomal HPV DNA or viral integration into the host's genome, and are closely and directly related to the development of cancers.16 There are numerous studies that have diagnosed HPV and the occurrence of CaCx via E6/E7 mRNA detection,20 but only a few studies have determined E6, but not E6/E7, expression at the protein level.21,22 After cervical cells have been infected with HPV, the presence of HPV DNA or mRNA is only a relatively low risk factor for CaCx progression because the major cause of pre-CaCx lesions is the functional expression of high-risk E6/E7 proteins.23 So E6/E7 protein expression might be more directly associated with cervical cancer risk. A pilot clinical study that used the OncoE6 assay demonstrated that it had higher specificity than the HPV DNA test for CIN3+ detection.21 In a previous study, the sensitivity of single E6 protein detection (OncoE6 testing) was only 42.4%,24 which was lower than the 65.5% that was observed for the HPV E6/E7 oncoprotein test used in this present study. This new technology for cervical cancer screening was evaluated in another clinical trial and it demonstrated higher specificity compared with HC2 (98.9% [95% confidence interval (CI) 98.6%, 99.2%] versus 86.8% [95% CI 85.9%, 87.7%], respectively), but lower sensitivity (67.3% [95% CI 52.5%, 80.1%] versus 98.0% [95% CI 89.1%, 99.9%], respectively) for CIN3+ detection only.22
This current study provided further evidence to show that the E6/E7 oncoprotein test is a potential new biomarker that has a satisfactory diagnostic value for CIN2+ CaCx screening. It demonstrated a better sensitivity than TCT and a better specificity than HC2 HPV DNA testing. For diagnosing ASC-US or LSIL, the sensitivity and specificity of the E6/E7 oncoprotein test were similar to E6/E7 mRNA for the same HPV types, but better with a higher PPV.25–27 These novel indicators (E6/E7 oncoproteins) could be quantified using standard detection methods established in the clinical screening of cervical cancer and this would reduce the limitations of other recent technologies used in this current study.
This current study had several strengths and limitations. The strengths included the prospective design and contemporaneous data collection with a large sample size. One limitation of this study was that it did not use all of the E6/E7 monoclonal antibodies with high specificity that are currently commercially available.28 In addition, the study only included Asian women from mainland China, so the usefulness of this screening test for pre-cancerous cervical changes in different human populations remains to be evaluated. Further multi-centre clinical studies with larger samples sizes are needed to evaluate the effectiveness of E6/E7 oncoprotein detection as a new biomarker for the screening and diagnosis of cervical pre-cancer and cancer.
Regarding HPV-related cancers, the overexpression of E6/E7 oncoproteins is closely associated with the malignant transformation of tumours.29 E6/E7 oncoproteins can inactivate two tumour suppressor proteins p53 and pRB and thereby promote atypical cell growth.30 The expression level of E6/E7 oncoproteins might differentiate the types of HPV infection; with HPV types with low E6/E7 expression being transient with low-risk of cancer induction.16 In contrast, HPV types with a high level or long-standing expression of E6/E7 oncoproteins are probably associated with a higher risk of inducing cancer.31 Based on this hypothesis, the screening threshold of E6/E7 oncoproteins should be quantified and used to test abnormal females in the future.32
In conclusion, this current study provided further evidence that measuring the levels of E6/E7 oncoproteins might be a potential new biomarker with satisfactory diagnostic values for HPV type 16. The relative diagnostic value might be further improved if the number of HPV oncogenic types was increased.
Acknowledgements
We would like to thank all of the people involved in this study including the patients, healthy control subjects, clinicians and healthcare professionals.
Author contributions
The authors were involved as follows: J.J.Z., H.Y.W., Y.W.L. – conception and design of the study; J.J.Z., H.Y.W., X.C.C, X.Y.Z., Y.W.L. – performance; H.Y.W., X.C.C – analysis and interpretation; J.J.Z., Y.W.L.– input into drafting the manuscript; J.J.Z., H.Y.W., X.C.C, X.Y.Z., Y.W.L. – responsible for revision of the manuscript.
Declaration of conflicting interests
The authors declare that there are no conflicts of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
References
- 1.Singh GK Azuine RE andSiahpush M.. Global inequalities in cervical cancer incidence and mortality are linked to deprivation, low socioeconomic status, and human development. Int J MCH AIDS 2012; 1: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knaul FM, Bhadelia A, Gralow J, et al. Meeting the emerging challenge of breast and cervical cancer in low- and middle-income countries. Int J Gynaecol Obstet 2012; 119(Suppl 1): S85–S88. [DOI] [PubMed] [Google Scholar]
- 3.Thun MJ, DeLancey JO, Center MM, et al. The global burden of cancer: priorities for prevention. Carcinogenesis 2010; 31: 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J Kang LN andQiao YL.. Review of the cervical cancer disease burden in mainland China. Asian Pac J Cancer Prev 2011; 12: 1149–1153. [PubMed] [Google Scholar]
- 5.Lee CH, Peng CY, Li RN, et al. Risk evaluation for the development of cervical intraepithelial neoplasia: development and validation of risk-scoring schemes. Int J Cancer 2015; 136: 340–349. [DOI] [PubMed] [Google Scholar]
- 6.Chen D Xue W andXiang J.. The intra-nucleus integration of mitochondrial DNA (mtDNA) in cervical mucosa cells and its relation with c-myc expression. J Exp Clin Cancer Res 2008; 27: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng ZM andBaker CC.. Papillomavirus genome structure, expression, and post-transcriptional regulation. Front Biosci 2006; 11: 2286–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheurer ME Tortolero-Luna G andAdler-Storthz K.. Human papillomavirus infection: biology, epidemiology, and prevention. Int J Gynecol Cancer 2005; 15: 727–746. [DOI] [PubMed] [Google Scholar]
- 9.Marth C, Landoni F, Mahner S, et al. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017; 28: iv72–iv83. [Mismatch] [DOI] [PubMed] [Google Scholar]
- 10.Long S, Lei W, Feng Y, et al. The feasibilities of TruScreen for primary cervical cancer screening: a self-controlled study. Arch Gynecol Obstet 2013; 288: 113–118. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. Cervical cancer, human papillomavirus (HPV) and HPV vaccines: key points for policy-makers and health professionals 2007; http://whqlibdoc.who.int/hq/2008/WHO_RHR_08.14_eng.pdf (2007, accessed 05 October 2017).
- 12.Apgar BS, Kittendorf AL, Bettcher CM, et al. Update on ASCCP consensus guidelines for abnormal cervical screening tests and cervical histology. Am Fam Physician 2009; 80: 147–155. [PubMed] [Google Scholar]
- 13.Boulet G, Horvath C, Vanden Broeck D, et al. Human papillomavirus: E6 and E7 oncogenes. Int J Biochem Cell Biol 2007; 39: 2006–2011. [DOI] [PubMed] [Google Scholar]
- 14.American Society of Cytopathology. The Bethesda System for Reporting Cervical Cytology, www.cytopathology.org/bethesda (accessed 15 Oct 2017).
- 15.Schiffman M, Herrero R, Hildesheim A, et al. HPV DNA testing in cervical cancer screening: results from women in a high-risk province of Costa Rica. JAMA 2000; 283: 87–93. [DOI] [PubMed] [Google Scholar]
- 16.Munagala R, Kausar H, Munjal C, et al. Withaferin A induces p53-dependent apoptosis by repression of HPV oncogenes and upregulation of tumor suppressor proteins in human cervical cancer cells. Carcinogenesis 2011; 32: 1697–1705. [DOI] [PubMed] [Google Scholar]
- 17.Chen CL, Hsieh FC, Lieblein JC, et al. Stat3 activation in human endometrial and cervical cancers. Br J Cancer 2007; 96: 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abramoff MD Magalhaes PJ andRam SJ.. Image Processing with ImageJ. Biophotonics International 2004; 11: 36–42. [Google Scholar]
- 19.Huang Y, Zhang J, Cui ZM, et al. Expression of the CXCL12/CXCR4 and CXCL16/CXCR6 axes in cervical intraepithelial neoplasia and cervical cancer. Chin J Cancer 2013; 32: 289–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zappacosta R, Caraceni D, Ciccocioppo L, et al. Implementing specificity of HPV-DNA primary screening in a successful organised cervical cancer prevention programme. Gynecol Oncol 2013; 128: 427–432. [DOI] [PubMed] [Google Scholar]
- 21.Schweizer J Lu PS andMahoney CW et al. . Feasibility study of a human papillomavirus E6 oncoprotein test for diagnosis of cervical precancer and cancer. J Clin Microbiol 2010; 48: 4646–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuzick J, Bergeron C, von Knebel Doeberitz M, et al. New technologies and procedures for cervical cancer screening. Vaccine 2012; 30(Suppl 5): F107–F116. [DOI] [PubMed] [Google Scholar]
- 23.Yoshimatsu Y, Nakahara T, Tanaka K, et al. Roles of the PDZ-binding motif of HPV 16 E6 protein in oncogenic transformation of human cervical keratinocytes. Cancer Sci 2017; 108: 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao FH, Jeronimo J, Qiao YL, et al. An evaluation of novel, lower-cost molecular screening tests for human papillomavirus in rural China. Cancer Prev Res (Phila) 2013; 6: 938–948. [DOI] [PubMed] [Google Scholar]
- 25.Benevolo M, Vocaturo A, Caraceni D, et al. Sensitivity, specificity, and clinical value of human papillomavirus (HPV) E6/E7 mRNA assay as a triage test for cervical cytology and HPV DNA test. J Clin Microbiol 2011; 49: 2643–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoler MH, Wright TC, Jr, Cuzick J, et al. APTIMA HPV assay performance in women with atypical squamous cells of undetermined significance cytology results. Am J Obstet Gynecol 2013; 208: 144.e1–8. [DOI] [PubMed] [Google Scholar]
- 27.Perez Castro S, Iñarrea Fernández A, Lamas González MJ, et al. Human papillomavirus (HPV) E6/E7 mRNA as a triage test after detection of HPV 16 and HPV 18 DNA. J Med Virol 2013; 85: 1063–1068. [DOI] [PubMed] [Google Scholar]
- 28.Zumbach K, Kisseljov F, Sacharova O, et al. Antibodies against oncoproteins E6 and E7 of human papillomavirus types 16 and 18 in cervical-carcinoma patients from Russia. Int J Cancer 2000; 85: 313–318. [DOI] [PubMed] [Google Scholar]
- 29.Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002; 2: 342–350. [DOI] [PubMed] [Google Scholar]
- 30.Godefroy N, Lemaire C, Mignotte B, et al. p53 and retinoblastoma protein (pRb): a complex network of interactions. Apoptosis 2006; 11: 659–661. [DOI] [PubMed] [Google Scholar]
- 31.Hong D, Liu J, Hu Y, et al. Viral E6 is overexpressed via high viral load in invasive cervical cancer with episomal HPV16. BMC Cancer 2017; 17: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wise-Draper TM andWells SI.. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci 2008; 13: 1003–1017. [DOI] [PubMed] [Google Scholar]