Abstract
The apolipoprotein mRNA editing enzyme catalytic polypeptide-like 3 (APOBEC3; A3) proteins are a family of seven cytidine deaminases (A3A, A3B, A3C, A3D, A3F, A3G and A3H) that restrict certain viral infections. These innate defence factors are best known for their ability to restrict the replication of human immunodeficiency virus type 1 (HIV-1) lacking a functional Vif protein (HIV-1Δvif) through the deamination of cytidine residues to uridines during reverse transcription, ultimately leading to lethal G → A changes in the viral genome. The best studied of the A3 proteins has been APOBEC3G because of its potent activity against HIV-1Δvif. However, one member of this family, A3A, has biological properties that make it unique among the A3 proteins. In this review, we will focus on the structural and phylogenetic features of the human and non-human primate A3A proteins, their role in the restriction of retroviruses and other viruses, and current findings on other biological properties affected by this protein.
Introduction
Several innate host restriction factors have been described that inhibit retroviruses such as human immunodeficiency virus type 1 (HIV-1). These innate restriction factors include the apolipoprotein B mRNA editing enzyme catalytic-polypeptide-like proteins 3 (APOBEC3; A3), bone marrow stromal antigen 2 (BST-2, also known as CD317, HM1.24 or tetherin) and tripartite motif (TRIM) proteins (Sheehy et al., 2002; Stremlau et al., 2004; Neil et al., 2008; Van Damme et al., 2008). The discovery of human APOBEC3G (hA3G) occurred during the search for a protein that restricted the replication of HIV lacking a functional Vif protein (HIV-1Δvif; Sheehy et al., 2002) and has been the most extensively studied of the three restriction factors mentioned above. It is now known that A3G is one of seven members of the A3 family in the human genome (hA3A, hA3B, hA3C, hA3D, hA3F, hA3G and hA3H) (Jarmuz et al., 2002; OhAinle et al., 2006). The A3 proteins, along with activation-induced deaminase (AID), APOBEC1 (A1) and APOBEC2 (A2) are cytidine deaminases. An 11th member of this family has been identified in humans, APOBEC4 (A4), but has no reported deaminase activity (Rogozin et al., 2005). This review aims to provide a summary of the studies on A3A including the restriction of lentiviruses from non-human primates and other retroviruses, restriction of select DNA viruses, inhibition of retrotransposition, editing of mRNA transcripts, its role in degradation of foreign DNA and its potential role in the genesis of cancer.
A3 proteins and the structure of hA3A
The seven genes for the A3 proteins are arranged tandemly on chromosomes 22 and 10 of the human and rhesus macaque genomes, respectively (Jarmuz et al., 2002; Schmitt et al., 2011). The A3 proteins are characterized by one or two zinc-coordinating domains (Z domains) with the sequence H-x-E-x23–28-PC-x2–4-C. hA3A, hA3C and hA3H have one Z domain, while A3B, A3D, A3F and A3G have two Z domains (Fig. 1). Phylogenetic analysis revealed a clustering among A3 zinc-finger motifs, further classified as A3Z1, A3Z2 or A3Z3 (Fig. 1) (Conticello et al., 2003; LaRue et al., 2009). This diversification probably preceded the split between placental mammals and marsupials (125–150 million years ago) (Bininda-Emonds et al., 2007; Münk et al., 2012). The structure of hA3A (aa 10–194 of 199 aa) was solved by nuclear magnetic resonance (NMR) spectroscopy (Byeon et al., 2013). Similar to other APOBEC (A2 and some A3 members) proteins for which a structure has been solved, the hA3A structure consists of six helices surrounding a central β-sheet of five strands. In another study, A3G was found to form high-order multimers as a function of protein concentration (Li et al., 2014). In contrast, A3A was only found as monomers at all concentrations tested. These investigators found a correlation with those A3 proteins with a propensity to multimerize (A3B, A3D, A3F, A3G and A3H) and restriction activities of various A3 proteins. Recently, however, the crystal structure of A3A E72A–C171A was solved at 2.85 Å resolution, which showed that hA3A exists as a homodimer (Logue et al., 2014; Bohn et al., 2015). A salient finding of this study was that A3A dimerization forms a highly positive groove that connects the active site of both monomers. These investigators identified two sets of residues that might contribute to dimerization, H11/H56 and H16/K30. Substitution of these residues showed that both sets of interactions were critical to cooperative DNA binding. These investigators speculated that A3A dimerization may provide a reason why A3 might form two domain fusions. They hypothesized that A3A monomers would not act on the target cytidine but rather were involved in substrate binding. They further stated that the evolution of two domain enzymes may have allowed the separation of binding and enzymatic activities, which resulted in less active proteins that were more specific to their targets. Comparison of structures of hA3A with other available APOBEC structures revealed that the A3A NMR structure is most similar to the X-ray crystallographic structure of the C-terminal domain of hA3G (Holden et al., 2008; Kitamura et al., 2012). An interesting feature of hA3A that is not found in the other hA3 proteins is the presence of 4 aa between the two cysteine residues of the canonical deaminase domain. Byeon et al. (2013) found that hA3A has a slightly higher binding affinity for ssRNA than ssDNA and displayed a similar catalytic activity for TTCA and CCCA substrates.
Fig. 1.
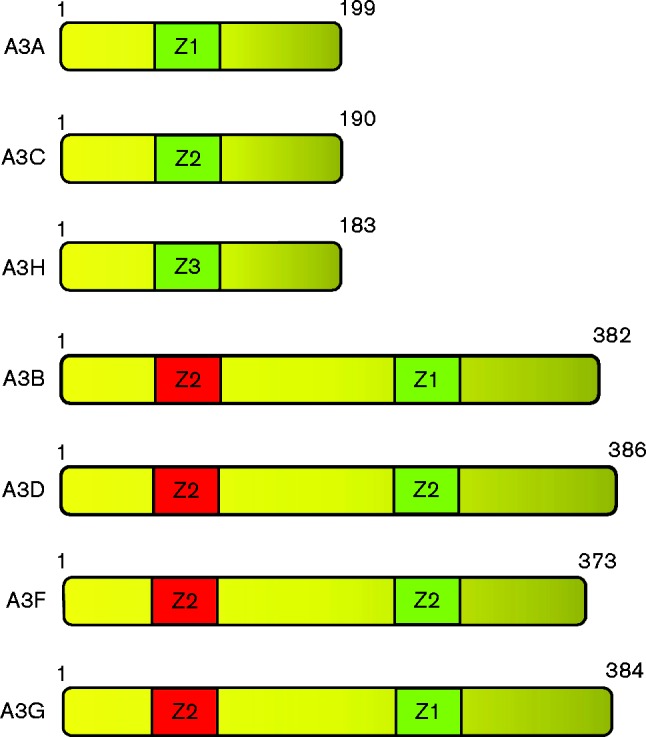
Diagram of the human APOBEC3A–H (A3A–A3H) proteins. The cytidine deaminase domains (Z domains) are indicated as active (green) or inactive (red). The numbers indicate the number of amino acids for each protein.
A3A sequence diversity among primates and tissue expression
Lentiviruses are endemic in multiple mammalian species, but the closest ancestors of HIV-1 and HIV-2 are the simian immunodeficiency viruses (SIVs) that infect west-central African chimpanzees (Keele et al., 2006) and sooty mangabeys (Santiago et al., 2005), respectively. Over 30 Old World monkey species also harbour endemic SIV infections (Hahn et al., 2000), but these infections are limited to sub-Saharan Africa. For example, Asian rhesus macaques are considered Old World monkeys but do not have endemic SIV infections. New World monkeys have also not been found to harbour endemic lentivirus infections. Notably, infection of rhesus macaques with SIV from sooty mangabeys (SIVsm) resulted in an AIDS phenotype (Letvin et al., 1985). To date, the SIV/rhesus macaque model remains the gold standard for understanding lentivirus pathogenesis in vivo.
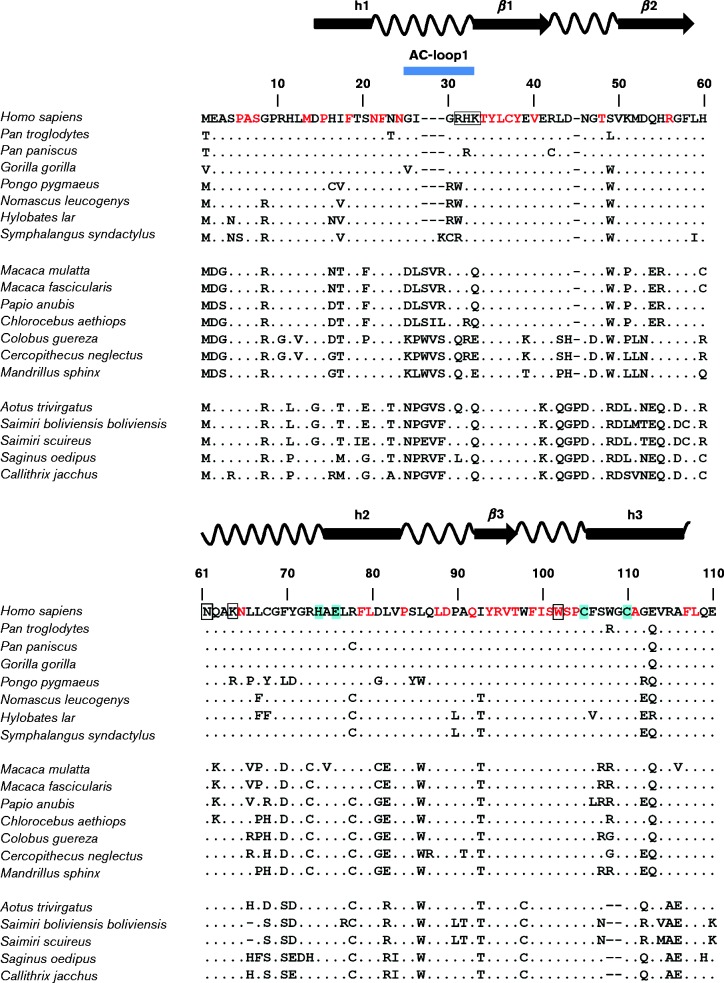
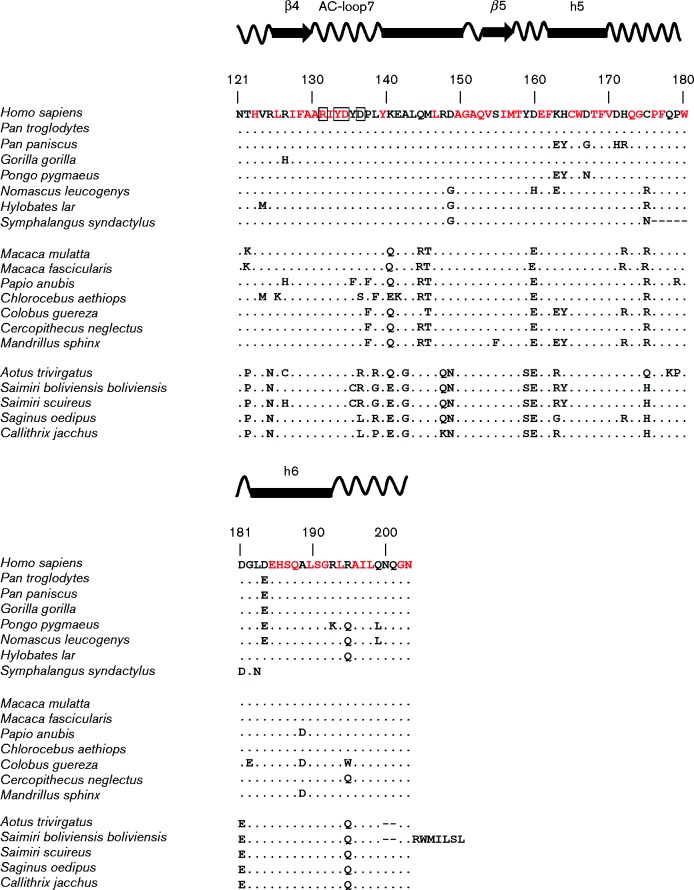
Full-length genomes of non-human primates are limited, thereby precluding an in-depth evolutionary analysis of the A3 locus during primate evolution. In fact, the draft genome of the rhesus macaque had significant gaps in the A3 locus, leading to an early hypothesis that rhesus macaques do not encode A3A (Virgen & Hatziioannou, 2007). However, by piecing together fragments from unassembled sequence contigs, our group cloned the rhesus macaque A3A (rhA3A) gene (Schmitt et al., 2011). The sequence of the A3A proteins from 20 primate species from the hominids (eight sequences), Old World monkeys (seven sequences) and New World monkeys (five sequences) are compared in Fig. 2. The primate A3A proteins vary in length from 199 to 206 aa with amino acid identity at 88 positions (∼45 %) and conserved amino acids at an additional 23 positions. The most variable region of the primate A3A proteins is the N-terminal region (Fig. 2). Notably, all hominid A3A proteins (with the exception of siamang) have a 3 aa deletion (positions 27–29) that is not observed in A3A proteins from Old or New World monkeys (Fig. 2). This indel will be discussed in greater detail later. Additionally, the New World monkey A3A proteins have a proline insertion at position 45 and a 2 aa deletion at positions 108–109 (Fig. 2).
Fig. 2.
Alignment of the amino acid sequence of hA3A with other hominid (top group) and Old (middle group) and New World (bottom group) monkey A3A proteins. Residues in red indicate invariant residues found in hominids and Old and New World monkeys with the exception of Symphalangus syndactylus (siaming) whose sequence appears to be truncated. A dot represents identity to the hA3A sequence and – represents gaps introduced into the sequence for purposes of alignment. The amino acids in red are identical in all species analysed. The species analysed were: Homo sapiens (human), Pan troglodytes (chimpanzee), Pan paniscus (bonobo), Gorilla gorilla (gorilla), Pongo pygmaeus (Bornean orangutan), Nomascus leucogenys (northern white-cheeked gibbon), Hylobates lar (common gibbon), Symphalangus syndactylus (siamang), Macaca mulatta (rhesus macaque), Macaca fascicularis (cynomolgus monkey), Papio anubis (baboon), Chlorocebus aethiops (African green monkey), Colobus guereza (black-and-white colobus), Cercopithecus neglectus (De Brazza's monkey), Mandrillus sphinx (mandrill), Aotus trivirgatus (northern owl monkey), Saimiri boliviensis bolivienis (black-capped squirrel monkey), Siamiri scuireus (common squirrel monkey), Saginus oedipus (cotton-tailed tamarin), Callithrix jacchus (common marmoset). Catalytic residues are highlighted in cyan, while residues of the polynucleotide-binding groove are indicated with an open box. Predicted α-helices (cylinders) and β-sheets (arrows) are shown.
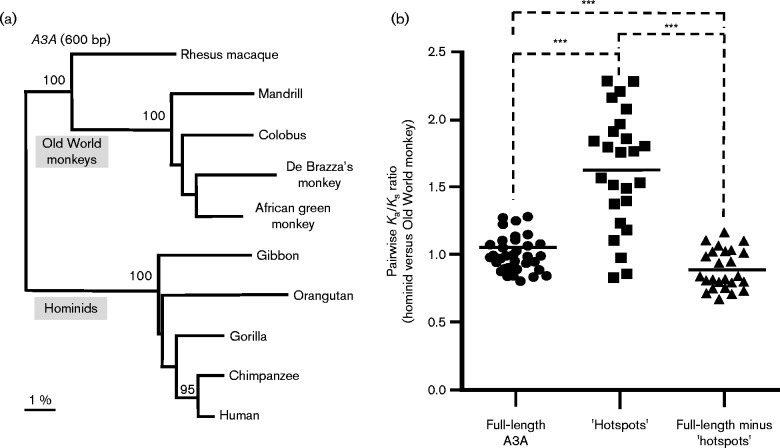
The interaction between host proteins and pathogens may leave a genetic mark on the host protein known as a positive-selection signature. This manifests as an increase in DNA sequence mutations that change the protein sequence (non-synonymous mutations, Ka) compared with mutations that preserve the protein sequence (synonymous mutations, Ks). A3G, A3C, A3D, A3F and A3H were noted to have undergone positive selection, with Ka/Ks values >1 (Sawyer et al., 2004; Zhang & Webb, 2004; OhAinle et al., 2006; Ortiz et al., 2009; Cagliani et al., 2011; Duggal et al., 2011). Lentivirus restriction factors such as BST-2, TRIM5α and SAMHD1 also exhibited a positive-selection signature. In contrast, A3A was not under positive selection (Sawyer et al., 2004), consistent with the notion that it may not have played a significant role in retrovirus restriction. We revisited this question using primate A3A sequences from five Old World monkeys and five hominids (Fig. 3a). Indeed, 25 pairwise comparisons of Ka/Ks ratios between hominid versus monkey A3A revealed that A3A was under neutral selection (Fig. 3b). However, when we looked at specific regions of A3A that were close to the predicted polynucleotide-binding groove (Bulliard et al., 2011), particularly the AC-loop1 (minus the indels), AC-loop3, AC-loop5 and α-helix4, there was a positive-selection signature. These data revealed that, while A3A does not exhibit signals of positive selection as do the other A3 genes, the regions surrounding the nucleic acid-binding groove are hotspots of genetic conflict. Another report found that A3A exhibited positive-selection signatures only on select branches of the primate A3A phylogenetic tree (Henry et al., 2012). Overall, the data suggest that A3A proteins from non-human primates may have intriguing antiretroviral properties. The antiretroviral properties of rhesus macaque as well as other Old and New World primate A3A proteins will be discussed later in this review.
Fig. 3.
Selection pressure in A3A following the evolutionary split between hominids and Old World monkeys. (a) A3A phylogenetic tree. A neighbour-joining (unrooted) tree for nucleotide sequences of five Old World monkey and five hominid species was reconstructed with gaps excluded and correcting for multiple substitutions using clustal_x. Support (>80 %) for specific clades is shown following 1000 subreplicates. (b) Identification of a positive-selection ‘hotspot’ in A3A. Pairwise ratios of Ka/Ks substitutions between hominid and Old World monkey A3A sequences were computed using the K-estimator program. Full-length A3A was essentially neutral. However, when sequences encoding only the AC-loop1, AC-loop3, AC-loop5 and α-helix4 regions were analysed, Ka/Ks ratios that were significantly greater than 1 were obtained. These regions are referred to as ‘hotspots’ of positive selection in A3A. The 3 aa indel was excluded from this analysis, and thus the level of positive selection may be higher. In contrast, residues outside the ‘hotspots’ had Ka/Ks values indicative of low-level purifying selection. Each dot corresponds to a pairwise comparison. Differences were evaluated using a two-tailed Student's t-test (***P < 0.0001).
Expression profile of A3A in tissues and cells
It is generally accepted that hA3G is expressed in the cytoplasm of cells, due to encoded cytoplasmic retention signals (Bennett et al., 2008; Stenglein et al., 2008),while hA3A has been shown to be expressed in both the cytoplasm and nucleus of cells (Muckenfuss et al., 2006). hA3A is known to be expressed in differentiated cell populations including monocyte/macrophages, dendritic cells and keratinocytes (Peng et al., 2007; Vartanian et al., 2008). To probe the localization of endogenous hA3A in differentiated macrophages, a rabbit polyclonal antibody was generated against the peptide CPFQPWDGLEEHSQALSGRLRAILQNQGN corresponding to the C terminus of A3A (Carpenter et al., 2012). Using this antibody, it was found that endogenous hA3A was expressed only in the cytoplasm of cells (Land et al., 2013). However, the C terminus of hA3A shows significant amino acid identity to hA3B and hA3G. The hA3A-derived peptide used to generate an antibody was 100 % identical to hA3B and only differed at three amino acid positions from A3G, which has the sequence CPFQPWDGLDEHSQDLSGRLRAILQNQEN (differences in bold). Thus, the detecting antibody for endogenous hA3A was probably not specific to hA3A. Convincing proof that hA3A has exclusive cytoplasmic localization may also require the identification of a cytoplasmic retention signal similar to that of hA3G.
A number of studies have comprehensively examined the presence of A3 mRNA species in human cells and tissues. In the first study, investigators found that hA3A mRNA was detected at low levels in T-cells and at higher levels in monocytes (Koning et al., 2009). They showed that IFN-α treatment of isolated CD4+T-cell populations resulted in the induction of A3A mRNA, which was followed by the appearance of protein detectable in Western blots. In a second study, hA3A mRNA was not detected in CD4+T-cells, and the addition of IFN-α did not induce hA3A (Refsland et al., 2010). In a third study, these same investigators found that neither hA3A nor rhA3A expression was increased in stimulated CD4+ T-cells from the respective hosts. We found that stimulated rhesus macaque CD4+T-cells isolated from PBMCs expressed rhA3A at robust levels comparable to those of rhA3G (Schmitt et al., 2011). The exact reasons for these discrepancies are unclear. However, it should be noted that in the two studies on rhesus macaque CD4+T-cells, the cells were stimulated with either phytohaemagglutinin (Hultquist et al., 2011) or Staphylococcus enterotoxin B (Schmitt et al., 2011). Thus, the method of stimulation may be critical to the induction of rhA3A.
Mechanisms of Vif-mediated degradation of A3 proteins
As the mechanism of Vif-mediated degradation of hA3G has been the most extensively studied, any discussion should begin with this protein. The HIV-1 Vif protein acts as an adaptor protein that results in the polyubiquitination and proteasomal degradation of hA3G. Vif accomplishes this by binding to hA3G and CBF-β, an allosteric regulator of Vif. Vif then recruits the E3 ubiquitin ligase composed of Cullin 5, Elongin B, Elongin C and ring box subunit 2 (RBX2), resulting in the polyubiquitination of hA3G and targeting the complex to the proteasome for destruction (Conticello et al., 2003; Marin et al., 2003; Sheehy et al., 2003; Stopak et al., 2003; Dussart et al., 2004; Liu et al., 2004; Mehle et al., 2004b; Yu et al., 2004; Kobayashi et al., 2005; Zhang et al.., 2011; Jäger et al., 2012). In terms of virus maturation, the end result of Vif-mediated ubiquitination and degradation of hA3G is that hA3G is not incorporated (or not to any meaningful extent) into budding virions. Several deletion and site-directed mutagenesis studies have identified regions on HIV-1 Vif that are important for the interactions with hA3G. One study showed that deletion of aa 43–59 abolished interactions with A3G, while another study using overlapping peptides found that a region from aa 33 to 88 formed a non-linear binding site for A3G (Wichroski et al., 2005; Mehle et al., 2007). Aa 14–17 and 40–44 of the HIV-1 Vif were specifically found to affect the interactions with human A3F and A3G, respectively (Russell & Pathak, 2007). Other investigators showed that domains 23SLVx4Yx9Y38, 69YXXL72, 81LGxGxxIxW89 and 171EDRW174 were also involved in neutralizing hA3G and hA3F (Chen et al., 2009; Pery et al., 2009; Dang et al., 2010). Highly conserved sites for the interaction of Vif with the proteasome machinery have also been identified. The viral BC box (SLQ(Y/F) LA) and the Zn2+-binding HCCH domains are necessary for the interactions with Elongin C and Cullin 5, respectively (Stopak et al., 2003; Yu et al., 2003, 2004; Mehle et al., 2004a, b, 2006; Luo et al., 2005). Finally, several studies have shown the importance of the amino acid at position 128 in hA3G. Chimaeric proteins between the hA3G and African green monkey A3G (agmA3G) and subsequently site-directed mutants revealed that the aspartic acid at position 128 of hA3G (a lysine in agmA3A) was necessary to specify interactions with HIV-1 Vif (Bogerd et al., 2004; Schröfelbauer et al., 2004).
Comparison of deamination of the HIV-1 genome in the presence of hA3G and hA3A
Upon transfection of HIV-1Δvif and hA3G into producer cells that do not express hA3G (e.g. 293 cells or SupT1 cells), hA3G is not degraded and is incorporated into the viral core of nascent virions released from producer cells. In the next round of replication, hA3G causes deamination of cytidine residues to uracils in the minus strand of ssDNA during reverse transcription (Yu et al., 2004). The mechanism of deamination involves the glutamic acid protonation of a water molecule, which acts as the catalytic nucleophile in the oxidation of a cytidine amino group, resulting in conversion to uridine and release of NH3 (Jarmuz et al., 2002). The nucleotide context in which deamination occurs is also important, with 5′-CC being the preferred context for hA3G, and deamination by hA3G is processive in nature (Chelico et al., 2006). This results in G → A hypermutation of the viral genome, generation of premature termination codons in the viral sequence, or inhibition of replication or degradation of the ssDNA by apurinic apyrimidinic endonuclease (Sheehy et al., 2002; Lecossier et al., 2003; Mangeat et al., 2003; Zhang et al., 2003; Yu et al., 2004). For hA3G, it is known that the Z3 domain has the catalytic activity hA3G (Fig. 1). Mutation of Glu67 in the active site of the Z1 domain of hA3G was found to be dispensable for antiviral activity, while mutation of Glu259 in the Z3 domain abolished antiviral activity (Navarro et al., 2005). In contrast, another group showed that Glu259Gln, lacking deaminase activity, still retained antiviral activity (Newman et al., 2005). This study has been challenged by other studies showing that the mutation of the active site of the Z3 domain had little or no antiviral activity (Miyagi et al., 2007; Schumacher et al., 2008; Browne et al., 2009). This indicates that deaminase activity is important for hA3G antiviral activity.
While hA3A is incorporated into nascent HIV-1 or HIV-1Δvif virions, studies have shown that hA3A does not restrict HIV-1 or HIV-1Δvif (Bishop et al., 2004; Goila-Gaur et al., 2007; Aguiar et al., 2008; Schmitt et al., 2011, 2013). These studies were all performed in 293 or HeLa cells. The studies indicated that little or no hA3A is incorporated into the nucleoprotein complex of virions compared with hA3G (Goila-Gaur et al., 2007). Two groups later showed that fusion of the hA3A gene to either the N-terminal domain hA3G or to Vpr resulted in proteins that were incorporated into the nucleoprotein complex and restricted virus replication (Goila-Gaur et al., 2007; Aguiar et al., 2008). Deamination of cytosine residues does occur with hA3A, and this process prefers cytosines in the nucleotide context of 5′-TC (Love et al., 2012; Logue et al., 2014). However, hA3A has been shown to induce fewer mutations in the HIV-1 genome and, unlike hA3G, is not processive in nature (Love et al., 2012). A recent study showed that the recognition loop 1(RL1; previously designated AC-loop1), which is truncated in hominid species, is a determinant of target specificity of hA3A (i.e. 5′-TC). These investigators showed that a chimaeric hA3A/A3G protein having the RL1 region from hA3G was more flexible with respect to the nucleotide 5′ to the deaminated C, deaminating AC, CC and GC with increased frequency. These investigators suggested that the presence of glycine and isoleucine residues at amino acid positions 25 and 26 in the RL1 of hA3A specified the 5′ nucleotide (Logue et al., 2014). This will be important in the discussion of the differences in virus restriction activity by different hominid and Old and New World monkey A3A proteins.
Virus restriction by A3 proteins by deaminase-independent mechanisms
In addition to deaminase-dependent activity of hA3G, other studies have shown that hA3G can restrict virus replication through deaminase-independent mechanisms. Different investigators have reported that hA3G can disrupt reverse transcription. Reports of inhibition of primer binding, strand transfer, transcript accumulation and integration have been described (Bishop et al., 2006; Iwatani et al., 2007; Li et al., 2007; Luo et al., 2007; Wang et al., 2012; Adolph et al., 2013; Bélanger et al., 2013). A major question that remains to be resolved using animal model systems is whether A3-mediated virus restriction of HIV-1 [or SIV/simian-human immunodeficiency virus (SHIV)] occurs through deaminase-dependent or -independent mechanisms.
Restriction of retroviruses by hA3A
As discussed above, it appears that A3A has little effect on the replication of HIV-1Δvif when expressed in 293 or HeLa cells. However, hA3A is expressed at high levels in macrophages and monocytes in response to IFN-α and was shown to inhibit incoming virus without incorporation (Peng et al., 2007; Berger et al., 2011; Koning et al., 2011). These studies suggest that hA3A could restrict replication in the producer cell and without being incorporated into the virion. A recent biochemical study showed that the pH can affect both the specificity and the deaminase activity of hA3A (Pham et al., 2013). These investigators found that purified A3A had optimal cytidine deaminase activity at a pH of 5.8–6.1 and a strict YYCR (where Y is a pyrimidine; R is a purine). In contrast, deaminase activity was reduced by between 13- and 30-fold and had a relaxed cytidine deamination specificity at a pH of 7.4–7.8 (Pham et al., 2013). This is potentially relevant to HIV-1-infected monocyte-derived macrophages (MDMs) because induction of autophagy has been reported to be necessary for HIV-1 replication in MDMs (Espert et al., 2009; Kyei et al., 2009). MDMs, cultured in the presence of 3-methyladenine, which inhibits class III phosphatidylinositol 3-kinase (PI3K) and autophagosome formation, exhibited substantial reductions in HIV-1 production of both R5 and X4 viruses (Espert et al., 2009).
Retroviruses other than lentiviruses can be restricted by A3A. One study reported that Rous sarcoma virus was moderately susceptible to hA3A (Wiegand & Cullen, 2007). A3A, A3B and A3H haplotype II proteins were shown to restrict human T-cell lymphotropic virus type 1 (HTLV-1) (Ooms et al., 2012). In this study, A3A was incorporated into virions, and a catalytic-site mutant abolished HTLV-1 restriction, suggesting that restriction may be dependent on deaminase activity (Ooms et al., 2012). These investigators also showed that A3A could mutate viral genomes using a technique known as differential DNA denaturation (3D)-PCR, biased to yield AT-rich, low-denaturation amplicons. These authors concluded that multiple independently mutated HTLV-I proviral genomes could be detected in HTLV-I cell lines. However, the 3D-PCR findings are not quantitative in nature, and thus the true editing frequency is unknown and may not significantly impact HTLV-I replication. Human endogenous retroviruses (HERVs) are a class of transposable elements that comprise over 8 % of the human genome (Lander et al., 2001; Bannert & Kurth, 2006; Jern & Coffin, 2008). HERVs most likely originated from germline infections by exogenous retroviruses during primate evolution. Most groups of HERVs are also present in Old World monkeys and apes, which suggests that their introduction into the germline probably occurred more than 30 million years ago (Barbulescu et al., 1999; Bock & Stoye, 2000). HERVs have accumulated numerous point mutations, deletions and insertions that have rendered them non-infectious. One such HERV [HERV-K(MHL-2)] was reconstituted to an infectious virus (Lee & Bieniasz, 2007). This resurrected virus was shown to be restricted by several A3 proteins including hA3A (Lee et al., 2008). This suggests that, during primate evolution, A3 proteins including A3A may have evolved to help combat the onslaught of retroviruses that eventually accumulated in the human germline. More recently, a study analysed the restriction of the retrovirus murine leukemia virus in hA3A and hA3G transgenic mice in a mouse A3-knockout background (Stravou et al., 2014). These investigators showed that hA3G and hA3A restricted murine retroviruses by different mechanisms. They observed that hA3G was packaged into virions and caused extensive deamination of the viral genome, while hA3A was not packaged into virions and restricted infection when expressed in target cells (Stravou et al., 2014). This study also found that hA3A can restrict murine retroviruses by a deamination-independent mechanism.
Virus restriction by non-human primate A3A proteins
In contrast to hA3A, a study in rhesus macaques showed that rhA3A was capable of restricting the replication of Vif-deficient SHIVΔvif at similar levels as rhA3G (Schmitt et al., 2011). Another study observed that rhA3A did not restrict replication (Hultquist et al., 2011). However, the reason for this discrepancy is probably due to where the haemagglutinin tag was fused to the rhA3A (Schmitt et al., 2013). Furthermore, hA3A was capable of significantly restricting SHIVΔvif although not to the same extent as rhA3G (Schmitt et al., 2011). These results indicate that hA3A is capable of restricting lentiviruses other than HIV-1Δvif. Similar to conclusions observed above for hA3A (Love et al., 2012), we observed that rhA3A is capable of cytidine deamination but not at the same frequency as rhA3G (Schmitt et al., 2011).
Our results with rhA3A raised the question of whether the A3A proteins from other non-human primates restrict HIV-1/SIV/SHIV and the importance of the 3 aa deletion in hominid A3A proteins. Recently, we examined the restriction properties of A3A proteins from an additional hominid (Lars gibbon; gibA3A), four additional Old World monkeys [African green monkey (agmA3A) and black and white colobus monkey (colA3A)] and two New World monkeys [squirrel monkey (sqmA3A) and northern owl monkey (nomA3A)] (Schmitt et al., 2013). Similar to hA3A, gibA3A did not restrict HIV-1Δvif or SHIVΔvif, indicating that the lack of HIV-1Δvif restriction is probably conserved among hominids. Similar to rhA3A, we found that agmA3A and colA3A restricted the replication of SHIVΔvif and that all three Old World monkey A3A proteins (agmA3A, colA3A, and rhA3A) restricted HIV-1Δvif to some extent, with mndA3A being the most effective. Among the New World monkeys, sqmA3A restricted SHIVΔvif but not HIV-1Δvif, whereas nomA3A restricted neither SHIVΔvif nor HIV-1Δvif (Schmitt et al., 2013). As the A3A proteins from select Old World monkeys could restrict HIV-1Δvif, we investigated the molecular determinants that define restriction. Using the distinct phenotypes of rhA3A (virus restriction) and hA3A (no virus restriction) in epithelial cells, a series of hA3A/rhA3A chimaeric proteins were constructed and their restriction properties on HIV-1Δvif were analysed (Schmitt et al., 2013). Our results showed that a chimaeric A3A protein, designated rh25–33hA3A, which had the 3 aa deletion of hA3A replaced by the amino acids of rhA3A (27SVR29) plus three additional amino acid substitutions in the AC-loop1, restored the restriction properties of hA3A to a similar level as rhA3A restriction of SHIVΔvif (Schmitt et al., 2013). Interestingly, rhA3A aa 25–33 map to the AC-loop1, and molecular modelling suggests that the AC-loop1 of rhA3A has a greater molecular surface compared with hA3A. Due to its predicted solvent accessibility, this additional molecular surface may be an important region for protein–protein interactions that do not occur with hA3A.
The finding that sqmA3A restricted SHIVΔvif is an interesting result, as the New and Old World monkeys branched approximately 30–35 million years ago with the separation of South America from Africa. To date, no lentiviruses have been isolated from New World monkeys. This brings up the question of whether some New World monkey A3A proteins may have evolved to restrict other viruses. One potential candidate is the foamy viruses (FVs), which are exogenous, persistent and non-pathogenic retroviruses in the subfamily Spumaretrovirinae (Switzer et al., 2005). FVs have been isolated from a broad range of mammals including non-human primates (both New and Old World monkeys), horses, cows and cats (Meiering & Linial, 2001; Hussain et al., 2003; Rethwilm, 2010). Previous studies have shown that the FVs have co-speciated with their mammalian hosts. For example, FVs express the Bet protein, which antagonizes A3 restriction and may play a role in both particle release and virus persistence (Saïb et al., 1995; Alke et al., 2001; Löchelt et al., 2005; Russell et al., 2005; Chareza et al., 2012). A remarkable feature of the FVs is that they have the capacity to cross species barriers and pose a significant risk of interspecies transmission to humans (Heneine et al., 2003; Switzer et al., 2005). While no disease has been associated with the transmission of FVs to humans, the ease of primate-to-primate transmission implies the co-evolution of host restriction factors, as viral disease mechanisms are not sufficient to prevent cross-species transmission (Leendertz et al., 2008; Khan, 2009). As the FVs are promising candidates for the development of viral vectors for gene delivery and vaccination, it will be of interest to determine whether New and Old World monkey A3A proteins are capable of restricting the replication of spumaviruses (Schwantes et al., 2003; Trobridge, 2009; Rethwilm, 2010).
A novel mechanism of virus restriction of HIV-1 by mndA3A, colA3A and debA3A
A significant finding from our analysis of non-human primate A3A proteins was that colA3A, mndA3A and debA3A restricted the replication of both HIV-1 and HIV-1Δvif (Schmitt et al., 2013; Katuwal et al., 2014). Thus, deciphering the mechanism of restriction could yield insight into potential novel antiviral strategies. Attempts to recover HIV-1 released from cells expressing colA3A and the HIV-1 genome resulted in very little Gag p24 (∼100-fold reduction) and infectious virus released from the producer cells. This was not observed with the other A3A or A3G proteins examined (Schmitt et al., 2013). We also analysed whether transfection of the viral genome into cells 24 h prior to transfection of the vector expressing colA3A would restrict the replication of HIV-1. Under these conditions, colA3A still restricted replication of HIV-1, suggesting that the early steps of virus replication (such as entry and reverse transcription) were not affected by the presence of colA3A (Schmitt et al., 2013). A recent study showed that both reverse transcription and integration were not affected by the presence of colA3A (Katuwal et al., 2014). Thus, these observations indicate that the mechanism of restriction occurs in the absence of being incorporated into virions and suggests that restriction may be at the level of transcription or translation.
As the substitution of the human A3A AC-loop 1 with that of rhesus A3A restored restriction activities of hA3A, we have also used a chimaeric protein approach to interrogate the structural basis for the restriction of HIV-1 by colA3A. A chimaeric protein with the N-terminal 100 aa fused to the C-terminal half of hA3A (col1–100hA3A) restricted HIV-1 replication, while the reverse chimaera (h1–100colA3A) had no effect on HIV-1 replication (Katuwal et al., 2014). Additional hA3A/colA3A chimaeric proteins revealed that aa 25–33 were critical to colA3A restriction of HIV-1. Comparison of the AC-loop 1 sequence from the A3A proteins of several Old World monkeys revealed that the sequences for three monkey species (rhesus macaque, African green monkey and baboon) were 25DLS(V/I) RGR(H/R)Q33, which is quite different from the AC-loop 1 sequence of the colobus monkey (25KPWVSGQRE33). We identified two additional Old World monkey A3A proteins with a similar or exact sequence. These were from Cercopithecus neglectus (De Brazza's monkey; debA3A; 25KPWVSGQRE33) and Mandrillus sphinx (mandrill; mndA3A; 25KLWVSGQHE33) (Fig. 2). Similar to colA3A, we observed that debA3A and mndA3A restricted HIV-1, with mndA3A consistently being more restrictive than colA3A and debA3A. Additionally, restriction also occurred in the producer cell (Katuwal et al., 2014). While speculative at this point, it will be of interest to determine whether the leucine at position 26 of mndA3A (both colA3A and debA3A have a proline at this position) is responsible for the slightly better restriction activity of mndA3A. As the mndA3A restricted HIV1 to higher levels than colA3A or debA3A, we constructed a chimaeric hA3A having the AC-loop1 region of mndA3A (mnd25–34hA3A) and determined whether it could restrict HIV-1. Our results indicated that this chimaeric A3A did indeed restrict HIV-1, confirming that this region is necessary and sufficient for hA3A to restrict HIV-1 in producer cells (Katuwal et al., 2014). The Old World monkeys (family Cercopithecidae) are composed of two subfamilies (Cercopithecinae and Colobinae) and four tribes (Papionini, Cercopithecini, Presbytini and Colobini) (Schrago & Russo, 2003; Schrago, 2007). Our findings indicate that restriction of WT HIV-1 by A3A is represented by species in three tribes (Colobini: colA3A; Cercopithecini: debA3A; and Papionini: mndA3A). These results also reinforce our previous findings that the sequence of the AC-loop1 region of A3A is critical for restriction of WT HIV-1 by select Old World monkey species. It will be of interest to determine whether other closely related species also express A3A proteins that restrict HIV-1 and whether the hA3A protein can potently inhibit HIV-1 with a few amino acid substitutions in the AC-loop 1 region.
As the data above suggested that the N-terminal region and more specifically the AC-loop1 region is very important for HIV-1 restriction in producer cells, we determined whether we could truncate colA3A or mndA3A to the N-terminal 100 aa and retain restriction activity. Our results indicated that truncation of these two proteins still restricted HIV-1 replication (Katuwal et al., 2014). As the truncation disrupted the Z domain of this protein, we can conclude that deamination is not required for colA3A or mndA3A restriction of HIV-1 (Katuwal et al., 2014). To the best of our knowledge, this is the first demonstration that a single deaminase domain A3 protein could be significantly truncated and retain virus restriction activity. Together with our data on mnd25–34hA3A, it will be of interest to determine whether a truncated mnd25–34hA3A is also active against HIV-1.
APOBEC3A and mRNA editing
Wilms Tumour 1 (WT1) mutations and variants are implicated in several diseases, including Wilms tumour and acute myeloid leukaemia. WT1 is a regulatory protein with dual tumour suppressor/oncogene activity depending on the isoforms expressed, including the Lys-Thr-Ser (KTS) variant. Recently, two G → A changes were detected (G1303A and G1586A) in cDNA clones from non-progenitor cord but not progenitor blood mononuclear cells (Niavarani et al., 2015). These investigators showed that A3A was expressed at high levels in non-progenitor cells compared with progenitor cells. Using small interfering RNA, they showed that knockdown of A3A expression, but not of A3B, A3D or A3F, led to near complete reversal of WT1 c.1303G → A. Furthermore, overexpression of A3A in a Fujioka cell line resulted in a significant increase in the WT1 c.1303G → A change. This represents the first report detailing a specific G → A mRNA editing of transcripts by hA3A. This was followed by a second study that showed that hA3A, which is expressed at high levels in monocytes and macrophages, induced RNA editing during the polarization of M1 macrophages and in monocytes under hypoxic conditions or in response to IFN (Sharma et al., 2015). It will be of interest to determine whether hA3A can edit viral mRNA from retroviruses, especially within infected macrophages where hA3A is expressed at high levels.
A3A restriction of DNA viruses
hA3A has been reported to restrict certain DNA viruses (Chen et al., 2006; Vartanian et al., 2008; Henry et al., 2009; Narvaiza et al., 2009; Suspène et al., 2011b; Ahasan et al., 2015; Warren et al., 2015). The parvovirus adeno-associated virus 2 (AAV-2) was shown to be restricted by hA3A (Chen et al., 2006). These investigators found that mutation of the cysteine at position 106 of the canonical deaminase motif (H-x-E-x23–28-PC-x2–4-C) to a serine did not affect its nuclear localization but did not restrict AAV-2, suggesting a role of the enzymatic site in restriction. In a subsequent study by the same group of investigators using comprehensive mutagenesis of A3A, they found that regions outside the catalytic site also contributed to A3A antiviral activities. They also found that deaminase-defective A3A mutants blocked the replication of both AAV-2 and the autonomous parvovirus minute virus of mice (Narvaiza et al., 2009). In a study on herpes simplex virus type 1 (HSV-1), investigators found that A3A could edit a small fraction of HSV-1 genomes without seriously impacting viral titres (Suspène et al., 2011b). This suggests that A3A probably does not play a significant role in restricting the herpesviruses tested. Another study analysed the ability of the seven A3 proteins to edit the hepatitis B virus (HBV) genome (Henry et al., 2009). These investigators, using 3D-PCR, showed that with the exception of A3D, the other A3 proteins edited the HBV genome, with A3A being the most efficient editor (Henry et al., 2009). In a subsequent study, it was shown that the overexpression of A3A induced hypermutation in the HBV genome, although the levels of hypermutants were less than those introduced by A3G (Abe et al., 2009). A third study examined the correlation between deletion of the A3B gene and chronic HBV infections (Ezzikouri et al., 2013). These investigators observed that patients with the A3B-deleted genotype had lower virus burdens than those patients without the A3B-deletion phenotype (Ezzikouri et al., 2013). A3A has also been shown to inhibit the replication of papillomaviruses (Ahasan et al., 2015; Warren et al., 2015). In one study, using a human papillomavirus 16 (HPV-16) pseudovirion (PsV) production system, in which PsVs are assembled by expression of HPV L1/L2 proteins and a reporter plasmid in 293FT cells, the investigators found that co-expression of hA3A in 293FT cells decreased the infectivity of the PsVs (Ahasan et al., 2015). These investigators presented evidence that the decreased infectivity of the PsVs was due to decreased copy number of the encapsidated reporter plasmid. In another study, HPV assembled in the presence of hA3A resulted in significantly decreased infectivity compared with a catalytically inactive hA3A/E/72Q or in the absence of hA3A (Warren et al., 2015). Using a sensitive next-generation sequencing approach, however, hA3A mutational signatures were not detected above the background in the HPV16 LCR and E2 regions of the viral genome. These investigators found that IFN-β induced the expression of hA3A, and this correlated with the ability to inhibit HPV-16 replication (Warren et al., 2014, 2015). Finally, these investigators showed that upregulation of hA3A in normal immortalized keratinocytes was dependent on the presence of the HPV oncoprotein E7, and that knockdown of hA3A in keratinocytes resulted in an increase in the infectivity of HPV (Warren et al., 2015).
A3A inhibition of retrotransposition
The retroelements, which includes the long interspersed nuclear elements (LINE-1), small interspersed nuclear elements, Alu elements and LTR retroelements (including human endogenous retroviruses and retrotransposons), comprise a large family of transposable genetic elements that make up approximately 42 % of the human genome and have shaped the genomes of humans and other mammals over millions of years. Some retroelements can cause genetic diseases through retrotransposition events that occur not only in germ cells but also in somatic cells, posing a threat to genomic stability throughout all cellular populations. In an effort to combat retroelements, mammals have developed an innate mechanism that provides resistance against the deleterious effects of retrotransposition. Within this, seven members of the A3 family of cytidine deaminases serve as highly active, intrinsic, anti-retroviral host factors. Based on their preferential expression in germ cells, in which retrotransposons may be active, it is likely that A3 proteins were acquired through mammalian evolution primarily to inhibit retrotransposition and thereby maintain genomic stability in these cells. The A3A protein, like the other six A3 proteins, has been shown to inhibit retrotransposition. In vitro experiments have demonstrated that ectopically expressed hA3A effectively inhibits the retrotransposition of LINE-1, Alu, intracisternal A particle (IAP) and MusD (Bogerd et al., 2006a, b; Chen et al., 2006; Muckenfuss et al., 2006; Kinomoto et al., 2007; Niewiadomska et al., 2007; Koito & Ikeda, 2011) through a deaminase-independent mechanism.
A3A and degradation of foreign DNA
hA3A has been reported to degrade foreign DNA introduced into cells (Stenglein et al., 2010). These investigators used an assay that was based on the number of fluorescent cells following the transfection of plasmids expressing GFP and hA3A. They found a decrease in the level of GFP-positive cells at 5 days but not at 2 days post-transfection, and proposed a model in which the foreign DNA is ‘degraded’, which leads to an IFN response and expression of A3A. This results in DNA deamination and excision of uracils by UNG2, and degradation. However, the mechanism for how foreign DNA is recognized is unknown. They concluded that related proteins exist in all vertebrates and that it may be a conserved innate immune mechanism. It could be argued that DNA viruses or viruses with DNA intermediates most likely constitute the single most common introduction of foreign DNA. Furthermore, many DNA viruses (e.g. poxviruses, herpesviruses, adenoviruses and some parvoviruses) will have entered, replicated, assembled and been released from cells within 48 h of infection, which may be concomitant with death of the cell. Similarly, most retroviruses will have entered, reverse transcribed their genomes and been integrated within 48 h. This study raises the question of why, if this is such an important innate immune mechanism, does it not occur early after transfection? Additionally, if this is evolutionarily conserved, it should also occur in other hominids and other non-human primates. The ability of A3A proteins from rhesus macaques and black-and-white colobus monkey to degrade foreign DNA has also been examined (Schmitt et al., 2011, 2013). In our assays, we introduced a vector expressing A3A protein 24 h prior to transfection of a vector expressing GFP. In these assays, we obtained more than 80 % of cells expressing GFP. While we observed a decrease in the level of fluorescent cells transfected with hA3A, neither rhA3A nor colA3A caused a decrease in the percentage of fluorescent cells (Schmitt et al., 2011, 2013).
DNA damage and A3A
Of the seven A3 proteins, three (A3A, A3C and A3H) have a nucleocytoplasmic localization (Muckenfuss et al., 2006). Thus, the potential exists for these proteins to deaminate cytidine residues of chromosomal DNA. The overexpression of A3A in cultured cells has been shown by some investigators to result in mutation of both nuclear and mitochondrial DNA (Landry et al., 2011; Suspène et al., 2011a; Aynaud et al., 2012; Lackey et al., 2013; Mussil et al., 2013). Moreover, ecotopic expression of A3A in HEK293 cells is generally toxic to cells.
Histone H2AX is a variant histone that represents approximately 10 % of the total H2A histone protein in normal human fibroblasts (Rokagou et al.., 1998, 1999). H2AX is required for checkpoint-mediated cell-cycle arrest and DNA repair following dsDNA breaks (Yuan et al., 2010). DNA damage, caused by ionizing radiation, UV light or radiomimetic agents, results in rapid phosphorylation of H2AX at Ser139 by PI3K-like kinases, including ATM, ATR and DNA-PK (Rokagou et al., 1998; Burma et al., 2001). Within minutes following DNA damage, H2AX is phosphorylated at Ser139 at sites of DNA damage and this variant is known as γH2AX (Rokagou et al., 1999). Human A3A was shown to induce a DNA damage response and phosphorylation of H2AX (Landry et al., 2011). More recently, using an antibody raised against an A3A-specific peptide, other investigators found that A3A expression in terminally differentiated macrophages was predominantly cytoplasmic and was not genotoxic (Land et al., 2013). However, as mentioned earlier, the peptide to which this antiserum was raised is identical in hA3B and 90 % identical to hA3G. However, as A3G is expressed in macrophages and is localized to the cytoplasm, this could lead to predominant cytoplasmic staining. Finally, a recent study showed that, like hA3A, A3A proteins from different mammalian species are also capable of hypermutating genomic DNA, suggesting that this activity has been conserved over approximately 148 million years of evolution (Caval et al., 2014a).
Possible roles of A3A in cancer
As the A3 proteins have a specificity for ssDNA, which is available during DNA replication and transcription of the genome, these proteins have the potential to be mutagens that could in turn lead to cancer. One study examined the editing of the HPV genome and found that hA3A, hA3C and hA3H were expressed in keratinocytes (Vartanian et al., 2008). They found that HPV-1a genomes from one of six plantar warts had consistent G → A and C → T edits, suggesting that both DNA strands were edited. Similar results were obtained in vitro when 293T cells were co-transfected with the HPV-1a genome and either hA3A, hA3C or hA3H. HPVs are known to cause cervical cancer with subtypes 16 and 18 most commonly associated with this cancer (Guan et al., 2012). These investigators found that HPV-16 sequences were edited in pre-cancerous cervical biopsies and concluded that stochastic or transient overexpression of these A3 proteins could expose the viral genome to mutations that could influence the development of tumours. Other studies have associated the A3A/A3B deletion and breast cancer of women of both Chinese and European ancestry, which is one of the strongest common genetic risks identified to date (Kidd et al., 2007; Long et al., 2013; Xuan et al., 2013). These results appear to be discordant from those published recently suggesting that hA3B overexpression might contribute to breast cancer (Burns et al., 2013). The deletion removes genomic sequences from exon 5 of hA3A transcripts to exon 8 of hA3B, resulting in a 29.5 kb deletion and fusion of A3A to the last exon of A3B. While the deletion did not affect the coding sequences of A3A, this transcript would contain the hA3A 5′ upstream regulatory sequences and the 3′ UTR from hA3B (A3A-UTRA3B) (Long et al., 2013). Recently, it was shown that A3A protein expression from the A3A-UTRA3B transcripts was 20 times more active than A3A-UTRA3A or A3A-ΔUTR (Caval et al., 2014b). Deletion of the entire A3B gene correlates with a higher risk of developing some cancers and a higher overall mutation burden per cancer genome (Nik-Zainal et al., 2014). These authors have suggested that because A3A expression correlates with inflammatory environments (such as type 1 and 2 IFNs) and chronic inflammation is usually associated with the genesis of cancer, A3A-induced damage may contribute to somatic mutation selection in different cancers. Genome-wide association studies have identified small nucleotide polymorphisms in a non-genic region approximately 25 kb centromeric of hA3A that were associated with bladder cancer (Rothman et al., 2010; Golka et al., 2011). Finally, a study revealed a homozygous deletion in the A3A and A3B genes in pancreatic cancer (Liang et al., 2014). Thus, whether hA3A overexpression leads to an increased incidence of cancer is still highly controversial.
Conclusions and future studies
While the A3A protein shares many structural properties with the other members of the A3 family of proteins, it has unique properties that could be either beneficial or detrimental to the host. The exact function of A3A in the host is still unclear. Similar to several other A3 proteins, A3A has probably contributed to the restriction and inactivation of HERVs, which comprise 8 % of the human genome (Jern & Coffin, 2008; Magiorkinis et al., 2013). Unlike the better-studied A3G, which readily restricts HIV-1Δvif by incorporation of hA3G into virus from the producer cell and restriction by cytidine deamination of the viral genome in target cells, hA3A does not significantly restrict the replication of HIV-1Δvif and its cytidine deamination does not appear to be processive in nature (Goila-Gaur et al., 2007). However, the findings that hA3A can restrict HIV-1 in macrophages and that the A3A proteins from other Old World monkey species such as the rhesus macaque (rhA3A) can restrict SHIVΔvif provides evidence that these proteins can restrict lentiviruses (Peng et al., 2007; Berger et al., 2011; Schmitt et al., 2011). Structure–function studies suggest that the AC-loop1 region (also referred to as RL1) may contain determinants for the restriction of HIV-1Δvif. Interestingly, the A3A proteins from hominid species have a 3 aa indel in this region (see Fig. 2). Thus, from an evolutionary standpoint, it will be of interest to learn why the hominids have this 3 aa deletion in their A3A proteins. hA3A can also restrict other retroviruses such as HTLV-1, although the mechanism is probably different from A3A restriction of SHIV/SIV. This protein can also restrict other viruses such as parvoviruses (both AAV-2 and autonomously replicating parvoviruses) and papillomaviruses. The available evidence indicates that this probably occurs through a deaminase-independent mechanism (Narvaiza et al., 2009; Warren et al., 2015). Currently, the mechanism used by hA3A to restrict these viruses is unknown. However, it was shown that HPV upregulation of A3A in keratinocytes required the expression of the E7 oncoprotein (Warren et al., 2015). This raises questions of whether hA3A can interact with one or more viral proteins from these viruses to restrict replication and whether an hA3A protein containing the AC-loop1 region of an Old World monkey can still restrict these viruses. Recent studies have shown that hA3A can edit at the RNA level. If these results are confirmed, it will be of interest to decipher whether A3A can contribute to sequence variation in the genomes of RNA viruses. Overexpression of hA3A has been shown by several investigators to be toxic to cells, and others have suggested that, because of its expression in the nucleus, it may cause cytidine deamination of the host cell chromosome. Whether the upregulation of hA3A during times of immune activation leads to genomic mutations and cancer is currently unknown. However, the finding that certain cancers are associated with a 25 kb deletion in the A3 locus that results in the deletion of hA3B and an upregulation in hA3A expression requires further study.
Acknowledgements
This work was supported by NIH grant R21 AI108391 to E. B. S. and NIH R01 AI090795 to M. L. S.
References
- Abe et al.., 2009.Abe H., Ochi H., Maekawa T., Hatakeyama T., Tsuge M., Kitamura S., Kimura T., Miki D., Mitsui F., other authors (2009). Effects of structural variations of APOBEC3A and APOBEC3B genes in chronic hepatitis B virus infection Hepatol Res 391159–1168 10.1111/j.1872-034X.2009.00566.x. [DOI] [PubMed] [Google Scholar]
- Adolph et al.., 2013.Adolph M. B., Webb J., Chelico L. (2013). Retroviral restriction factor APOBEC3G delays the initiation of DNA synthesis by HIV-1 reverse transcriptase PLoS One 8e64196. 10.1371/journal.pone.0064196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguiar et al.., 2008.Aguiar R. S., Lovsin N., Tanuri A., Peterlin B. M. (2008). Vpr.A3A chimera inhibits HIV replication J Biol Chem 2832518–2525 10.1074/jbc.M706436200. [DOI] [PubMed] [Google Scholar]
- Ahasan et al.., 2015.Ahasan M. M., Wakae K., Wang Z., Kitamura K., Liu G., Koura M., Imayasu M., Sakamoto N., Hanaoka K., other authors (2015). APOBEC3A and 3C decrease human papillomavirus 16 pseudovirion infectivity Biochem Biophys Res Commun 457295–299 10.1016/j.bbrc.2014.12.103. [DOI] [PubMed] [Google Scholar]
- Alke et al.., 2001.Alke A., Schwantes A., Kido K., Flötenmeyer M., Flügel R. M., Löchelt M. (2001). The bet gene of feline foamy virus is required for virus replication Virology 287310–320 10.1006/viro.2001.1065. [DOI] [PubMed] [Google Scholar]
- Aynaud et al.., 2012.Aynaud M. M., Suspène R., Vidalain P. O., Mussil B., Guétard D., Tangy F., Wain-Hobson S., Vartanian J. P. (2012). Human Tribbles 3 protects nuclear DNA from cytidine deamination by APOBEC3A J Biol Chem 28739182–39192 10.1074/jbc.M112.372722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N., Kurth R. (2006). The evolutionary dynamics of human endogenous retroviral families Annu Rev Genomics Hum Genet 7149–173 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Barbulescu et al.., 1999.Barbulescu M., Turner G., Seaman M. I., Deinard A. S., Kidd K. K., Lenz J. (1999). Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans Curr Biol 9861–868 10.1016/S0960-9822(99)80390-X. [DOI] [PubMed] [Google Scholar]
- Bélanger et al.., 2013.Bélanger K., Savoie M., Rosales Gerpe M. C., Couture J. F., Langlois M. A. (2013). Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses Nucleic Acids Res 417438–7452 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett et al.., 2008.Bennett R. P., Presnyak V., Wedekind J. E., Smith H. C. (2008). Nuclear exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding J Biol Chem 2837320–7327 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- Berger et al.., 2011.Berger G., Durand S., Fargier G., Nguyen X. N., Cordeil S., Bouaziz S., Muriaux D., Darlix J. L., Cimarelli A. (2011). APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells PLoS Pathog 7e1002221. 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds et al.., 2007.Bininda-Emonds O. R., Cardillo M., Jones K. E., MacPhee R. D., Beck R. M., Grenyer R., Price S. A., Vos R. A., Gittleman J. L., Purvis A. (2007). The delayed rise of present-day mammals Nature 446507–512 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Bishop et al.., 2004.Bishop K. N., Holmes R. K., Sheehy A. M., Davidson N. O., Cho S. J., Malim M. H. (2004). Cytidine deamination of retroviral DNA by diverse APOBEC proteins Curr Biol 141392–1396 10.1016/j.cub.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Bishop et al.., 2006.Bishop K. N., Holmes R. K., Malim M. H. (2006). Antiviral potency of APOBEC proteins does not correlate with cytidine deamination J Virol 808450–8458 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock M., Stoye J. P. (2000). Endogenous retroviruses and the human germline Curr Opin Genet Dev 10651–655 10.1016/S0959-437X(00)00138-6. [DOI] [PubMed] [Google Scholar]
- Bogerd et al.., 2004.Bogerd H. P., Doehle B. P., Wiegand H. L., Cullen B. R. (2004). A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor Proc Natl Acad Sci U S A 1013770–3774 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd et al.., 2006a.Bogerd H. P., Wiegand H. L., Doehle B. P., Lueders K. K., Cullen B. R. (2006a). APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells Nucleic Acids Res 3489–95 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd et al.., 2006b.Bogerd H. P., Wiegand H. L., Hulme A. E., Garcia-Perez J. L., O'Shea K. S., Moran J. V., Cullen B. R. (2006b). Cellular inhibitors of long interspersed element 1 and Alu retrotransposition Proc Natl Acad Sci U S A 1038780–8785 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn et al.., 2015.Bohn M. F., Shandilya S. M., Silvas T. V., Nalivaika E. A., Kouno T., Kelch B. A., Ryder S. P., Kurt-Yilmaz N., Somasundaran M., Schiffer C. A. (2015). The ssDNA mutator APOBEC3A is regulated by cooperative dimerization Structure 23903–911 10.1016/j.str.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne et al.., 2009.Browne E. P., Allers C., Landau N. R. (2009). Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent Virology 387313–321 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulliard et al.., 2011.Bulliard Y., Narvaiza I., Bertero A., Peddi S., Röhrig U. F., Ortiz M., Zoete V., Castro-Díaz N., Turelli P., other authors (2011). Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities J Virol 851765–1776 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma et al.., 2001.Burma S., Chen B. P., Murphy M., Kurimasa A., Chen D. J. (2001). ATM phosphorylates histone H2AX in response to DNA double-strand breaks J Biol Chem 27642462–42467 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- Burns et al.., 2013.Burns M. B., Lackey L., Carpenter M. A., Rathore A., Land A. M., Leonard B., Refsland E. W., Kotandeniya D., Tretyakova N., other authors (2013). APOBEC3B is an enzymatic source of mutation in breast cancer Nature 494366–370 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon et al.., 2013.Byeon I. J., Ahn J., Mitra M., Byeon C. H., Hercík K., Hritz J., Charlton L. M., Levin J. G., Gronenborn A. M. (2013). NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity Nat Commun 41890. 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani et al.., 2011.Cagliani R., Riva S., Fumagalli M., Biasin M., Caputo S. L., Mazzotta F., Piacentini L., Pozzoli U., Bresolin N., other authors (2011). A positively selected APOBEC3H haplotype is associated with natural resistance to HIV-1 infection Evolution 653311–3322 10.1111/j.1558-5646.2011.01368.x. [DOI] [PubMed] [Google Scholar]
- Carpenter et al.., 2012.Carpenter M. A., Li M., Rathore A., Lackey L., Law E. K., Land A. M., Leonard B., Shandilya S. M., Bohn M. F., other authors (2012). Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A J Biol Chem 28734801–34808 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caval et al.., 2014a.Caval V., Suspène R., Vartanian J.-P., Wain-Hobson S. (2014a). Orthologous mammalian APOBEC3A cytidine deaminases hypermutate nuclear DNA Mol Biol Evol 31330–340 10.1093/molbev/mst195. [DOI] [PubMed] [Google Scholar]
- Caval et al.., 2014b.Caval V., Suspène R., Shapira M., Vartanian J. P., Wain-Hobson S. (2014b). A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3′UTR enhances chromosomal DNA damage Nat Commun 55129. 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- Chareza et al.., 2012.Chareza S., Slavkovic Lukic D., Liu Y., Räthe A. M., Münk C., Zabogli E., Pistello M., Löchelt M. (2012). Molecular and functional interactions of cat APOBEC3 and feline foamy and immunodeficiency virus proteins: different ways to counteract host-encoded restriction Virology 424138–146 10.1016/j.virol.2011.12.017. [DOI] [PubMed] [Google Scholar]
- Chelico et al.., 2006.Chelico L., Pham P., Calabrese P., Goodman M. F. (2006). APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA Nat Struct Mol Biol 13392–399 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- Chen et al.., 2006.Chen H., Lilley C. E., Yu Q., Lee D. V., Chou J., Narvaiza I., Landau N. R., Weitzman M. D. (2006). APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons Curr Biol 16480–485 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen et al.., 2009.Chen G., He Z., Wang T., Xu R., Yu X. F. (2009). A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G J Virol 838674–8682 10.1128/JVI.00653-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello et al.., 2003.Conticello S. G., Harris R. S., Neuberger M. S. (2003). The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G Curr Biol 132009–2013 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- Dang et al.., 2010.Dang Y., Wang X., York I. A., Zheng Y. H. (2010). Identification of a critical T(Q/D/E)x5ADx2(I/L) motif from primate lentivirus Vif proteins that regulate APOBEC3G and APOBEC3F neutralizing activity J Virol 848561–8570 10.1128/JVI.00960-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal et al.., 2011.Duggal N. K., Malik H. S., Emerman M. (2011). The breadth of antiviral activity of Apobec3DE in chimpanzees has been driven by positive selection J Virol 8511361–11371 10.1128/JVI.05046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussart et al.., 2004.Dussart S., Courcoul M., Bessou G., Douaisi M., Duverger Y., Vigne R., Decroly E. (2004). The Vif protein of human immunodeficiency virus type 1 is posttranslationally modified by ubiquitin Biochem Biophys Res Commun 31566–72 10.1016/j.bbrc.2004.01.023. [DOI] [PubMed] [Google Scholar]
- Espert et al.., 2009.Espert L., Varbanov M., Robert-Hebmann V., Sagnier S., Robbins I., Sanchez F., Lafont V., Biard-Piechaczyk M. (2009). Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection PLoS One 4e5787. 10.1371/journal.pone.0005787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzikouri et al.., 2013.Ezzikouri S., Kitab B., Rebbani K., Marchio A., Wain-Hobson S., Dejean A., Vartanian J. P., Pineau P., Benjelloun S. (2013). Polymorphic APOBEC3 modulates chronic hepatitis B in Moroccan population J Viral Hepat 20678–686 10.1111/jvh.12042. [DOI] [PubMed] [Google Scholar]
- Goila-Gaur et al.., 2007.Goila-Gaur R., Khan M. A., Miyagi E., Kao S., Strebel K. (2007). Targeting APOBEC3A to the viral nucleoprotein complex confers antiviral activity Retrovirology 461. 10.1186/1742-4690-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golka et al.., 2011.Golka K., Selinski S., Lehmann M. L., Blaszkewicz M., Marchan R., Ickstadt K., Schwender H., Bolt H. M., Hengstler J. G. (2011). Genetic variants in urinary bladder cancer: collective power of the wimp SNPs Arch Toxicol 85539–554 10.1007/s00204-011-0676-3. [DOI] [PubMed] [Google Scholar]
- Guan et al.., 2012.Guan P., Howell-Jones R., Li N., Bruni L., de Sanjosé S., Franceschi S., Clifford G. M. (2012). Human papillomavirus types in 115,789 HPV-positive women: a meta-analysis from cervical infection to cancer Int J Cancer 1312349–2359 10.1002/ijc.27485. [DOI] [PubMed] [Google Scholar]
- Hahn et al.., 2000.Hahn B. H., Shaw G. M., De Cock K. M., Sharp P. M. (2000). AIDS as a zoonosis: scientific and public health implications Science 287607–614 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Heneine et al.., 2003.Heneine W., Schweizer M., Sandstrom P., Folks T. (2003). Human infection with foamy viruses Curr Top Microbiol Immunol 277181–196. [DOI] [PubMed] [Google Scholar]
- Henry et al.., 2009.Henry M., Guétard D., Suspène R., Rusniok C., Wain-Hobson S., Vartanian J. P. (2009). Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G PLoS One 4e4277. 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry et al.., 2012.Henry M., Terzian C., Peeters M., Wain-Hobson S., Vartanian J. P. (2012). Evolution of the primate APOBEC3A cytidine deaminase gene and identification of related coding regions PLoS One 7e30036. 10.1371/journal.pone.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden et al.., 2008.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008). Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications Nature 456121–124 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultquist et al.., 2011.Hultquist J. F., Lengyel J. A., Refsland E. W., LaRue R. S., Lackey L., Brown W. L., Harris R. S. (2011). Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1 J Virol 8511220–11234 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain et al.., 2003.Hussain A. I., Shanmugam V., Bhullar V. B., Beer B. E., Vallet D., Gautier-Hion A., Wolfe N. D., Karesh W. B., Kilbourn A. M., other authors (2003). Screening for simian foamy virus infection by using a combined antigen Western blot assay: evidence for a wide distribution among Old World primates and identification of four new divergent viruses Virology 309248–257 10.1016/S0042-6822(03)00070-9. [DOI] [PubMed] [Google Scholar]
- Iwatani et al.., 2007.Iwatani Y., Chan D. S., Wang F., Maynard K. S., Sugiura W., Gronenborn A. M., Rouzina I., Williams M. C., Musier-Forsyth K., Levin J. G. (2007). Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G Nucleic Acids Res 357096–7108 10.1093/nar/gkm750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger et al.., 2012.Jäger S., Kim D. Y., Hultquist J. F., Shindo K., LaRue R. S., Kwon E., Li M., Anderson B. D., Yen L., other authors (2012). Vif hijacks CBF-β to degrade APOBEC3G and promote HIV-1 infection Nature 481371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz et al.., 2002.Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002). An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22 Genomics 79285–296 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- Jern P., Coffin J. M. (2008). Effects of retroviruses on host genome function Annu Rev Genet 42709–732 10.1146/annurev.genet.42.110807.091501. [DOI] [PubMed] [Google Scholar]
- Katuwal et al.., 2014.Katuwal M., Wang Y., Schmitt K., Guo K., Halemano K., Santiago M. L., Stephens E. B. (2014). Cellular HIV-1 inhibition by truncated Old World primate APOBEC3A proteins lacking a complete deaminase domain Virology 468-470532–544 10.1016/j.virol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele et al.., 2006.Keele B. F., Van Heuverswyn F., Li Y., Bailes E., Takehisa J., Santiago M. L., Bibollet-Ruche F., Chen Y., Wain L. V., other authors (2006). Chimpanzee reservoirs of pandemic and nonpandemic HIV-1 Science 313523–526 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S. (2009). Simian foamy virus infection in humans: prevalence and management Expert Rev Anti Infect Ther 7569–580 10.1586/eri.09.39. [DOI] [PubMed] [Google Scholar]
- Kidd et al.., 2007.Kidd J. M., Newman T. L., Tuzun E., Kaul R., Eichler E. E. (2007). Population stratification of a common APOBEC gene deletion polymorphism PLoS Genet 3e63. 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomoto et al.., 2007.Kinomoto M., Kanno T., Shimura M., Ishizaka Y., Kojima A., Kurata T., Sata T., Tokunaga K. (2007). All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition Nucleic Acids Res 352955–2964 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura et al.., 2012.Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., other authors (2012). The APOBEC3C crystal structure and the interface for HIV-1 Vif binding Nat Struct Mol Biol 191005–1010 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- Kobayashi et al.., 2005.Kobayashi M., Takaori-Kondo A., Miyauchi Y., Iwai K., Uchiyama T. (2005). Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function J Biol Chem 28018573–18578 10.1074/jbc.C500082200. [DOI] [PubMed] [Google Scholar]
- Koito A., Ikeda T. (2011). Intrinsic restriction activity by AID/APOBEC family of enzymes against the mobility of retroelements Mob Genet Elements 1197–202 10.4161/mge.1.3.17430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning et al.., 2009.Koning F. A., Newman E. N., Kim E. Y., Kunstman K. J., Wolinsky S. M., Malim M. H. (2009). Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets J Virol 839474–9485 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning et al.., 2011.Koning F. A., Goujon C., Bauby H., Malim M. H. (2011). Target cell-mediated editing of HIV-1 cDNA by APOBEC3 proteins in human macrophages J Virol 8513448–13452 10.1128/JVI.00775-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyei et al.., 2009.Kyei G. B., Dinkins C., Davis A. S., Roberts E., Singh S. B., Dong C., Wu L., Kominami E., Ueno T., other authors (2009). Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages J Cell Biol 186255–268 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackey et al.., 2013.Lackey L., Law E. K., Brown W. L., Harris R. S. (2013). Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination Cell Cycle 12762–772 10.4161/cc.23713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land et al.., 2013.Land A. M., Law E. K., Carpenter M. A., Lackey L., Brown W. L., Harris R. S. (2013). Endogenous APOBEC3A is cytoplasmic and non-genotoxic J Biol Chem 28817253–17260 10.1074/jbc.M113.458661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander et al.., 2001.Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., other authors (2001). Initial sequencing and analysis of the human genome Nature 409860–921 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Landry et al.., 2011.Landry S., Narvaiza I., Linfesty D. C., Weitzman M. D. (2011). APOBEC3A can activate the DNA damage response and cause cell-cycle arrest EMBO Rep 12444–450 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRue et al.., 2009.LaRue R. S., Andrésdóttir V., Blanchard Y., Conticello S. G., Derse D., Emerman M., Greene W. C., Jónsson S. R., Landau N. R., other authors (2009). Guidelines for naming nonprimate APOBEC3 genes and proteins J Virol 83494–497 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier et al.., 2003.Lecossier D., Bouchonnet F., Clavel F., Hance A. J. (2003). Hypermutation of HIV-1 DNA in the absence of the Vif protein Science 3001112. 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lee Y. N., Bieniasz P. D. (2007). Reconstitution of an infectious human endogenous retrovirus PLoS Pathog 3e10. 10.1371/journal.ppat.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee et al.., 2008.Lee Y. N., Malim M. H., Bieniasz P. D. (2008). Hypermutation of an ancient human retrovirus by APOBEC3G J Virol 828762–8770 10.1128/JVI.00751-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leendertz et al.., 2008.Leendertz F. H., Zirkel F., Couacy-Hymann E., Ellerbrok H., Morozov V. A., Pauli G., Hedemann C., Formenty P., Jensen S. A., other authors (2008). Interspecies transmission of simian foamy virus in a natural predator-prey system J Virol 827741–7744 10.1128/JVI.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letvin et al.., 1985.Letvin N. L., Daniel M. D., Sehgal P. K., Desrosiers R. C., Hunt R. D., Waldron L. M., MacKey J. J., Schmidt D. K., Chalifoux L. V., King N. W. (1985). Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III Science 23071–73. [DOI] [PubMed] [Google Scholar]
- Li et al.., 2007.Li X. Y., Guo F., Zhang L., Kleiman L., Cen S. (2007). APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription J Biol Chem 28232065–32074 10.1074/jbc.M703423200. [DOI] [PubMed] [Google Scholar]
- Li et al.., 2014.Li J., Chen Y., Li M., Carpenter M. A., McDougle R. M., Luengas E. M., Macdonald P. J., Harris R. S., Mueller J. D. (2014). APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells J Mol Biol 4261296–1307 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang et al.., 2014.Liang J. W., Shi Z. Z., Shen T. Y., Che X., Wang Z., Shi S. S., Xu X., Cai Y., Zhao P., other authors (2014). Identification of genomic alterations in pancreatic cancer using array-based comparative genomic hybridization PLoS One 9e114616. 10.1371/journal.pone.0114616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al.., 2004.Liu B., Yu X., Luo K., Yu Y., Yu X. F. (2004). Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G J Virol 782072–2081 10.1128/JVI.78.4.2072-2081.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löchelt et al.., 2005.Löchelt M., Romen F., Bastone P., Muckenfuss H., Kirchner N., Kim Y. B., Truyen U., Rösler U., Battenberg M., other authors (2005). The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein Proc Natl Acad Sci U S A 1027982–7987 10.1073/pnas.0501445102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue et al.., 2014.Logue E. C., Bloch N., Dhuey E., Zhang R., Cao P., Herate C., Chauveau L., Hubbard S. R., Landau N. R. (2014). A DNA sequence recognition loop on APOBEC3A controls substrate specificity PLoS One 9e97062. 10.1371/journal.pone.0097062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long et al.., 2013.Long J., Delahanty R. J., Li G., Gao Y. T., Lu W., Cai Q., Xiang Y. B., Li C., Ji B. T., other authors (2013). A common deletion in the APOBEC3 genes and breast cancer risk J Natl Cancer Inst 105573–579 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love et al.., 2012.Love R. P., Xu H., Chelico L. (2012). Biochemical analysis of hypermutation by the deoxycytidine deaminase APOBEC3A J Biol Chem 28730812–30822 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo et al.., 2005.Luo K., Xiao Z., Ehrlich E., Yu Y., Liu B., Zheng S., Yu X. F. (2005). Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G Proc Natl Acad Sci U S A 10211444–11449 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo et al.., 2007.Luo K., Wang T., Liu B., Tian C., Xiao Z., Kappes J., Yu X. F. (2007). Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation J Virol 817238–7248 10.1128/JVI.02584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorkinis et al.., 2013.Magiorkinis G., Belshaw R., Katzourakis A. (2013). ‘There and back again’: revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era Philos Trans R Soc Lond B Biol Sci 36820120504. 10.1098/rstb.2012.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat et al.., 2003.Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. (2003). Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts Nature 42499–103 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Marin et al.., 2003.Marin M., Rose K. M., Kozak S. L., Kabat D. (2003). HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation Nat Med 91398–1403 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Mehle et al.., 2004a.Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. (2004a). Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation Genes Dev 182861–2866 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle et al.., 2004b.Mehle A., Strack B., Ancuta P., Zhang C., McPike M., Gabuzda D. (2004b). Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway J Biol Chem 2797792–7798 10.1074/jbc.M313093200. [DOI] [PubMed] [Google Scholar]
- Mehle et al.., 2006.Mehle A., Thomas E. R., Rajendran K. S., Gabuzda D. (2006). A zinc-binding region in Vif binds Cul5 and determines cullin selection J Biol Chem 28117259–17265 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- Mehle et al.., 2007.Mehle A., Wilson H., Zhang C., Brazier A. J., McPike M., Pery E., Gabuzda D. (2007). Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding J Virol 8113235–13241 10.1128/JVI.00204-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiering C. D., Linial M. L. (2001). Historical perspective of foamy virus epidemiology and infection Clin Microbiol Rev 14165–176 10.1128/CMR.14.1.165-176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi et al.., 2007.Miyagi E., Opi S., Takeuchi H., Khan M., Goila-Gaur R., Kao S., Strebel K. (2007). Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1 J Virol 8113346–13353 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenfuss et al.., 2006.Muckenfuss H., Hamdorf M., Held U., Perkovic M., Löwer J., Cichutek K., Flory E., Schumann G. G., Münk C. (2006). APOBEC3 proteins inhibit human LINE-1 retrotransposition J Biol Chem 28122161–22172 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- Münk et al.., 2012.Münk C., Willemsen A., Bravo I. G. (2012). An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals BMC Evol Biol 1271. 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussil et al.., 2013.Mussil B., Suspène R., Aynaud M. M., Gauvrit A., Vartanian J. P., Wain-Hobson S. (2013). Human APOBEC3A isoforms translocate to the nucleus and induce DNA double strand breaks leading to cell stress and death PLoS One 8e73641. 10.1371/journal.pone.0073641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaiza et al.., 2009.Narvaiza I., Linfesty D. C., Greener B. N., Hakata Y., Pintel D. J., Logue E., Landau N. R., Weitzman M. D. (2009). Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase PLoS Pathog 5e1000439. 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil et al.., 2008.Neil S. J., Zang T., Bieniasz P. D. (2008). Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu Nature 451425–430 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- Newman et al.., 2005.Newman E. N., Holmes R. K., Craig H. M., Klein K. C., Lingappa J. R., Malim M. H., Sheehy A. M. (2005). Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity Curr Biol 15166–170 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Niavarani et al.., 2015.Niavarani A., Currie E., Reyal Y., Anjos-Afonso F., Horswell S., Griessinger E., Luis Sardina J., Bonnet D. (2015). APOBEC3A is implicated in a novel class of G-to-A mRNA editing in WT1 transcripts PLoS One 10e0120089. 10.1371/journal.pone.0120089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska et al.., 2007.Niewiadomska A. M., Tian C., Tan L., Wang T., Sarkis P. T., Yu X. F. (2007). Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association J Virol 819577–9583 10.1128/JVI.02800-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal et al.., 2014.Nik-Zainal S., Wedge D. C., Alexandrov L. B., Petljak M., Butler A. P., Bolli N., Davies H. R., Knappskog S., Martin S., other authors (2014). Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer Nat Genet 46487–491 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle et al.., 2006.OhAinle M., Kerns J. A., Malik H. S., Emerman M. (2006). Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H J Virol 803853–3862 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms et al.., 2012.Ooms M., Krikoni A., Kress A. K., Simon V., Münk C. (2012). APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1 J Virol 866097–6108 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz et al.., 2009.Ortiz M., Guex N., Patin E., Martin O., Xenarios I., Ciuffi A., Quintana-Murci L., Telenti A. (2009). Evolutionary trajectories of primate genes involved in HIV pathogenesis Mol Biol Evol 262865–2875 10.1093/molbev/msp197. [DOI] [PubMed] [Google Scholar]
- Peng et al.., 2007.Peng G., Greenwell-Wild T., Nares S., Jin W., Lei K. J., Rangel Z. G., Munson P. J., Wahl S. M. (2007). Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression Blood 110393–400 10.1182/blood-2006-10-051763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pery et al.., 2009.Pery E., Rajendran K. S., Brazier A. J., Gabuzda D. (2009). Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif J Virol 832374–2381 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham et al.., 2013.Pham P., Landolph A., Mendez C., Li N., Goodman M. F. (2013). A biochemical analysis linking APOBEC3A to disparate HIV-1 restriction and skin cancer J Biol Chem 28829294–29304 10.1074/jbc.M113.504175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland et al.., 2010.Refsland E. W., Stenglein M. D., Shindo K., Albin J. S., Brown W. L., Harris R. S. (2010). Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction Nucleic Acids Res 384274–4284 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rethwilm A. (2010). Molecular biology of foamy viruses Med Microbiol Immunol (Berl) 199197–207 10.1007/s00430-010-0158-x. [DOI] [PubMed] [Google Scholar]
- Rogakou et al.., 1998.Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139 J Biol Chem 2735858–5868 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rogakou et al.., 1999.Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999). Megabase chromatin domains involved in DNA double-strand breaks in vivo J Cell Biol 146905–916 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin et al.., 2005.Rogozin I. B., Basu M. K., Jordan I. K., Pavlov Y. I., Koonin E. V. (2005). APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis Cell Cycle 41281–1285 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- Rothman et al.., 2010.Rothman N., Garcia-Closas M., Chatterjee N., Malats N., Wu X., Figueroa J. D., Real F. X., Van Den Berg D., Matullo G., other authors (2010). A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci Nat Genet 42978–984 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. A., Pathak V. K. (2007). Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F J Virol 818201–8210 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell et al.., 2005.Russell R. A., Wiegand H. L., Moore M. D., Schäfer A., McClure M. O., Cullen B. R. (2005). Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors J Virol 798724–8731 10.1128/JVI.79.14.8724-8731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saïb et al.., 1995.Saïb A., Koken M. H., van der Spek P., Périès J., de Thé H. (1995). Involvement of a spliced and defective human foamy virus in the establishment of chronic infection J Virol 695261–5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago et al.., 2005.Santiago M. L., Range F., Keele B. F., Li Y., Bailes E., Bibollet-Ruche F., Fruteau C., Noë R., Peeters M., other authors (2005). Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Taï Forest, Côte d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2 J Virol 7912515–12527 10.1128/JVI.79.19.12515-12527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer et al.., 2004.Sawyer S. L., Emerman M., Malik H. S. (2004). Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G PLoS Biol 2e275. 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt et al.., 2011.Schmitt K., Guo K., Algaier M., Ruiz A., Cheng F., Qiu J., Wissing S., Santiago M. L., Stephens E. B. (2011). Differential virus restriction patterns of rhesus macaque and human APOBEC3A: implications for lentivirus evolution Virology 41924–42 10.1016/j.virol.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt et al.., 2013.Schmitt K., Guo K., Katuwal M., Wilson D., Prochnow C., Bransteitter R., Chen X. S., Santiago M. L., Stephens E. B. (2013). Lentivirus restriction by diverse primate APOBEC3A proteins Virology 44282–96 10.1016/j.virol.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrago C. G. (2007). On the time scale of New World primate diversification Am J Phys Anthropol 132344–354 10.1002/ajpa.20459. [DOI] [PubMed] [Google Scholar]
- Schrago C. G., Russo C. A. (2003). Timing the origin of New World monkeys Mol Biol Evol 201620–1625 10.1093/molbev/msg172. [DOI] [PubMed] [Google Scholar]
- Schröfelbauer et al.., 2004.Schröfelbauer B., Chen D., Landau N. R. (2004). A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc Natl Acad Sci U S A 1013927–3932 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher et al.., 2008.Schumacher A. J., Haché G., Macduff D. A., Brown W. L., Harris R. S. (2008). The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction J Virol 822652–2660 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwantes et al.., 2003.Schwantes A., Truyen U., Weikel J., Weiss C., Löchelt M. (2003). Application of chimeric feline foamy virus-based retroviral vectors for the induction of antiviral immunity in cats J Virol 777830–7842 10.1128/JVI.77.14.7830-7842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma et al.., 2015.Sharma S., Patnaik S. K., Taggart R. T., Kannisto E. D., Enriquez S. M., Gollnick P., Baysal B. E. (2015). APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages Nat Commun 66881. 10.1038/ncomms7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy et al.., 2002.Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein Nature 418646–650 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy et al.., 2003.Sheehy A. M., Gaddis N. C., Malim M. H. (2003). The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif Nat Med 91404–1407 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Stavrou et al.., 2014.Stavrou S., Crawford D., Blouch K., Browne E. P., Kohli R. M., Ross S. R. (2014). Different modes of retrovirus restriction by human APOBEC3A and APOBEC3G in vivo PLoS Pathog 10e1004145. 10.1371/journal.ppat.1004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein et al.., 2008.Stenglein M. D., Matsuo H., Harris R. S. (2008). Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization J Virol 829591–9599 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein et al.., 2010.Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010). APOBEC3 proteins mediate the clearance of foreign DNA from human cells Nat Struct Mol Biol 17222–229 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak et al.., 2003.Stopak K., de Noronha C., Yonemoto W., Greene W. C. (2003). HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability Mol Cell 12591–601 10.1016/S1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Stremlau et al.., 2004.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. (2004). The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys Nature 427848–853 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- Suspène et al.., 2011a.Suspène R., Aynaud M. M., Guétard D., Henry M., Eckhoff G., Marchio A., Pineau P., Dejean A., Vartanian J. P., Wain-Hobson S. (2011a). Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism Proc Natl Acad Sci U S A 1084858–4863 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspène et al.., 2011b.Suspène R., Aynaud M. M., Koch S., Pasdeloup D., Labetoulle M., Gaertner B., Vartanian J. P., Meyerhans A., Wain-Hobson S. (2011b). Genetic editing of herpes simplex virus 1 and Epstein–Barr herpesvirus genomes by human APOBEC3 cytidine deaminases in culture and in vivo J Virol 857594–7602 10.1128/JVI.00290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Switzer et al.., 2005.Switzer W. M., Salemi M., Shanmugam V., Gao F., Cong M. E., Kuiken C., Bhullar V., Beer B. E., Vallet D., other authors (2005). Ancient co-speciation of simian foamy viruses and primates Nature 434376–380 10.1038/nature03341. [DOI] [PubMed] [Google Scholar]
- Trobridge G. D. (2009). Foamy virus vectors for gene transfer Expert Opin Biol Ther 91427–1436 10.1517/14712590903246388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme et al.., 2008.Van Damme N., Goff D., Katsura C., Jorgenson R. L., Mitchell R., Johnson M. C., Stephens E. B., Guatelli J. (2008). The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein Cell Host Microbe 3245–252 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian et al.., 2008.Vartanian J. P., Guétard D., Henry M., Wain-Hobson S. (2008). Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions Science 320230–233 10.1126/science.1153201. [DOI] [PubMed] [Google Scholar]
- Virgen C. A., Hatziioannou T. (2007). Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins J Virol 8113932–13937 10.1128/JVI.01760-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al.., 2012.Wang X., Ao Z., Chen L., Kobinger G., Peng J., Yao X. (2012). The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication J Virol 863777–3786 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren et al.., 2014.Warren C. J., Griffin L. M., Little A. S., Huang I. C., Farzan M., Pyeon D. (2014). The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus PLoS One 9e96579. 10.1371/journal.pone.0096579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren et al.., 2015.Warren C. J., Xu T., Guo K., Griffin L. M., Westrich J. A., Lee D., Lambert P. F., Santiago M. L., Pyeon D. (2015). APOBEC3A functions as a restriction factor of human papillomavirus J Virol 89688–702 10.1128/JVI.02383-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski et al.., 2005.Wichroski M. J., Ichiyama K., Rana T. M. (2005). Analysis of HIV-1 viral infectivity factor-mediated proteasome-dependent depletion of APOBEC3G: correlating function and subcellular localization J Biol Chem 2808387–8396 10.1074/jbc.M408048200. [DOI] [PubMed] [Google Scholar]
- Wiegand H. L., Cullen B. R. (2007). Inhibition of alpharetrovirus replication by a range of human APOBEC3 proteins J Virol 8113694–13699 10.1128/JVI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan et al.., 2013.Xuan D., Li G., Cai Q., Deming-Halverson S., Shrubsole M. J., Shu X. O., Kelley M. C., Zheng W., Long J. (2013). APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry Carcinogenesis 342240–2243 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al.., 2003.Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003). Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex Science 3021056–1060 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Yu et al.., 2004.Yu Q., König R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J. M., Landau N. R. (2004). Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome Nat Struct Mol Biol 11435–442 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- Yuan et al.., 2010.Yuan J., Adamski R., Chen J. (2010). Focus on histone variant H2AX: to be or not to be FEBS Lett 5843717–3724 10.1016/j.febslet.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Webb D. M. (2004). Rapid evolution of primate antiviral enzyme APOBEC3G Hum Mol Genet 131785–1791 10.1093/hmg/ddh183. [DOI] [PubMed] [Google Scholar]
- Zhang et al.., 2003.Zhang H., Yang B., Pomerantz R. J., Zhang C., Arunachalam S. C., Gao L. (2003). The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA Nature 42494–98 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al.., 2012.Zhang W., Du J., Evans S. L., Yu Y., Yu X. F. (2012). T-cell differentiation factor CBF-β regulates HIV-1 Vif-mediated evasion of host restriction Nature 481376–379. [DOI] [PubMed] [Google Scholar]